Abstract
The sfp cluster, encoding Sfp fimbriae and located in the large plasmid of sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157 (pSFO157), has been considered a unique characteristic of this organism. We discovered and then characterized the sfp cluster in EHEC O165:H25/NM (nonmotile) isolates of human and bovine origin. All seven strains investigated harbored a complete sfp cluster (carrying sfpA, sfpH, sfpC, sfpD, sfpJ, sfpF, and sfpG) of 6,838 bp with >99% nucleotide sequence homology to the sfp cluster of SF EHEC O157:NM. The sfp cluster in EHEC O165:H25/NM strains was located in an ∼80-kb (six strains) or ∼120-kb (one strain) plasmid which differed in structure, virulence genes, and sfp flanks from pSFO157. All O165:H25/NM strains belonged to the same multilocus sequence type (ST119) and were only distantly phylogenetically related to SF EHEC O157:NM (ST11). The highly conserved sfp cluster in different clonal backgrounds suggests that this segment was acquired independently by EHEC O165:H25 and SF EHEC O157:NM. Its presence in an additional EHEC serotype extends the diagnostic utility of PCR targeting sfpA as an easy and efficient approach to seek EHEC in patients' stools. The reasons for the convergence of pathogenic EHEC strains on a suite of virulence loci remain unknown.
Escherichia coli O157:H7 is the most commonly isolated serotype of enterohemorrhagic E. coli (EHEC) worldwide (29, 48). However, an increasing number of non-O157:H7 EHEC serotypes have been isolated from patients (3, 5, 9, 19, 27, 28, 29, 31, 32, 33, 51). Different serotypes possess various, often serotype-specific, combinations of virulence genes (5, 46, 49, 55) within genomic islands, bacteriophages, or large plasmids (4, 10, 11, 12, 13, 15, 16, 30, 37, 38, 39). Allelic variations of some genes, such as eae, encoding the adhesin intimin, and of course stx, encoding Shiga toxin (Stx), are present in most EHEC strains (3, 5, 9). However, other genes are restricted to particular serotypes. Such genes include, for example, the lpf operon (encoding long polar fimbriae) in O island (OI) 154 of E. coli O157:H7 EDL933 (50), which has been found only in EHEC O157:H7/NM (nonmotile) isolates and their progenitor, E. coli O55:H7 (5, 49, 50), and the sfp gene cluster, which we recently proposed to be restricted to the large plasmid of sorbitol-fermenting (SF) EHEC O157:NM, pSFO157 (11, 12). The sfp cluster is inserted into the region of pSFO157 where katP and espP (encoding a catalase-peroxidase and a serine protease, respectively) reside in the large plasmid of EHEC O157:H7, pO157 (13). The cluster is flanked by insertion sequences and an origin of plasmid replication (11, 12), indicating that horizontal transfer gave rise to its presence in SF EHEC O157:NM.
The sfp cluster (carrying sfpA, sfpH, sfpC, sfpD, sfpJ, sfpF, and sfpG) encodes Sfp fimbriae that mediate mannose-resistant hemagglutination (11) and possibly also adherence of SF EHEC O157:NM organisms to human intestinal epithelial cells (34). The absence of this locus in thousands of Stx-producing E. coli strains belonging to many different serotypes led us to assume that the sfp cluster is unique to SF EHEC O157:NM (23). However, using PCR to target sfpA (encoding the major fimbrial subunit) in stool samples (23), we unexpectedly isolated an sfpA-positive EHEC strain of serotype O165:H25 from a patient with hemolytic-uremic syndrome (HUS). Here we report the extended analysis of this serotype, focusing on conservation of the sfp cluster.
MATERIALS AND METHODS
Bacterial strains.
sfpA-positive EHEC O165:H25 strain 820/08 from a patient with HUS was isolated at the National Consulting Laboratory for HUS, Münster, Germany, during routine diagnostic efforts by use of a published protocol (23). Additionally, five EHEC O165:H25/NM strains, four from epidemiologically unrelated patients with diarrhea and one from a healthy bull (Table 1), were from the collection of the Robert Koch Institute, Wernigerode, Germany. Three of these strains were described previously (25). EHEC O165:H25 strain MT52 (Table 1) was from a patient with bloody diarrhea in Montana (27). SF EHEC O157:NM HUS isolate 3072/96, in which the sfp cluster was originally identified (11), and EHEC O157:H7 strain EDL933 (13, 35) were used as positive and negative controls, respectively.
TABLE 1.
EHEC O165:H25/NM strains and controls investigated in this study, with plasmid characteristics
| Strain no. | Yr, country of isolationa | Clinical diagnosisb or source | Serotypec | fliC RFLP type | Plasmid characteristics
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plasmid profiled | Southern blot hybridization with probee
|
|||||||||
| sfpA | EHEC hlyA | espPf | katP | etpD | ||||||
| 820/08 | 2008, G | HUS | O165:H25 | H25 | 80 | 80 | 80 | 80 | 80 | — |
| 98-08419-1 | 1998, G | D | O165:NM | H25 | 80, 120 | 80 | 80 | 80 | 80 | — |
| 99-02258 | 1999, G | D | O165:H25 | H25 | 80, 120 | 80 | 80 | 80 | 80 | — |
| 02-11228 | 2002, G | D | O165:H25 | H25 | 80, 120 | 120 | 80, 120 | 80 | 80, 120 | — |
| 04-07734 | 2004, G | D | O165:H25 | H25 | 80 | 80 | 80 | 80 | 80 | — |
| MT52 | 2000, US | BD | O165:H25 | H25 | 80 | 80 | 80 | 80 | 80 | — |
| 00-09087 | 2000, G | Cattle | O165:H25 | H25 | 60, 80 | 80 | 80 | 80 | 80 | — |
| 3072/96 | 1996, G | HUS | O157:NM | H7 | 121 | 121 | 121 | — | — | 121 |
| EDL933 | 1982, US | Hamburger | O157:H7 | H7 | 92 | — | 92 | 92 | 92 | 92 |
G, Germany; US, United States.
HUS, hemolytic-uremic syndrome (defined as the triad of hemolytic anemia, thrombocytopenia, and acute renal failure) (48); BD, bloody diarrhea; D, diarrhea.
