Abstract
Accurate modeling of the infectious aerosol risk associated with the land application of biosolids requires an in-depth knowledge of the magnitudes and changes in pathogen concentrations for a variety of class A and class B stabilization methods. The following survey used quantitative PCR (qPCR) and culture assays to detect environmentally resistant bacterial and viral pathogens and biosolid indicator organisms for 36 biosolid grab samples. Biosolids were collected from 14 U.S. states and included 16 class B mesophilic anaerobic digestion (MAD) samples and 20 class A biosolid samples from temperature-phased anaerobic digestion (TPAD), MAD plus composting (COM), and MAD plus heat pelletization processes. The indicator concentrations of fecal coliforms and male-specific coliphages as well as pathogen genome concentrations for human adenovirus species, Legionella pneumophila, Staphylococcus aureus, and Clostridium difficile were significantly lower in the class A samples, and a multivariate analysis of variance ranked the stabilization processes from the lowest pathogen/indicator load to the highest as (i) class A COM, (ii) class A TPAD, and (iii) class B MAD. Human adenovirus genomes were found in 88% of the class B samples and 70 to 100% of the class A samples. L. pneumophila, S. aureus, and C. difficile genomes were detected at the qPCR assay detection limits in 19 to 50% of the class B and class A anaerobic digestion samples, while L. pneumophila was detected in 50% of the class A compost samples. When considering all the stabilization methods, both the fecal coliform and the male-specific coliphage concentrations show a significant linear correlation with the pathogen genome concentrations. This survey provides the necessary pathogen concentrations to add to biosolid aerosol risk and pathogen exposure analyses and clarifies the effectiveness of class A stabilization methods with the pathogen and indicator loads in biosolids.
Domestic sewage sludges that have been stabilized to reduce pathogen content are termed biosolids; their beneficial reuse for agricultural applications constitutes approximately 60% of the 7 million dry tons generated each year in the United States (6). Driven by health complaints from residents living near application sites, concern has been expressed that a hazardous exposure is created when biosolid aerosols are spread onto fields, incorporated into soils, and released during high-wind events (23, 36). Research in the last 5 years has made significant progress in estimating the human exposure to total biosolid aerosols and biosolid indicator microorganisms. Advances include both the development and calibration of theoretical and empirically based microbial aerosol models to assess off-site exposure during land application operations (8, 16, 31, 54) and the application of culture and DNA-based microbial-source tracking methods to these aerosols (5, 14). However, the paucity of information on the viral and bacterial pathogen content of biosolids has limited model use and effectiveness.
The U.S. Environmental Protection Agency (EPA) classifies end-product biosolids as class A (pathogen free) or class B (contains pathogens) based on indicator content and/or technologies used for stabilization (48). It is therefore important to distinguish between regulatory classes and treatment classes when describing the pathogen load in biosolids. In large U.S. municipalities, biosolids are most commonly stabilized with mesophilic anaerobic digestion (MAD) to produce a class B product, but current trends show that U.S. utilities have or are considering options for upgrading stabilization technology to produce class A biosolids. This includes the conversion to temperature-phased anaerobic digestion (TPAD) operations and/or composting the biosolids after anaerobic digestion (COM) (20). Meeting class A status requires the monitoring of fecal coliforms for estimating pathogen content, while the monitoring of enteric viruses, helminth ova, and Salmonella spp. is not required as long as “time and temperature” requirements are met. The majority of U.S. class A operations choose to meet class A status with time and temperature treatment-based alternatives. Based on these regulatory monitoring targets, most culture- or PCR-based biosolid studies have focused on these microorganisms and similar enteric pathogens (i.e., Listeria monocytogenes, enterovirus, and Clostridium perfringens) but have not diversified to other relevant airborne pathogens, like Legionella pneumophila, that may proliferate during stabilization where enteric pathogens cannot (1, 9, 22, 28, 34, 40, 41, 53). Only recently, and in limited sample sizes, have researchers begun to directly compare the diverse sludge stabilization methods available, including MAD, liming, and composting, to understand how selected enteric pathogens and indicators are removed (18, 22, 32, 34).
Comprehensive surveys that include dominant class A and class B stabilization practices have not been undertaken, and relevant airborne pathogens such as Legionella pneumophila and adenovirus have not been quantified in biosolids. Furthermore, the relationships between resistant-pathogen concentrations and fecal indicators have not been studied. This lack of information precludes meaningful aerosol pathogen exposure analysis, and subsequent regulatory and infrastructural decisions (such as requiring class A standards for land application) cannot be accurately evaluated. The production of such information has therefore been placed as a top priority among concerned citizens, biosolid researchers, and practitioners (4, 19, 21, 38).
