Abstract
Among 165 Spanish Haemophilus influenzae isolates with mutations in the ftsI gene (ftsI+) (2005 to 2007), 73% were β-lactamase negative and 26.7% were positive. The proportion of β-lactamase-negative isolates to β-lactamase-positive isolates was 2:1 to 4:1 in general, versus 1:3 in pediatric hospitals. Among 44 β-lactamase-positive strains, 8 strains produced ROB-1 (5 from the pediatric hospital). β-Lactamase-positive ftsI+ strains were phylogenetically closer than were β-lactamase-negative strains.
Since previous studies showed that ampicillin-susceptible β-lactamase-negative Haemophilus influenzae strains showing an ampicillin MIC of 1 μg/ml should be interpreted with caution because they may carry ftsI gene mutations (5), the presence of these mutations in β-lactamase-positive strains susceptible to amoxicillin-clavulanic acid exhibiting MICs of 2/1 to 4/2 μg/ml can be suspected. The aim of this study was to genotypically and phenotypically characterize (with respect to β-lactam susceptibility) H. influenzae isolates with ampicillin MIC of ≥1 μg/ml for β-lactamase-negative or amoxicillin-clavulanic acid MIC of ≥2/1 μg/ml for β-lactamase-positive strains. Spanish hospitals were contacted with a request for isolates with these susceptibility characteristics that were collected from March 2005 to March 2007. Six hospitals sent isolates that were retested in triplicate, and those showing the required or 1-dilution-lower modal MICs were included. Of the 252 strains received, 199 were recovered and exhibited the susceptibility requirements. Susceptibility to β-lactams was determined by microdilution (1). The susceptibility breakpoints considered were ≤1 μg/ml for ampicillin, ≤4/2 μg/ml for amoxicillin-clavulanic acid, ≤8 μg/ml for cefaclor, ≤4 μg/ml for cefuroxime, ≤1 μg/ml for cefdinir, and ≤2 μg/ml for cefotaxime (2). Nonsusceptibility was considered when MICs were above the susceptibility breakpoints. β-Lactamase production was determined by the chromogenic cephalosporin test (7).
For amplification and sequencing of the ftsI gene, DNA was obtained using the QIAamp DNA kit (Qiagen, Hilden, Germany). PCR amplification of the ftsI, acrR, blaTEM, and blaROB genes was performed using referenced primers (6, 8, 9). Strains with mutations in the ftsI gene were genotypically defined as BLNAR (β-lactamase negative, ampicillin resistant) or BLPACR (β-lactamase positive, amoxicillin-clavulanic acid resistant) that, when possible, were classified into the groups and subgroups proposed by Dabernat et al. (3) and Ubukata et al. (9). The ClustalW2 program (http://www.ebi.ac.uk) was used to construct phylogenetic trees of a 1,030-bp sequence from the ftsI gene.
Of the 199 strains exhibiting the susceptibility requirements, amplification failed in three isolates excluded for further analysis (all were β-lactamase negative; two strains had an ampicillin MIC of 2 μg/ml and one of 1 μg/ml).
Of the 196 strains tested, 31 (15.8%) did not present mutation in the ftsI gene, 10 were β-lactamase negative, and 21 were β-lactamase positive by the chromogenic cephalosporin test. Of the 165 strains showing ftsI mutations, 121 (73%) were β-lactamase negative (BLNAR), and 44 (26.7%) were β-lactamase positive (BLPACR). The proportion of BLNAR to BLPACR strains was approximately 2:1 to 4:1 in all general hospitals, but the proportion was reversed (approximately 1:3) in the pediatric hospital (H. S. Joan de Deu) (Table 1).
TABLE 1.