NM, nonmotile.
Sizes of plasmids (in kb). The sizes of plasmids in strains 3072/96 and EDL933 are based on published sequences (GenBank accession no. AF401292 and AF074613, respectively) (12, 13); the sizes of the other plasmids were estimated according to their mobility by agarose gel electrophoresis (Fig. 2A).
The sfpA, EHEC hlyA, espP, katP, and etpD genes encode the major subunit of Sfp fimbriae, EHEC hemolysin, a serine protease, a catalase-peroxidase, and a type II secretion system, respectively (12, 13). The data are sizes (in kb) of plasmids that hybridized with the respective probes. —, no signal. The presence or absence of the respective genes was confirmed by PCR.
All EHEC O165:H25/NM strains contain espP subtype δ, whereas strain EDL933 contains espP subtype α, as demonstrated using the espP subtyping scheme (8).
Phenotyping.
Isolates were confirmed as E. coli (API 20 E; bioMérieux, Nürtingen, Germany) and serotyped (27, 40). Fermentation of sorbitol was determined on sorbitol MacConkey agar (SMAC), β-d-glucuronidase activity was assessed using nutrient agar with 4-methylumbelliferyl-β-d-glucuronide (Becton Dickinson, Sparks, MD), EHEC hemolysin production was identified using enterohemolysin agar (Sifin, Berlin, Germany), and production of α-hemolysin was sought with Columbia blood agar (Heipha, Heidelberg, Germany). Resistance to tellurite was determined by assessing the ability of strains to grow on cefixime-tellurite SMAC (CT-SMAC). Stx production was determined using a Vero cell assay (3).
Analysis of the sfp cluster.
The presence of the complete sfp cluster was sought using concatenated PCRs in which each sfp gene and linkages between contiguous genes were amplified (Fig. 1 and Table 2). Purified PCR products (PCR purification kit; Qiagen, Hilden, Germany) were sequenced using an automated ABI Prism 3130xl genetic analyzer and an ABI Prism BigDye Terminator ready reaction cycle sequencing kit (version 3.1; Applied Biosystems, Darmstadt, Germany). Sequences were analyzed with Ridom TraceEditPro software (Ridom GmbH, Würzburg, Germany), and homology was sought using the EMBL-GenBank database (http://www.ncbi.nlm.nih.gov/BLAST).
FIG. 1.
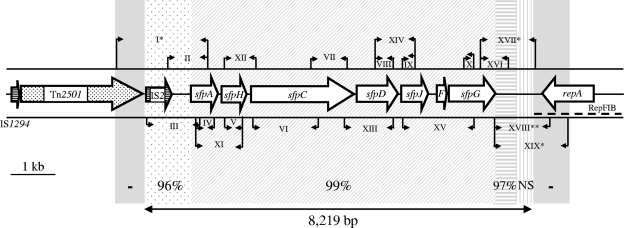
Physical map of the sfp cluster and its flanks in SF EHEC O157:NM strain 3072/96 and PCR strategy used for analysis of the sfp cluster in EHEC O165:H25/NM. The large arrows indicate sfp genes (F, sfpF) and genes adjacent to the sfp cluster in strain 3072/96. A transposon-like element is dotted, and insertion sequences are horizontally striped. The dashed line below repA depicts the RepFIB origin of replication of pSFO157. The small arrows indicate the positions of PCR primers (details for PCRs I to XIX are given in Table 2). In all but four PCRs, amplicons of identical sizes were obtained from each of seven EHEC O165 strains and strain 3072/96. In the remaining PCRs, no amplicons (*) or amplicons of different sizes (**) than those from strain 3072/96 (∼600 bp instead of 1,343 bp) were elicited from EHEC O165 strains. The double arrow below the graph depicts the 8,219-bp plasmid region which was sequenced from EHEC O165:H25 strains 820/08 and 04-07734 and which comprises the sfp cluster (6,838 bp) and its upstream (828 bp) and downstream (553 bp) flanks. The numbers above this arrow indicate nucleotide sequence homologies of the corresponding regions between EHEC O165:H25 strains (which were 100% identical) and strain 3072/96. NS, no significant homology was found using BLAST algorithms (http://www.ncbi.nlm.nih.gov/BLAST). The gray regions (−) adjacent on both sides to the sequenced stretch were not detected in EHEC O165 by the PCR strategy used.
TABLE 2.
