Abstract
Fluoroquinolone MICs are increased through the acquisition of chromosomal mutations in the genes encoding gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE), increased levels of the multidrug efflux pump AcrAB, and the plasmid-borne genes aac(6′)-Ib-cr and the qnr variants in Escherichia coli. In the accompanying report, we found that ciprofloxacin, gatifloxacin, levofloxacin, and norfloxacin MICs for fluoroquinolone-resistant E. coli clinical isolates were very high and widely varied (L. Becnel Boyd, M. J. Maynard, S. K. Morgan-Linnell, L. B. Horton, R. Sucgang, R. J. Hamill, J. Rojo Jimenez, J. Versalovic, D. Steffen, and L. Zechiedrich, Antimicrob. Agents Chemother. 53:229-234, 2009). Here, we sequenced gyrA, gyrB, parC, and parE; screened for aac(6′)-Ib-cr and qnrA; and quantified AcrA levels in E. coli isolates for which patient sex, age, location, and site of infection were known. We found that (i) all fluoroquinolone-resistant isolates had gyrA mutations; (ii) ∼85% of gyrA mutants also had parC mutations; (iii) the ciprofloxacin and norfloxacin MICs for isolates harboring aac(6′)-Ib-cr (∼23%) were significantly higher, but the gatifloxacin and levofloxacin MICs were not; (iv) no isolate had qnrA; and (v) ∼33% of the fluoroquinolone-resistant isolates had increased AcrA levels. Increased AcrA correlated with nonsusceptibility to the fluoroquinolones but did not correlate with nonsusceptibility to any other antimicrobial agents reported from hospital antibiograms. Known mechanisms accounted for the fluoroquinolone MICs of 50 to 70% of the isolates; the remaining included isolates for which the MICs were up to 1,500-fold higher than expected. Thus, additional, unknown fluoroquinolone resistance mechanisms must be present in some clinical isolates.
Fluoroquinolones have become some of the most frequently prescribed antimicrobial agents worldwide. Fluoroquinolone MICs can be increased by mutations in the genes encoding the targets gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE), by increased levels of the multidrug efflux pump AcrAB (12, 27, 29), and by the presence of plasmid-borne mechanisms QnrA, QnrB, QnrS, and Aac(6′)-Ib-cr (9, 23, 28). However, these mechanisms do not equally affect the MICs of all fluoroquinolones; in general, ciprofloxacin and norfloxacin MICs are affected by all of these mechanisms, and gatifloxacin and levofloxacin MICs are less affected or are not affected (14, 23, 26). In a companion study, we found tremendous variation among the MICs of ciprofloxacin, gatifloxacin, levofloxacin, and norfloxacin, such that the four fluoroquinolones were affected to the same extent only in a small subset of isolates (∼5%) (4). In addition to the tremendous diversity in MICs of the four tested fluoroquinolones, we recently uncovered clear correlations between fluoroquinolone resistance in Escherichia coli isolates and patient age, patient sex, hospital location, and culture site (3). It is possible that these patient factors correlate with some of the MIC relationships or with some resistance genotype in the fluoroquinolone-resistant isolates, but such potential relationships have not yet been explored.
Here, we determined topoisomerase quinolone resistance-determining region (QRDR) sequences, AcrAB levels, and whether or not isolates contained aac(6′)-Ib-cr or qnrA. We analyzed the results in light of the fluoroquinolone MICs and patient factors (age, sex, hospital ward, culture site, and time, or date of culture). Altered topoisomerase gene sequences, increased AcrAB levels, and/or the prevalence of plasmid-encoded resistance mechanisms have been well documented in fluoroquinolone-resistant clinical isolates (12, 17, 22, 28); however, no study, to our knowledge, has examined all of these mechanisms simultaneously, and in large part because too few isolates were studied, potential relationships between these resistance mechanisms and how they correlate with patient data are unknown.
The results presented here showed correlations between resistance mechanisms and fluoroquinolone MICs, with important implications for clinical treatment. We found that all fluoroquinolone-resistant isolates encoded topoisomerase mutations, while only one-third of these isolates had increased levels of AcrA. Additionally, aac(6′)-Ib-cr was present in approximately 20% of fluoroquinolone-resistant isolates; however, qnrA was not present in any of the isolates. The presence of the resistance mechanisms could account for the fluoroquinolone MICs for the majority of isolates; however, the MICs for some isolates were 1,500-fold higher than anticipated, given the resistance mechanisms present. This work presents new insights into the development of fluoroquinolone resistance and the role of individual resistance mechanisms to resistance.
MATERIALS AND METHODS
Chemicals and reagents.
Mueller-Hinton (MH) and Luria-Bertani (LB) agar and broth were from Difco (Sparks, MD); tryptone, yeast extract, and agar were from Becton Dickinson and Company (San Jose, CA). Arabinose, chloramphenicol, ciprofloxacin, levofloxacin, and norfloxacin were from Sigma Aldrich (St. Louis, MO), and gatifloxacin was from Bristol-Myers Squibb (New York, NY). Etest strips were from AB Biodisk (Solna, Sweden). Wizard genomic DNA isolation kits were from Promega (Madison, WI). Oligonucleotide primers were from IDT DNA (Coralville, IA). Recombinant Taq polymerase was from Invitrogen (Carlsbad, CA). AcrA antibodies were a generous gift from Helen I. Zgurskaya (34), and pHSH-J53 10-2 CCR+, encoding aac(6′)-Ib-cr and qnrA, was a kind gift from David C. Hooper (28).
