Abstract
Bacterial biofilm formation is thought to enhance survival in natural environments and during interaction with hosts. A robust colonizer of the human gastrointestinal tract, Escherichia coli Nissle 1917, is widely employed in probiotic therapy. In this study, we performed a genetic screen to identify genes that are involved in Nissle biofilm formation. We found that F1C fimbriae are required for biofilm formation on an inert surface. In addition, these structures are also important for adherence to epithelial cells and persistence in infant mouse colonization. The data suggest a possible connection between Nissle biofilm formation and the survival of this commensal within the host. Further study of the requirements for robust biofilm formation may improve the therapeutic efficacy of Nissle 1917.
A biofilm is defined as a community of microorganisms attached to an inert or living surface by a self-produced polymeric matrix. Biofilms have been shown to protect encased bacteria from natural immune defenses such as antimicrobial compounds. This protection may be due to the inability of these compounds to penetrate the complex biofilm architecture or to the physiology of the bacterial cells within the biofilm (3). Several studies have shown that environmental conditions can trigger changes in gene expression that enable bacteria to adhere to a surface (23). Additional regulatory changes make this attachment intimate and irreversible (24, 33).
For many bacteria, adherence to epithelial cells is essential for the colonization or invasion of host tissues (5). This event is typically mediated by fibrillar structures such as fimbriae or pili (12). Fimbrial adhesins can mediate the binding of the bacteria to the host target cell either directly or indirectly by forming cross-link liaisons with the natural microbiota. Flagella, curli fibers, phase-modulated type 1 fimbriae, and antigen 43 are examples of adhesins that play a role in Escherichia coli K-12 biofilm formation (33).
Non-K-12 E. coli strains may present other biofilm-promoting factors which make them better suited to the colonization of specific niches. Uropathogenic E. coli (UPEC) strains, for example, express P, S, and F1C fimbriae in addition to type 1 fimbriae, the most ubiquitous adhesin structure among E. coli strains (8, 14, 18, 21). All of these structures are postulated to be important in the ascent of UPEC to the bladder via various surfaces in the urinary tract. In addition, MR/P fimbriae have been shown previously to contribute to biofilm formation by uropathogenic Proteus mirabilis (10) and are most commonly associated with the virulence of UPEC isolates (2, 11). Type 3 and F9 fimbriae, also present in UPEC, have been shown to confer increased biofilm capacity on K-12 strains (20, 32).
E. coli Nissle 1917 is a robust colonizer of the human intestinal tract and a widely employed probiotic. Under the registered trade name Mutaflor, this microorganism has shown promising results in the treatment of Crohn's disease, ulcerative colitis, and other inflammatory bowel diseases as part of a probiotic therapy (15, 19, 27). The partial genome sequence of Nissle 1917 has revealed several factors which may distinguish the colonization and biofilm-forming capacities of this strain from those of a closely related UPEC strain: Nissle 1917 has a K5 as opposed to a K2 capsule, carries an inactive P-fimbrial operon, and lacks S fimbriae (7, 30). Nissle does express type 1 fimbriae and F1C fimbriae, although the expression of the latter has traditionally been associated with uropathogens coexpressing P fimbriae (21).
Several studies have shown previously that only 50 to 80% of patients treated daily with Nissle 1917 present detectable levels of the probiotic strain in stool samples. However, the number of Nissle bacteria recovered from stool declines after administration is ceased (22). It remains a mystery why Nissle's apparently robust colonization phenotype does not enable long-term persistence in many patients. The basis of Nissle's probiotic action has been investigated; however, the molecular mechanisms involved in Nissle colonization of host epithelia are not well-known. There is a lack of information concerning the genetic requirements for Nissle biofilm formation. In this study, we investigated the molecular mechanisms involved in E. coli Nissle 1917 biofilm formation and the correlation among adherence factors involved in adherence to an inert surface and host epithelial cells and in vivo colonization. We found that F1C fimbriae are important for biofilm formation, adhesiveness to abiotic and biotic surfaces, and murine intestinal colonization.
Transposon mutagenesis screen to identify genes required for Nissle biofilm formation.