PCR primers and conditions used for analysis of the sfp cluster and its flanking regions in EHEC O165:H25/NM
| PCR no. | Primera | Sequence (5′-3′) | Target | PCR conditionsb
|
PCR product size (bp) | ||
|---|---|---|---|---|---|---|---|
| Denaturing | Annealing | Extension | |||||
| I | wprom-3 | GCT GGA CGC CGG TGC TTA TT | Tn2501-sfpA | 94°C, 30 s | 61°C, 60 s | 72°C, 150 s | 1,900 |
| wprom-4 | CCA GAT TGC CGC TCG CTT CAG | ||||||
| II | IS2-1 | GTG ATA ACA AGC GTC TGG AAA | IS2-sfpA | 94°C, 30 s | 57°C, 60 s | 72°C, 60 s | 867 |
| wprom-4 | CCA GAT TGC CGC TCG CTT CAG | ||||||
| III | IS2-3 | CGA TGA CTG GAA GGA TAG | IS2-sfpA | 94°C, 30 s | 50°C, 60 s | 72°C, 90 s | 1,161 |
| sfpA-2 | AGC CAG TTC CTT GAC ATC | ||||||
| IV | sfpA-U | AGC CAA GGC CAA GGG ATT ATT A | sfpA | 94°C, 30 s | 59°C, 60 s | 72°C, 60 s | 440 |
| sfpA-L | TTA GCA ACA GCA GTG AAG TCT C | ||||||
| V | sfpH-1 | CCA TCG CCC TCC TTT CCT CT | sfpH | 94°C, 30 s | 58°C, 60 s | 72°C, 60 s | 425 |
| sfpH-2 | TCC AGC CCC TCT TCA TTT CC | ||||||
| VI | sfpC-5 | TAT GCG TTG TTG TTT TCC | sfpC 5′ end | 94°C, 30 s | 55°C, 60 s | 72°C, 150 s | 1,643 |
| sfpC-6 | TCC CGT TTC TGG TAT TTC | ||||||
| VII | sfpC-1 | CGG GCA TCG GAC TAT TAC AC | sfpC 3′ end | 94°C, 30s | 58°C, 60 s | 72°C, 60 s | 787 |
| sfpC-2 | CCA CCA TCC CCA GTT CAC G | ||||||
| VIII | sfpD-1 | CGC CAA TGA TAA CAA ACA G | sfpD | 94°C, 30 s | 52°C, 60 s | 72°C, 60 s | 410 |
| sfpD-2 | TAC CAC CAA TGC CAA TCA C | ||||||
| IX | sfpJ-1 | ATG AGC AGG ACA ACA ACA GG | sfpJ | 94°C, 30 s | 52°C, 60 s | 72°C, 60 s | 251 |
| sfpJ-2 | TAT TTT CCG TTA TCC TCC AG | ||||||
| X | sfpG-1 | CGT AGT GAC TTG GAT GCT G | sfpG | 94°C, 30s | 52°C, 60s | 72°C, 60s | 267 |
| sfpG-2 | TTG CCA TCC ATT ACC ATT C | ||||||
| XI | sfpA-U | AGC CAA GGC CAA GGG ATT ATT A | sfpA-sfpH | 94°C, 30s | 59°C, 60s | 72°C, 60s | 1,026 |
| sfpH-2 | TCC AGC CCC TCT TCA TTT CC | ||||||
| XII | sfpH-1 | CCA TCG CCC TCC TTT CCT CT | sfpH-sfpC | 94°C, 30s | 59°C, 60s | 72°C, 60s | 733 |
| sfpC-3 | CGC CAG CCA GGG AAA ACA ACA A | ||||||
| XIII | sfpC-4 | GTG GGG AGT TGG TGT AGT GA | sfpC-sfpD | 94°C, 30s | 56°C, 60s | 72°C, 60s | 1,030 |
| sfpD-2 | TAC CAC CAA TGC CAA TCA C | ||||||
| XIV | sfpD-1 | CGC CAA TGA TAA CAA ACA G | sfpD-sfpJ | 94°C, 30s | 52°C, 60s | 72°C, 60s | 875 |
| sfpJ-2 | TAT TTT CCG TTA TCC TCC AG | ||||||
| XV | sfpJ-1 | ATG AGC AGG ACA ACA ACA GG | sfpJ-sfpG | 94°C, 30s | 52°C, 60s | 72°C, 150s | 1,748 |
| sfpG-2 | TTG CCA TCC ATT ACC ATT C | ||||||
| XVI | sfpG-3 | GCA TAA CAA TAA ACT CCA AA | sfpG | 94°C, 30 s | 48°C, 60 s | 72°C, 60 s | 588 |
| Gfla-1 | TGA CAA GCA AAC TGA TTA CC | Flank 1 | |||||
| XVII | sfpG-3 | GCA TAA CAA TAA ACT CCA AA | sfpG | 94°C, 30 s | 50°C, 60 s | 72°C, 120 s | 1,325 |
| Gfla-3 | CAT TGC TCC CTT CAT AAA A | Flank 2 | |||||
| XVIII | sfpG-5 | GGA TCG TAA TGC TGT TTC A | sfpG-repA | 94°C, 30 s | 51°C, 60 s | 72°C, 150 s | 1,343 |
| Gfla-4 | AAG CAC CGA ATG ATG AGA G | ||||||
| XIX | sfpG-5 | GGA TCG TAA TGC TGT TTC A | sfpG-repA | 94°C, 30 s | 51°C, 60 s | 72°C, 180 s | 1,768 |
| Gfla-5 | TAC CGT GAT CTC CTT TTC C | ||||||
The primers were derived from the respective genes of the sfp cluster and from the sfp flanking regions in SF EHEC O157:NM strain 3072/96 (GenBank accession no. AF401292). Except for primer pairs wprom-3/wprom-4 and sfpA-U/sfpA-L, which were described previously (11), all primers were designed for this study.
All PCRs included 30 cycles as given in the table, preceded by an initial denaturation (94°C, 5 min) and followed by a final extension (72°C, 5 min).
Genotypic characterization.
Putative virulence genes, ter genes encoding tellurite resistance (Table 3), and plasmid genes (Table 1) were sought using PCR (1, 2, 5, 8, 11, 18, 30, 46, 49). fliC, encoding the flagellin subunit, and eae, espP, and stx genes were subtyped (3, 8, 46, 54, 55). The stx2d activatable allele (3, 56) was sought using a specific PCR (56).
TABLE 3.
Characteristics of EHEC O165:H25/NM and O157:NM strains harboring the sfp cluster
| Characteristica | Presence of gene, value, or description
|
|
|---|---|---|
| O165:H25/NM (n = 7)b | O157:NM (3072/96) | |
| stx genotype | stx2 plus stx2cc | stx2 |
| cdtd | − | + |
| eae type | ɛ | γ |
| lpfAO157/OI 154 | − | + |
| lpfAO113 | + | − |
| OI 122 (efa1/sen/nleE/nleB/pagC)e | +/+/+/+/− | +/+/+/+/+ |
| ter clusterf | − | − |
| Sorbitol fermentation | −/+g | + |
| β-d-Glucuronidase | + | + |
| Growth on CT-SMAC | − | − |
| EHEC hemolysin production | + | −h |
| α-Hemolysin production | − | − |
| Shiga toxin titeri | 256-1,024 | 512 |
| Phylogenetic group | A | D |
The genes mentioned encode the following proteins or phenotypes: stx, Shiga toxin; cdt, cytolethal distending toxin; eae, intimin; lpfAO113 and lpfAO157/OI 154, major fimbrial subunits of long polar fimbriae of EHEC O113 and EHEC O157, respectively, encoded on OI 154; efa1, EHEC factor for adherence; sen, homologue of Shigella flexneri enterotoxin 2; nleE and nleB, non-locus-of-enterocyte-effacement effector proteins NleE and NleB, respectively; pagC, homologue of PagC protein of Salmonella enterica serovar Typhimurium; ter cluster, tellurite resistance. +, the characteristic is present; −, the characteristic is absent.