Number of isolates exhibiting mutations in the ftsI gene, by institution
| Center | No. of isolates with the ftsI gene sequenced | No. (%) of isolates with mutations in the ftsI gene
|
||
|---|---|---|---|---|
| Total | β-Lactamase negativea | β-Lactamase positivea | ||
| H. General Univ. Gregorio Marañon | 115 | 103 (89.6) | 82 (79.6) | 21 (20.4) |
| H. S. Joan de Deu | 31 | 21 (67.7) | 8 (26.6) | 13 (73.4) |
| H. Clinic | 26 | 22 (84.6) | 18 (81.8) | 4 (18.2) |
| H. Univ. Marques de Valdecilla | 20 | 19 (95.0) | 13 (68.4) | 6 (31.6) |
| Other | 4 | 0.0 (0) | ||
| Total | 196 | 165 (84.2) | 121 (73.3) | 44 (26.7) |
Percentage values in parentheses represent the proportion of the number of the indicated isolates to the total number of isolates with mutations in the ftsI gene.
Table 2 shows the MIC50, MIC90, and nonsusceptibility rates to the different antibiotics tested. Susceptibility problems with cefaclor, cefuroxime, and cefdinir are present even in strains without mutations in the ftsI gene, with nonsusceptibility rates of 20% to cefuroxime, 33% to 40% to cefdinir, and 40% to 76% to cefaclor. It is remarkable that among isolates without ftsI gene mutations, three β-lactamase-negative strains were nonsusceptible to ampicillin, and one β-lactamase-positive strain was nonsusceptible to amoxicillin-clavulanic acid.
TABLE 2.
Antimicrobial susceptibility of strains with (ftsI+) and without mutations in the ftsI gene distributed by β-lactamase production
| Antibiotic | Value (μg/ml) for strains without mutations
|
Value(μg/ml) for ftsI+ strains
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Lactamase negative (n = 10)
|
β-Lactamase positive (n = 21)
|
β-Lactamase negative (n = 121)
|
β-Lactamase positive (n = 44)
|
|||||||||
| MIC50 | MIC90 | %NS | MIC50 | MIC90 | %NS | MIC50 | MIC90 | %NS | MIC50 | MIC90 | %NS | |
| AMP | 1 | 2 | 30.0 | ≥128 | ≥128 | 100 | 2 | 4 | 98.3 | ≥128 | ≥128 | 95.5 |
| AMC | 0.5 | 4 | 0.0 | 2 | 4 | 4.8 | 2 | 4 | 9.9 | 4 | 8 | 25.0 |
| CEC | 8 | 32 | 40.0 | 32 | ≥128 | 76.2 | 32 | ≥128 | 70.2 | 32 | 64 | 86.4 |
| CXM | 1 | 8 | 20.0 | 2 | 8 | 19 | 4 | 16 | 48.8 | 4 | 16 | 45.4 |
| CDR | 1 | 4 | 40.0 | 0.5 | 4 | 33.3 | 2 | 8 | 57.0 | 2 | 4 | 52.3 |
| CTX | 0.03 | 0.25 | 0.0 | 0.03 | 0.06 | 0.0 | 0.06 | 0.5 | 0.8 | 0.03 | 0.25 | 0.0 |
| CDN | 0.03 | 0.06 | NA | 0.03 | 0.06 | NA | 0.06 | 0.06 | NA | 0.03 | 0.06 | NA |
aNonsusceptibility (NS) defined as MICs above susceptibility breakpoints. Susceptibility breakpoints (μg/ml) ≤1 μg/ml for ampicillin, ≤4/2 μg/ml for amoxicillin-clavulanic acid, ≤8 μg/ml for cefaclor, ≤4 μg/ml for cefuroxime, ≤1 μg/ml for cefdinir, and ≤2 μg/ml for cefotaxime (2). NA, no CLSI breakpoints available; AMP, ampicillin; AMC, amoxicillin-clavulanic acid; CEC, cefaclor; CXM, cefuroxime; CDR, cefdinir; CTX, cefotaxime; CDN, cefditoren.
Among the 21 β-lactamase-positive isolates without mutations in the ftsI gene, 19 (90.5%) isolates produced TEM-1, concomitantly with ROB-1 in two strains. The remaining two strains were positive by the chromogenic cephalosporin test, but TEM and ROB β-lactamases were not detected. Among the 44 β-lactamase-positive isolates with mutations in the ftsI gene (BLPACR isolates), 37 (84.1%) strains produced TEM-1, and 8 (18.2%) produced ROB-1, with two strains (4.5%) producing both β-lactamases. The remaining strain was positive by the chromogenic cephalosporin test, but TEM and ROB β-lactamases were not detected. Of the eight BLPACR strains producing ROB-1 β-lactamase, five were from the pediatric hospital (H. S. Joan de Deu), representing 38.5% (5 out of 13) of the BLPACR strains from that center.