E. coli K-12 strains and plasmids.
The isogenic C600-based E. coli strains and the AcrAB overexpression and empty vector control plasmids used in this study are listed in Table 1. For all comparative analyses, isogenic strains and plasmids were used.
TABLE 1.
E. coli K-12 strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant feature | Reference |
|---|---|---|
| E. coli strain | ||
| C600 | F−thr-1 leu-6 thi-1 lacY1 supE44 tonA21 | 1 |
| SKM18 | C600 except gyrAL83, Y87zei-723::Tn10 parCI80, G84 | 14 |
| 1596 | C600 except gyrAL83zei-723::Tn10 parCL80; kanr | 10 |
| 1982 | C600 except gyrAL83zei-723::Tn10 parCK84; kanr | 10 |
| Plasmid | ||
| pAB | PBAD-acrAB in pACYC184 | 34 |
| pACYC184 | Camr; p15A origin | 5 |
aAbbreviations: r, resistant; kan, kanamycin; cam, chloramphenicol.
Numbers of isolates.
From our accompanying study of 214 fluoroquinolone-resistant and 27 fluoroquinolone-susceptible E. coli clinical isolates (4), we chose 153 isolates for Western blotting to determine AcrA levels. These isolates represented the full range of ciprofloxacin, gatifloxacin, levofloxacin, and norfloxacin MICs and included isolates ELZ4004, ELZ4032, ELZ4601, and ELZ4686, which had the unprecedented fluoroquinolone MIC relationships (4). For 78 of the isolates with quantified AcrA levels (including the four isolates with unusual resistance phenotypes), we sequenced the topoisomerase QRDRs and screened for the plasmid-located genes aac(6′)-Ib-cr and qnrA.
Fluoroquinolone susceptibility determinations.
The drug susceptibility of E. coli K-12 strains containing pAB or pACYC184 was assessed by Etest according to the manufacturer's instructions as described previously for strains C600, 1596, and 1982 (31) and by broth dilution according to CLSI guidelines for strain SKM18 (15). Overnight standing cultures were grown in LB broth with chloramphenicol (30 μg/ml). Cultures were diluted 1:50 in 10 ml LB broth with chloramphenicol and 0.2% arabinose for either the Etest or broth dilution assay. For strains C600, 1596, and 1982, once the culture reached an optical density at 600 nm (OD600) of 0.4, the cells were spread by sterile cotton swabs onto LB agar with 0.2% arabinose for the Etest. For strain SKM18, broth dilution assays were performed in LB broth with 0.2% arabinose.
MICs for the clinical isolates were determined using the agar dilution method according to CLSI guidelines (15) except with a modified dilution scheme, as described previously (4). When the MIC was <1 μg/ml by the agar dilution method, at least two Etest measurements were performed according to the manufacturer's instructions to quantify MICs.
Because susceptible MICs vary by up to 3.5-fold for different fluoroquinolones, direct comparisons of MIC data for different fluoroquinolones are difficult. Therefore, for some comparisons shown here, we normalized the data by determining the increase (n-fold) in MICs compared to the MICs for the CLSI standard strain ATCC 25922, which were (within error) 0.015 μg/ml for ciprofloxacin and gatifloxacin, 0.03 μg/ml for levofloxacin, and 0.06 μg/ml for norfloxacin.
Gene amplification and sequencing.
The QRDRs of gyrA, gyrB, parC, and parE were amplified by PCR twice independently and sequenced by Agencourt Biosciences (Beverly, MA). gyrA was amplified by nested PCR with outer (gyrA_QRDR_F or gyrA_QRDR_R) and inner (gyrA_QRDR2_F or gyrA_QRDR2_R) primers (Table 2). The ATCC 25922 reference strain with wild-type topoisomerase genes and the strains with the gyrAL83 (LZ1319) (33) or parCK84 (LZ1596) (10) mutation served as sequencing controls.
TABLE 2.
Primers used in this study
| Name | Sequence |
|---|---|
| gyrA_QRDR_F | 5′-TACACCGGTCAACATTGAGG |
| gyrA_QRDR_R | 5′-TTAATGATTGCCGCCTCGG |
| gyrA_QRDR2_F | 5′-CCGTCCCGTACTTTACGC |
| gyrA_QRDR2_R | 5′-CCGTATAACGCATTGCCGC |
| gyrB_QRDR_F | 5′-CTCCTCCCAGACCAAAGACA |
| gyrB_QRDR_R | 5′TCACGACCGATACCACAGC |
| parC_QRDR_F | 5′GTACGTGATCATGGACCGTGCG |
| parC_QRDR_R | 5′GCTCGCTCAATAGCAGCTCGG |
| parE_QRDR_F | 5′-GCCTGGCAAACTGGCTGATGTA |
| parE_QRDR_R | 5′-GATCAATACGGTAGAGCGGTGGC |
aac(6′)-Ib-cr and qnrA screens.