All seven EHEC O165:H25/NM strains had identical characteristics, except for sorbitol fermentation (see below).
One strain (bovine isolate 00-09087) lost stx before it could be subtyped.
cdt I, cdt II, cdt III, cdt IV, and cdt V genes were targeted (1); strain 3072/96 contains the cdt V cluster.
A complete OI 122 which contains all genes representative of its three modules (efa1, sen, nleE, nleB, and pagC) (30) was present in EHEC O157, whereas all EHEC O165 strains contained OI 122 without pagC (incomplete OI 122) (30).
All ter genes (terZ, terA, terB, terC, terD, terE, and terF) were absent.
All six EHEC O165:H25/NM strains from Germany are non-SF, whereas the U.S. strain MT52 is SF.
The EHEC hlyA gene is present but not expressed.
The highest dilution of a culture supernatant which caused cytotoxicity in 50% of the Vero cell monolayer after 3 days of incubation. The range of Stx titers for the seven EHEC O165:H25/NM strains is shown.
Plasmid analysis.
Plasmid profiles were determined (41), and plasmid restriction fragment length polymorphism (RFLP) patterns were produced using plasmid-extracted DNA (Plasmid Midi kit; Qiagen) digested with EcoRI (New England Biolabs, Frankfurt, Germany) and separated in a 0.6% agarose gel. For Southern hybridization, undigested or EcoRI-digested plasmid DNA was transferred to a nylon membrane (Roche Molecular Biochemicals, Mannheim, Germany) and hybridized with digoxigenin-labeled (DIG High Prime kit; Roche Molecular Biochemicals) sfpA, EHEC hlyA, katP, espP, and etpD probes. The sfpA probe was derived from SF EHEC O157:NM strain 3072/96 by PCR with primers sfpA-U and sfpA-L (Table 2). All other probes were derived from E. coli O157:H7 strain EDL933, using primers HlyA1 and HlyA4 (EHEC hlyA), esp-A and esp-B (espP), D1 and D13R (etpD) (46), and kat-1 (5′-GGCGGAAGAGAAGATGACTG-3′) and kat-2 (5′-GCCACAGTCTCCTCATCATC-3′) (katP). Labeled probes were detected using a DIG luminescence detection kit (Roche Molecular Biochemicals).
Multilocus sequence typing (MLST) and PCR phylogrouping.
Internal regions of seven housekeeping genes were sequenced (4, 53) to assign alleles and sequence types (ST) (http://web.mpiib-berlin.mpg.de/mlst/dbs/Ecoli). Relationships between strains harboring the sfp cluster were characterized using the eBURST algorithm (21), which groups strains sharing six or more identical alleles into the same clonal complex (CC). Similarities were calculated using MEGA software v. 4.0 (47), and a minimum spanning tree was generated with SeqSphere software v. 0.7 beta (Ridom GmbH). In addition, isolates were classified into E. coli reference phylogenetic groups A, B1, B2, and D (43), using PCR (14).
Strain accession number.
The EHEC O165:H25 HUS isolate 820/08 characterized in this study is included in our recently created HUSEC reference collection (33; www.ehec.org) as HUSEC042.
Nucleotide sequence accession numbers.
The 6,838-bp sfp clusters and their 828-bp upstream and 553-bp downstream flanks in the large plasmids of O165:H25 strains 820/08 and 04-07734 were deposited in the EMBL-GenBank database under accession no. EU980314 and EU980315, respectively.
RESULTS
EHEC O165:H25/NM strains contain a complete sfp cluster.
All seven EHEC O165:H25/NM isolates yielded amplicons of the same sizes as those elicited from the control SF EHEC O157:NM strain 3072/96 in all PCRs targeting sfp genes and their links (Fig. 1). This demonstrates the resemblance between the sfp clusters of EHEC O165 and SF O157 strains.
The sfp cluster is conserved in EHEC O165:H25/NM and SF EHEC O157:NM.
The sequences of the sfp clusters (6,838 bp) in EHEC O165:H25 strains 820/08 and 04-07734 (chosen to represent severe and mild disease phenotypes, respectively) were identical to each other and >99% identical to the sfp cluster of SF EHEC O157:NM strain 3072/96 (GenBank accession no. AF401292). The 23 nucleotide differences between the sfp genes of EHEC O165:H25 and strain 3072/96 consisted of 10 nonsynonymous and 13 synonymous changes.
sfp clusters in EHEC O165:H25/NM are in large plasmids.
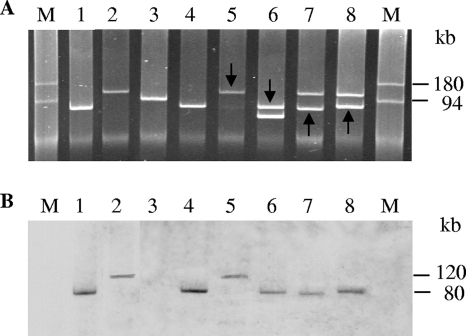
To determine if the sfp cluster in EHEC O165 is located on a plasmid, plasmids were isolated from the seven EHEC O165:H25/NM strains and hybridized with the sfpA probe. All strains contained a plasmid of ∼80 kb, either alone or with a larger (∼120 kb) or smaller (∼60 kb) plasmid (Fig. 2A; Table 1). For six strains, the sfpA probe hybridized to the 80-kb plasmid (Fig. 2B, lanes 1, 4, and 6 to 8; Table 1), and for one strain, the probe bound to a 120-kb plasmid (Fig. 2B, lane 5). The latter strain was the only one in which the size of the sfp-harboring plasmid was comparable to that in SF EHEC O157:NM strain 3072/96 (Fig. 2B, lane 2).
FIG. 2.
Plasmid profiles (A) and plasmid hybridization with the sfpA probe (B) of EHEC O165 and control strains. In lanes 1 and 4 to 8, the following EHEC O165:H25/NM strains are shown: lanes 1, 820/08; lanes 4, 04-07734; lanes 5, 02-11228; lanes 6, 00-09087; lanes 7, 99-02258; and lanes 8, 98-08419-1. In lanes 2 and 3, SF EHEC O157:NM strain 3072/96 and EHEC O157:H7 strain EDL933, respectively, are shown for comparison. For strains which contained two plasmids, the plasmid which hybridized with the sfpA probe is marked by an arrow (A); the sizes of the sfpA-hybridizing plasmids are indicated in panel B. M, molecular size marker (DNAs of plasmids R27 [180 kb] and R100 [94 kb]) (45; http://www.biotech.bham.ac.uk/Plasmids/default.htm).