To address these needs, the following study evaluates selected indicator and human pathogen levels in a library of class A and class B biosolids. A total of 16 class B and 20 class A samples were collected from treatment plants across the United States; these biosolids were produced using the common class B stabilization method of MAD and class A stabilization methods, including TPAD, COM, and MAD, plus heat pelletization (MH). The relationship of culturable bacterial and viral indicators with quantitative PCR (qPCR)-derived pathogen concentrations was investigated for each class and stabilization method to ascertain how indicators could be used to estimate human pathogen levels. The pathogens considered include the respiratory- and gastrointestinal-related human adenovirus species, the airborne pathogen Legionella pneumophila, and two environmentally resistant pathogens, Staphylococcus aureus and Clostridium difficile, which are capable of extended survival under desiccation, UV irradiation, and oxidative-stress conditions (13).
MATERIALS AND METHODS
Sampling.
Biosolid samples were collected from 29 wastewater utilities across the United States in states where agricultural application is the primary biosolid end-use regimen. The targeted stabilization processes included MAD (class B; 16 samples), TPAD (the time and temperature treatment option for class A; 8 samples), class A COM (10 samples), and class A MH (2 samples). For six treatment locations, both an anaerobically digested and a compost sample were obtained, and two compost samples were taken from one facility as two different end products, one having a longer curing time. All facilities treated a mixture of primary and secondary sludge. Table 1 provides a summary of the solid retention times, stabilization temperatures, and dewatering processes used for the biosolid samples in this survey.
TABLE 1.
Operational parameters for biosolid stabilization processes
| Stabilization process and configuration | Parametersa
|
Solid content (%)b | |
|---|---|---|---|
| 1 | 2 | ||
| MAD | 35-37°C for <20 days (5), for 20-25 days (6), and for >25 days (4) | High-solids centrifuge (6), belt filter press (5), solid-bowl centrifuge (2), and none (2) | 22 ± 2.9, 17 ± 3.0, 22, 1 |
| TPAD | |||
| Meso-Thermo | 38-41°C for 15-20 days, 55°C for 5-15 days (2) | High-solids centrifuge (4), belt filter press (3), none (1) | 29 ± 3.2, 19 ± 1.3, 3 |
| Thermo-Meso | 55°C for 15-30 days, 35-37°C for 15-30 days (3); 55°C aerobic for 5-10 days, 32-37°C for 15-30 days (1) | ||
| Thermo-Thermo | 54-56°C for 20 days, 48-56°C for 3-5 days (2) | ||
| MH | 35-37°C for 10-20 days, dewatering, followed by a low-pressure oxidation drying system (2) | High-solids centrifuge (1), solid bowl centrifuge (1) | 93, 91 |
| COM | Agitated windrow method (8) | Sawdust, green waste, woodchips, and paper (6); no amendment (2) | 45 ± 12.4; 80 ± 2.2 |
| In-vessel composting (1 short and 1 long curing) | Sawdust, woodchips, paper (2) | 38 ± 1.4 | |
Parameters for the MAD, TPAD, and MH stabilization processes are temperatures and solid retention times (no. of samples) in column 1 and the dewatering processes (no. of samples) in column 2. Parameters for the COM stabilization process are composting configurations (no. of samples) in column 1 and amendments in column 2.
Solid content percentages correspond to the averages ± standard deviations for the respective dewatering methods and amendments.
The anaerobically digested samples were aseptically collected at conveyor belts at the end of the treatment process or the truck loading area in accordance with U.S. EPA-recommended procedures in method 1680 (50). The compost samples were collected from curing piles that were ready for agriculture application. Five 100-g samples were shipped overnight on ice to the laboratory and processed within 24 h of collection. Upon arrival, the five 100-g samples were mixed together to form a composite biosolid sample, and the sample was subsequently split into fractions for immediate culture analysis and gravimetric solid content analysis (15) or frozen at −80°C for qPCR analysis.
Culture enumeration.
Biosolid indicators, including fecal coliforms, male-specific coliphages, and sulfite-reducing Clostridia, were enumerated by culturing. For bacterial indicator analyses, duplicate 15 to 25 g (wet) samples were eluted in sterile 0.1% peptone water with 0.05% Tween 80 (29) followed by mixing for 1 h at 250 rpm. Serial dilutions were made in sterile 1× phosphate-buffered saline (0.14 M NaCl, 0.01 M phosphate, 0.03 M KCl [pH 7.4]), and indicator bacteria were quantified using membrane filtration with 0.45-μm-pore-size, 47-mm-diameter filters (Millipore Co., Billerica, MA). Fecal coliforms were cultured for 24 h at 44.5°C on mFC agar (Difco, Inc., Detroit, MI) with 0.01% rosolic acid and identified as dark blue colonies (15). Sulfite-reducing clostridia were cultured with egg-yolk-free tryptose sulfite cycloserine agar (Difco, Inc., Detroit, MI) in anaerobic chambers (BBL GasPak; Becton Dickinson, Inc., Sparks, MD) for 48 h at 37°C (42). Black colonies were identified as sulfite-reducing clostridia.