Clinical isolates were screened in duplicate for the plasmid-mediated resistance genes aac(6′)-Ib-cr and qnrA using PCR primers described previously (19), and each PCR product was sequenced. ATCC 25922 and pHSH-J53 10-2 CCR+ (28) were negative and positive control strains, respectively.
Western blots.
Cultures were grown in MH broth to an OD600 of 0.7 to 0.9 and normalized by the OD600. Cells then were pelleted and resuspended in 100 μl cell lysis buffer (167 mM Tris base, pH 6.8, 19% glycerol, 1.9% sodium dodecyl sulfate, 9.5% β-mercaptoethanol [final concentrations]). Whole-cell extracts were separated by polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Membranes were probed using anti-AcrA antibodies as described previously (34). Each strain was analyzed in duplicate, and ATCC 25922 was included on each gel for the normalization of the AcrA levels. Isolates with AcrA levels more than two standard deviations greater than the average level for fluoroquinolone-susceptible isolates were considered significantly increased (≥1.9-fold).
Electronic data storage.
An electronic file containing all E. coli antibiograms from 1 July 1999 to 31 December 2004 (n > 17,000) obtained from Ben Taub General Hospital was parsed and stored in an Oracle database that could be queried electronically (3). The file included data from the patient (age, sex, and hospital location) and the susceptibility status of two or more aminoglycosides (amikacin, gentamicin, and/or tobramycin), carbapenems (imipenem and meropenem), cephalosporins (ceftriaxone, ceftazidime, cefepime, cefotaxime, cefoxitin, cefazolin, cefpodoxime, cefuroxime, and cefotetan), fluoroquinolones (ciprofloxacin, gatifloxacin, levofloxacin, norfloxacin, and/or ofloxacin), and penicillins (ampicillin, augmentin, piperacillin, piperacillin-tazobactam, and ticarcillin-clavulanic acid), as well as aztreonam, nitrofurantoin, and sulfamethoxazole-trimethoprim. All molecular characterization data from the clinical isolates in spreadsheets were imported into the database. Thus, the antibiogram data described above, together with all clinical isolate and MIC data, are housed in our custom-designed, evolving Oracle database.
Statistical methods.
All parameters were analyzed by paired t tests. t test values with P < 0.05 (95% confidence interval) were considered significant.
RESULTS
Topoisomerase mutations in clinical isolates.
At least three mutations, two of which must be in gyrA, are required to achieve CLSI-classified clinical resistance (14, 26), and the MICs for quadruple mutants are ∼10-fold higher than those for triple mutants (14). MICs differed for individual fluoroquinolones such that ciprofloxacin and norfloxacin MICs were increased 10-fold more by the topoisomerase gene mutations than were gatifloxacin or levofloxacin MICs (14, 26).
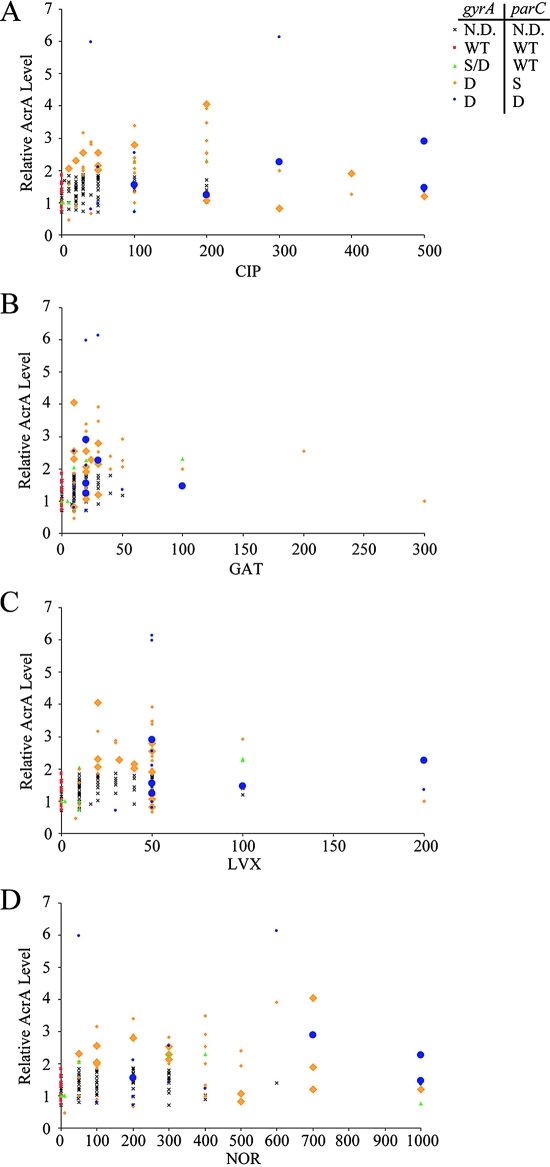
The simultaneous presence of two mutations in gyrA and one or two mutations in parC could account for some of the high MICs observed for the fluoroquinolone-resistant clinical isolates. Thus, we sequenced the gyrA, gyrB, parC, and parE QRDRs. Seventy-eight isolates were chosen to represent the full range of fluoroquinolone MICs and the four strains with unique phenotypes (4). All of the fluoroquinolone-resistant isolates (n = 58) had gyrA QRDR mutations, and two of these isolates also had gyrB QRDR mutations (Table 3). Approximately 85% of the fluoroquinolone-resistant isolates with gyrA QRDR mutations also had parC QRDR mutations, and ∼35% of strains encoding mutations in gyrA and parC also had parE QRDR mutations. No isolate had the wild-type gyrA sequence and mutations in the QRDR of parC. All of the gyrA and parC QRDR mutations that we uncovered have been documented previously for clinical isolates and are known to increase fluoroquinolone MICs (reviewed in references 11, 14, and 24). The mutations in gyrB and parE may increase fluoroquinolone MICs somewhat. We found, however, that the MICs for clinical isolates encoding gyrA and parC mutations with wild-type gyrB and parE sequences were greater than or equal to those for strains with gyrB or parE mutations. Therefore, it does not appear that these gyrB and parE mutations discernibly affect fluoroquinolone MICs. Consequently, we show the data only by the number of gyrA and parC mutations in Fig. 1. Fifteen isolates had wild-type gyrA and parC, 6 had either one or two gyrA mutations and wild-type parC, 42 had two mutations in gyrA and one in parC, and 12 had two mutations in both gyrA and parC. In general, the fluoroquinolone MICs for clinical isolates with increased numbers of gyrA and parC mutations were higher (Fig. 1, Table 3).