Comparison of sfp-harboring plasmids of EHEC O165:H25/NM strains and pSFO157.
sfp-harboring plasmids from EHEC O165:H25/NM strains (pO165) were further hybridized with probes representing a panel of putative virulence plasmid-carried genes of E. coli O157:H7 (EHEC hlyA, espP, katP, and etpD) (Table 1). In each instance where the sfp cluster was in the 80-kb plasmid (Fig. 2B, lanes 1, 4, and 6 to 8; Table 1), the same plasmid also hybridized with the EHEC hlyA, espP, and katP probes (Table 1). In contrast, in the only strain that contained the sfp cluster in the 120-kb plasmid (Fig. 2B, lane 5), the same plasmid also contained EHEC hlyA and katP sequences, but espP was located in the second (80-kb) plasmid (Table 1); the latter plasmid also harbored additional copies of EHEC hlyA and katP (Table 1). None of the pO165 plasmids hybridized with the etpD probe (Table 1). In contrast to pO165, and in accordance with its sequence (GenBank accession no. AF401292), the 121-kb pSFO157 plasmid from strain 3072/96 hybridized with EHEC hlyA and etpD but not with the katP and espP probes (Table 1). Thus, pO165 and pSFO157 plasmids differ by gene composition in addition to (in most cases) size.
Plasmid RFLP and sfpA Southern blot hybridization.
For three EHEC O165:H25 strains that contained a single plasmid (Table 1), the plasmid RFLP patterns of the two German strains were identical and differed from that of the U.S. strain; moreover, all three EHEC O165:H25 strains differed in plasmid RFLP pattern from strain 3072/96. For EHEC O165, the sfpA probe hybridized to either an ∼50-kb (both German strains) or ∼15-kb (MT52) fragment of plasmid DNA, whereas in strain 3072/96, an ∼11-kb DNA fragment reacted with the probe (data not shown).
sfp-flanking regions in EHEC O165:H25/NM.
A fragment of the insertion sequence IS2, but not a transposon-like sequence (Tn2501), is located upstream of the sfp cluster in EHEC O165 strains (Fig. 1). Furthermore, a region homologous to the RepFIB origin of replication of pSFO157, which starts 686 bp downstream of sfpG in strain 3072/96 and includes an open reading frame for the replication protein RepA (11) (GenBank accession no. AF401292), was not found at this position in EHEC O165:H25/NM (Fig. 1). In these strains, a 205-bp fragment of the insertion sequence IS1 is located 333 bp downstream of sfpG (GenBank accession no. EU980314 and EU980315), displaying no significant homology to this region in pSFO157 (Fig. 1).
Virulence genes and phenotypes of EHEC O165:H25/NM strains containing an sfp cluster.
In addition to the sfp cluster and other plasmid genes (EHEC hlyA, espP, and katP) (Table 1), the EHEC O165:H25/NM strains also shared chromosomal virulence loci and displayed, except for sorbitol fermentation by the U.S. isolate MT52, identical phenotypes (Table 3). However, they differed from SF EHEC O157:NM with respect to most of these characteristics (Table 3).
MLST analysis and phylogeny PCR.
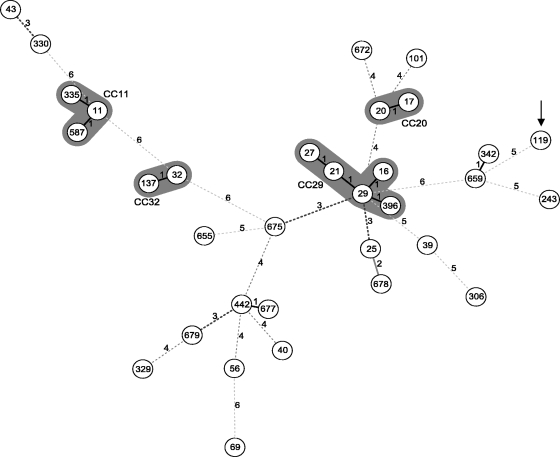
The phylogenetic relationships between EHEC O165:H25/NM strains and representative HUS-associated EHEC strains (HUSEC collection) (33) are shown in Fig. 3. All EHEC O165:H25/NM strains with the sfp cluster belonged to the same sequence type (ST119), which is quite distinct from ST11 of SF EHEC O157:NM strain 3072/96. Furthermore, EHEC O165:H25/NM strains share only two of the seven loci with the next most closely related organisms (ST342 and ST659). At the nucleotide level, ST119 and ST11 differ in 48 of the 3,423 bp analyzed by MLST in the seven housekeeping genes (1.40% pairwise distance). In contrast, the overall average pairwise distance within the HUSEC collection is only 0.89%. The sfp-positive EHEC O165:H25/NM isolates differ at only 24 sites (0.70%) from Shigella dysenteriae strain M1354 (ST243) (http://web.mpiib-berlin.mpg.de/mlst/dbs/Ecoli), which was used as an outgroup for the minimum spanning tree (Fig. 3).
FIG. 3.
Minimum spanning tree based on MLST of the sfp-positive EHEC O165:H25/NM strains (ST119; depicted by arrow) in comparison to the HUSEC collection (33) and Shigella dysenteriae strain M1354 as an outgroup (ST243) (data are from http://web.mpiib-berlin.mpg.de/mlst/dbs/Ecoli). Each circle represents a unique ST (with the designation given in the circle). The numbers on the connecting lines display the numbers of differing alleles. Clonal complexes (CC) of the major EHEC serogroups (33) are highlighted in gray.
Phylogenetic grouping (43) further supports the distant relationship between the O165 and O157 EHEC strains carrying sfp clusters. Whereas SF EHEC O157:NM strains belong to E. coli reference group D, all seven O165:H25/NM strains were classified as group A (Table 3). These groups represent the two extremes of the E. coli population (43, 53).
