Abstract
A detailed kinetic characterization of echinocandin inhibition was performed for mutant 1,3-β-d-glucan synthase enzymes from clinical isolates of Candida albicans with nine different FKS1 mutations resulting in high MICs. Among 14 mutant Fks1p enzymes studied, the kinetic parameters 50% inhibitory concentration and Ki increased 50-fold to several thousandfold relative to those for the wild type. Enzymes with mutations at Ser645 (S645P, S645Y, and S645F) within hot spot 1 showed the most prominent decrease in sensitivity, while those with mutations at the N- and C-terminal ends of hot spot 1 generally retained greater sensitivity to all three drugs. Kinetic inhibitions by caspofungin, micafungin, and anidulafungin were comparable among the fks1 mutant enzymes, although absolute values did vary with specific mutations. Amino acid substitutions in Fks1p did not alter Km values, although some mutations decreased the Vmax. Given the association of FKS1 mutations with clinical resistance, an evaluation of the kinetic parameters for the inhibition of mutant 1,3-β-d-glucan synthase as a function of the MIC enabled an independent evaluation of the recently adopted susceptibility breakpoint for echinocandin drugs. Overall, a breakpoint MIC of ≥2 μg/ml for caspofungin captured nearly 100% of fks1 C. albicans strains when a kinetic inhibition rise threshold of ≤50-fold for the Ki was used as a measure of susceptibility. A similar MIC breakpoint for micafungin and anidulafungin was less inclusive, and a projected MIC of ≥0.5 μg/ml was required for >95% coverage of clinical isolates. However, when MIC determinations were performed in the presence of 50% serum, all fks1 mutants showed MIC values of ≥2 μg/ml for the three echinocandin drugs. The 1,3-β-d-glucan synthase kinetic inhibition data support the proposed susceptibility breakpoint for caspofungin in C. albicans, but a lower susceptibility breakpoint (≤0.5 μg/ml) may be more appropriate for anidulafungin and micafungin. Overall, the data indicate that MIC testing with caspofungin may serve as a surrogate marker for resistance among the class of echinocandin drugs.
The introduction of echinocandin class drugs was an important advance in the treatment of systemic fungal infections by reducing the problems of cross-resistance to azoles and toxicity associated with polyene drugs (45, 46, 51). Echinocandins inhibit 1,3-β-d-glucan synthase (EC 2.4.1.34), which catalyzes the biosynthesis of 1,3-β-d-glucan, the major glucan component of fungal cell walls (10, 31). 1,3-β-d-Glucan synthase is an enzyme complex with at least two subunits, Fksp and Rho1p. The latter is a regulatory element involved in a number of cellular processes (23, 44). Fksp, encoded by three related genes, FKS1, FKS2, and FKS3, contains the active site, which catalyzes the transfer of sugar moieties from activated donor molecules (UDP glucose [UDPG]) to specific acceptor molecules [(1,3)-β-d-glucosyl(N)] forming glycosidic bonds (23, 47, 49). The inhibition of 1,3-β-d-glucan synthesis by echinocandin drugs disrupts the structure of the growing cell wall, resulting in osmotic instability and the death of susceptible yeast cells (5, 21). In recent years, a great deal has been learned about echinocandin pharmacokinetics, safety, spectrum, efficacy, and resistance (2, 4, 16, 17, 27, 35, 48, 50, 52). In contrast, the biochemistry and kinetics of 1,3-β-d-glucan synthase are poorly understood, with limited data obtained from crude lysates, detergent-activated enzymes, or microsomal fractions (11, 12). Furthermore, the biochemical mechanism of inhibition of 1,3-β-d-glucan synthase by echinocandin drugs remains largely unknown.
Echinocandin resistance in susceptible species like Candida albicans is uncommon, but it has been associated with amino acid substitutions in two conserved regions of Fks1p (35). These mutations, which result in elevated MICs, reduce the sensitivity of 1,3-β-d-glucan synthase to drug by several hundredfold to a thousandfold (33). The biochemical mechanism of this reduced sensitivity is not understood. There is a complex relationship between elevated MIC and clinical success (20, 39), largely because cells can induce a variety of adaptive pathways that lead to reduced susceptibility without directly impacting the behavior of the drug on 1,3-β-d-glucan synthase (20, 25, 26). Furthermore, in vitro MIC assays of echinocandin susceptibility according to CLSI protocol M27-A3 do not take into account the strong effects of serum protein binding, which influences the relative efficacies of echinocandin drugs (30, 32, 53). Nevertheless, the CLSI Antifungal Subcommittee proposed an echinocandin MIC breakpoint for susceptibility of ≤2 μg/ml for Candida spp. (40). To assess the relationship between the new breakpoint and the inherent sensitivity of 1,3-β-d-glucan synthase from fks1 mutants to echinocandin drugs, a detailed kinetic analysis of mutant and wild-type enzymes was undertaken to compare kinetic inhibition parameters with the MICs for clinical and laboratory isolates of C. albicans.
MATERIALS AND METHODS
Strains and compounds.
Fifteen C. albicans clinical strains, three control C. albicans strains, and two spontaneous mutants isolated with anidulafungin (ANF) (Pfizer, New York, NY) were used in this study (Table 1). Moreover, three laboratory-generated fks1 mutant strains of C. albicans reported previously were utilized only for FKS expression profiling (3). Twelve of the clinical isolates and the spontaneous mutants showed reduced echinocandin susceptibility (MIC ≥ 2 μg/ml), and three strains (strains 1002, 3107, and 3795) were echinocandin susceptible. Two of the echinocandin-susceptible clinical strains were fluconazole resistant (strain 3107 harbors the Y132F amino acid change in Erg11p, and strain 3795 showed CDR2 and MDR2 overexpression) (34). C. albicans strains SC5314, ATCC 90028, and ATCC 36082 were used as control strains. ANF, caspofungin (CSF) (Merck & Co., Inc., Rahway, NJ), and micafungin (MCF) (Astellas Pharma USA, Inc., Deerfield, IL) were obtained as standard powders from their manufacturers. CSF and MCF were dissolved in sterile distilled water, while ANF was dissolved in 100% dimethyl sulfoxide (Sigma-Aldrich). Stock solutions of each drug were kept at −86°C.
TABLE 1.