Male-specific coliphages were eluted from biosolids following a modified U.S. EPA method for the recovery of viruses from sludge (35, 51). Briefly, 25 ml of sterile 10% beef extract buffer was added to duplicate 5- to 9-g (wet) biosolid samples and adjusted to a pH of 9. The samples were stirred for 2 h at 250 rpm, followed by centrifugation in sterile Teflon tubes at 10,000 × g for 30 min at 4°C. The supernatant was collected, adjusted to a pH of 7.2, 0.2-μm filter sterilized, and analyzed by the double agar layer method (49). For the double agar layer method, serial dilutions were made in sterile phosphate-buffered saline, and duplicate 1-ml aliquots of each biosolid dilution were combined with 4 ml molten tryptic soy agar (0.7%) and 1 ml of log-phase Escherichia coli ATCC 15597 (American Type Culture Collection, Manassas, VA) and poured onto tryptic soy agar plates. The plates were incubated for 24 h at 37°C, and the plaques in the E. coli lawn were identified as male-specific coliphages. Escherichia coli phage MS2 (ATCC 15597-B1) was used as a positive control.
For the culture-based bacteria and bacteriophage indicator assays, detection limits were calculated based on the wet sample weight and the sample's biosolid content, which varied primarily by the stabilization treatment. For each biosolid treatment, the ranges of detection limits (geometric average, maximum, and minimum, respectively, in CFU/dry g) calculated for the bacterial assays were 17.6, 139.8, and 7.1 for MAD; 12.0, 19.3, and 7.9 for TPAD; 6.8, 13.5, and 3.4 for COM; and 3.9, 4.8, and 3.1 for MH. The ranges of detection limits (in PFU/dry g) for the bacteriophage assays were 26.1, 321.8, and 14.2 for MAD; 20.0, 60.6, and 11.7 for TPAD; 9.5, 17.1, and 5.2 for COM; and 4.6, 4.8, and 4.4 for MH.
Pathogen genome enumeration with qPCR.
The quantification of human pathogen genomes in biosolids was performed using TaqMan real-time qPCR. Prior to the qPCR analyses, experiments were conducted to optimize biosolid inhibitor removal during DNA extraction and the qPCR assay. An exogenous external control was also used to control for inhibition in each biosolid DNA extraction prior to the qPCR analyses.
(i) Extraction of nucleic acids.
DNA was extracted from 0.05 to 0.3 g wet biosolids with the Mo Bio PowerSoil DNA kit (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer's instructions for high yields of DNA. For the composted biosolids, greater inhibition and brown discoloration were initially noted, so the amount of starting material was decreased in the composting matrix (<0.15 g). To increase the DNA yields, the following modifications were used. The initial cell lysis utilized an additional heat step at 70°C for 10 min followed by bead beating (Mini-Beadbeater; Biospec Products, Bartlesville, OK) at 2,500 rpm for 3 min instead of vortex agitation. The incubation with buffers S2 and S3 was increased to 10 min at 4°C, and a second centrifugation step was used to maximize the supernatant recovery at each step. Also, the final DNA elution from the silica filter was performed by adding Tris-EDTA buffer heated to 50°C and incubating for 5 min at room temperature before the final centrifugation.
DNA extraction efficiencies were determined for each biosolid matrix and used in the final calculation of the pathogen genomes/dry g biosolids. To calculate the DNA extraction efficiency of the Mo Bio PowerSoil DNA kit, Enterococcus faecalis pure cultures were spiked at a concentration of 107 total cells/0.25 g biosolids and extracted using the previously described extraction protocol. The E. faecalis genomes were then diluted and quantified using the Enterococcus genus qPCR assay specified in Table 2. E. faecalis was chosen for its extraction efficiency due to its gram-positive cell wall and relatively high resistance to cell lysis. Parallel DNA extractions were performed on biosolid samples that were not spiked to determine the background qPCR Enterococcus levels and to ensure that the levels were at least 3 orders of magnitude less than the spiked E. faecalis concentration. The average biosolid DNA extraction efficiency was calculated by dividing the amount of E. faecalis genomes recovered from the biosolid matrix by the amount recovered from E. faecalis spiked in tubes with no biosolids. The final extraction efficiencies were based on the average of five biosolid samples for each biosolid matrix (MAD, TPAD, and COM).
TABLE 2.