To study biofilm formation by Nissle 1917, we first examined the capacity of the strain to form biofilms on two abiotic surfaces under different growth conditions. Wild-type cells were grown aerobically or anaerobically in rich Luria-Bertani (LB) broth or minimal M9 medium in glass or polystyrene tubes until reaching stationary phase. After the cultures were removed, tubes were rinsed with distilled water and stained with 1% crystal violet. The wild-type Nissle strain formed heavy biofilms that were easily detected with crystal violet staining under all conditions tested (Fig. 1 and data not shown). In order to explore the genetic basis of Nissle biofilm formation, we performed transposon mutagenesis using the mini-Tn5 derivative pRL27 (16) and screened bacteria grown in glass tubes containing LB broth under aerobic conditions for deficiency in biofilm formation. Transposon insertions were evaluated by arbitrary PCR and DNA sequencing. Because the Nissle 1917 sequenced genome is not yet publicly available, we analyzed our sequence data based on Nissle homology to the most closely related sequenced strain, the UPEC strain CFT073 (34). Approximately 10,000 mini-Tn5 transposon insertions in Nissle were screened, and 13 independent insertion mutants presented reduced abilities to form biofilms compared to that of the wild-type strain grown under the same conditions (Table 1).
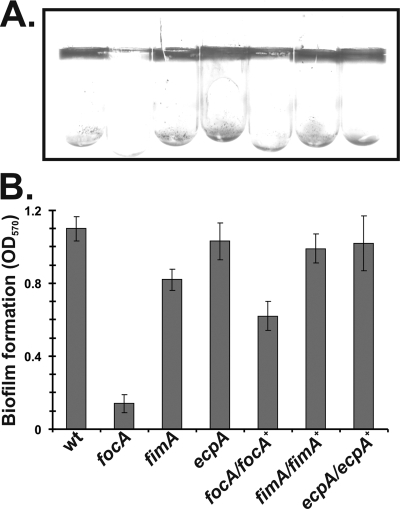
FIG. 1.
Role of surface adhesins in Nissle 1917 biofilm formation. (A) Crystal violet staining of biofilms formed by overnight cultures inside borosilicate glass tubes. (B) Quantitation of biofilm formation. Stained biofilms were dissolved in dimethyl sulfoxide, and the optical density at 570 nm (OD570) was recorded. The results are averages from three experiments. Error bars indicate the standard deviations. Deletion mutant strains are indicated by the relevant gene(s). wt, wild type.
TABLE 1.
Transposon mutagenesis screen of Nissle 1917 biofilm-deficient mutants
| Mutant | Gene (ORF no.) with transposon insertiona | Putative gene product | Biofilm formation (% of wild-type level) (SD)b |
|---|---|---|---|
| Nutrient metabolism mutants | |||
| J43A4 | aspA (c0522) | Aspartate ammonia-lyase | 24.1 (4.1) |
| J54B6 | ilvI (c0095) | Acetolactate synthase 3 catalytic subunit | 54.6 (13.7) |
| J10E1 | ybiW (c0908) | Formate acetyltransferase | 10.3 (1.4) |
| J2A7 | cmk (c1048) | Cytidylate kinase | 4.2 (0.54) |
| W4H2 | pyrD (c1081) | Dihydroorotate dehydrogenase | 24.2 (6.6) |
| Energy metabolism mutants | |||
| J17B9 | mdh (c3991) | Malate dehydrogenase | 22.6 (1.6) |
| W28B11 | gltA (c0796) | Type II citrate synthase | 24.3 (3.4) |
| W30G11 | sdhA (c0801) | Succinate dehydrogenase flavoprotein subunit | 21.1 (2.9) |
| W21E1 | sdhC (c0798) | Succinate dehydrogenase cytochrome subunit | 19.2 (5.1) |
| Transport and envelope-associated protein mutants | |||
| J57F3 | fucP (c0332) | l-Fucose permease | 26.0 (5.6) |
| W8D10 | dsbA (c4804) | Periplasmic protein disulfide isomerase | 37.2 (1.8) |
| J58C1 | mppA (c1803) | Periplasmic murein peptide binding protein precursor | 36.7 (1.8) |
| J59H1 | focC (c1241) | F1C periplasmic chaperone | 25.0 (4.0) |
Open reading frame (ORF) numbers are based on the homologous E. coli CFT073 genome sequence.
Biofilm formation on glass surfaces was quantitated by crystal violet staining. Averages and standard deviations of results for three replicates are shown.
