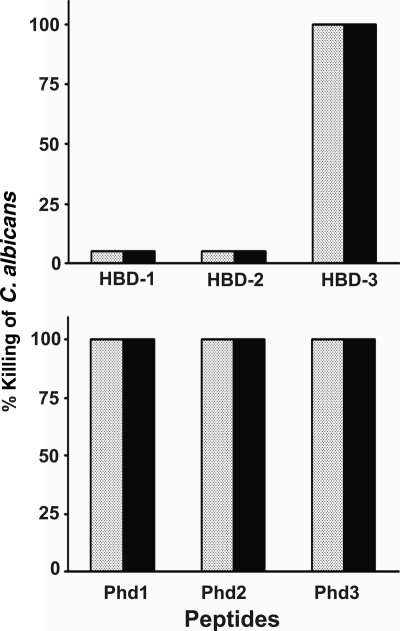
Abstract
The activities of defensins HBD-1, HBD-2, and HBD-3 and their C-terminal analogs Phd1, Phd2, and Phd3 against Candida albicans were investigated. Phd1 to Phd3 showed lower-level activities than HBD-1 to HBD-3, although metabolic inhibitors did not render Phd1 to Phd3 inactive. Their activities were also less salt sensitive than those of HBD-1 to HBD-3. Confocal microscope images indicated that the initial site of action was the fungal membrane.
Mammalian defensins comprising the alpha and beta families are important components of the innate immune system (1, 8, 17, 18, 24, 25, 29, 30). HBD-1 and HBD-2 are active against gram-negative bacteria. Their activities are attenuated by increasing concentrations of NaCl (2, 9, 10). HBD-3 is active against both gram-negative and gram-positive bacteria and is not affected by NaCl (3, 11). The findings of extensive studies have indicated that native disulfide bridges are not essential for antibacterial activity and that segments of HBD-1 to HBD-3 shorter than the full-length defensins also exhibit antibacterial activities (12-16, 21, 22, 26, 28, 35, 36). In recent years, there has been considerable interest in the antifungal activities of beta-defensins, as Candida albicans is responsible for causing oral candidiasis, particularly in patients infected with human immunodeficiency virus (5, 20). HBD-1 to HBD-3 have been detected previously in salivary glands and salivary secretions (4, 6, 7, 23, 27). The killing of C. albicans by HBD-2 and HBD-3 is salt sensitive and energy dependent (33). We have shown that single disulfide peptides spanning the C-terminal segments of HBD-1 to HBD-3, i.e., Phd1 (ACPIFTKIQGTYRGKAKCK), Phd2 (FCPRRYKQIGTGLPGTKCK), and Phd3 (SCLPKEEQIGKSTRGRKCRRKK) (disulfide bridges indicated by underlining), exhibit antibacterial activities (16). In this report, we describe their activities against C. albicans and compare the effects of salts and metabolic inhibitors on these peptides with the effects on HBD-1 to HBD-3.
HBD-1, HBD-2, and HBD-3 were purchased from Peptides International (Louisville, KY). Phd1, Phd2, and Phd3 were synthesized as described earlier using 4-(hydroxymethyl)phenoxyacetamidomethyl resin and 9-fluorenylmethoxy carbonyl chemistry (16). The formation of disulfide bonds was accomplished by air oxidation at a peptide concentration of 0.5 mg/ml for 24 h at room temperature. Purified peptides were characterized by matrix-assisted laser desorption ionization-time of flight mass spectrometry on an ABI Voyager DE STR matrix-assisted laser desorption ionization-time of flight mass spectrometer (PerSeptive Biosystems) using recrystallized α-cyano-4-hydroxycinnamic acid as a matrix (16). Peptide labeling with carboxyfluorescein (CF) at a free amino group of the N-terminal amino acid was carried out by treating 10 mg of resin-bound peptide with 0.8 ml of dimethylformamide containing CF and activating agents as described earlier (34). The deprotection of CF-labeled peptides (CF-Phd1 to CF-Phd3) from the resin, purification, and characterization by mass spectrometry were carried out as described earlier (16).