In vitro whole-cell susceptibility (MIC) and GS inhibition profiles (IC50) for ECD of the C. albicans strains included in the studyh
| Strain | FKS1 genotype | FKS1 HS1a | FKS1 HS2b | MIC (μg/ml)c
|
IC50 (ng/ml)d
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANF
|
CSF
|
MCF
|
ANF | CSF | MCF | |||||||
| No serum | 50% serum | No serum | 50% serum | No serum | 50% serum | |||||||
| SC5314e | WT | WT | WT | 0.08 | 0.50 | 0.42 | 0.50 | 0.05 | 0.50 | 0.89 (11.41) | 3.88 (64.13) | 58.2 (198.6) |
| 36082e | WT | WT | WT | 0.03 | 0.12 | 0.21 | 0.25 | 0.03 | 1.00 | 1.51 (20.1) | 0.9 (16.5) | 10.2 (31.3) |
| 90028e | WT | WT | WT | 0.03 | 0.25 | 0.21 | 0.25 | 0.03 | 0.50 | 1.83 (25.4) | 0.5 (9.4) | 18.8 (60.1) |
| 1002f | WT | WT | WT | 0.06 | 0.44 | 0.33 | 0.50 | 0.04 | 0.50 | 7.96 | 1.75 | 53.34 |
| 3107f | WT | WT | WT | 0.03 | 0.25 | 0.25 | 0.33 | 0.03 | 0.50 | 3.12 | 3.62 | 14.72 |
| 3795f | WT | WT | WT | 0.06 | 0.44 | 0.33 | 0.50 | 0.03 | 0.50 | 6.76 | 1.44 | 38.89 |
| 119f | T1921C | F641L | WT | 0.16 | 2.00 | 2.00 | 8.00 | 0.33 | 8.00 | 11.3 | 10.03 | 183.6 |
| 177f | T1922C | F641S | WT | 0.83 | 4.00 | 4.00 | 16.0 | 1.00 | 16.0 | 2622 | 1091 | 1441 |
| 2762f | T1922C | F641S | WT | 1.00 | 4.00 | 3.33 | 8.00 | 1.00 | 16.0 | 1085 | 910 | 1693 |
| 205f | T1933C | S645P | WT | 1.33 | 8.00 | 8.00 | 8.00 | 4.00 | 8.00 | 989 | 245.4 | 1085 |
| 5415f | T1933C | S645P | WT | 2.00 | 8.00 | 8.00 | 8.00 | 4.00 | 8.00 | 1152 | 698.1 | 1254 |
| 89f | C1934A | S645Y | WT | 2.00 | 8.00 | 8.00 | 8.00 | 4.00 | 8.00 | 2739 | 2075 | 2533 |
| 85f | C1934T | S645F | WT | 2.00 | 8.00 | 4.00 | 8.00 | 2.67 | 8.00 | 787 | 459.1 | 449.8 |
| 149f | G1942T | D648Y | WT | 0.83 | 4.00 | 2.67 | 4.00 | 0.83 | 4.00 | 93.3 | 68.22 | 375.5 |
| 122f | C1946A | P649H | WT | 0.67 | 8.00 | 4.00 | 4.00 | 0.83 | 4.00 | 491 | 82.66 | 924.6 |
| 121f | G4082A | WT | R1361H | 0.25 | 8.00 | 2.00 | 8.00 | 0.25 | 8.00 | 75.99 | 33.72 | 498.2 |
| 194f | C1934T/G4082R | S645F | R1361R/H | 4.00 | 8.00 | 4.00 | 8.00 | 2.67 | 8.00 | 782 | 530.7 | 1765 |
| 90f | G4082R | WT | R1361R/H | 0.25 | 4.00 | 1.00 | 4.00 | 0.50 | 4.00 | 40.6 (1.01) | 20.43 (2.05) | 77.39 (22.5) |
| A15g | T1933Y | S645S/P | WT | 0.50 | 2.00 | 4.00 | 8.00 | 0.50 | 2.00 | 128.7 (0.39) | 218.1 (4.19) | 177.3 (35.5) |
| A15-10g | T1933C | S645P | WT | 4.00 | 8.00 | 8.00 | 16.0 | 4.00 | 8.00 | 901 | 322.4 | 1194 |
HS1, hot spot 1 (FKS1 amino acids 641 and 649). The wild-type sequence is 41-FLTLSLRDP-649.
HS2, hot spot 2 (FKS1 amino acids 1357 and 1364). The wild-type sequence is 1357-DWIRRTYL-1364.
Shown are geometric means (three repetitions on three separate days).
IC50 values obtained using trapped 1,3-β-d-glucan synthase enzyme. Shown are arithmetic means (three repetitions on three separate days). IC50 values obtained using crude membranes (strains SC5314, 36082, and 90028) and IC501 (susceptible allele) using a two-site competition-fitting algorithm (strains 90 and A15) are shown in parentheses.
Control strains.
Clinical strains.
Spontaneous mutants derived from strain SC5314. A15 were obtained by selection with 0.5 μg/ml ANF (paternal strain SC5314), and A15-10 was selected using 8 μg/ml ANF (A15 paternal strain).
WT, wild type.
Isolation of echinocandin-resistant spontaneous C. albicans mutants.
Spontaneous echinocandin-resistant mutants of C. albicans strain SC5314 were isolated using a two-step methodology. In the first step, 108 yeast cells were plated onto YPD agar plates (2% yeast extract, 4% Bacto peptone, and 4% dextrose) containing 0.5 μg/ml of ANF. The mutants isolated were subsequently plated onto YPD agar with 8 μg/ml of ANF. Selection plates were incubated for 7 days at 37°C. Resistant colonies were selected and regrown on fresh ANF-containing plates (0.5 or 8 μg/ml) to confirm the phenotype. Echinocandin MICs were obtained as described hereafter.
Antifungal susceptibility testing.
Susceptibility testing was performed in triplicate in accordance with CLSI document M27-A3 (7) in the presence or absence of 50% human serum (Sigma) (32). Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019 were used as quality control strains.
FKS gene sequence analysis.
The C. albicans FKS1 and FKS2 genes were amplified and sequenced from genomic DNA extracted from C. albicans cells grown overnight in YPD broth medium with the Q-Biogene (Irvine, CA) FastDNA kit. PCR and sequencing primers were designed based on the C. albicans FKS1 and FKS2 sequences (GenBank accession no. XM_716336 and XM_712867, respectively). DNA sequencing was performed with a CEQ dye terminator sequencing kit (Beckman Coulter, Fullerton, CA) according to the manufacturer's recommendations. Sequence analyses were performed with CEQ 8000 genetic analysis system software (Beckman Coulter, Fullerton, CA) and BioEdit Sequence Alignment Editor (Ibis Therapeutics, Carlsbad, CA).
C. albicans 1,3-β-d-glucan synthase isolation and inhibition assay.
All the isolates used in this work were grown with vigorous shaking at 37°C to early stationary phase in YPD broth, and cells were collected by centrifugation. Cell disruption, membrane protein extraction, and 1,3-β-d-glucan synthase enrichment by-product entrapment were done as described previously (33). Sensitivity to echinocandin drugs was measured in a polymerization assay using a 96-well multiscreen HTS filtration system (Millipore Corporation, Bedford, MA) in a final reaction mixture volume of 100 μl, as previously described (33). Serial dilutions of ANF, CSF, and MCF were added (1 μl/well). Control reactions were performed in the presence of 1% dimethyl sulfoxide when ANF was used. All reactions were initiated by the addition of the product-entrapped 1,3-β-d-glucan synthase enzyme to the reaction mixture. Inhibition curves and 50% inhibitory concentrations (IC50s) were determined using a sigmoidal response (variable-slope) curve and a two-site competition-fitting algorithm with GraphPad Prism, version 4.0, software (Prism Software, Irvine, CA).
Characterization of the 1,3-β-d-glucan product.
The product of the reaction mixtures was characterized as 1,3-β-d-glucan using a Glucatell kit (Associates of Cape Cod Inc., Falmouth, MA) according to the manufacturer's instructions. The addition of the 1,3-β-d-glucan synthase-trapped enzyme was used to initiate the reaction in a 100-μl final volume. Reaction mixtures were incubated at 25°C for 60 min and were then stopped by rapid cooling on ice. Using the end-point assay Glucatell kit and comparing the results obtained with those using the [3H]UDPG incorporation assay, it was established that 2.4 × 10−2 nmol of glucose was incorporated (∼10 pg of 1,3-β-d-glucan/ml).
Kinetic analyses.
All reactions were run with a 96-well multiscreen HTS filtration system (Millipore) in a final volume of 100 μl. Each well contained 50 mM HEPES (pH 7.5), 10% (wt/vol) glycerol, 1.5 mg/ml bovine serum albumin, 25 mM KF, 1 mM EDTA, 25 μM GTP-γ-S, 1 μg 1,3-β-d-glucan synthase enzyme, [3H]UDPG (7,000 dpm/nmol glucose), and echinocandin drugs, as indicated below. The plates were incubated for 60 min at 25°C. All reactions were initiated by the addition of product-entrapped 1,3-β-d-glucan synthase to the mixture. [3H]UDPG was used as the substrate in concentrations varying from 0.015 to 2 mM to determine the different kinetic parameters, which were analyzed by linear regression to obtain slopes in dpm/min. This value was then converted to nM glucose incorporated per minute. The maximum velocity (Vmax) and the Michaelis-Menten constant (Km) were determined for product-entrapped 1,3-β-d-glucan synthase enzyme by varying the amount of UDPG (between 0.015 and 2 mM) using Lineweaver-Burke plots. Furthermore, Dixon plots were used to determine the inhibition constant (Ki) and the nature of kinetic inhibition by echinocandin drugs. Ki values were calculated by varying the echinocandin drug concentration (between 0.01 and 50 ng/ml for wild-type GS and between 10 and 10,000 ng/ml for mutant GS) at different fixed substrate concentrations ranging from 0.125 to 0.5 mM UDPG. All data were analyzed with GraphPad Prism 4.0 software (Prism Software).