Nonsusceptibility rates in strains showing mutations in the ftsI gene were similar regardless of β-lactamase production, except in the case of amoxicillin-clavulanic acid, where nonsusceptibility rates (MIC ≥ 8/4 μg/ml; resistance, in this case, since no intermediate category is CLSI defined) increased from 9.9% in β-lactamase-negative strains with mutations in the ftsI gene (ftsI+) to 25% in ftsI+ β-lactamase-positive strains (Table 2). More than 50% of these ftsI+ β-lactamase-positive strains (MIC range, 8/4 to 32/16 μg/ml) were from the pediatric hospital H. S. Joan de Deu. This hospital also showed the highest prevalence of β-lactamase production among ftsI+ strains (13/31 [41.9%], versus 31/165 [18.8%] in general hospitals) (Table 1) and the highest prevalence of amoxicillin-clavulanic acid resistance (MIC ≥ 8/4 μg/ml) among ftsI+ β-lactamase-positive strains (6/13 [46.2%], versus 5/31 [16.1%] in general hospitals). This may be related to different patterns of antibiotic consumption in children and adults. It has been suggested to be due to a relationship between amoxicillin-clavulanic acid consumption and the evolution of BLNAR strains (4), which can be extended to BLPACR strains and children, with higher amoxicillin-clavulanic acid consumption.
Cefotaxime exhibited MIC50/MIC90s of ≤0.06/≤0.5 μg/ml and cefditoren values of ≤0.06/≤0.06 μg/ml regardless the presence of ftsI mutations and/or β-lactamase production. Only one BLNAR strain was nonsusceptible to cefotaxime (cefotaxime MIC = 4 μg/ml, cefditoren MIC = 0.06 μg/ml).
Eleven strains presented changes that predicted early termination of the acrR reading frame. Of them, 10 strains had ftsI mutations (one of them is a β-lactamase producer). These strains, presumably hyperproducers of an AcrAB efflux pump, did not present higher ampicillin MICs, suggesting, as in previous studies (4), unrelatedness to high-level ampicillin resistance.
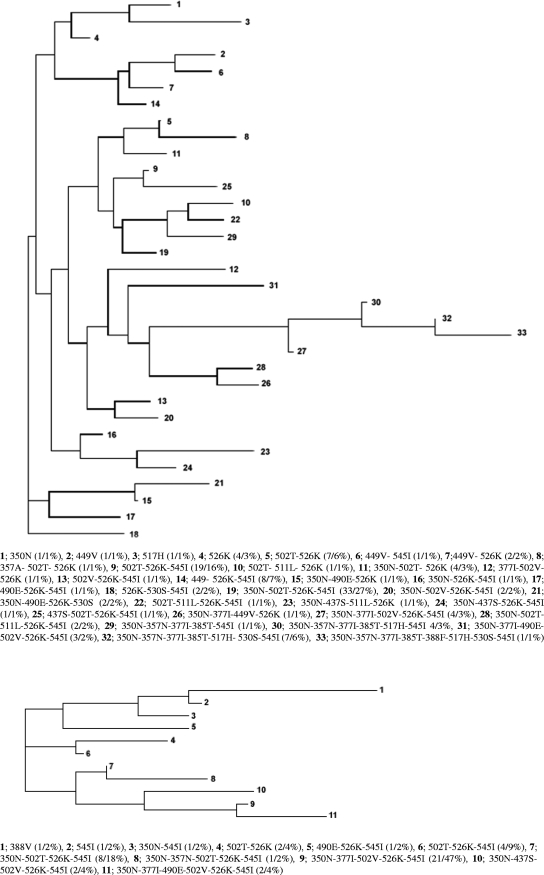
Figure 1 shows phylogenetic trees of a 1,030-bp sequence of the ftsI gene. The most frequent group was IIc in BLNAR (62/121 strains [51.2%]) and IIb in BLPACR (25/44 strains [56.8%]). Among BLPACR strains, the number of patterns of amino acid substitutions in the ftsI gene was 11, with 47% of isolates belonging to one single pattern (350N, 377I, 502V, 526K, 545I [phylogenetic no. 9]) that represented only 3% (phylogenetic no. 27) of BLNAR strains. Among BLNAR strains, the amino acid substitution profile 350N, 502T, 526K, 545I was the most prevalent pattern (27%) that was also represented in BLPACR strains (18%). BLPACR strains showed a closer phylogenetic relationship than did BLNAR strains, among which 33 patterns were found, only two of them (different than the prevalent pattern in BLPACR strains) showing >10% prevalence.