The activities of HBD-1 to HBD-3 and Phd1 to Phd3 in final volumes of 50 μl against C. albicans (ATCC 18804) in sterile 96-well plates were determined as described previously (33), with slight modifications. Briefly, minimum fungicidal concentrations (MFC) of the peptides were determined by growing C. albicans aerobically in yeast extract-peptone-dextrose (YEPD) medium at 30°C. After 20 h, 0.5 ml from this suspension was subcultured for 2 h in 20 ml of YEPD broth to obtain a mid-log-phase culture. Cells were harvested by centrifugation, washed with 10 mM phosphate buffer (PB), pH 7.4, and resuspended in the same buffer, and the concentration was adjusted to 106 cells/ml. Aliquots of diluted cells were incubated with peptides in 50-μl volumes at 30°C for 2 h. Cell suspensions were diluted and plated onto YEPD agar plates, and the plates were incubated for 24 h at 30°C. Colonies were counted, and the concentrations of the peptides at which no viable colonies were formed were taken as the MFC. The averages of results from three independent experiments done with duplicate samples were taken for the calculation of MFC. In order to determine ion specificity, various concentrations of NaCl, CaCl2, and MgCl2 were added to the incubation buffer. For the experiments evaluating the energy requirements, mid-log-phase cells (106/ml) in PB were preincubated with 5 mM sodium azide or 50 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) (19) for 2 h at 30°C with shaking before being treated with peptides.
Intracellular localization was analyzed by treating C. albicans with CF-Phd1, CF-Phd2, and CF-Phd3 (at 50% of the MFC) and propidium iodide (PI) for 15 min at 30°C. The cells were examined with a Zeiss LSM 510 META confocal microscope. Optical sectioning was done at 1 airy unit by using the 488- and 543-nm-wavelength laser lines with a 63× water lens objective. Emission data were collected using 500- to 530-nm band-pass and 565- to 615-nm band-pass filters for CF and PI, respectively, in the multitrack mode. Z-sections were acquired at 0.35-μm intervals and projected using the LSM-FCS software version 3.2. The bright-field images were obtained simultaneously using the transmitted-light detector. The images were assembled using Adobe Photoshop version 6.
Membrane permeabilization of C. albicans was determined using the fluorescent dye Sytox green (Molecular Probes, Eugene, OR) (31). Mid-log-phase C. albicans cells (107 CFU per ml) were washed and resuspended in PB containing 1 μM Sytox green. A greater number of organisms were used in this experiment than in the antifungal assays in order to detect changes in fluorescence. Aliquots of diluted cells were mixed with the peptide concentrations specified in the figure legends in 0.5-ml cuvettes held at 30°C. All measurements were carried out on a FluoroLog model 3-22 fluorescence spectrophotometer (Jobin Yvon) at an excitation wavelength of 488 nm (slit width, 2 nm) and an emission wavelength of 540 nm (slit width, 5 nm).
The hemolytic activities of Phd1 to Phd3 were determined using human erythrocytes as described earlier (32). Briefly, erythrocytes were obtained by the centrifugation (800 × g) of heparinized blood and were washed three times with 5 mM HEPES (pH 7.4) containing 150 mM NaCl. Aliquots containing 107 red blood cells/ml were incubated in the presence of different peptide concentrations in 0.5-ml tubes containing a final volume of 100 μl for 30 min at 37°C with gentle mixing. The samples were centrifuged, and the absorbance of the supernatants at 540 nm was measured. The level of erythrocyte lysis occurring with 0.1% Triton X-100 was taken as the maximal level of lysis.
The antifungal activities of HBD-1 to HBD-3 and Phd1 to Phd3 are summarized in Table 1. We observed that HBD-1, obtained from Peptides International, showed substantial activity. Phd1 to Phd3 showed activities against C. albicans, but with lower potencies than those of HBD-1 to HBD-3.
TABLE 1.