RNA isolation and expression profiling.
C. albicans strains were grown in YPD broth and incubated at 37°C with shaking (150 rpm) for 16 h. Total RNA was extracted using the RNeasy minikit (Qiagen), and gene expression profiles were performed using the one-step Sybr green QRT-PCR kit (Stratagene, La Jolla, CA) using the Stratagene Mx3005P multiplex quantitative PCR system. Differential expression was analyzed for the three C. albicans FKS genes. FKS1, FKS2, and FKS3 expression profiling primers were designed using GenBank accession numbers XM_716336, XM_712867, and XM_713421, respectively (Table 2). Relative expression was evaluated using a method described previously by Pfaffl (36). The C. albicans URA3 gene (GenBank accession no. XP_721787.1) was used for the normalization of expression (13).
TABLE 2.
Primers used in this study
| Primer | Orientation (5′→3′) | Sequence (5′→3′) |
|---|---|---|
| CaFKS1expF | Sense | TGATACTGGTAATCATAGACCAAAAA |
| CaFKS1expR | Antisense | AACTCTGAATGGATTTGTAGAATAAGG |
| CaFKS2expF | Sense | ACTTGCTAGCAGTCGCCAAT |
| CaFKS2expR | Antisense | ACCACCATGAGCGGTTAGAC |
| CaFKS3expF | Sense | ACCTCAATATTCAGCTTGGTGCCC |
| CaFKS3expR | Antisense | GGACAACTCATTCGACTTGACCGT |
| CaURA3F | Sense | CAACACTAAGACCTATAGTGAGAGAGC |
| CaURA3R | Antisense | TGCACATAAATTGGTTTTCTTCA |
Statistical and kinetic breakpoint analysis.
Kinetic and susceptibility testing was performed in triplicate. Arithmetic means and standard deviations were used to statistically analyze all the continuous variables studied (IC50s, Km, Vmax, and Ki). Geometric means were used to statistically compare MIC results. Significant differences in MICs and kinetic parameters were determined by a Student's t test (unpaired and unequal variance); a P value of <0.05 was considered to be significant. In order to approximate a normal distribution, the MICs were transformed to log2 values to establish susceptibility differences between strains. Both on-scale and off-scale results were included in the analysis. The off-scale MICs were converted to the next concentration up or down. MICs were compared with Ki values using plots of the log2 MIC versus log10 Ki. The MIC-Ki breakpoint was defined as the MIC for which ≥90% of the strains had Ki values for 1,3-β-d-glucan synthase at least 100-fold higher than that for the wild-type enzyme. Statistical analysis was done with the Statistical Package for the Social Sciences (version 13.0; SPSS Inc., Chicago, IL).
RESULTS
Correlation between MICs and FKS1 mutations.
All strains (wild type and fks1 mutants) exhibited statistically significantly higher MICs for CSF than for the other drugs in the absence of serum (P = 0.0001 for CSF versus ANF and P = 0.0006 for CSF versus MCF). However, the MICs of each drug varied with the specific FKS1 mutations (Table 1). The fks1 mutant isolates showed 4- to 30-fold MIC increases for CSF, while ANF and MCF showed 90-fold and 110-fold increases, respectively, relative to those of standard laboratory and clinical wild-type strains (n = 6). The most significant MIC increases (8- to >100-fold) were related to amino acid changes at Ser645 (S645P, S645F, and S645Y), whereas the other mutations accounted for smaller increases (4- to 30-fold). The average increases in MIC for the fks1 mutant strains (n = 14) were 52.57, 26.41-, and 14.57-fold for MFG, ANF, and CSF, respectively. The spontaneous ANF mutants isolated showed MICs comparable to those of clinical isolates harboring the same mutations (Table 1). The heterozygous mutant A15 (S645S/P) showed intermediate MICs comparable to those obtained from clinical strains with F641L, D648Y, P649H, R1361R/H, and R1361H Fks1p amino acid changes. The efficacy of echinocandin drugs is influenced by human serum, which is absent in standard in vitro assays (30, 32, 53). To evaluate the antifungal efficacies of echinocandin drugs in the presence of human serum, MICs were also determined in the presence of 50% serum. As previously reported (30, 32, 53), serum neutralized most potency differences between drugs observed in vitro assays. The CSF MIC in the presence of 50% serum increased 2.3-fold on average for mutant strains. On the other hand, serum had a more pronounced and statistically significant effect on the other drugs showing an MIC increase ranging from 4- to 32-fold for ANF and MCF (P < 0.001).
Effect of Fks1p substitutions on 1,3-β-d-glucan synthase inhibition.
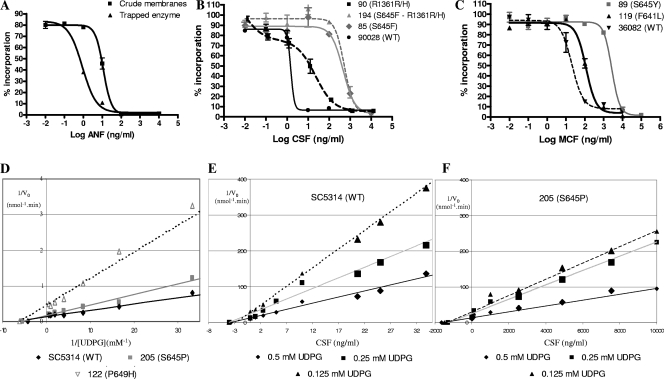
Evaluation of kinetic inhibition (IC50) for wild-type crude microsomes and product-entrapped enzyme demonstrated that the entrapped enzyme was approximately 10-fold more sensitive to all echinocandin drugs (Fig. 1A and Table 1). Moreover, the IC50 values obtained using different crude microsome preparations were highly variable (variation coefficient of 34.5% on average) (data not shown), making this preparation less reliable for kinetic studies. 1,3-β-d-Glucan synthase enzymes isolated from fks1 mutant strains showed higher IC50 values for ANF (243-fold) than for the other drugs (29- and 14-fold for CSF and MCF, respectively). Among the 14 mutant Fks1p enzymes studied, mutations at Ser645 (S645P, S645Y, and S645F) showed the highest IC50 increases relative to the wild type (P < 0.01). On the other hand, F641S mutants showed higher IC50 values than did F641L mutants (Fig. 1B). As demonstrated previously (33), the heterozygous R1361H mutants (strains 90 and A15) showed two IC50 values (two inflection points in the inhibition curve), demonstrating that the two isoenzymes (mutant and wild type) are present in the extracts (Fig. 1C). The spontaneous ANF mutants showed IC50 values comparable to those of clinical isolates carrying the same mutations (Table 1).
FIG. 1.
(A) ANF titration curves for strain SC5314 (wild type) 1,3-β-glucan synthase in crude microsomal membranes or in a trapped-enzyme preparation. (B) CSF titration curves for strains ATCC 90028 (wild type [WT]), 90 (R1361R/H), 85(S645F), and 194 (S645F and R1361R/H). (C) MCF titration curves for strains ATCC 36082 (wild type), 119 (F641L), and 177 (F641S). (D) Lineweaver-Burke double-reciprocal plotting for wild-type strain SC5314, Fks1p mutant S645P strain 205, and Fks1p mutant P649H strain 122. (E and F) Dixon plotting using 0.5 mM, 0.25 mM, and 0.125 mM UDPG for the 1,3-β-glucan synthase-trapped enzyme isolated from strains SC5314 (E) and strain 205 (F).
Enzyme kinetics.