PCR primers, probes, and parameters used in the study
| Target group (gene) | ID | Sequence (5′-3′) | Size (bp) | Tempanneala (oC), time | Standard curve slope, intercept (R2)b | Reference(s) |
|---|---|---|---|---|---|---|
| Adenovirus spp. (all 51 types) (hexon) | 18895F | GGA CGC CTC GGA GTA CCT GAG | 120 | 55, 60 s | −3.380, 43.187 (0.992) | 27 |
| 18990R | ACN GTG GGG TTT CTG AAC TTG TT | |||||
| 18923P | CTGGTGCAGTTCGCCCGTGCCA | |||||
| Staphylococcus aureus (nuc) | F | CGCTACTAGTTGCTTAGTGTTAACTTTAGTTG | 124 | 60, 60 s | −3.572, 42.635 (0.979) | 2 |
| R | TGCACTATATACTGTTGGATCTTCAGAA | |||||
| P | TGCATCACAAACAGATAACGGCGTAAATAGAAG | |||||
| Clostridium difficile (tcdB) | F | GAAAGTCCAAGTTTACGCTCAAT | 177 | 57, 60 s | −3.571, 42.456 (0.998) | 52 |
| R | GCTGCACCTAAACTTACACCA | |||||
| P | ACAGATGCAGCCAAAGTTGTTGAATT | |||||
| Legionella pneumophila (mip) | 409F | AATGGTGTTAAACCTGGTAAATCGG | 114 | 60, 60 s | −3.550, 44.161 (0.989) | 17 |
| 523R | CCTGAAAAGTAGCTGGCTTACCAGT | |||||
| 460P | CGTCTGATTGATGGTACCGTTTTTGACAG | |||||
| Enterococcus spp. (23S)c | 748F | AGAAATTCCAAACGAACTTG | 96 | 60, 120 s | −3.379, 39.750 (0.976) | 24 |
| 854R | CAGTGCTCTACCTCCATCATT | |||||
| 813P | TGGTTCTCTCCGAAATAGCTTTAGGGCTA | |||||
| puc19 exogenous control | 49F | CGG AGA CGG TCA CAG CT | 94 | 55, 120 s | N/A | 46, this study |
| 448R | TTG CAT GCC TGC AGG T | |||||
| 200F | ATTCAGGCTGCGCAACTGTTG | |||||
| 294R | TTAATCGCCTTGCAGCACAT | |||||
| 231P | TCGGTGCGGGCCTCTTCGCTATTAC |
Tempanneal, annealing temperature.
Standard curve values represent the average of the results from at least three independent standard dilutions. Concentrations (CT) were determined with the following formula: CT = slope × log(concentration) + y intercept. N/A, not applicable.
Standard curve calculations account for the four 23S gene copies in each Enterococcus cell.
(ii) qPCR assays.
Previously published qPCR primers and TaqMan probes were used for the qPCR amplification of pathogen genomes; the sequences used are listed in Table 2. Commercial TaqMan probes were labeled with 6-carboxyfluorescein and butylhydroquinone (Biosearch Technologies, Novato, CA). For all the TaqMan qPCR analyses, triplicate 20-μl qPCRs were prepared with 2× TaqMan PCR master mix with ROX passive reference dye (Roche Diagnostics, Indianapolis, IN), 0.4 mg/ml bovine serum albumin, 1.5% polyvinylpyrrolidone (molecular weight, 25,000; Sigma-Aldrich, St. Louis, MO), and 5 μl DNA template (34). Adenovirus species and internal control reactions used 250 nM of each primer and 125 nM of TaqMan probe, while all bacterial analyses were conducted with 300 nM of each primer and 100 nM of TaqMan probe. The qPCRs were run on an ABI Prism 7500 sequence detector under standard thermal-cycling conditions, consisting of an initial 10 min of denaturation at 95°C, followed by 45 cycles of 15 s of denaturation at 95°C and 60 s of annealing/extension (the assay-specific temperatures are listed in Table 2).
Standard curves were developed for each qPCR bacterial pathogen assay using genomic DNA. Starting nucleic acid concentrations were determined by a Nanodrop ND-1000 UV-Vis spectrophotometer (Nanodrop Technologies, Wilmington, DE). Three to five independent dilution series were aliquoted from 100 to 106 genomic units (GU)/reaction with the genomic DNA of Legionella pneumophila subsp. pneumophila (ATCC 33152), Staphylococcus aureus subsp. aureus strain MU50 (ATCC 700699), Clostridium difficile (ATCC 90556-M6S), and Enterococcus faecalis (ATCC 19433). For adenovirus species standard curves, a 301-bp region of the hexon gene from adenovirus serotype 40 was PCR amplified with previously published primers (3). The PCR amplicon was subsequently cloned into a pCR4-TOPO plasmid vector, transformed into competent E. coli following the manufacturer's instructions (TOPO TA cloning kit; Invitrogen, Carlsbad, CA), and purified with a QIAprep spin miniprep kit (Qiagen, Inc., Valencia, CA). The hexon gene plasmid DNA was then quantified and diluted as previously described for genomic DNA. Standard curves were used to relate a threshold cycle value to genome concentrations in biosolid samples, and standard log regression parameters are listed for each assay in Table 2. Negative controls and a standard curve were included with every assay. Each qPCR pathogen assay could consistently detect 10 pathogen genomes per reaction, while cases with one pathogen genome were typically detected in only one out of three replicates.