Antifungal activities of human beta-defensins HBD-1 to HBD-3 and C-terminal analogs Phd1 to Phd3
| Peptide | MFC (μM)a for C. albicans |
|---|---|
| HBD-1 | 7 |
| Phd1 | 18 |
| HBD-2 | 8 |
| Phd2 | 18 |
| HBD-3 | 2.5 |
| Phd3 | 16 |
The values reported are averages of results from three different experiments done with duplicate samples, and variations were 3%.
The data shown in Fig. 1 compare the candidacidal activities of the peptides at the MFC in the presence of CCCP and sodium azide. HBD-3 and analogs Phd1 to Phd3 were active in the presence of sodium azide and CCCP, whereas HBD-1 and HBD-2 were inactive. The results indicate that HBD-3 and Phd1 to Phd3 kill C. albicans by energy-independent mechanisms, unlike HBD-1 and HBD-2.
FIG. 1.
Effect of metabolic inhibitors on the candidacidal activities of HBD-1 to HBD-3 and Phd1 to Phd3 against C. albicans. Cells pretreated with 50 μM CCCP or 5 mM sodium azide for 2 h at 30°C were further incubated with peptides at the MFC in PB for 2 h at 30°C. Suitably diluted aliquots were plated onto YEPD agar plates, which were incubated for 24 h. The colonies formed were counted, and the percent killing of C. albicans cells was determined. Shaded bars and dark bars represent peptide activities in the presence of 50 μM CCCP and 5 mM sodium azide, respectively. The values represent averages of results from three independent experiments done with duplicate samples, and variations were 3%.
The effects of different concentrations of salts on the candidacidal activities of HBD-1 to HBD-3 and Phd1 to Phd3 at their MFC are indicated in Fig. 2. The data in Fig. 2A show that Phd1 and Phd2 exhibited activities at 25 mM NaCl, unlike HBD-1 and HBD-2, which were inactive. HBD-3 and Phd3 showed comparable activities at 25 mM NaCl. At 100 mM NaCl, all peptides at their MFC showed very little activity whereas 50% killing was observed at double their MFC (data not shown). As summarized in Fig. 2B and C, Phd1 and Phd2 showed considerably greater activities than HBD-1 and HBD-2 in the presence of 0.5 mM Ca2+ or Mg2+ whereas HBD-3 and Phd3 showed comparable activities. At a 5 mM CaCl2 concentration, Phd1 and Phd2 were inactive, like HBD-1 and HBD-2, while HBD-3 exhibited greater activity than Phd3. Unlike the full-length peptides HBD-1 to HBD-3, Phd1 and Phd2 were active at 0.5 mM MgCl2 while Phd3 was active even at 25 mM MgCl2.
FIG. 2.
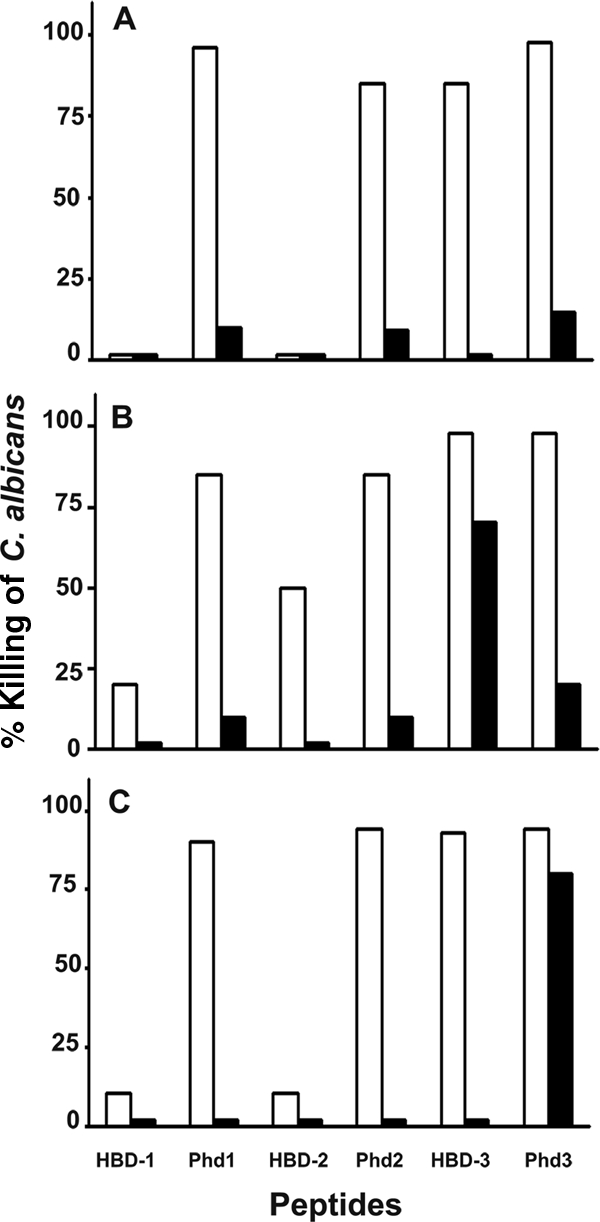