A comparison of the kinetic properties of 1,3-β-d-glucan synthases from the wild-type strain (SC5314) to those from an isogenic Fks1-S645P mutant (A15-10) shows that this mutation decreases echinocandin susceptibility but also decreases the catalytic Vmax approximately 50% relative to that of the wild type (Table 3). The Km and Vmax values for the wild-type enzyme are comparable to average values of 0.099 ± 0.022 mM and 5.962 ± 0.723 nmol/min, respectively, obtained for six wild-type (sensitive) enzymes (Table 4), highlighting the consistency of the various enzyme preparations. The Km values obtained for wild-type and Fks1p mutant enzymes were not statistically different (P = 0.47), indicating that amino acid substitutions decreasing sensitivity do not alter substrate binding. In contrast, the Vmax values of all Fks1p mutant enzymes were significantly lower than those of wild-type Fks1s (P = 0.0013) (Table 4). Among the mutant enzymes, the amino acid substitutions F641S and S645P showed the highest relative Vmax values (Fig. 1D).
TABLE 3.
In vitro properties of the trapped 1,3-β-glucan synthase enzyme from the C. albicans strain SC5314 and its spontaneous echinocandin-resistant mutant A15-10
| Kinetic property | Value(s)
|
|
|---|---|---|
| SC5314 (parental) | A15-10 (S645P) | |
| Mean Km (mM) ± SD | 0.125 ± 0.009 | 0.091 ± 0.021 |
| Mean Vmax (nmol·min−1) ± SD | 6.717 ± 0.246 | 3.263 ± 0.115 |
| Mean Ki (ng/ml) ± SD | ||
| ANF | 1.33 ± 0.56 | 2563.7 ± 11.1 |
| CSF | 2.97 ± 1.58 | 1358.0 ± 25.4 |
| MCF | 27.47 ± 1.82 | 2249.5 ± 54.5 |
| Linear with time (min) | 30-90 | 30-90 |
| Linear with protein (μg) | 0.25-2.5 | 0.25-2.5 |
TABLE 4.
Kinetic properties and Ki values of strains included in the studye
| Strain | Genotype | FKS1
|
Mean Vmax (nmol·min−1)c ± SD | Mean Km (mM)c ± SD | Mean Ki (ng/ml)c ± SD
|
|||
|---|---|---|---|---|---|---|---|---|
| HS1a | HS2b | ANF | CSF | MCF | ||||
| 5314 | WT | WT | WT | 6.717 ± 0.246 | 0.125 ± 0.009 | 1.33 ± 0.56 | 2.97 ± 1.58 | 27.47 ± 1.82 |
| 36082 | WT | WT | WT | 5.872 ± 0.199 | 0.089 ± 0.005 | 1.43 ± 0.14 | 0.95 ± 0.05 | 23.53 ± 2.37 |
| 90028 | WT | WT | WT | 4.715 ± 0.479 | 0.067 ± 0.005 | 0.63 ± 0.18 | 0.46 ± 0.04 | 15.47 ± 2.05 |
| 1002 | WTd | WT | WT | 5.798 ± 0.264 | 0.122 ± 0.004 | 2.39 ± 0.30 | 6.26 ± 0.90 | 34.52 ± 1.32 |
| 3107 | Erg11 (Y132F)d | WT | WT | 6.038 ± 0.117 | 0.102 ± 0.004 | 1.67 ± 0.26 | 8.79 ± 2.54 | 41.87 ± 12.86 |
| 3795 | CDR2 and MDR2d | WT | WT | 6.630 ± 0.085 | 0.088 ± 0.003 | 16.70 ± 7.27 | 4.67 ± 0.51 | 58.64 ± 11.06 |
| 119 | T1921C | F641L | WT | 2.545 ± 0.163 | 0.054 ± 0.003 | 6.77 ± 0.22 | 4.92 ± 1.00 | 155.90 ± 14.53 |
| 177 | T1922C | F641S | WT | 4.401 ± 0.240 | 0.050 ± 0.004 | 4342.33 ± 229.61 | 1817.60 ± 3.48 | 4404.23 ± 42.10 |
| 2762 | T1922C | F641S | WT | 3.571 ± 0.403 | 0.108 ± 0.011 | 4481.27 ± 81.83 | 498.93 ± 53.18 | 1447.88 ± 108.66 |
| 205 | T1933C | S645P | WT | 5.540 ± 0.184 | 0.116 ± 0.005 | 3223.10 ± 99.65 | 1480.00 ± 172.55 | 2673.63 ± 118.85 |
| 5415 | T1933C | S645P | WT | 3.121 ± 0.118 | 0.098 ± 0.009 | 4154.11 ± 55.78 | 2122.49 ± 109.32 | 2753.01 ± 25.57 |
| 89 | C1934A | S645Y | WT | 2.507 ± 0.177 | 0.061 ± 0.002 | 1163.37 ± 52.76 | 860.00 ± 34.12 | 867.13 ± 10.95 |
| 85 | C1934T | S645F | WT | 1.826 ± 0.153 | 0.025 ± 0.005 | 445.93 ± 20.07 | 519.33 ± 17.21 | 1289.19 ± 42.60 |
| 149 | G1942T | D648Y | WT | 1.220 ± 0.150 | 0.032 ± 0.003 | 170.55 ± 26.19 | 200.00 ± 12.53 | 489.23 ± 2.14 |
| 122 | C1946A | P649H | WT | 1.951 ± 0.292 | 0.152 ± 0.012 | 646.07 ± 26.72 | 291.67 ± 50.16 | 2588.53 ± 32.25 |
| 121 | G4082A | WT | R1361H | 4.405 ± 0.262 | 0.129 ± 0.013 | 84.57 ± 4.08 | 308.00 ± 19.67 | 840.48 ± 28.74 |
| 194 | C1934T/G4082R | S645F | R1361R/H | 5.810 ± 1.018 | 0.061 ± 0.006 | 299.24 ± 11.94 | 466.89 ± 8.80 | 493.83 ± 27.42 |
| 90 | G4082R | WT | R1361R/H | 2.358 ± 0.245 | 0.031 ± 0.006 | 48.50 ± 6.46 | 148.15 ± 3.67 | 264.73 ± 11.97 |
| A15-10 | T1933C | S645P | WT | 3.263 ± 0.115 | 0.091 ± 0.021 | 2563.7 ± 11.1 | 1358.0 ± 25.4 | 2249.5 ± 54.5 |
HS1, hot spot 1 (FKS1 amino acids 641 and 649). The wild-type sequence is 641-FLTLSLRDP-649.
HS2, hot spot 2 (FKS1 amino acids 1357 and 1364). The wild-type sequence is 1357-DWIRRTYL-1364.
Arithmetic means ± standard deviations (three repetitions).
See reference 34.
WT, wild type.
Kinetic inhibition by the three echinocandin drugs was assessed against wild-type and fks1 mutant enzymes (Table 4). Dixon plots of enzyme inhibition were linear, and all drugs showed noncompetitive inhibition (9, 49) (Fig. 1E and F). The inhibition constant (Ki) values for all enzymes and each drug were evaluated. The ANF Ki values were the highest among all fks1 mutant 1,3-β-d-glucan synthases, showing a ≥100-fold increase compared with wild-type enzymes. The mutations conferring the highest Ki values for all three echinocandin drugs were F641S, S645P, and S645Y. For CSF, the highest Ki values were observed in mutants with amino acid changes at Ser645 (S645P, S645F, and S645Y). Mutations at the C-terminal end of Fks1 hot spot 1 (D648Y and P649H) confer higher MCF Ki values than those for the other drugs tested. For Fks1 hot spot 2 amino acid substitutions, the R1361H 1,3-β-d-glucan synthase mutant showed statistically significantly higher Ki values than wild-type 1,3-β-d-glucan synthases (P < 0.001). However, the CSF Ki was three times higher than those of ANF and MCF. As expected, the R1361H homozygous mutant (strain 121) showed higher Ki values than did the heterozygous mutant (strain 90) for all three echinocandin drugs tested. Finally, in compound mutant strain 194 (S645F-R1361R/H), ANF and MCF Ki values were lower than that of the S645F mutant alone (strain 85), indicating that the two resistance sites do not synergize.