Several disruptions of genes involved in nutrient metabolism produced biofilm deficiency, but this observation was probably the result of general growth deficiency rather than effects on specific determinants of adhesion or biofilm formation. For example, J2A7 (Table 1) contained an insertion in cmk, which encodes a cytidylate kinase. This mutant exhibited a growth defect compared to the wild-type strain under the same conditions (data not shown), confirming the previous finding that a cmk deletion mutant has a lower replication elongation rate than that of a wild-type strain due to a reduced pool of dCTP and dTTP nucleotides (6). Four of 13 biofilm-deficient mutants isolated during the screen revealed insertions in genes coding for tricarboxylic acid cycle enzymes. The genes mdh, gltA, and sdhA and sdhC, involved in the production of oxaloacetate, citrate, and fumarate, respectively, have been reported previously to stimulate biofilm formation by Staphylococcus aureus (28). The mechanism through which these compounds promote biofilm formation in this bacterium is not yet known, but it is apparently dependent on fibronectin binding proteins and the GraRS two-component sensor kinase system (28). Another category of genes identified in the screen encode envelope-associated proteins, some of which are involved in transport. DsbA is a periplasmic disulfide isomerase which plays an important role in the correct folding of periplasmic proteins. A dsbA deletion mutant of a Shiga toxin-producing enterohemorrhagic E. coli strain showed weaker biofilm-forming ability and was less virulent than the parent strain, probably as a result of the lack of efficient interaction of the bacterium with the host cell in a Caenorhabditis elegans infection model (17).
Another mutant exhibiting a dramatic change in biofilm formation capacity contained an insertion in the focC gene, part of the F1C fimbria-encoding foc operon. The product of focC is a periplasmic chaperone protein responsible for the stabilization of F1C fimbriae in the periplasm and/or their transport to the cell surface. A focC congenic mutant of UPEC strains has been characterized previously as defective in fimbria production due to a decreased level of expression of focA (encoding the major F1C fimbrial subunit) compared to that in the parent strains (26). Since fimbrial adhesins often play important roles in colonization and biofilm formation (31), we decided to test the adherence properties of Nissle F1C fimbriae involved in biofilm formation and in vivo colonization.
Biofilm formation and epithelial cell adherence by Nissle require F1C fimbriae.
In order to confirm that F1C fimbriae are important for biofilm formation, we constructed an in-frame deletion of the gene encoding the major F1C fimbrial subunit, focA, by the λ-Red recombination method (4). For comparison, we also constructed mutations in fimA, encoding the major subunit of type 1 fimbriae, known to be important for E. coli K-12 biofilm formation (30), and yagZ, also known as ecpA, encoding the major subunit of the E. coli common pilus, which is present in all E. coli genome sequences determined to date and known to aid the in vitro adherence of enterohemorrhagic E. coli (25). The fimA and ecpA mutant strains were generated by moving the deletion mutant constructs from the Keio collection (1) into Nissle by P1 transduction. The focA mutant showed a profound defect in the ability to form a biofilm, while wild-type Nissle formed a robust film on the glass tube wall when cells were grown statically in LB broth at 37°C until stationary phase (Fig. 1). The fimA mutant produced a slightly less extensive biofilm than that produced by the wild type, whereas the ecpA deletion mutant did not differ significantly from the wild type. The wild-type strain showed strong type 1 fimbria production in the static cultures used in our experiments, as judged by yeast agglutination (data not shown) (29). In contrast, the fimA mutant showed no evidence of yeast agglutination. To ensure that it was the focA mutation, and not some other mutation on the chromosome, that affected biofilm formation, we also performed complementation assays. We integrated the focA gene under the control of its native promoter into the chromosome of the Nissle focA deletion mutant at the φ80 phage attachment site attφ80. The expression of focA from the attφ80 locus largely restored biofilm formation (Fig. 1). Our data suggest that under the in vitro conditions used in this study, F1C fimbriae play a major role in mediating the ability of Nissle to adhere to inert surfaces.