TABLE 3.
Distribution of gyrA, gyrB, parC, and parE sequences in clinical E. coli isolates
| Topoisomerase sequencea
|
No. of isolatesb | MIC range (median), in μg/ml
|
||||||
|---|---|---|---|---|---|---|---|---|
| gyrA | gyrB | parC | parE | CIP | GAT | LVX | NOR | |
| WT | WT | WT | WT | 15 | 0.004-0.094 (0.012) | 0.007-0.094 (0.016) | 0.016-0.125 (0.034) | 0.023-0.115 (0.064) |
| WT | WT | WT | A458 | 1 | 0.010 | 0.010 | 0.034 | 0.047 |
| A83 | WT | WT | WT | 1 | 0.094 | 0.094 | 0.168 | 0.194 |
| L83, N87 | WT | WT | WT | 2 | 10, 100 | 10 | 10 | 10, 1000 |
| L83, N87 | ND | WT | ND | 2 | 10, 200 | 10, 100 | 10, 100 | 50, 400 |
| L83, N87 | WT | K84 | WT | 1 | 100 | 30 | 50 | 200 |
| L83, N87 | ND | K84 | ND | 3 | 20-30 (30) | 10-20 (20) | 10-50 (20) | 50-100 (100) |
| L83, N87 | WT | I80 | WT | 9 | 10-400 (100) | 10-40 (20) | 8-50 (50) | 10-500 (100) |
| L83, N87 | ND | I80 | ND | 13 | 30-300 (50) | 10-200 (20) | 20-100 (45) | 50-400 (100) |
| L83, N87 | WT | I80 | M464 | 1 | 100 | 20 | 50 | 400 |
| L83, N87 | WT | I80 | A458 | 8 | 100-200 (200) | 10-300 (25) | 20-200 (50) | 50-700 (500) |
| L83, N87 | WT | I80, G84 | WT | 5 | 50-500 (250) | 10-100 (25) | 50-200 (75) | 200-1,000 (500) |
| L83, N87 | WT | R80, V84 | WT | 1 | 500 | 20 | 50 | 700 |
| L83, N87 | T471 | I80, G84 | WT | 1 | 100 | 20 | 30 | 200 |
| L83, N87 | WT | I80, K84 | D460 | 1 | 200 | 20 | 50 | 400 |
| L83, N87 | V456 | I80 | A458 | 1 | 200 | 20 | 50 | 700 |
| L83, Y87 | WT | WT | WT | 2 | 2, 16 | 0.500, 20 | 2, 100 | 8, 300 |
| L83, Y87 | WT | K84 | WT | 1 | 500 | 30 | 50 | 1000 |
| L83, Y87 | WT | I80 | WT | 5 | 10-300 (50) | 10-50 (20) | 10-50 (20) | 50-400 (100) |
| L83, Y87 | ND | I80 | ND | 3 | 40-50 (45) | 20-30 (24) | 20-32 (32) | 200-300 (250) |
| L83, Y87 | WT | I80 | A458 | 2 | 100 | 20 | 50 | 300, 400 |
| L83, Y87 | WT | I80, G84 | WT | 3 | 40-100 (100) | 10 | 50 | 100-300 (300) |
| L83, Y87 | WT | I80, G84 | A458 | 1 | 300 | 30 | 200 | 1,000 |
WT, wild type; ND, not determined.
The total no. of isolates was 75.
FIG. 1.