Prior to any qPCR pathogen analyses, DNA inhibition was explored in each biosolid DNA extract using an exogenous external control (the pUC19 plasmid) assay. The pUC19 plasmid was spiked into each biosolid sample with no dilution, a 1:2 dilution, and a 1:5 dilution at 104 copies puc19/reaction. The percent difference in threshold cycle values from each biosolid dilution was compared to the control, and in cases where inhibition was observed (Student's t test, P ≤ 0.05), the biosolid DNA extract was diluted until inhibition was removed (12). The inhibitory effects were also checked for each pathogen assay by spiking known concentrations of genomic DNA for the relevant pathogen and comparing to the standard curve with at least two biosolid samples from each treatment.
(iii) Pathogen confirmation with DNA sequencing.
Pathogen serotypes were determined for a subset of the positive biosolid qPCRs with DNA sequencing. After qPCR, positive reactions were purified with the MinElute reaction cleanup kit (Qiagen, Inc., Valencia, CA). Sequencing was performed on an AB 3730xl DNA analyzer, and phylogenetic analyses were conducted using MEGA version 4 (45). To ascertain further differences in the sequences for L. pneumophila and adenovirus species, nested PCR followed by sequencing was also performed using previously described assays (3, 10).
Statistical interpretation.
All culturable indicator and pathogen genome data were log10 transformed and evaluated for lognormality using the Andersen-Darling test. Analysis of variance (ANOVA) followed by Tukey's postcomparison tests were used to show the differences between the concentrations of pathogens and indicators (P ≤ 0.05) for each biosolid stabilization treatment. Multivariate ANOVA (MANOVA) was used to rank biosolids based on all indicator and pathogen data. Lognormal data were also analyzed with Pearson's correlation coefficient and linear regression to understand the relationships between culturable indicator and human pathogen concentrations. Statistics were calculated in Minitab version 15.0.
RESULTS
The following results present a comparison of biosolid indicators and human pathogen genomes in biosolid samples originating from domestic wastewater sludges stabilized via the following methods: MAD (class B), TPAD (class A), COM (class A), and MH (class A). Since the goal of the survey was to capture trends between different treatment processes and because the concentration data were found to be lognormally distributed, the results are presented as the geometric (log) average and standard deviation for each treatment type rather than for individual biosolid samples. The class A MH results are shown for comparison purposes but are not statistically evaluated because of the limited sample size (n = 2).
Culturable indicators in biosolids.
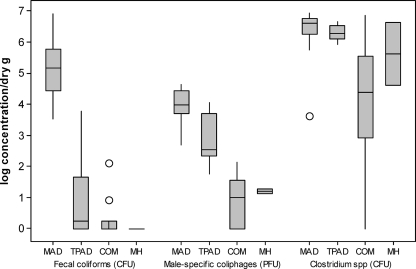
Figure 1 depicts the concentration of culturable indicators in biosolids from the four stabilization methods using a box-plot format. Comparisons between each biosolid stabilization treatment show that indicator concentrations in class A biosolids are lower than class B biosolids for fecal coliforms, male-specific coliphages, and sulfite-reducing clostridia (ANOVA, P ≤ 0.005). Mean fecal coliform concentrations corresponded to U.S. EPA regulatory limits for each biosolid class (class B, 2 × 106 CFU/dry g; class A, 103 CFU/dry g), with class B MAD samples averaging 1.5 × 105 CFU/dry g and all but seven class A samples with fecal coliform concentrations below the detection limits. Two of 16 class B MAD samples exceeded the allowable U.S. EPA fecal coliform limits but were not higher than 107 CFU/dry g, while only one TPAD sample surpassed the U.S. EPA class A allowable limit of 103 CFU/dry g. Male-specific coliphages were detected in the class A treatments when fecal coliforms were not, especially in the TPAD samples, which averaged 102 to 103 PFU/dry g. Since sulfite-reducing clostridia are known to grow well in anaerobic digesters, their presence was expected in the high concentrations seen here (4.5 × 106 CFU/dry g for MAD samples and 2.0 × 106 CFU/dry g for TPAD samples). Sulfite-reducing clostridia were also present in the class A COM samples but at two logs lower (1.3 × 104 CFU/dry g) than both the MAD and TPAD biosolids (ANOVA, P ≤ 0.005). Overall, the magnitude differences between class B and class A treatments were highest for fecal coliforms (5-log difference), while 1- to 3-log differences were observed for male-specific coliphages. Male-specific coliphage concentrations can be used to rank the stabilization methods using Tukey's postcomparison tests, with the rank order as follows: class B MAD (8.7 × 103 PFU/dry g) > class A TPAD (6.4 × 102 PFU/dry g) ≫ class A COM (8 PFU/dry g). On the other hand, Tukey's postcomparison tests for fecal coliforms show only that class B biosolids ≫ class A TPAD and COM, while no differences are revealed between the class A treatments.