Expression profiling.
Real-time PCR was used to evaluate transcription levels for the three different C. albicans FKS genes. Table 5 indicates that FKS1 is expressed 2.92- and 26.9-fold more on average than FKS2 and FKS3, respectively. The level of expression for FKS2 and FKS3 increased when FKS1 harbored a resistance mutation. The spontaneous mutants selected from strain SC5314 using ANF (this study) (Table 1) and CSF (3, 33) showed similar decreases in expressed FKS1/FKS2 and FKS1/FKS3 ratios, indicating that mutations in FKS1 affect the regulation of all three genes. Heterozygous mutants (S645S/P) showed FKS1/FKS2 and FKS1/FKS3 ratios intermediate to those obtained using their parental (S645) and homozygous (S645P) counterpart strains. This phenomenon likely reflects the notion that the expression of the FKS family of genes is coordinately regulated with the level of the 1,3-β-d-glucan-synthesizing capacity in the cell.
TABLE 5.
Relative expression ratios between FKS genes
| Strain | Genotypec
|
Expression ratio
|
||
|---|---|---|---|---|
| FKS1-HS1 | FKS1-HS2 | FKS1/FKS2 | FKS1/FKS3 | |
| 5314 | WT | WT | 3.33 | 20.00 |
| A15a | S645S/P | WT | 1.41 | 7.14 |
| A15-10a | S645P | WT | 1.15 | 3.57 |
| 36082 | WT | WT | 2.56 | 33.33 |
| 90028 | WT | WT | 2.75 | 25.00 |
| 70b | WT | WT | 3.04 | 27.14 |
| S1b | S645S/P | WT | 1.28 | 12.56 |
| S36b | S645P | WT | 0.74 | 2.17 |
| 119 | F641L | WT | 1.46 | 8.33 |
| 177 | F641S | WT | 0.38 | 4.76 |
| 205 | S645P | WT | 0.97 | 3.85 |
| 89 | S645Y | WT | 0.53 | 4.76 |
| 85 | S645F | WT | 1.47 | 12.50 |
| 149 | D648Y | WT | 2.13 | 25.00 |
| 122 | P649H | WT | 3.92 | 25.00 |
| 121 | WT | R1361H | 1.29 | 9.09 |
| 194 | S645F | R1361R/H | 0.49 | 4.35 |
| 90 | WT | R1361R/H | 1.79 | 33.33 |
Spontaneous mutants selected using ANF and SC5314 as the paternal strain.
Spontaneous mutants selected using CSF and strain 70 as the paternal strain (3).
WT, wild type.
Correlation between Ki and MIC.
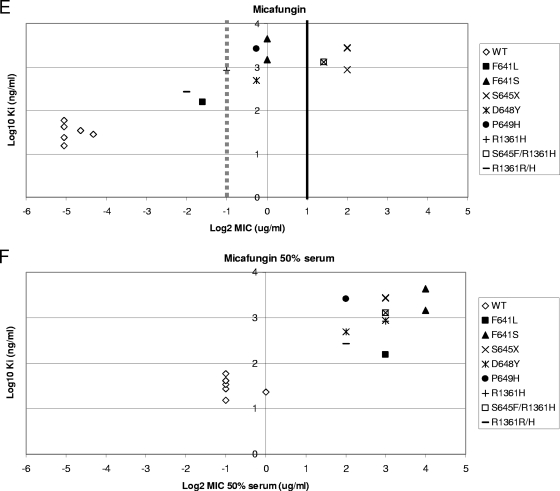
The Ki values established for wild-type and mutant enzymes (Table 4) provide a quantitative measure of the inhibitor potency of the echinocandin drugs. In principle, increased or decreased drug potency at the target should directly correlate with in vitro growth inhibition. Figure 1 illustrates that FKS1 mutations resulting in elevated MICs correlate with several-log increases in Ki. This linear-log relationship suggests that 1,3-β-d-glucan synthase can sustain a wide range of affinity changes for echinocandins without impacting relative susceptibility in in vitro growth. However, above a certain threshold (50- to 1,000-fold) of changes in Ki values, C. albicans cells grow at echinocandin concentrations higher than the proposed breakpoint (>2 μg/ml). As illustrated for MCF, substitutions at Fks1p Ser645 showed the highest MIC and Ki increases, while the lowest increases were found for mutants that carried changes in the Fks1p hot spot 1 C-terminal edge (D648 and P649).
Echinocandin susceptibility breakpoint relative to Ki.
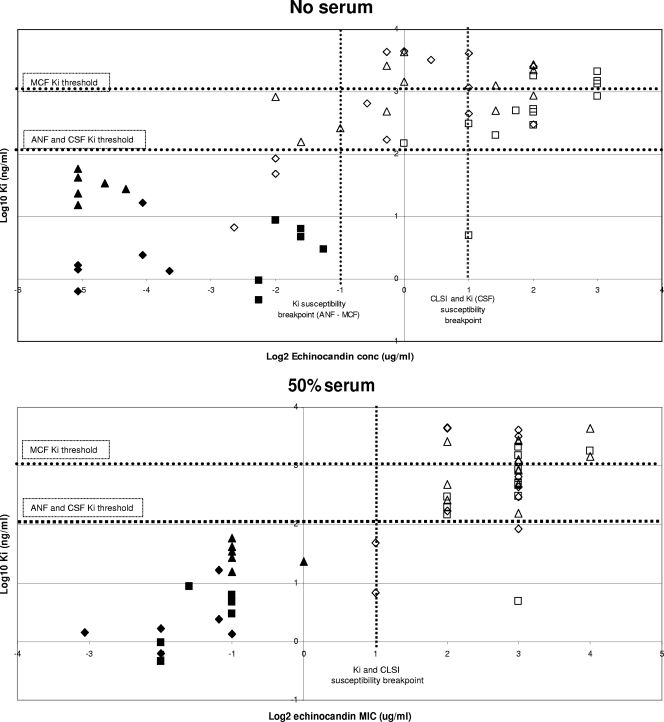
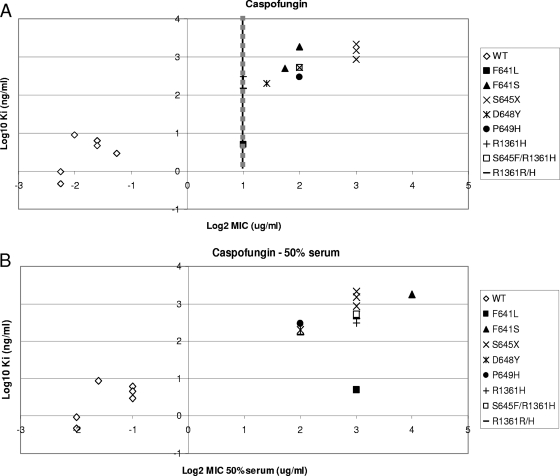
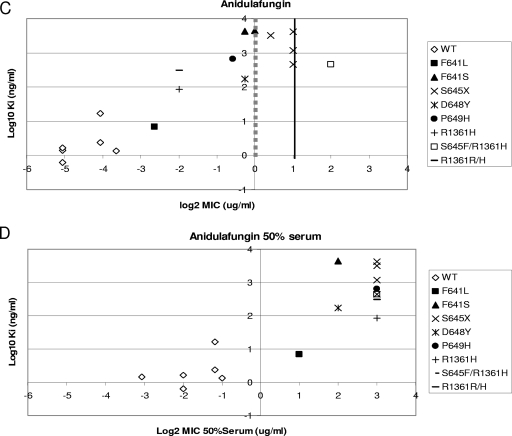
The correlations established in Fig. 2 indicate that Ki values can be used to distinguish between susceptible and resistant strains. In order to determine if the echinocandin breakpoint includes all the strains with 1,3-β-d-glucan synthase Ki values at least 50-fold higher than the average wild-type values (ANF and CSF Ki values of >200 ng/ml and MCF Ki values of >1,500 ng/ml), a quantitative evaluation of Ki and MIC values was performed for the wild type and fks1 mutants. An evaluation of log2 MICs, in the presence and absence of 50% human serum, and the log10 Ki demonstrated for CSF that all high-Ki isolates were captured at an MIC of ≥2 μg/ml (Fig. 3A). The separation between susceptible and resistant strains was maximized in the presence of 50% serum (Fig. 3B). An MIC of 0.5 μg/ml was sufficient to distinguish between most strains with high-Ki 1,3-β-d-glucan synthases for ANF (Fig. 3C) and MCF (Fig. 3E). Typically, these strains at the margin showed changes in Ki values of <100-fold relative to that of wild type (Table 4). A cutoff value of 2 μg/ml for these drugs, as proposed by the new CLSI recommendation, missed several strains with fks1 mutations except those containing substitutions at Ser645 (S645P, S645Y, S645F, and S645F-R1361R/H). However, a cutoff value of 2 μg/ml was sufficient to distinguish between all resistant fks1 mutant strains and susceptible strains when MICs were obtained in the presence of 50% serum (Fig. 3D and F).