To determine if F1C fimbriae play a role in adhering to biotic surfaces, we examined the binding of the wild-type Nissle strain and the focA, fimA, and ecpA Nissle mutants to epithelial cells. The human larynx epithelial cell line HEp-2 (ATCC CCL-23) was grown in a 5% CO2 incubator at 37°C in RPMI 1640 medium supplemented with 10% calf serum. Once confluent on coverslips, cells were washed with plain RPMI 1640 medium to remove serum. Overnight (stationary-phase) cultures were then inoculated at 100:1 (E. coli cell/host cell ratio) and incubated for 12 h at 37°C. At 4 and 8 h, the medium containing unbound bacteria was aspirated and replaced with fresh medium. After the cells were fixed with 1% paraformaldehyde, slides were visualized using an Olympus IX81 microscope at a magnification of ×100 and representative field images were acquired with a SensiCam QE camera (Cooke Corporation, Romulus, MI) and IPLab version 4.0 software (Scanalytics, Fairfax, VA). Figure 2A shows that wild-type Nissle adhered extensively to HEp-2 cells, with most bacteria immobilized in sheets coating the eukaryotic cells. The focA mutant, however, did not attach to HEp-2 cells, while the mutant strain complemented with focA did (Fig. 2B), suggesting that F1C fimbriae are critical for adherence to epithelial cell surfaces. The ecpA common pilus mutant was also deficient in binding to HEp-2 cells (Fig. 2C), confirming previous findings that this pilus structure is important for E. coli adherence to eukaryotic cell surfaces (25). The fimA type 1 fimbria mutant strain showed patterns of binding to epithelial cells similar to those of the focA and ecpA mutants (Fig. 2D) under these assay conditions, suggesting that the Nissle type 1 fimbriae may be as important as F1C fimbriae and common pili for adherence to epithelial cells. However, the reintroduction of the fimA gene by integration at the phage φ80 attachment site did not enhance the binding to HEp-2 cells (data not shown). In addition, we did not observe any evidence of yeast agglutination for this strain (data not shown). It is possible that the fimA deletion may affect downstream genes involving type 1 fimbria synthesis or that the fimA upstream regions of K-12 and Nissle are different. We also note that all mutants had growth rates in tissue culture medium that were similar to that of the wild-type strain (data not shown).
FIG. 2.
Nissle 1917 adherence to HEp-2 cells in culture. Shown are representative micrographs of the Nissle wild-type strain (A); the focA (B), ecpA (C), and fimA (D) deletion mutants; and the focA φ80(Pfoc::focA) (E) and ecpA φ80(PecpA::ecpA) (F) complemented strains after a 12-h incubation with periodic washing to remove unbound bacteria. Examples of bacteria, which appear black in the phase-contrast images, are indicated with arrows. Scale bars, 10 μm.
F1C fimbriae are important for Nissle colonization of infant mice.
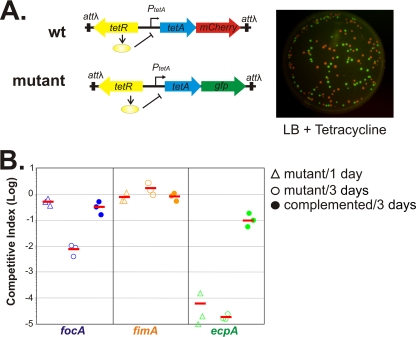
To study Nissle's colonization properties within the host, we tested the ability of Nissle to colonize infant mice. The use of infant mice is particularly attractive as it is convenient and inexpensive to perform assays (13). We orally inoculated 107 CFU of a wild-type Nissle strain that was marked with the gene for a fluorescent protein under inducible control (described below) into 6-day-old suckling CD-1 mice. At different time points, mouse intestines were harvested and the Nissle bacterial number was determined by serial dilution and plating onto selective medium agar plates. We found that approximately 106 to 108 CFU of Nissle could be recovered from intestines up to 12 days postinoculation (data not shown), indicating that Nissle bacteria can colonize infant mice. To investigate whether fimbriae and the E. coli common pilus are important for colonization, we performed competition assays by coinoculating wild-type and mutant strains into infant mice. We constructed inducible fluorescent reporters in Nissle strains to easily differentiate wild-type and mutant bacteria on plates. A tetR-tetA-mCherry construct was integrated into the chromosome of the Nissle wild type at the phage lambda attachment site attλ. Similarly, a tetR-tetA-gfp construct was used as a reporter for Nissle mutants (Fig. 3A). The expression of fluorescent reporter genes remains silent until bacteria are exposed to treatment with tetracycline in LB plates after recovery from mouse intestines, thus avoiding potential effects on cell physiology from fluorescent protein overexpression during colonization (Fig. 3A). The competition assay revealed that the focA mutant has the same ability to colonize infant mice at 1 day postinoculation as wild-type Nissle; however, levels of the same mutant were reduced nearly 100-fold at 3 days (Fig. 3B). In contrast, the fimA mutant was detected at wild-type levels throughout the experiment, while the ecpA mutant was almost undetectable at both 1 and 3 days postinoculation. Both focA and ecpA complementation restored colonization ability fully or partially. These data suggest that the E. coli common pilus is absolutely essential for the initial stages of colonization, while F1C fimbriae may play a role in Nissle persistence in the infant mouse intestinal tract. Type 1 fimbriae are not necessary for Nissle colonization in our murine infant model; however, the type 1 fimbriae may be important for adhesion to abiotic and biotic surfaces in other contexts (Fig. 1 and 2). The expression of type 1 fimbriae is phase modulated by the inversion of a 314-bp DNA segment by the FimB and FimE recombinases in E. coli. The switching frequency is known to vary from strain to strain and under different environmental conditions (9). Additionally, Nissle is the only known example of an E. coli strain with a non-FimB-mediated off-to-on switch (29). It is possible that this second mechanism of phase switching was upregulated under our in vitro biofilm and cell adherence conditions but not in vivo.