FIG. 1.
Box-and-whisker plots of the log biosolid indicator concentrations after four stabilization treatments (pooled data for 16 MAD, 8 TPAD, 10 COM, and 2 MH samples). The inner box lines represent the geometric medians, while the outer box lines represent the 25th and 75th data percentiles (inner quartile range [IQR], the whiskers extend to 1.5 times the IQR, and the unfilled circles indicate data outliers.
Human pathogen genomes in biosolids.
Before human pathogen genome detection was performed with qPCR, both the inhibitory effects of the biosolid DNA extracts and the qPCR detection limits were evaluated. This evaluation compared common laboratory DNA extraction methods (5) and commercial kits used for environmental matrices (Qiagen Miniprep, Mo Bio PowerSoil DNA, and Mo Bio UltraSoil DNA kits) and found that the Mo Bio PowerSoil DNA kit effectively removed inhibitors in all biosolid treatment types, whereas other kits did not (data not shown). Next, to ensure that there was no inhibition during qPCR, the qPCR amplification for an exogenous external control (the puc19 plasmid) in each biosolid DNA extract was compared to a control standard in a noninhibitory matrix. No significant decreases in amplification efficiency (<5%) were noted in most biosolid samples, and in those with a greater-than-5% decrease in efficiency, a 1:5 dilution of the biosolid DNA extract removed this effect. The qPCR detection limits were next determined as shown in Table 3; these detection limits varied from sample to sample due to differences in the biosolid moisture content, the need to dilute templates, and the DNA extraction efficiency for a particular treatment. Average DNA extraction efficiencies were used to calculate both the final pathogen concentrations and detection limits for each biosolid treatment (Table 3). Based on the calculated qPCR assay sensitivity, pathogen concentrations should be at least 3.8 × 103 GU/dry g for MAD, 6.3 × 102 GU/dry g for TPAD, and 5 × 102 GU/dry g for COM to be detected by the qPCR method.
TABLE 3.
DNA extraction efficiency and quantitative PCR assay detection limits for each biosolid matrixa
| Treatment | Solid content (%)
|
DNA extraction efficiency (%)
|
Log detection limit/dry gb
|
||||
|---|---|---|---|---|---|---|---|
| Avg | SD | Avg | SD | Min | Avg | Max | |
| MAD | 17 | 7.4 | 10.8 | 8.6 | 3.38 | 3.58 | 4.29 |
| TPAD | 22 | 9.5 | 45.9 | 34.3 | 2.65 | 2.87 | 3.36 |
| COM | 51 | 18.2 | 55.5 | 39.3 | 2.48 | 2.71 | 2.89 |
| MHEAT | 92 | NA | ND | ND | 2.52 | 2.54 | 2.55 |
Avg, average; SD, standard deviation; Min, minimum; Max, maximum; NA, not applicable; ND, not determined.
Detection limits were calculated as (1 genome/qPCR volume) × DNA elution volume/(wet weight × solid content × % DNA extraction efficiency).
Human pathogen genomes were detected by qPCR in both class A and class B biosolids. Table 4 shows the detection frequencies for genomes of adenovirus species, Staphylococcus aureus, Clostridium difficile, and Legionella pneumophila for each treatment. The detection frequencies were highest for adenovirus genomes because concentrations were detected well above the detection limits. On the other hand, bacterial pathogen genomes were observed at or near the detection limits, so the higher detection frequencies for the class A treatments in Table 4 likely resulted from the ability to detect lower log concentrations than for the class B biosolids. In most samples, genomes from at least one pathogen were detected. The exceptions were samples within the class A treatments and included 2 COM samples, one with the longest curing time, and a TPAD sample that utilized both aerobic and anaerobic digestion. Genomes from multiple pathogens were detected in 16/36 biosolid samples; concurrent encounters of L. pneumophila and adenovirus species were most frequently observed (10/36 samples).
TABLE 4.
Percentage of biosolid samples with detectable pathogen genomes
| Human pathogen | % of positive samples
|
|||
|---|---|---|---|---|
| Class B MAD (n = 16) | Class A
|
|||
| TPAD (n = 8) | COM (n = 10) | MH (n = 2) | ||
| Virus | ||||
| Adenovirus spp. | 88 | 75 | 70 | 100 |
| Bacteria | ||||
| Legionella pneumophila | 31 | 25 | 50 | 0 |
| Staphylococcus aureus | 19 | 50 | 0 | 0 |
| Clostridium difficile | 25 | 38 | 0 | 0 |
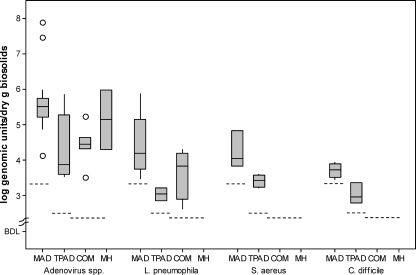
Figure 2 provides qPCR results for the selected viral and bacterial pathogen genomes. When pathogen genomes were present, the geometric average concentration for all pathogen genomes was lower for the class A biosolids relative to that of the class B biosolids. The differences were greatest for adenovirus species genomes in class B MAD biosolids (5.0 × 105 GU/dry g) compared to class A COM biosolids (2.5 × 104 GU/dry g) (ANOVA, P ≤ 0.05). While S. aureus and C. difficile genomes were found more frequently in class A TPAD samples (Table 4), their concentrations were significantly lower than class B MAD concentrations (ANOVA, P ≤ 0.05).