FIG. 1.
Phylogenetic trees of a 1,030-bp sequence from the ftsI gene of ftsI+ β-lactamase-negative isolates (top tree) and of ftsI+ β-lactamase-positive isolates (bottom tree). Phylogenetic numbers, amino acid substitutions, numbers and percentages of strains are shown below each phylogenetic tree.
The study of the prevalence of resistance mechanisms (β-lactamase and ftsI gene mutations) to β-lactams in H. influenzae and its genetic relatedness may help to establish adequate therapeutic and preventive measures to counter their selection/diffusion.
Acknowledgments
L.M.-M. is supported by Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III, Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008). This study was supported by an unrestricted grant from Tedec-Meiji Farma S.A., Madrid, Spain.
We thank M. Casal and J. L. Gómez-Garcés for participating in the study and M. Gimeno for her collaboration.
Footnotes
Published ahead of print on 27 October 2008.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard. CLSI document M7-A7. Clinical Laboratory Standards Institute, Wayne, PA.
- 2.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Dabernat, H., C. Delmas, M. Seguy, R. Pelissier, G. Faucon, S. Bennamani, and C. Pasquier. 2002. Diversity of beta-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae. Antimicrob. Agents Chemother. 46:2208-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Cobos, S., J. Campos, E. Lázaro, F. Román, E. Cercenado, C. García-Rey, M. Pérez-Vázquez, J. Oteo, and F. de Abajo. 2007. Ampicillin-resistant non-beta-lactamase-producing Haemophilus influenzae in Spain: recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob. Agents Chemother. 51:2564-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.García-de-Lomas, J., M. Lerma, L. Cebrián, J. L. Juan-Bañón, P. Coronel, M. J. Giménez, and L. Aguilar. 2007. Influence of Haemophilus influenzae beta-lactamase production and/or ftsI gene mutations on in vitro activity of and susceptibility rates to aminopenicillins and second- and third-generation cephalosporins. Int. J. Antimicrob. Agents 30:190-192. [DOI] [PubMed] [Google Scholar]
- 6.Kaczmarek, F. S., T. D. Gootz, F. Dib-Hajj, W. Shang, S. Hallowell, and M. Cronan. 2004. Genetic and molecular characterization of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae with unusually high resistance to ampicillin. Antimicrob. Agents Chemother. 48:1630-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Callaghan, C. H., S. M. Kirby, A. Morris, R. E. Waller, and R. E. Duncombe. 1972. Correlation between hydrolysis of the β-lactam bond of the cephalosporin nucleus and expulsion of the 3-substituent. J. Bacteriol. 110:988-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scriver, S. R., S. L. Walmsley, C. L. Kau, D. J. Hoban, J. Brunton, A. McGeer, T. C. Moore, E. Witwicki, et al. 1994. Determination of antimicrobial susceptibilities of Canadian isolates of Haemophilus influenzae and characterization of their beta-lactamases. Antimicrob. Agents Chemother. 38:1678-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ubukata, K., Y. Shibasaki, K. Yamamoto, N. Chiba, K. Hasegawa, Y. Takeuchi, K. Sunakawa, M. Inoue, and M. Konno. 2001. Association of amino acid substitutions in penicillin-binding protein 3 with beta-lactam resistance in beta-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 45:1693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]