FIG. 3.
Results from infant mouse competition colonization assays. (A) Construction of fluorescence reporters and detection of a mixture of wild-type (wt; red) and mutant (green) colonies. (B) Abilities of focA (blue), fimA (orange), and ecpA (green) mutants to colonize infant mice. The competitive index (the total number of CFU of the mutant in the whole intestinal homogenate divided by the number of CFU of the wild type) is shown for each mouse at different time points (open triangles, 1 day postinoculation, and open circles, 3 days postinoculation) and for the three mutants complemented with a copy of the respective deleted gene (closed circles). Short horizontal lines represent the averages of the competitive indexes for each mutant.
In this study, we have shown that F1C fimbriae are required for Nissle to adhere avidly to both abiotic and biotic surfaces and for persistence during the colonization of mouse intestine. Although the Nissle focA mutant was able to colonize at a level comparable to that of the wild-type strain at 1 day postinoculation, the abundance of focA mutant bacteria steeply declined by 3 days postinoculation. The loss of the E. coli common pilus, in contrast, caused the complete and immediate loss of the colonization ability of Nissle. These data suggest that there are distinct requirements for the initial colonization by versus the long-term persistence of this commensal. In addition, it is possible that the importance of F1C fimbriae for commensal colonization is through the role of these structures in promoting biofilms. Further research on the regulatory mechanisms controlling biofilm formation by E. coli commensals, and their role in intestinal colonization, may aid our understanding of how natural microbiota survive adverse conditions inside the human intestines, as well as elucidate new methods of combating persistent infections caused by pathogenic biofilms.
Acknowledgments
We thank Erin Frey for preparing HEp-2 cell cultures.
This work is supported by NIH grants R01AI072479 (to J.Z.) and R01GM080279 (to M.G.).
Footnotes
Published ahead of print on 7 November 2008.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahrani, F. K., G. Massad, C. V. Lockatell, D. E. Johnson, R. G. Russell, J. W. Warren, and H. L. Mobley. 1994. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 62:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fricke, J., J. Neuhard, R. A. Kelln, and S. Pedersen. 1995. The cmk gene encoding cytidine monophosphate kinase is located in the rpsA operon and is required for normal replication rate in Escherichia coli. J. Bacteriol. 177:517-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grozdanov, L., C. Raasch, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, and U. Dobrindt. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hull, R. A., R. E. Gill, P. Hsu, B. H. Minshew, and S. Falkow. 1981. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun. 33:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iida, K., Y. Mizunoe, S. N. Wai, and S. Yoshida. 2001. Type 1 fimbriation and its phase switching in diarrheagenic Escherichia coli strains. Clin. Diagn. Lab. Immunol. 8:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansen, A. M., V. Lockatell, D. E. Johnson, and H. L. Mobley. 2004. Mannose-resistant Proteus-like fimbriae are produced by most Proteus mirabilis strains infecting the urinary tract, dictate the in vivo localization of bacteria, and contribute to biofilm formation. Infect. Immun. 72:7294-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallenius, G., R. Mollby, S. B. Svenson, I. Helin, H. Hultberg, B. Cedergren, and J. Winberg. 1981. Occurrence of P-fimbriated Escherichia coli in urinary tract infections. Lancet 2:1369-1372. [DOI] [PubMed] [Google Scholar]
- 12.Klemm, P., and M. A. Schembri. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 290:27-35. [DOI] [PubMed] [Google Scholar]
- 13.Klose, K. E. 2000. The suckling mouse model of cholera. Trends Microbiol. 8:189-191. [DOI] [PubMed] [Google Scholar]