FIG. 2.
Box-and-whisker plots of the log pathogen genome concentrations of positive samples for each biosolid stabilization method. The inner box lines represent the geometric medians, while the outer box lines represent the 25th and 75th data percentiles (IQR), the whiskers extend to 1.5 times the IQR, and the unfilled circles indicate data outliers. For each treatment, the dashed line represents the minimum detection limit within the treatment.
To confirm the qPCR results, a subset of positive samples was sequenced. For the adenovirus species, types 31 (subgenus A), 3 (subgenus B:1), and 41 (subgenus F) were isolated; these strains correspond to human adenovirus causing gastrointestinal (subgenus F) and acute respiratory infections (subgenera B:1 and A) (3). L. pneumophila sequences were closely related to serotypes L. pneumophila subsp. Dallas and L. pneumophila subsp. Corby in 5 MAD, 2 TPAD, and 3 COM samples but not to the control, which was L. pneumophila subsp. Philadelphia. S. aureus was also confirmed for 100% of the MAD and TPAD samples tested, but the 124-bp amplified region of the nuc gene used in the qPCR did not allow for differentiation among S. aureus serotypes. Similarly, C. difficile-positive biosolids were confirmed at the species level for all positive MAD and TPAD samples.
Finally, MANOVAs were used to rank the biosolid stabilization methods by giving equal weight to the pathogen genome and indicator concentrations. From the method with the lowest to the highest pathogen and indicator loads, the biosolid stabilization methods can be ranked as follows: (i) class A COM, (ii) class A TPAD, and (iii) class B MAD (MANOVA, P ≤ 0.005).
Statistical relationships between indicators and pathogen genomes.
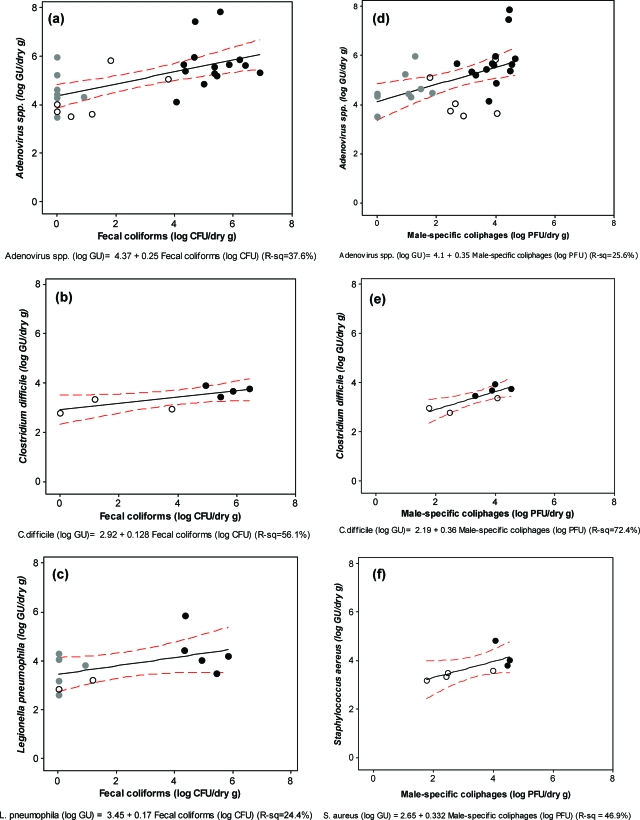
Linear correlations between culturable indicators and pathogen genomes were investigated for both the pooled data set (all treatments) and data from each stabilization treatment. For the bacterial pathogen genomes, only results with pathogen presence above detection limits are included in this analysis. Since each biological parameter was lognormally distributed, parametric statistics were used first to determine statistically significant correlations (Pearson's correlation coefficient [rp] and P value) and to then understand the behavior and variance of correlated pathogen-indicator pairs (linear regression equation [R2]). In general, an rp of >0.6 shows a strong positive correlation, and an rp of >0.3 but <0.6 shows a weak to moderate positive relationship. When the biosolids data were analyzed together, strong positive correlations were noted between both indicator-indicator and pathogen-indicator pairs. Fecal coliforms, the chosen U.S. EPA indicator, were positively correlated to human pathogen genomes, including adenovirus species genomes (rp = 0.613; P ≤ 0.005) and C. difficile genomes (rp = 0.749; P ≤ 0.05), and weakly correlated to L. pneumophila genomes (rp = 0.493; P = 0.10). Correlations were also observed between male-specific coliphages and adenovirus species genomes (rp = 0.506; P ≤ 0.005), C. difficile genomes (rp = 0.851; P ≤ 0.05), and S. aureus genomes (rp = 0.685; P ≤ 0.10). All indicator-indicator relationships were strongly significant (e.g., fecal coliforms to male-specific coliphages, rp = 0.773 and P ≤ 0.005), but no pathogen genome-pathogen genome relationships were found. The sulfite-reducing Clostridium indicator was only correlated to other indicators but not to the bacterial and viral pathogens in this study. The data analyzed within a single treatment did not reveal significant pathogen-indicator relationships, likely due to the limited number of points and small spread of concentrations found for each treatment.