FIG. 2.
Correlation between ANF (diamonds), CSF (squares), and MCF (triangles) Ki values and MICs with and without 50% serum. Filled shapes represent wild-type glucan synthases, and open shapes represent mutants. Ki threshold lines represent >50-fold-higher-than-average wild-type glucan synthase Ki values.
FIG. 3.
Ki-MIC correlations arranged by Fks1p amino acid changes. Ki values are expressed in ng/ml (log10), and MICs are expressed in μg/ml (log2). Black lines represent the CLSI susceptibility breakpoint (2 μg/ml). Gray dotted lines represent the breakpoint suggested in this work. (A, C, and E) MICs were obtained using RPMI 1640 medium (CLSI document M27-A3) (7). (B, D, and F) MICs were obtained using RPMI 1640 medium with 50% serum. WT, wild type.
DISCUSSION
Mutations in Fks1 help define resistance.
It was proposed previously that mutations conferring echinocandin resistance reside in two hot spot regions of FKS1p (CaFks1 F641-P649 and D1357-L1364), which are highly conserved among FKS genes in different fungal species (33, 35). The whole-cell and biochemical data obtained in this study provide strong evidence that a relatively narrow spectrum of FKS1 mutations in strains of C. albicans confer reduced susceptibility across the entire class of echinocandin drugs even though there are subtle differences in drug behaviors among the various mutants (Table 1 and 4). At the biochemical level, mutations that decrease enzyme sensitivity (IC50 or Ki) to drug by 50-fold to several thousandfold do so across the entire class of drugs (Tables 1, 3, and 4). Whole-cell growth inhibition studies (MIC) demonstrate that mutants harboring amino acid changes at Ser645 in Fks1p have the highest MICs, while amino acid changes in the C-terminal portion of hot spot 1 (Asp648 and Pro649) and in Arg1361 (hot spot 2) have less pronounced effects on susceptibility to echinocandins (Table 1). The F641L mutations are somewhat different, as they show elevated CSF MICs but low IC50 values. However, the relative effect depends on the substituted amino acid (Tables 1 and 4). Many of the phenotypic differences observed with fks mutants are most acutely manifested in standard testing medium and largely disappear in the presence of 50% human serum, as previously reported (30, 32, 53). Similarly, as reported previously by Paderu et al. (32) for wild-type enzymes, serum also shifts the relative efficacy (IC50) of each drug on mutant enzymes containing Fks1p substitutions (Table 1). In serum, it is apparent that all FKS1 mutants show a clustering of high MICs and high Ki values (Fig. 2). As the echinocandins are heavily serum protein bound in vivo, MICs of 4 to 16 μg/ml in the presence of serum (Table 1) may place the response of such mutants on the edge of therapeutic efficacy, depending on the pharmacokinetic properties of the echinocandin drugs (1).
Relationship of Ki to MIC and CLSI breakpoint.
The susceptibility of common yeasts to the three echinocandin drugs has remained largely constant since the introduction of CSF in 2001 (37). It was determined for a wide range of species that greater than 99% of isolates were inhibited by ≤2 μg/ml of either CSF, MCF, or ANF. Therefore, the CLSI Antifungal Subcommittee recently established an MIC of ≤2 μg/ml as an interpretive MIC breakpoint for susceptibility of Candida spp. to the three echinocandin drugs (7). An MIC predictive of resistance could not be assigned because of the paucity of isolates available above the proposed breakpoint (40). In this work, we have provided a detailed molecular and kinetic characterization of clinical isolates of C. albicans with FKS1 mutations resulting in high MICs. It is now well recognized that high-MIC isolates of Candida spp. from patients failing therapy often contain amino acid substitutions in Fks subunits (Fks1p and/or Fks2p) of the 1,3-β-d-glucan synthase complex (6, 8, 14, 15, 19, 22, 24, 28, 33). The elevated MIC phenotype linked to mutations in FKS is a direct consequence of a decrease in the biochemical sensitivity of 1,3-β-d-glucan synthase to drug, which is reflected as an increase of 50-fold to several thousandfold for the related enzyme inhibition constants IC50 and Ki (Tables 1 and 4). The decrease in enzyme sensitivity (IC50 or Ki) does not correlate strictly with an equivalent rise in MIC (Tables 1 and 4). However, there is an unambiguous linkage between increasing Ki and MIC values (Fig. 2). This tightly coupled interrelationship between amino acid substitutions in Fks1p, 1,3-β-d-glucan synthase sensitivity, and MIC (Tables 1 to 4 and Fig. 2) provides a window to examine resistance and, in particular, the relationship between MIC and the potential for successful clinical outcome, as proposed by the new CLSI guidelines. In principle, a plot of the inherent biochemical parameter Ki as a function of MIC should establish a drug level that correlates with enzyme insensitivity to drug, which can be operationally defined as a Ki level at least 50 times that of the average sensitive wild-type enzyme (Table 4). Using this threshold criterion for enzyme-mediated resistance, it is apparent for CSF that all clinical isolates containing FKS1 mutations, except F641L, are captured at MICs of ≥2 μg/ml (Fig. 3C), which fully supports the CLSI breakpoint. When MICs are obtained in the presence of serum, there is a 100% capture of all fks1 mutants above the MIC breakpoint (Fig. 2 and 3B, D, and F). However, a threshold MIC of ≥2 μg/ml is less inclusive when similar relationships for ANF (Fig. 3C) and MCF (Fig. 3E) in the absence of serum were evaluated. For those drugs, an MIC of >0.5 μg/ml would permit >95% capture of mutant isolates with fks1 mutations. Again, if serum is included in the growth medium, the MICs shift sufficiently for both MCF and ANF, which enables the complete capture of fks1 mutants above an MIC of ≥2 μg/ml. The inclusion of serum in routine testing medium would simplify matters, but in practice, it would be difficult to standardize. Nevertheless, even under CLSI testing methodology standard M27-A3, echinocandin susceptibility surveillance studies showed that more than 99% of the C. albicans strains have ANF and MCF MICs of ≤0.5 μg/ml, which were considered to be susceptible (38, 40). Lower breakpoints would therefore not result in false resistance detection. However, while attractive, such a view does not take into account the full range of considerations for the breakpoint determination, including the tenant that the threshold value should not bisect MIC distributions of wild-type populations such as Candida parapsilosis or Candida guillermondii. In this context, this work has immediate relevance to strains with fks1 mutant backgrounds; further studies would be required to support lower breakpoints for ANF and MCF.
FKS1 mutations influence 1,3-β-d-glucan synthase kinetics.