Effect of salts on candidacidal activities of HBD-1 to HBD-3 and Phd1 to Phd3 against C. albicans. Cells were incubated with peptides at their MFC in PB containing different concentrations of salts for 2 h at 30°C. Suitably diluted aliquots were incubated for 24 h on YEPD agar plates. The colonies formed were counted, and the percent killing of C. albicans cells was determined. Panels: A, 25 mM NaCl and 100 mM NaCl; B, 0.5 mM CaCl2 and 5 mM CaCl2; and C, 0.5 mM MgCl2 and 25 mM MgCl2. Light bars and dark bars represent lower and higher concentrations of salts, respectively. The values represent averages of results from three independent experiments done with duplicate samples.
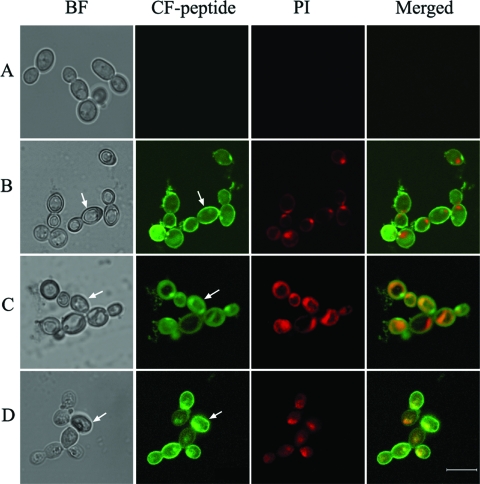
The cellular localization of Phd1 to Phd3 in C. albicans was investigated using CF-Phd1 to CF-Phd3 and confocal microscopy analysis as presented in Fig. 3. The cells exhibited intense fluorescence at the locations indicated in the fluorescence images and the corresponding bright-field images. The data indicate that the peptides were localized on the membrane. A diffuse intracellular staining pattern was also observed, which indicates the translocation of the peptides into the cells. These cells showed intense PI staining, indicating membrane damage (Fig. 3B, C, and D). Control cells showed negative staining for PI (Fig. 3A).
FIG. 3.
Confocal microscope images showing the localization of CF-Phd1, CF-Phd2, and CF-Phd3 incubated with C. albicans. Cells were treated with 50% MFC of peptide and 4 μg/ml of PI. Arrows in the panels show membranes in the fluorescence images (CF-peptide) and also in the corresponding bright-field (BF) images. Panels: A, control cells without peptide; B, CF-Phd1; C, CF-Phd2; and D, CF-Phd3. The bar represents 5 μm.
Membrane damage was also assessed by an increase in fluorescence due to the influx of Sytox green, a high-affinity nucleic acid stain that does not cross the membranes of live cells. However, it penetrates cells with damaged plasma membranes and binds to nucleic acids, resulting in the enhancement of its fluorescence intensity (31). The data shown in Fig. 4 indicate that Phd1 and Phd2 caused greater membrane permeabilization than the parent peptides HBD-1 and HBD-2. However, HBD-3 showed more fluorescence enhancement than Phd3. Although the data shown in Fig. 4 correspond to changes in fluorescence at one concentration, the increase in fluorescence was concentration dependent. HBD-3 and Phd3 caused greater membrane permeabilization than HBD-1 and HBD-2 and Phd1 and Phd2, respectively.