Topoisomerase gene mutations, the presence of aac(6′) Ib-cr, and relative levels of AcrA as a function of fluoroquinolone MICs. Ciprofloxacin (A), gatifloxacin (B), levofloxacin (C), and norfloxacin (D) MICs (in micrograms/milliliter) were determined by agar dilution or Etest and are displayed on the x axis. The y axis denotes the relative level of AcrA compared to that of the CLSI control strain ATCC 25922, as determined by Western blotting. Each point represents one clinical isolate. Symbols represent gyrA and/or parC sequences: wild-type gyrA and parC are indicated by red squares (WT, WT), gyrA single or double mutations and wild-type parC by green triangles (S/D, WT), double gyrA and single parC mutations by orange diamonds (D, S), and double gyrA and double parC mutations by blue circles (D, D). Larger symbols represent that strains had aac(6′) Ib-cr. An X indicates isolates with no topoisomerase sequence or plasmid-borne gene data. CIP, ciprofloxacin; GAT, gatifloxacin; LVX, levofloxacin; NOR, norfloxacin; N.D., not determined; S, single mutation; and D, double mutation.
Plasmid-mediated resistance genes.
To determine whether known plasmid-borne resistance genes were present, we screened all of the isolates with sequenced topoisomerase QRDRs. Approximately 25% carried aac(6′)-Ib-cr (Fig. 1), and all of these isolates also had at least three topoisomerase mutations (Fig. 1). Three isolates harbored the parental aac(6′)-Ib gene, which does not affect fluoroquinolone resistance (23). None of the isolates harbored qnrA. Because qnrB occurs with the same low frequency as qnrA (2%) and qnrS has yet to be found in E. coli clinical isolates in the United States (22), we did not screen for these genes.
Because aac(6′)-Ib-cr increases ciprofloxacin and norfloxacin MICs (23), we expected that it would be present concurrently in isolates for which the normalized MICs of these two fluoroquinolones were high (4). Indeed, ∼60% of the isolates for which the normalized ciprofloxacin and norfloxacin MICs were high had aac(6′)-Ib-cr. Paired t tests showed that the ciprofloxacin and norfloxacin MICs for isolates with aac(6′)-Ib-cr were significantly higher (P < 0.01) than those for strains without aac(6′)-Ib-cr. Like ciprofloxacin and norfloxacin, gatifloxacin lacks a piperazinyl amine substitution, but the effect of Aac(6′)-Ib-cr on gatifloxacin has not been determined (23). Paired t tests revealed no significant difference between gatifloxacin (or levofloxacin) MICs for isolates with or without the plasmid-borne gene. Thus, the presence of aac(6′)-Ib-cr is very well correlated with increased MICs of ciprofloxacin and norfloxacin but not of gatifloxacin or levofloxacin.
AcrAB and fluoroquinolone resistance in isogenic laboratory strains.
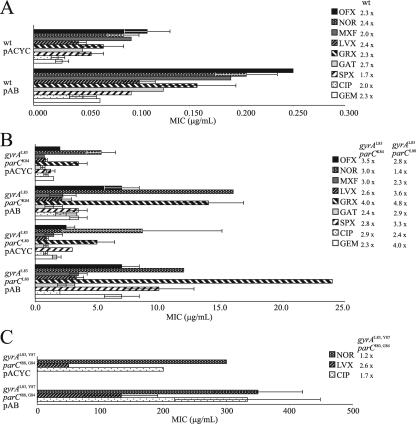
Previous studies have shown that increased levels of AcrAB can increase ciprofloxacin and norfloxacin MICs up to fivefold for a wild-type laboratory E. coli K-12 strain (16); however, the effect of increased AcrAB levels on other fluoroquinolone MICs or when combined with topoisomerase mutations has not been reported. We used the AcrAB overexpression plasmid pAB (16) to measure fluoroquinolone MICs for isogenic strains based upon the parental C600 strain (15) encoding defined gyrA and/or parC mutations to determine whether there were synergistic effects of topoisomerase mutations and increased AcrAB (Fig. 2). Western blotting confirmed that AcrA levels were increased ∼20-fold in strains with pAB in these experiments (data not shown).
FIG. 2.
Fluoroquinolone MICs for strains with the AcrAB overexpression plasmid and the empty vector control plasmid. Fluoroquinolone MICs for strains containing either the AcrAB overexpression plasmid pAB or the empty vector control plasmid pACYC184 are shown on the x axis. The y axis denotes the isogenic strains. (A) C600 parental wild-type (wt) strains; (B) C600-based strains encoding single gyrA and single parC mutations; and (C) C600-based strains encoding double gyrA and double parC mutations. The bars are filled according to the fluoroquinolone tested as indicated in the legend, and the increases (n-fold) in MICs for each strain are shown next to the legend. OFX, ofloxacin; NOR, norfloxacin; MXF, moxifloxacin; LVX, levofloxacin; GRX, grepafloxacin; GAT, gatifloxacin; SPX, sparfloxacin; CIP, ciprofloxacin; and GEM, gemifloxacin.
We found that all of the tested fluoroquinolone MICs increased 1.7- to 2.6-fold for the parental strain with wild-type topoisomerase genes (Fig. 2A). Fluoroquinolone MICs also increased for strains encoding a single mutation in gyrA combined with a single mutation in parC (Fig. 2B). For the gyrAL83, Y87 parCI80, G84 quadruple topoisomerase mutant strain, the ciprofloxacin and norfloxacin MICs were not significantly increased, and the levofloxacin MIC increased approximately threefold (Fig. 2C). Thus, there is not a synergistic effect on fluoroquinolone MICs by combining AcrAB and topoisomerase mutations. In addition, increased levels of AcrAB did not always increase fluoroquinolone MICs, and when they did it was by less than threefold.