Little is known about the inhibition kinetics of 1,3-β-d-glucan synthase and how echinocandin drugs interact with the enzyme complex. Genetic analysis of resistance can help define structural binding/interaction domains, especially when mutations are clustered in limited regions, which tend to favor direct or short-range interactions over long-range conformational rearrangements. Kinetic analysis of mutant enzymes indicates that amino acid substitutions conferring reduced drug susceptibility have little or no effect on the affinity of enzyme for substrate (Km) (Tables 3 and 4), while certain mutations alter the catalytic capacity of the enzyme (Vmax) (Tables 3 and 4). These kinetic findings suggest that the binding site for echinocandin drugs is apart from the binding site for substrate, which is also consistent with the noncompetitive inhibition kinetics observed (Fig. 1E and F) (9, 49). The reduced catalytic capacity of some mutations suggests a possible effect on a transition state intermediate in the reaction. The structural location of the binding site is not known, although the hot spot regions are predicted from topology models (29) to lie near the extracytoplasmic surface of the enzyme. The clustering of mutations in two structurally defined hot spot regions (F641-P649 and D1357-L1364) may help define a binding site for echinocandin drugs. Such a notion would imply that amino acids Phe641, Ser645, Asp648, Pro649, and Arg1361 promote echinocandin-enzyme interactions. Moreover, Ser645 would be most important in this interaction since all substitutions have the most pronounced effect on MIC for both CSF (3) and ANF, as observed in this study.
Certain FKS1 mutations are associated with decreases in Vmax, which were observed in mutant Fks1p isolated from isogenic strains (SC5314 and A15-10) (Table 3). The decreased catalytic capacity may have a fitness cost to the cell. However, the relative expression data (Table 5) presented here show a possible way for the cell to compensate for this effect, since C. albicans is able to increase the expression of the other nonmutated FKS genes. This idea was confirmed when isogenic strains showing a moderate and more important increase in FKS2 and FKS3 expression levels for heterozygous and homozygous isogenic mutants, respectively, were studied. However, this phenomenon was not seen in D648Y (strain 149) and P649H (strain 122) mutants and in other Candida spp. with equivalent mutations (13). Coincidently, these strains showed the lowest MICs in this collection. Surprisingly, our kinetic assays did not reflect elevated levels of FKS2 and FKS3 expression, since smooth curves were obtained, representing a single kinetic species. Thus, the majority of the kinetic response for C. albicans GS appears to be dominated by one enzyme type (presumably Fks1p). This phenomenon could have several possible explanations. One explanation is that the expression of FKS2 and FKS3 does not translate into assembled enzyme due to posttranscriptional control (e.g., RNA or enzyme turnover), or the enzyme extraction was more selective for Fks1p, as stated previously (13). Conversely, when heterozygous FKS1 mutants (Fks1p/fks1p) were studied, double-inflection inhibition curves (two IC50 values) and nonlinear double reciprocal plots were seen, representing two isoenzymes (18, 33).
Overall, it is apparent that the three echinocandin drugs share target, mechanism of resistance, spectrum, and in vitro potency, allowing the possibility of using CSF as a surrogate marker to predict the echinocandin susceptibility of C. albicans strains. This concept was suggested previously by Pfaller et al. (41-43) using fluconazole MIC breakpoints to assess relative voriconazole (or other azole) susceptibility, and our data with CSF support an analogous application for echinocandin drugs.
Acknowledgments
This work was supported by NIH grant 1R01AI069397-01 to D.S.P.
Footnotes
Published ahead of print on 27 October 2008.
REFERENCES
- 1.Andes, D., D. J. Diekema, M. A. Pfaller, R. A. Prince, K. Marchillo, J. Ashbeck, and J. Hou. 2008. In vivo pharmacodynamic characterization of anidulafungin in a neutropenic murine candidiasis model. Antimicrob. Agents Chemother. 52:539-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andes, D. R., D. J. Diekema, M. A. Pfaller, K. Marchillo, and J. Bohrmueller. 2008. In vivo pharmacodynamic target investigation for micafungin against Candida albicans and C. glabrata in a neutropenic murine candidiasis model. Antimicrob. Agents Chemother. 52:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balashov, S. V., S. Park, and D. S. Perlin. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50:2058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjamin, D. K., Jr., T. Driscoll, N. L. Seibel, C. E. Gonzalez, M. M. Roden, R. Kilaru, K. Clark, J. A. Dowell, J. Schranz, and T. J. Walsh. 2006. Safety and pharmacokinetics of intravenous anidulafungin in children with neutropenia at high risk for invasive fungal infections. Antimicrob. Agents Chemother. 50:632-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, J. C., P. S. Hicks, M. B. Kurtz, H. Rosen, D. M. Schmatz, P. A. Liberator, and C. M. Douglas. 2002. The antifungal echinocandin caspofungin acetate kills growing cells of Aspergillus fumigatus in vitro. Antimicrob. Agents Chemother. 46:3001-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary, J. D., G. Garcia-Effron, S. W. Chapman, and D. S. Perlin. 2008. Reduced Candida glabrata susceptibility secondary to an FKS1 mutation developed during candidemia treatment. Antimicrob. Agents Chemother. 52:2263-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A3, 3rd ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Desnos-Ollivier, M., F. Dromer, and E. Dannaoui. 2008. Detection of caspofungin resistance in Candida spp. by Etest. J. Clin. Microbiol. 46:2389-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon, M. 1953. The determination of enzyme inhibitor constants. Biochem. J. 55:170-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas, C. M. 2001. Fungal beta(1,3)-D-glucan synthesis. Med. Mycol. 39(Suppl. 1):55-66. [DOI] [PubMed] [Google Scholar]
- 11.Frost, D., K. Brandt, C. Estill, and R. Goldman. 1997. Partial purification of (1,3)-beta-glucan synthase from Candida albicans. FEMS Microbiol. Lett. 146:255-261. [DOI] [PubMed] [Google Scholar]
- 12.Frost, D. J., K. Brandt, J. Capobianco, and R. Goldman. 1994. Characterization of (1,3)-beta-glucan synthase in Candida albicans: microsomal assay from the yeast or mycelial morphological forms and a permeabilized whole-cell assay. Microbiology 140:2239-2246. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Effron, G., S. K. Katiyar, S. Park, T. D. Edlind, and D. S. Perlin. 2008. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 52:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha, Y. S., S. F. Covert, and M. Momany. 2006. FsFKS1, the 1,3-β-glucan synthase from the caspofungin-resistant fungus Fusarium solani. Eukaryot. Cell 5:1036-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakki, M., J. F. Staab, and K. A. Marr. 2006. Emergence of a Candida krusei isolate with reduced susceptibility to caspofungin during therapy. Antimicrob. Agents Chemother. 50:2522-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hope, W. W., S. Shoham, and T. J. Walsh. 2007. The pharmacology and clinical use of caspofungin. Expert Opin. Drug Metab. Toxicol. 3:263-274. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, M. D., and J. R. Perfect. 2003. Caspofungin: first approved agent in a new class of antifungals. Expert Opin. Pharmacother. 4:807-823. [DOI] [PubMed] [Google Scholar]