DISCUSSION
We identified the fimbrial sfp operon, which we previously proposed to be unique to SF E. coli O157:NM (23), in an additional EHEC serotype, O165:H25. Despite their host cells' distant relatedness, the sfp cluster is highly conserved in these two serotypes and resides on large plasmids. However, the sfp-harboring plasmids in the two serotypes differ in size, structure, gene composition, and position of the sfp cluster, as well as in the sequences flanking the sfp genes. Our data have diagnostic, epidemiological, and evolutionary implications.
EHEC O165 strains cause HUS and bloody diarrhea (7, 9, 17, 20, 22, 27, 28, 31, 44, 52). The need to reliably identify such strains is therefore obvious. Their possession of the sfp cluster offers an additional target, besides stx/Stx, to detect them in patients' stools. In work that led to the identification of the sfp cluster in EHEC O165, we were initially stymied by the presence of sfpA and the absence of rfbO157, which then prompted extensive efforts to isolate the strain of interest and its subsequent characterization. Thus, the PCR detection of sfpA and the inability to detect rfbO157 provided the impetus to identify an infecting yet unexpected pathogen in the primary stool culture. A systematic pursuit of such findings by isolation of the PCR-positive strains might lead in the future to identification of other E. coli serotypes that harbor the sfp cluster. An additional diagnostically useful marker of EHEC O165:H25/NM strains is that they mostly fail to ferment sorbitol after overnight incubation on SMAC (Table 3). This unusual phenotype, which is generally restricted to EHEC O157:H7 (29, 48) and other select EHEC isolates (44; www.ehec.org) and which was also reported for an EHEC O165:NM strain isolated from an HUS patient in Brazil (17), can assist in the isolation of such strains from primary cultures. However, in contrast to EHEC O157:H7, the EHEC O165:H25/NM strains produce β-d-glucuronidase, lack the ter cluster, and are accordingly susceptible to tellurite and do not grow on CT-SMAC (Table 3); this is in agreement with the tellurite susceptibility of EHEC O165 strains isolated from patients in Japan (44). The SF phenotype of the U.S. EHEC O165:H25 strain was also reported for an EHEC strain of this serotype isolated in Finland (31).
In studies that associated human diseases with EHEC O165, the source of the infection remained unknown (17). However, in one of these studies, a fresh, homemade cheese consumed by the patient was suspected as the source of the EHEC O165 (17), suggesting that cattle are a possible reservoir. Indeed, isolation of EHEC O165:H25/NM from cattle feces and from beef has been reported (6, 24, 25, 26, 36). Some of these studies demonstrated that bovine EHEC O165:H25/NM strains share virulence genes, such as eae ɛ, EHEC hlyA, and efa1, with human isolates of this serotype (6, 25). Moreover, as we show, such strains also share the sfp cluster, which encodes a putative adhesin (34). Taken together, these data suggest that cattle can harbor EHEC O165:H25/NM and can thus be sources of human infections.
EHEC O165:H25 bovine isolates can lose their large plasmids (and presumably the sfp cluster) during laboratory processing (25). We believe that such an event might explain why an EHEC O165:NM (fliCH25) strain we isolated from a patient in the 1990s (22) was sfp negative when it was screened several years later using sfpA PCR (23), thereby prompting us to consider that this locus was not found in strains belonging to the O165 serogroup (23). A recent more detailed analysis of this strain demonstrated that it lacks a large plasmid and all other genes typically located on pO165. This supports the hypothesis that this strain might have lost its sfp-harboring plasmid in vivo or during laboratory storage.
Most EHEC O157 and non-O157 strains possess large plasmids (37, 38, 39, 41, 46), which we also detected in EHEC O165:H25/NM. Analysis of colocalization of the sfp cluster with other plasmid genes (Table 1) and RFLP analysis of sfp-harboring plasmids demonstrated that the sfp cluster is located in EHEC O165 and O157 in several different plasmids, namely, (i) the 121-kb plasmid pSFO157 of strain 3072/96, harboring the sfp cluster together with EHEC hlyA and the etp cluster but lacking espP and katP (Table 1); (ii) the ∼80-kb plasmid pO165, which contains sfp in combination with EHEC hlyA, espP, and katP but lacks etp (Table 1) and structurally differs in strains from Germany and the United States (data not shown); and (iii) the ∼120-kb plasmid pO165, harboring the sfp cluster together with EHEC hlyA and katP but without espP and etp (Table 1). The precise mechanisms of acquisition of these loci, including sfp, by EHEC strains are unknown. The megaplasmids (10, 11, 12, 13, 37, 38) are clearly mosaics, and most putative virulence genes are flanked by mobile genetic determinants, which probably facilitate their transfer. However, partial deletions of these mobile elements, as shown with the regions that flank the sfp operon in pO165, can stabilize particular virulence loci. Because EHEC O157:NM and O165:H25/NM are unrelated, we hypothesize that the sfp genes in these two serotypes were acquired independently in different events and from different sources. However, we cannot exclude the possibility of a direct transmission of the sfp cluster from one serotype to the other via horizontal transfer. It is interesting that although the sfp locus is present in EHEC O165:H25 strains associated with human disease in the United States, SF EHEC O157:NM, which regularly possesses this cluster, has not to date been isolated in the New World.
In SF EHEC O157:NM, the expression of Sfp fimbriae is strongly influenced by environmental conditions, in particular by oxygen tension (34). Studies are under way in our laboratory to determine conditions that promote Sfp expression in EHEC O165. Such studies will provide a closer insight into the role of Sfp fimbriae in the virulence of EHEC strains of these two serotypes.
In conclusion, the finding of a highly conserved sfp locus in divergent phylogenetic backgrounds prompts the need to perform further molecular and evolutionary comparative analyses of these unrelated EHEC lineages. It is interesting that suites of virulence genes seem to be common to human-pathogenic EHEC strains, where the possession of stx genes, for example, is almost always accompanied by the presence of one of the eae alleles (5, 9). Such associations occur much more frequently than would be expected by chance, and indeed, intimin and Stx interact in their effects on epithelial cells (42). To facilitate further studies on such convergence of virulence loci in phylogenetically diverse EHEC strains (33), the EHEC O165:H25 HUS isolate 820/08 characterized in this study is included in our recently created HUSEC reference collection (33; www.ehec.org).