AcrAB and fluoroquinolone resistance in clinical isolates.
We first determined the AcrA levels, relative to that of the CLSI control strain, in a sample (n = 22) of fluoroquinolone-susceptible clinical isolates in order to establish the baseline level of AcrA. The data for the susceptible isolates can be seen as the points below the MICs for the CLSI susceptibility breakpoints in Fig. 1 (≤1 μg/ml ciprofloxacin, ≤2 μg/ml for gatifloxacin and levofloxacin, and ≤4 μg/ml for norfloxacin). The average relative AcrA level in the 22 fluoroquinolone-susceptible isolates was 1.18, with a standard deviation of 0.37, which was not statistically different from the value for the CLSI control strain (ATCC 25922). Isolates with AcrA levels that were >2 standard deviations above the average AcrA levels of susceptible isolates were considered significantly increased. No fluoroquinolone-susceptible isolates (based upon hospital antibiogram data and the MIC measurements determined in this study) had increased AcrA levels greater than 1.9-fold.
With the level of AcrA in drug-susceptible strains established, we then determined the relative AcrA levels in the 78 isolates from sequencing and screening experiments, as well as those of an additional 75 isolates for greater statistical significance. Approximately one-third of all fluoroquinolone-resistant clinical isolates had increased levels of AcrA (average increase [n-fold], 2.71 ± 0.98, ranging from 1.9 to 6.3). AcrA levels in isolates for which the norfloxacin MICs were ≥400 μg/ml had significantly higher AcrA levels (average, 1.9) than those for which the norfloxacin MICs were below 400 μg/ml (average, 1.6) by paired t test (P < 0.01).
To determine whether AcrA levels correlated with the hospital-determined susceptibility status of any antibiotic (see Materials and Methods for a complete list of included drugs), we analyzed the hospital antibiograms for all isolates with quantified AcrA levels. Increased levels of AcrA (>1.9-fold) were never observed in isolates that the hospitals already had deemed to be fluoroquinolone susceptible. For all other drugs included on the antibiograms, AcrA was increased in both susceptible and nonsusceptible isolates. Thus, increased AcrA clearly correlates with fluoroquinolone nonsusceptibility but not with the nonsusceptibility of any other antimicrobial agent included in the antibiograms.
Correlations between fluoroquinolone MICs and molecular resistance mechanisms.
In the companion paper (4), we found that the ciprofloxacin and levofloxacin MICs for most clinical isolates fell into two distinct groups (20 and 100 μg/ml for ciprofloxacin and 10 and 50 μg/ml for levofloxacin). The characterization of the molecular resistance mechanisms of these isolates showed that those in the lower-MIC groups (10 μg/ml levofloxacin and 20 μg/ml ciprofloxacin) generally had one or two gyrA mutations, lacked parC mutations, did not harbor aac(6′)-Ib-cr, and had normal AcrA levels (Fig. 1). Isolates in the higher-MIC groups (50 μg/ml levofloxacin and 100 μg/ml ciprofloxacin) had two gyrA mutations, one or two parC mutations, and occasionally the presence of aac(6′)-Ib-cr and increased levels of AcrA (Fig. 1). Thus, differences in known resistance genotypes likely account for the two groups of MICs for ciprofloxacin and levofloxacin.
Additionally, MICs of two other drug pairs (ciprofloxacin and norfloxacin as well as gatifloxacin and levofloxacin) were similar for the drugs in each pair but different from the other pair. In general, ciprofloxacin and norfloxacin MICs were higher than gatifloxacin and levofloxacin MICs. Because the accumulation of multiple topoisomerase gene mutations and Aac(6′)-Ib-cr affect ciprofloxacin and norfloxacin MICs more than gatifloxacin and levofloxacin MICs (14), the presence of multiple gyrA and parC mutations and Aac(6′)-Ib-cr in these clinical isolates likely explains these MIC data.
Patient factors and fluoroquinolone resistance.
We previously showed that E. coli isolates from males, older patients, urine, and hospitalized patients were more likely to be nonsusceptible to fluoroquinolones than those from females, younger patients, culture samples other than urine, and outpatients (3). Because the MICs for fluoroquinolone-resistant isolates were so high and widely variable among the different fluoroquinolones, it is possible that these MIC differences correlate with culture site or patient age, location, and/or sex. Therefore, we asked whether these patient factors correlated with MICs or any particular resistance mechanisms that affect fluoroquinolone MICs, as they did with fluoroquinolone nonsusceptibility in our previous work (3). Paired t tests showed that males and older patients had a distribution of fluoroquinolone MICs and resistance mechanisms that was indistinguishable from that of females and younger patients (data not shown). All patients, regardless of age, sex, or hospital location, were equally likely to have an isolate for which the fluoroquinolone MICs were high or had a particular resistance mechanism (P > 0.05).