FIG. 4.
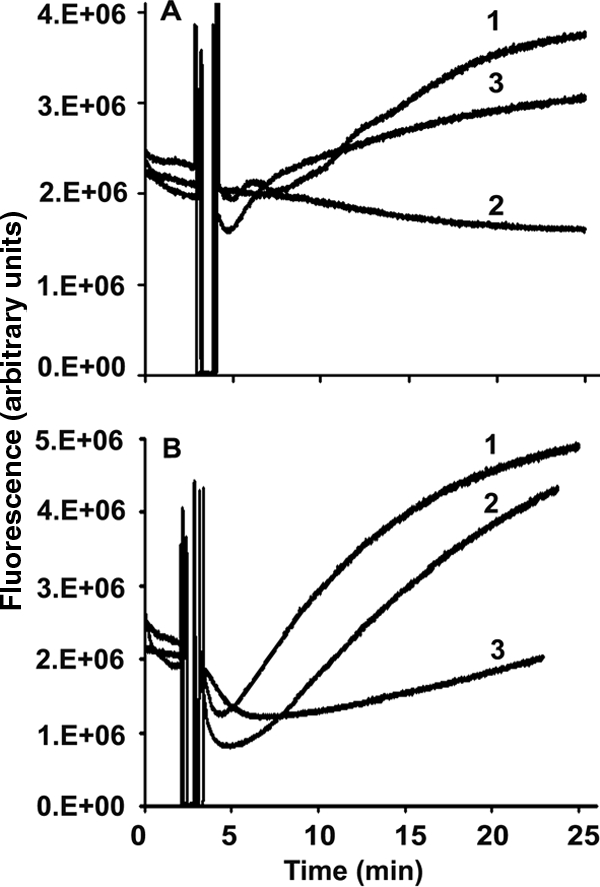
Membrane permeabilization with peptides was measured by the influx of Sytox green into C. albicans. Cells (107 CFU per ml) were incubated with 1 μM Sytox green. Once the basal fluorescence reached a constant value, peptides at the corresponding concentrations were added and the increase in fluorescence was monitored at 30°C with excitation at 488 nm and emission at 540 nm. (A) HBD-1 (1), HBD-2 (2), and HBD-3 (3); (B) C-terminal analogs Phd1 (1), Phd2 (2), and Phd3 (3). Concentrations of peptides were 4 μM for HBD-1, HBD-2, Phd1, and Phd2 and 1 μM for HBD-3 and Phd3.
Phd1 to Phd3 showed no hemolytic activities at concentrations of up to 75 μM, which exceeds the MFC by three- to fourfold. At 100 μM, 15% lysis was observed.
Although Phd1 to Phd3 were less active than HBD-1 to HBD-3, their activities were not lost in the presence of metabolic inhibitors. Also, the activities were less salt sensitive than those of the parent peptides HBD-1 and HBD-2. Hence, Phd1 to Phd3 and possibly the C-terminal regions of other defensins may be attractive candidates for development as therapeutic agents as well as for analysis to understand the mechanism of action.
Acknowledgments
Funding from CSIR Network project NWP-05 is gratefully acknowledged.
Footnotes
Published ahead of print on 22 September 2008.