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG)-funded International Graduate School project “Molecular Interactions of Pathogens with Biotic and Abiotic Surfaces” (GRK1409), by a grant from EU Network ERA-NET PathoGenomics (no. 0313937C), and by a grant from EU Network of Excellence EuroPathoGenomics (no. LSHB-CT-2005-512061). Strain MT52 was recovered and characterized using support from the Centers for Disease Control and Prevention (cooperative agreement U50/CCU814408) and NIH grant AI-47499. P.I.T. was also supported by NIH grant 5P30 DK052574 to the Washington University Digestive Diseases Research Core Center.
We thank Nadine Brandt and Margret Junge for excellent technical assistance.
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Bielaszewska, M., M. Fell, L. Greune, R. Prager, A. Fruth, H. Tschäpe, M. A. Schmidt, and H. Karch. 2004. Characterization of cytolethal distending toxin genes and expression in Shiga toxin-producing Escherichia coli strains of non-O157 serogroups. Infect. Immun. 72:1812-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielaszewska, M., P. I. Tarr, H. Karch, W. Zhang, and W. Mathys. 2005. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J. Clin. Microbiol. 43:452-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielaszewska, M., A. W. Friedrich, T. Aldick, R. Schurk-Bulgrin, and H. Karch. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160-1167. [DOI] [PubMed] [Google Scholar]
- 4.Bielaszewska, M., R. Prager, R. Köck, A. Mellmann, W. Zhang, H. Tschäpe, P. I. Tarr, and H. Karch. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl. Environ. Microbiol. 73:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielaszewska, M., R. Köck, A. W. Friedrich, C. von Eiff, L. B. Zimmerhackl, H. Karch, and A. Mellmann. 2007. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS ONE 2:e1024. doi: 10.1371/journal.pone.0001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco, M., J. E. Blanco, A. Mora, G. Dahbi, M. P. Alonso, E. A. González, M. I. Bernárdez, and J. Blanco. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eaeξ). J. Clin. Microbiol. 42:645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockmeyer, J., M. Bielaszewska, A. Fruth, M.-L. Bonn, A. Mellmann, H.-U. Humpf, and H. Karch. 2007. Subtypes of the plasmid-encoded serine protease EspP in Shiga toxin-producing Escherichia coli: distribution, secretion and proteolytic activity. Appl. Environ. Microbiol. 73:6351-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks, J. T., E. G. Sowers, J. G. Wells, K. D. Greene, P. M. Griffin, R. M. Hoekstra, and N. A. Strockbine. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192:1422-1429. [DOI] [PubMed] [Google Scholar]
- 10.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 11.Brunder, W., A. S. Khan, J. Hacker, and H. Karch. 2001. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H−. Infect. Immun. 69:4447-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunder, W., H. Karch, and H. Schmidt. 2006. Complete sequence of the large virulence plasmid pSFO157 of the sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H− strain 3072/96. Int. J. Med. Microbiol. 296:467-474. [DOI] [PubMed] [Google Scholar]
- 13.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombes, B. K., M. E. Wickham, M. Mascarenhas, S. Gruenheid, B. B. Finlay, and M. A. Karmali. 2008. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 74:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creuzburg, K., J. Recktenwald, V. Kuhle, S. Herold, M. Hensel, and H. Schmidt. 2005. The Shiga toxin 1-converting bacteriophage BP-4795 encodes an NleA-like type III effector protein. J. Bacteriol. 187:8494-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Souza, R. L., L. S. Nishimura, and B. E. C. Guth. 2007. Uncommon Shiga toxin-producing Escherichia coli serotype O165:HNM as cause of hemolytic uremic syndrome in São Paulo, Brazil. Diagn. Microbiol. Infect. Dis. 59:223-225. [DOI] [PubMed] [Google Scholar]
- 18.Doughty, S., J. Sloan, V. Bennett-Wood, M. Robertson, R. M. Robins-Browne, and E. L. Hartland. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott, E. J., R. M. Robins-Browne, E. V. O'Loughlin, V. Bennett-Wood, J. Bourke, P. Henning, G. G. Hogg, J. Knight, H. Powell, D. Redmond, et al. 2001. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch. Dis. Child. 85:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ethelberg, S., K. E. Olsen, F. Scheutz, C. Jensen, P. Schiellerup, J. Enberg, A. M. Petersen, B. Olesen, P. Gerner-Smidt, and K. Molbak. 2004. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg. Infect. Dis. 10:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 23.Friedrich, A. W., K. V. Nierhoff, M. Bielaszewska, A. Mellmann, and H. Karch. 2004. Phylogeny, clinical associations, and diagnostic utility of the pilin subunit gene (sfpA) of sorbitol-fermenting, enterohemorrhagic Escherichia coli O157:H−. J. Clin. Microbiol. 42:4697-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geue, L., M. Segura-Alvarez, F. J. Conraths, T. Kuczius, J. Bockemuhl, H. Karch, and P. Gallien. 2002. A long-term study on the prevalence of Shiga toxin-producing Escherichia coli (STEC) on four German cattle farms. Epidemiol. Infect. 129:173-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geue, L., T. Selhorst, C. Schnick, B. Mintel, and F. J. Conraths. 2006. Analysis of the clonal relationship of Shiga toxin-producing Escherichia coli serogroup O165:H25 isolated from cattle. Appl. Environ. Microbiol. 72:2254-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussein, H. S. 2007. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 85(Suppl. 13):E63-E72. [DOI] [PubMed] [Google Scholar]
- 27.Jelacic, J. K., T. Damrow, G. S. Chen, S. Jelacic, M. Bielaszewska, M. Ciol, H. M. Carvalho, A. R. Melton-Celsa, A. D. O'Brien, and P. I. Tarr. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719-729. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, K. E., C. M. Thorpe, and C. L. Sears. 2006. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 43:1587-1595. [DOI] [PubMed] [Google Scholar]