- 14.Korhonen, T. K., V. Vaisanen, H. Saxen, H. Hultberg, and S. B. Svenson. 1982. P-antigen-recognizing fimbriae from human uropathogenic Escherichia coli strains. Infect. Immun. 37:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruis, W., E. Schutz, P. Fric, B. Fixa, G. Judmaier, and M. Stolte. 1997. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 11:853-858. [DOI] [PubMed] [Google Scholar]
- 16.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 17.Lee, Y., Y. Kim, S. Yeom, S. Kim, S. Park, C. O. Jeon, and W. Park. 2008. The role of disulfide bond isomerase A (DsbA) of Escherichia coli O157:H7 in biofilm formation and virulence. FEMS Microbiol. Lett. 278:213-222. [DOI] [PubMed] [Google Scholar]
- 18.Lund, B., B. I. Marklund, N. Stromberg, F. Lindberg, K. A. Karlsson, and S. Normark. 1988. Uropathogenic Escherichia coli can express serologically identical pili of different receptor binding specificities. Mol. Microbiol. 2:255-263. [DOI] [PubMed] [Google Scholar]
- 19.Malchow, H. A. 1997. Crohn's disease and Escherichia coli. A new approach in therapy to maintain remission of colonic Crohn's disease? J. Clin. Gastroenterol. 25:653-658. [DOI] [PubMed] [Google Scholar]
- 20.Ong, C. L., G. C. Ulett, A. N. Mabbett, S. A. Beatson, R. I. Webb, W. Monaghan, G. R. Nimmo, D. F. Looke, A. G. McEwan, and M. A. Schembri. 2008. Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation. J. Bacteriol. 190:1054-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pere, A., M. Leinonen, V. Vaisanen-Rhen, M. Rhen, and T. K. Korhonen. 1985. Occurrence of type-1C fimbriae on Escherichia coli strains isolated from human extraintestinal infections. J. Gen. Microbiol. 131:1705-1711. [DOI] [PubMed] [Google Scholar]
- 22.Prilassnig, M., C. Wenisch, F. Daxboeck, and G. Feierl. 2007. Are probiotics detectable in human feces after oral uptake by healthy volunteers? Wien. Klin. Wochenschr. 119:456-462. [DOI] [PubMed] [Google Scholar]
- 23.Pruss, B. M., C. Besemann, A. Denton, and A. J. Wolfe. 2006. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J. Bacteriol. 188:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 25.Rendon, M. A., Z. Saldana, A. L. Erdem, V. Monteiro-Neto, A. Vazquez, J. B. Kaper, J. L. Puente, and J. A. Giron. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. USA 104:10637-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riegman, N., R. Kusters, H. Van Veggel, H. Bergmans, P. Van Bergen en Henegouwen, J. Hacker, and I. Van Die. 1990. F1C fimbriae of a uropathogenic Escherichia coli strain: genetic and functional organization of the foc gene cluster and identification of minor subunits. J. Bacteriol. 172:1114-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz, M. 2008. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm. Bowel Dis. 14:1012-1018. [DOI] [PubMed] [Google Scholar]
- 28.Shanks, R. M., M. A. Meehl, K. M. Brothers, R. M. Martinez, N. P. Donegan, M. L. Graber, A. L. Cheung, and G. A. O'Toole. 2008. Genetic evidence for an alternative citrate-dependent biofilm formation pathway in Staphylococcus aureus that is dependent on fibronectin binding proteins and the GraRS two-component regulatory system. Infect. Immun. 76:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stentebjerg-Olesen, B., T. Chakraborty, and P. Klemm. 1999. Type 1 fimbriation and phase switching in a natural Escherichia coli fimB null strain, Nissle 1917. J. Bacteriol. 181:7470-7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, J., F. Gunzer, A. M. Westendorf, J. Buer, M. Scharfe, M. Jarek, F. Gossling, H. Blocker, and A. P. Zeng. 2005. Genomic peculiarity of coding sequences and metabolic potential of probiotic Escherichia coli strain Nissle 1917 inferred from raw genome data. J. Biotechnol. 117:147-161. [DOI] [PubMed] [Google Scholar]
- 31.Torres, A. G., X. Zhou, and J. B. Kaper. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulett, G. C., A. N. Mabbett, K. C. Fung, R. I. Webb, and M. A. Schembri. 2007. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology 153:2321-2331. [DOI] [PubMed] [Google Scholar]
- 33.Van Houdt, R., and C. W. Michiels. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156:626-633. [DOI] [PubMed] [Google Scholar]
- 34.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]