REFERENCES
- 1.Agerberth, B., and G. H. Gudmundsson. 2006. Host antimicrobial defence peptides in human disease. Curr. Top. Microbiol. Immunol. 306:67-90. [DOI] [PubMed] [Google Scholar]
- 2.Bals, R., X. Wang, Z. Wu, T. Freeman, V. Bafna, M. Zasloff, and J. M. Wilson. 1998. Human beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J. Clin. Investig. 102:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batoni, G., G. Maisetta, S. Esin, and M. Campa. 2006. Human beta-defensin-3: a promising antimicrobial peptide. Mini Rev. Med. Chem. 6:1063-1073. [DOI] [PubMed] [Google Scholar]
- 4.Bonass, W. A., A. S. High, P. J. Owen, and D. A. Devine. 1999. Expression of beta-defensin genes by human salivary glands. Oral Microbiol. Immunol. 14:371-374. [DOI] [PubMed] [Google Scholar]
- 5.Coleman, D. C., D. E. Bennett, D. J. Sullivan, P. J. Gallagher, M. C. Henman, D. B. Shanley, and R. J. Russell. 1993. Oral Candida in HIV infection and AIDS: new perspectives/ new approaches. Crit. Rev. Microbiol. 19:61-82. [DOI] [PubMed] [Google Scholar]
- 6.Dale, B. A., and S. Krisanaprakornkit. 2001. Defensin antimicrobial peptide in the oral cavity. J. Oral Pathol. Med. 30:321-327. [DOI] [PubMed] [Google Scholar]
- 7.Dunsche, A., Y. Acil, R. Siebert, J. Harder, J. M. Schroder, and S. Jepsen. 2001. Expression profile of human defensins and antimicrobial proteins in oral tissues. J. Oral Pathol. Med. 30:154-158. [DOI] [PubMed] [Google Scholar]
- 8.Ganz, T. 2004. Defensins: antimicrobial peptides of vertebrates. C. R. Biol. 327:539-549. [DOI] [PubMed] [Google Scholar]
- 9.Goldman, M. J., G. M. Anderson, E. D. Stolzenberg, U. P. Kari, M. Zasloff, and J. M. Wilson. 1997. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553-560. [DOI] [PubMed] [Google Scholar]
- 10.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 1997. A peptide antibiotic from human skin. Nature 387:861-862. [DOI] [PubMed] [Google Scholar]
- 11.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 12.Hoover, D. M., Z. Wu, K. Tucker, W. Lu, and J. Lubkowski. 2003. Antimicrobial characterization of human beta-defensin 3 derivatives. Antimicrob. Agents Chemother. 47:2804-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kluver, E., K. Adermann, and A. Schulz. 2006. Synthesis and structure-activity relationship of β-defensins, multi-functional peptides of the immune system. J. Pept. Sci. 12:243-257. [DOI] [PubMed] [Google Scholar]
- 14.Kluver, E., S. Schulz-Maronde, S. Scheid, B. Meyer, W. G. Forssmann, and K. Adermann. 2005. Structure-activity relation of human beta-defensin 3: influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry 44:9804-9816. [DOI] [PubMed] [Google Scholar]
- 15.Krishnakumari, V., A. Sharadadevi, S. Singh, and R. Nagaraj. 2003. Single disulfide and linear analogues corresponding to the carboxy-terminal segment of bovine β-defensin-2: effects of introducing the beta hairpin nucleating sequence d-Pro-Gly on antibacterial activity and biophysical properties. Biochemistry 42:9307-9315. [DOI] [PubMed] [Google Scholar]
- 16.Krishnakumari, V., S. Singh, and R. Nagaraj. 2006. Antibacterial activities of synthetic peptides corresponding to the carboxy-terminal region of human beta-defensins 1-3. Peptides 27:2607-2613. [DOI] [PubMed] [Google Scholar]
- 17.Lehrer, R. I. 2004. Primate defensins. Nat. Rev. Microbiol. 2:727-738. [DOI] [PubMed] [Google Scholar]
- 18.Lehrer, R. I., and T. Ganz. 2002. Defensins of vertebrate animals. Curr. Opin. Immunol. 14:96-102. [DOI] [PubMed] [Google Scholar]