The majority of the isolates (∼80%) in this study originated from urine, where many fluoroquinolones concentrate. The MICs for E. coli strains isolated from urine, therefore, might be expected to be higher than those isolated from other tissues. Urine and nonurine culture sites, however, had statistically indistinguishable distributions of fluoroquinolone MICs (P > 0.05). Thus, whereas clinical fluoroquinolone nonsusceptibility strongly correlates with patient variables, MICs appear to be independent of these patient variables in fluoroquinolone- resistant clinical isolates. The analyses of fluoroquinolone resistance factors from more patients or knowledge of previous antibiotic exposure could provide additional insight into the complex relationships between the bacterium and the host.
DISCUSSION
The role of AcrAB in fluoroquinolone resistance.
The overproduction of AcrAB had minimal effects (1.7- to 2.6-fold), if any, on fluoroquinolone MICs (Fig. 2) (16, 31). At the same time, increased AcrAB levels were observed only in fluoroquinolone-resistant isolates and never in fluoroquinolone-susceptible isolates. This correlation did not exist for any other antibiotic tested in the antibiograms. It seems unlikely that the twofold increase in fluoroquinolone MICs that increased AcrAB caused in laboratory isolates plays a large role in directly increasing fluoroquinolone MICs. If AcrAB plays a small direct role in increasing fluoroquinolone MICs, then there must be additional mutations present to account for the high MICs observed for clinical isolates. Indeed, all of the isolates with increased AcrAB also contained at least two mutations in gyrA. Furthermore, the data presented here suggest that while there is a large additive effect in the accumulation of multiple topoisomerase mutations, increased AcrAB levels do not further increase fluoroquinolone MICs in already highly resistant strains. However, it is possible that increased levels of AcrAB affect fluoroquinolone resistance in a way that does not directly involve fluoroquinolone efflux.
Existing data support a model in which AcrAB plays a role in quorum sensing by emitting quorum-sensing signals (13, 20, 21, 32). In E. coli, the overexpression of the quorum-sensing regulator SdiA increases fluoroquinolone MICs and also increases AcrAB levels. Increased fluoroquinolone MICs mediated by this SdiA overexpression is dependent upon AcrAB (21). Bacteria grown in the presence of conditioned media from separate cultures of cells overexpressing AcrAB grew slower than those in conditioned media from cells not overexpressing AcrAB (13, 20, 21, 32). Thus, it is possible that clinical isolates increase levels of AcrAB because of its role in quorum sensing or because doing so confers some fitness advantage to the isolates.
MarA, RobA, SoxA, and SdiA are global regulators that increase fluoroquinolone MICs, and this increase is dependent upon AcrAB (18, 21, 25, 29, 30). Increased MarA levels not only increase fluoroquinolone MICs but also somehow protect bacteria from the bactericidal effect of fluoroquinolones at concentrations higher than the MIC (8). Whether this protection has clinical relevance is not known. Therefore, increased AcrAB levels in clinical isolates might have arisen because of alterations in MarA, RobA, or SoxA or from selection for MarA-regulated bactericidal protection.
Unexplained fluoroquinolone MICs.
We established an estimated range of expected MICs based upon the published MICs for strains encoding defined topoisomerase mutations (14), the MICs for strains with increased AcrAB, and published MICs for strains containing aac(6′)-Ib-cr. Based on their characterized genotypes, the MICs for 40 to 70% of isolates were consistent with those expected based upon data from isogenic laboratory E. coli strains. However, the MICs for 20 to 45% of the isolates (4) were lower than expected, and the MICs for 5 to 15% were higher than expected. Fluoroquinolone MICs for the majority of these isolates, including all with lower values than expected, deviated 10- to 50-fold in either direction. The MICs for three isolates (ELZ4013, ELZ4072, and ELZ4258), however, were 500- to 1,600-fold higher than could be explained by the resistance mechanisms present. The three isolates encoded only a single or double mutation in gyrA, and two of these isolates (ELZ4072 and ELZ4258) also had increased AcrA levels. The norfloxacin MIC for a defined laboratory strain encoding the same double gyrA mutation as these three isolates was 0.625 μg/ml, while the norfloxacin MICs for ELZ4013, ELZ4072, and ELZ4258 were 1,000, 400, and 300 μg/ml, respectively. It is possible that compensatory mutations that alter fluoroquinolone MICs occurred as a consequence of a loss of fitness from the original mutation, such as the reduced gyrase supercoiling activity caused by gyrA mutations (2). In addition, some unknown genotypes or aspects of the genetic backgrounds of the isolates may have affected the MICs. If additional, unknown genotypes exist, they could either confer relatively small (i.e., 2- to 16-fold, similarly to that of most of the known genotypes alone) or dramatic (i.e., 10,000-fold, unlike any known genotype) increases in fluoroquinolone MICs.
The isolates (ELZ4004, ELZ4132, ELZ4601, and ELZ4686) with unusual resistance phenotypes uncovered in the companion study (4) had three or four mutations in gyrA and parC and increased relative levels of AcrA or aac(6′)-Ib-cr. Other isolates with identical combinations of the known resistance genotypes did not share the MIC phenotypes. Thus, it seems likely that unknown mutations, genetic backgrounds, or novel fluoroquinolone resistance genotypes exist in these clinical isolates. Two known backgrounds include a hypermutator background or one that is particularly susceptible to SOS induction by the fluoroquinolones (6, 7), both of which have been shown to be important in the development of fluoroquinolone resistance.