- 18.Kahn, J. N., G. Garcia-Effron, M. J. Hsu, S. Park, K. A. Marr, and D. S. Perlin. 2007. Acquired echinocandin resistance in a Candida krusei isolate due to modification of glucan synthase. Antimicrob. Agents Chemother. 51:1876-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kahn, J. N., M. J. Hsu, F. Racine, R. Giacobbe, and M. Motyl. 2006. Caspofungin susceptibility in Aspergillus and non-Aspergillus molds: inhibition of glucan synthase and reduction of β-d-1,3 glucan levels in culture. Antimicrob. Agents Chemother. 50:2214-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kartsonis, N., J. Killar, L. Mixson, C. M. Hoe, C. Sable, K. Bartizal, and M. Motyl. 2005. Caspofungin susceptibility testing of isolates from patients with esophageal candidiasis or invasive candidiasis: relationship of MIC to treatment outcome. Antimicrob. Agents Chemother. 49:3616-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kartsonis, N. A., J. Nielsen, and C. M. Douglas. 2003. Caspofungin: the first in a new class of antifungal agents. Drug Resist. Updat. 6:197-218. [DOI] [PubMed] [Google Scholar]
- 22.Katiyar, S., M. Pfaller, and T. Edlind. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 50:2892-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondoh, O., Y. Tachibana, Y. Ohya, M. Arisawa, and T. Watanabe. 1997. Cloning of the RHO1 gene from Candida albicans and its regulation of β-1,3-glucan synthesis. J. Bacteriol. 179:7734-7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laverdiere, M., R. G. Lalonde, J. G. Baril, D. C. Sheppard, S. Park, and D. S. Perlin. 2006. Progressive loss of echinocandin activity following prolonged use for treatment of Candida albicans oesophagitis. J. Antimicrob. Chemother. 57:705-708. [DOI] [PubMed] [Google Scholar]
- 25.Lesage, G., J. Shapiro, C. A. Specht, A. M. Sdicu, P. Menard, S. Hussein, A. H. Tong, C. Boone, and H. Bussey. 2005. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markovich, S., A. Yekutiel, I. Shalit, Y. Shadkchan, and N. Osherov. 2004. Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 48:3871-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maschmeyer, G., and A. Glasmacher. 2005. Pharmacological properties and clinical efficacy of a recently licensed systemic antifungal, caspofungin. Mycoses 48:227-234. [DOI] [PubMed] [Google Scholar]
- 28.Miller, C. D., B. W. Lomaestro, S. Park, and D. S. Perlin. 2006. Progressive esophagitis caused by Candida albicans with reduced susceptibility to caspofungin. Pharmacotherapy 26:877-880. [DOI] [PubMed] [Google Scholar]
- 29.Mio, T., M. Adachi-Shimizu, Y. Tachibana, H. Tabuchi, S. B. Inoue, T. Yabe, T. Yamada-Okabe, M. Arisawa, T. Watanabe, and H. Yamada-Okabe. 1997. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in β-1,3-glucan synthesis. J. Bacteriol. 179:4096-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odabasi, Z., V. Paetznick, J. H. Rex, and L. Ostrosky-Zeichner. 2007. Effects of serum on in vitro susceptibility testing of echinocandins. Antimicrob. Agents Chemother. 51:4214-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onishi, J., M. Meinz, J. Thompson, J. Curotto, S. Dreikorn, M. Rosenbach, C. Douglas, G. Abruzzo, A. Flattery, L. Kong, A. Cabello, F. Vicente, F. Pelaez, M. T. Diez, I. Martin, G. Bills, R. Giacobbe, A. Dombrowski, R. Schwartz, S. Morris, G. Harris, A. Tsipouras, K. Wilson, and M. B. Kurtz. 2000. Discovery of novel antifungal (1,3)-β-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 44:368-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paderu, P., G. Garcia-Effron, S. Balashov, G. Delmas, S. Park, and D. S. Perlin. 2007. Serum differentially alters the antifungal properties of echinocandin drugs. Antimicrob. Agents Chemother. 51:2253-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, S., R. Kelly, J. N. Kahn, J. Robles, M. J. Hsu, E. Register, W. Li, V. Vyas, H. Fan, G. Abruzzo, A. Flattery, C. Gill, G. Chrebet, S. A. Parent, M. Kurtz, H. Teppler, C. M. Douglas, and D. S. Perlin. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49:3264-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlin, D. S. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaller, M. A., L. Boyken, R. J. Hollis, J. Kroeger, S. A. Messer, S. Tendolkar, and D. J. Diekema. 2008. In vitro susceptibility of invasive isolates of Candida spp. to anidulafungin, caspofungin, and micafungin: six years of global surveillance. J. Clin. Microbiol. 46:150-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaller, M. A., L. Boyken, R. J. Hollis, S. A. Messer, S. Tendolkar, and D. J. Diekema. 2006. In vitro susceptibilities of Candida spp. to caspofungin: four years of global surveillance. J. Clin. Microbiol. 44:760-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfaller, M. A., D. J. Diekema, L. Boyken, S. A. Messer, S. Tendolkar, R. J. Hollis, and B. P. Goldstein. 2005. Effectiveness of anidulafungin in eradicating Candida species in invasive candidiasis. Antimicrob. Agents Chemother. 49:4795-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaller, M. A., D. J. Diekema, L. Ostrosky-Zeichner, J. H. Rex, B. D. Alexander, D. Andes, S. D. Brown, V. Chaturvedi, M. A. Ghannoum, C. C. Knapp, D. J. Sheehan, and T. J. Walsh. 2008. Correlation of MIC with outcome for Candida species tested against caspofungin, anidulafungin, and micafungin: analysis and proposal for interpretive MIC breakpoints. J. Clin. Microbiol. 46:2620-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2004. Cross-resistance between fluconazole and ravuconazole and the use of fluconazole as a surrogate marker to predict susceptibility and resistance to ravuconazole among 12,796 clinical isolates of Candida spp. J. Clin. Microbiol. 42:3137-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2007. Use of fluconazole as a surrogate marker to predict susceptibility and resistance to voriconazole among 13,338 clinical isolates of Candida spp. tested by Clinical and Laboratory Standards Institute-recommended broth microdilution methods. J. Clin. Microbiol. 45:70-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfaller, M. A., S. A. Messer, L. Boyken, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2008. Selection of a surrogate agent (fluconazole or voriconazole) for initial susceptibility testing of posaconazole against Candida spp.: results from a global antifungal surveillance program. J. Clin. Microbiol. 46:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roh, D. H., B. Bowers, H. Riezman, and E. Cabib. 2002. Rho1p mutations specific for regulation of beta(1→3)glucan synthesis and the order of assembly of the yeast cell wall. Mol. Microbiol. 44:1167-1183. [DOI] [PubMed] [Google Scholar]
- 45.Sabra, R., and R. A. Branch. 1990. Amphotericin B nephrotoxicity. Drug Saf. 5:94-108. [DOI] [PubMed] [Google Scholar]
- 46.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 47.Sawistowska-Schroder, E. T., D. Kerridge, and H. Perry. 1984. Echinocandin inhibition of 1,3-beta-D-glucan synthase from Candida albicans. FEBS Lett. 173:134-138. [DOI] [PubMed] [Google Scholar]
- 48.Stone, J. A., X. Xu, G. A. Winchell, P. J. Deutsch, P. G. Pearson, E. M. Migoya, G. C. Mistry, L. Xi, A. Miller, P. Sandhu, R. Singh, F. deLuna, S. C. Dilzer, and K. C. Lasseter. 2004. Disposition of caspofungin: role of distribution in determining pharmacokinetics in plasma. Antimicrob. Agents Chemother. 48:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang, J. and T. R. Parr, Jr. 1991. W-1 solubilization and kinetics of inhibition by cilofungin of Candida albicans (1,3)-β-d-glucan synthase. Antimicrob. Agents Chemother. 35:99-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, C., W. Graninger, E. Presterl, and C. Joukhadar. 2006. The echinocandins: comparison of their pharmacokinetics, pharmacodynamics and clinical applications. Pharmacology 78:161-177. [DOI] [PubMed] [Google Scholar]
- 51.White, T. C., S. Holleman, F. Dy, L. F. Mirels, and D. A. Stevens. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiederhold, N. P., and J. S. Lewis. 2007. The echinocandin micafungin: a review of the pharmacology, spectrum of activity, clinical efficacy and safety. Expert Opin. Pharmacother. 8:1155-1166. [DOI] [PubMed] [Google Scholar]
- 53.Wiederhold, N. P., L. K. Najvar, R. Bocanegra, D. Molina, M. Olivo, and J. R. Graybill. 2007. In vivo efficacy of anidulafungin and caspofungin against Candida glabrata and association with in vitro potency in the presence of sera. Antimicrob. Agents Chemother. 51:1616-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]