- 29.Karch, H., P. I. Tarr, and M. Bielaszewska. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405-418. [DOI] [PubMed] [Google Scholar]
- 30.Karmali, M. A., M. Mascarenhas, S. Shen, K. Ziebell, S. Johnson, R. Reid-Smith, J. Isaac-Renton, C. Clark, K. Rahn, and J. B. Kaper. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keskimaki, M., M. Saari, T. Heiskanen, and A. Siitonen. 1998. Shiga toxin-producing Escherichia coli in Finland from 1990 through 1997: prevalence and characteristics of isolates. J. Clin. Microbiol. 36:3641-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellmann, A., M. Bielaszewska, L. B. Zimmerhackl, R. Prager, D. Harmsen, H. Tschäpe, and H. Karch. 2005. Enterohemorrhagic Escherichia coli in human infection: in vivo evolution of a bacterial pathogen. Clin. Infect. Dis. 41:785-792. [DOI] [PubMed] [Google Scholar]
- 33.Mellmann, A., M. Bielaszewska, R. Köck, A. W. Friedrich, A. Fruth, B. Middendorf, D. Harmsen, M. A. Schmidt, and H. Karch. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 14:1287-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müsken, A., M. Bielaszewska, L. Greune, C. H. Schweppe, J. Müthing, H. Schmidt, M. A. Schmidt, H. Karch, and W. Zhang. 2008. Anaerobic conditions promote expression of Sfp fimbriae and adherence of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM to human intestinal epithelial cells. Appl. Environ. Microbiol. 74:1087-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Brien, A. D., A. T. Lively, M. E. Chen, S. W. Rothman, and S. B. Formal. 1983. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 Shiga like cytotoxin. Lancet i:702. [DOI] [PubMed] [Google Scholar]
- 36.Parma, A. E., M. E. Sanz, J. E. Blanco, J. Blanco, M. R. Viñas, M. Blanco, N. L. Padola, and A. I. Etcheverría. 2000. Virulence genotypes and serotypes of verotoxigenic Escherichia coli isolated from cattle and foods in Argentina. Importance in public health. Eur. J. Epidemiol. 16:757-762. [DOI] [PubMed] [Google Scholar]
- 37.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paton, A. W., P. Srimanote, U. M. Talbot, H. Wang, and J. C. Paton. 2004. A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pradel, N., Y. Bertin, C. Martin, and V. Livrelli. 2008. Molecular analysis of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients and dairy samples in France. Appl. Environ. Microbiol. 74:2118-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prager, R., U. Strutz, A. Fruth, and H. Tschäpe. 2003. Subtyping of pathogenic Escherichia coli strains using flagellar H-antigens: serotyping versus fliC polymorphisms. Int. J. Med. Microbiol. 292:477-486. [DOI] [PubMed] [Google Scholar]
- 41.Prager, R., S. Annemuller, and H. Tschäpe. 2005. Diversity of virulence patterns among Shiga toxin-producing Escherichia coli from human clinical cases—need for more detailed diagnostics. Int. J. Med. Microbiol. 295:29-38. [DOI] [PubMed] [Google Scholar]
- 42.Robinson, C. M., J. F. Sinclair, M. J. Smith, and A. D. O'Brien. 2006. Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. USA 103:9667-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selander, R. K., D. A. Caugant, and T. S. Whittam. 1987. Genetic structure and variation in natural populations of Escherichia coli, p. 1625-1648. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 44.Seto, K., M. Taguchi, K. Kobayashi, and S. Kozaki. 2007. Biochemical and molecular characterization of minor serogroups of Shiga toxin-producing Escherichia coli isolated from humans in Osaka prefecture. J. Vet. Med. Sci. 69:1215-1222. [DOI] [PubMed] [Google Scholar]
- 45.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, V. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonntag, A. K., R. Prager, M. Bielaszewska, W. Zhang, A. Fruth, H. Tschäpe, and H. Karch. 2004. Phenotypic and genotypic analyses of enterohemorrhagic Escherichia coli O145 strains from patients in Germany. J. Clin. Microbiol. 42:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 48.Tarr, P. I., C. A. Gordon, and W. L. Chandler. 2005. Shiga toxin-producing Escherichia coli and the haemolytic uraemic syndrome. Lancet 365:1073-1086. [DOI] [PubMed] [Google Scholar]
- 49.Toma, C., E. Martinez Espinosa, T. Song, E. Miliwebsky, I. Chinen, S. Iyoda, M. Iwanaga, and M. Rivas. 2004. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 42:4937-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres, A. G., K. J. Kanack, C. B. Tutt, V. Popov, and J. B. Kaper. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol. Lett. 238:333-344. [DOI] [PubMed] [Google Scholar]
- 51.Tozzi, A. E., A. Caprioli, F. Minelli, A. Gianviti, L. De Petris, A. Edefonti, G. Montini, A. Ferretti, Z. De Palo, M. Gaido, G. Tizzoni, et al. 2003. Shiga toxin-producing Escherichia coli infections associated with hemolytic uremic syndrome in Italy 1988-2000. Emerg. Infect. Dis. 9:106-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uchida, H., H. Kanegane, K. Yoshiya, K. Kitamura, T. Ihara, H. Kamiya, Y. Kobayashi, H. Miyazawa, and T. Takeda. 1995. Four cases of hemolytic uremic syndrome (HUS) associated with serotype O165 verotoxin producing Escherichia coli (VTEC) identified by LPS-solid phase enzyme-linked immunosorbent assay (ELISA). Kansenshogaku Zasshi 69:678-683. [DOI] [PubMed] [Google Scholar]
- 53.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, W., B. Kohler, E. Oswald, L. Beutin, H. Karch, S. Morabito, A. Caprioli, S. Suerbaum, and H. Schmidt. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, W., A. Mellmann, A. K. Sonntag, L. Wieler, M. Bielaszewska, H. Tschäpe, H. Karch, and A. W. Friedrich. 2007. Structural and functional differences between disease-associated genes of enterohaemorrhagic Escherichia coli O111. Int. J. Med. Microbiol. 297:17-26. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, J., S. Cui, L. D. Teel, S. Zhao, R. Singh, A. D. O'Brien, and J. Meng. 2008. Identification and characterization of Shiga toxin type 2 variants in Escherichia coli isolated from animals, food, and humans. Appl. Environ. Microbiol. 74:5645-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]