Acknowledgments
We thank Robert L. Atmar and Charles E. Stager for collecting clinical isolates and patient data; Richard J. Hamill, James Versalovic, Richard Sucgang, Robert S. Atmar, Sheila I. C. Hull, Barbara E. Murray, Timothy G. Palzkill, and Joseph F. Petrosino for helpful advice; and Nicholas G. Brown and Carey B. Gardener for technical assistance.
S.K.M. was supported by the Houston Area Molecular Biophysics (NIH T32-GM008280) and L.B.B. by the Pharmacoinformatics (NIH T90 DK070109) training programs. L.Z. was funded by NIH R01-AI054830. A grant from The Burroughs Wellcome Fund was used to construct the Oracle database.
Footnotes
Published ahead of print on 6 October 2008.
REFERENCES
- 1.Appleyard, R. K. 1954. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagel, S., V. Hullen, B. Wiedemann, and P. Heisig. 1999. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 43:868-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becnel Boyd, L., R. L. Atmar, G. L. Randall, R. J. Hamill, D. Steffen, and L. Zechiedrich. 2008. Increased fluoroquinolone resistance with time in Escherichia coli from >17,000 patients at a large county hospital as a function of culture site, age, sex, and location. BMC Infect. Dis. 8:4. [DOI] [PMC free article] [PubMed]
- 4.Becnel Boyd, L., M. J. Maynard, S. K. Morgan-Linnell, L. B. Horton, R. Sucgang, R. J. Hamill, J. Rojo Jimenez, J. Versalovic, D. Steffen, and L. Zechiedrich. 2009. Relationships among ciprofloxacin, gatifloxacin, levofloxacin, and norfloxacin MICs for Escherichia coli clinical isolates. Antimicrob. Agents Chemother. 53:229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirz, R. T., J. K. Chin, D. R. Andes, V. de Crecy-Lagard, W. A. Craig, and F. E. Romesberg. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 3:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirz, R. T., and F. E. Romesberg. 2006. Induction and inhibition of ciprofloxacin resistance-conferring mutations in hypermutator bacteria. Antimicrob. Agents Chemother. 50:220-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman, J. D., D. G. White, and S. B. Levy. 1996. Multiple antibiotic resistance (mar) locus protects Escherichia coli from rapid cell killing by fluoroquinolones. Antimicrob. Agents Chemother. 40:1266-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khodursky, A. B., E. L. Zechiedrich, and N. R. Cozzarelli. 1995. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:11801-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu, T., X. Zhao, and K. Drlica. 1999. Gatifloxacin activity against quinolone-resistant gyrase: allele-specific enhancement of bacteriostatic and bactericidal activities by the C-8-methoxy group. Antimicrob. Agents Chemother. 43:2969-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazzariol, A., Y. Tokue, T. M. Kanegawa, G. Cornaglia, and H. Nikaido. 2000. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein acrA. Antimicrob. Agents Chemother. 44:3441-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan-Linnell, S. K., and L. Zechiedrich. 2007. Contributions of the combined effects of topoisomerase mutations toward fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 51:4205-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 6th edition, M7-A6. NCCLS, Wayne, PA.
- 16.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oethinger, M., W. V. Kern, A. S. Jellen-Ritter, L. M. McMurry, and S. B. Levy. 2000. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob. Agents Chemother. 44:10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pesci, E. C., J. B. J. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinlone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahmati, S., S. Yang, A. L. Davidson, and E. L. Zechiedrich. 2002. Control of the AcrAB multidrug efflux pump by quorum sensing regulator SdiA. Mol. Microbiol. 43:677-685. [DOI] [PubMed] [Google Scholar]
- 22.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 23.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 24.Ruiz, J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 51:1109-1117. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka, T., T. Horii, K. Shibayama, K. Sato, S. Ohsuka, Y. Arakawa, K. Yamaki, K. Takagi, and M. Ohta. 1997. RobA-induced multiple antibiotic resistance largely depends on the activation of the AcrAB efflux. Microbiol. Immunol. 41:697-702. [DOI] [PubMed] [Google Scholar]
- 26.Turner, A. K., S. Nair, and J. Wain. 2006. The acquisition of full fluoroquinolone resistance in Salmonella Typhi by accumulation of point mutations in the topoisomerase targets. J. Antimicrob. Chemother. 58:733-740. [DOI] [PubMed] [Google Scholar]
- 27.Wang, H., J. L. Dzink-Fox, M. Chen, and S. B. Levy. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webber, M. A., and L. J. Piddock. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob. Agents Chemother. 45:1550-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, S., S. Clayton Rahmati, and E. L. Zechiedrich. 2003. Relative contributions of the AcrAB, MdfA and NorE efflux pumps to quinolone resistance in Escherichia coli. J. Antimicrob. Chemother. 51:545-556. [DOI] [PubMed] [Google Scholar]
- 32.Yang, S., C. R. Lopez, and E. L. Zechiedrich. 2006. Quorum sensing and multidrug transporters in Escherichia coli. Proc. Natl. Acad. Sci. USA 103:2386-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida, H., T. Kojima, J.-I. Yamagishi, and S. Nakamura. 1988. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol. Gen. Genet. 211:1-7. [DOI] [PubMed] [Google Scholar]
- 34.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 182:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]