- 19.Lehrer, R. I., T. Ganz, D. Szklarek, and M. E. Selsted. 1988. Modulation of the in vitro candidacidal activity of human neutrophil defensins by target cell metabolism and divalent cations. J. Clin. Investig. 81:1829-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupetti, A., R. Danesi, J. W. van't Wout, J. T. van Dissel, S. Senesi, and P. H. Nibbering. 2002. Antimicrobial peptides: therapeutic potential for the treatment of Candida infections. Expert Opin. Investig. Drugs 11:309-318. [DOI] [PubMed] [Google Scholar]
- 21.Mandal, M., M. V. Jagannadham, and R. Nagaraj. 2002. Antibacterial activities and conformations of bovine beta-defensin BNBD-12 and analogs: structural and disulfide bridge requirements for activity. Peptides 23:413-418. [DOI] [PubMed] [Google Scholar]
- 22.Mandal, M., and R. Nagaraj. 2002. Antibacterial activities and conformations of synthetic alpha-defensin HNP-1 and analogs with one, two and three disulfide bridges. J. Pept. Res. 59:95-104. [DOI] [PubMed] [Google Scholar]
- 23.Mathews, M., H. P. Jia, J. M. Guthmiller, G. Losh, S. Graham, G. K. Johnson, B. F. Tack, and P. B. McCray, Jr. 1999. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oppenheim, J. J., A. Biragyn, L. W. Kwak, and D. Yang. 2003. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Ann. Rheum. Dis. 62:17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pazgier, M., D. M. Hoover, D. Yang, W. Lu, and J. Lubkowski. 2006. Human beta-defensins. Cell. Mol. Life Sci. 63:1294-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pazgier, M., X. Li, W. Lu, and J. Lubkowski. 2007. Human defensins: synthesis and structural properties. Curr. Pharm. Des. 13:3096-3118. [DOI] [PubMed] [Google Scholar]
- 27.Sahasrabudhe, K. S., J. R. Kimball, T. H. Morton, A. Weinberg, and B. A. Dale. 2000. Expression of the antimicrobial peptide, human beta-defensin-1, in duct cells of minor salivary glands and detection in saliva. J. Dent. Res. 79:1669-1674. [DOI] [PubMed] [Google Scholar]
- 28.Sahl, H. G., U. Pag, S. Bonness, S. Wagner, N. Antcheva, and A. Tossi. 2005. Mammalian defensins: structures and mechanism of antibiotic activity. J. Leukoc. Biol. 77:466-475. [DOI] [PubMed] [Google Scholar]
- 29.Schneider, J. J., A. Unholzer, M. Schaller, M. Schafer-Korting, and H. C. Korting. 2005. Human defensins. J. Mol. Med. 83:587-595. [DOI] [PubMed] [Google Scholar]
- 30.Selsted, M., and A. J. Ouellette. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551-557. [DOI] [PubMed] [Google Scholar]
- 31.Thevissen, K., F. R. Terras, and W. F. Broekaert. 1999. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl. Environ. Microbiol. 65:5451-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varkey, J., and R. Nagaraj. 2005. Antibacterial activity of human neutrophil defensin HNP-1 analogs without cysteines. Antimicrob. Agents Chemother. 49:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vylkova, S., N. Nayyar, W. Li, and M. Edgerton. 2007. Human beta-defensins kill Candida albicans in an energy-dependent and salt-sensitive manner without causing membrane disruption. Antimicrob. Agents Chemother. 51:154-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber, P. J. A., J. E. Bader, G. Folkers, and A. G. Beck-Sickinger. 1998. A fast and inexpensive method for N-terminal fluorescein-labeling of peptides. Bioorg. Med. Chem. Lett. 8:597-600. [DOI] [PubMed] [Google Scholar]
- 35.Wu, Z., D. M. Hoover, D. Yang, C. Boulegue, F. Santamaria, J. J. Oppenheim, J. Lubkowski, and W. Lu. 2003. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc. Natl. Acad. Sci. USA 100:8880-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou, G., E. de Leeuw, C. Li, M. Pazgier, C. Li, P. Zeng, W.-Y. Lu, J. Lubkowski, and W. Lu. 2007. Toward understanding the cationicity of defensins: Arg and Lys versus their noncoded analogs. J. Biol. Chem. 282:19653-19665. [DOI] [PubMed] [Google Scholar]