Positive correlations between pathogen genomes and indicators are shown in Fig. 3, which shows the fitted-line plots for each significantly related pathogen-indicator pair. Both fecal coliform and male-specific coliphage indicators can predict weak to moderate linear increases in pathogen genomes (R2, 24 to 72%). The major trend that emerges within the linear regression equations is that the slope of the line is both positive and similar for a specific indicator, regardless of the pathogen. The culturable fecal coliforms (Fig. 3a to c) increase 5.4 logs for each l-log change in pathogen genomes, while the culturable male-specific coliphages (Fig. 3d to f) average a 2.8-log increase for every 1-log increase in pathogen genomes.
FIG. 3.
Linear regression analyses between significantly correlated pathogen genomes (GU) and culturable indicators. Only positive samples were used for the linear regression of bacterial pathogen genomes. Stabilization methods are represented by the following symbols: black circles, MAD; unfilled circles, TPAD; and gray circles, COM. For each plot, the best-fit linear regression line (solid line) and 95% confidence intervals (dashed lines) are shown for the linear equation below the y axis. R2 values are presented to indicate how much variance is described through the linear regression analyses.
DISCUSSION
Biosolids are increasingly diverted for agricultural application in the U.S. and worldwide. The concerns over infectious risk, coupled with recent trends to upgrade stabilization technologies to produce class A biosolids, underscore the need to understand how indicator and pathogen contents vary for commonly used stabilization processes. The survey results presented here allow for an enhanced understanding of biosolids and associated biosolid aerosol pathogen loads as a function of class and stabilization, demonstrate the utility of quantifying pathogen genomes in biosolids, and provide new information on the effectiveness of indicators used in the U.S. EPA land application regulations.
Reduced pathogen genome and indicator loads in class A biosolids.
Both culturable indicator and pathogen genome concentrations were consistently lower in class A biosolids compared to class B biosolids. These concentration differences were greater for culturable indicators (Fig. 1) than pathogen genomes (Fig. 2). Of the four biosolid stabilization technologies targeted here, class A COM and class A MH provided the lowest levels of both culturable indicators and pathogen genomes, followed by class A TPAD and finally class B MAD. Indicator concentrations were comparable to previously reported values (1, 18, 22, 32, 33, 40) and were homogenous within treatment types, even with highly diverse operation parameters (Table 1). For the pathogens considered, only adenovirus species genomes have been previously quantified in biosolids. In this previous study, which was limited to MAD samples (7), adenovirus genome concentrations were 1 to 2 logs lower than those reported for MAD samples in our study. One difference that partially explains the concentration difference is the inclusion of DNA extraction efficiencies in our study.
While our approach was to identify trends using averaged treatment values, an analysis of individual plant data showed one notable exception. In contrast to the average decreasing trend from class B to class A, three of the five plants that provided influent (MAD) and effluent (COM) samples had an increase in Legionella pneumophila genomes. Of the three indicators and four pathogens considered, L. pneumophila genomes were the only ones to show an increase during composting. L. pneumophila is associated with warm, aerobic environments similar to that of a compost heap and appears to have the potential to proliferate during composting. The transmission of Legionnaires' disease from garden compost has been documented (11), and L. pneumophila has also been shown to grow on heat-killed microbial cells in thermophilic environments (47). It is important to note that our research focus was not on L. pneumophila inactivation in compost and we are not aware of other reports detailing the inactivation or growth of L. pneumophila in compost heaps. We therefore list this as a noteworthy observation that merits further investigation rather than as a firm conclusion.
This study also reports the first detection (by culture or PCR) of Staphylococcus aureus and Clostridium difficile in biosolids. Special attention should be given to the detection of S. aureus genomes here as a link has been suggested between this organism and infection in residents living near agricultural class B land application sites (30). Rusin et al. (41) previously applied culture-based methods to detect S. aureus in 2 raw sewage sludge, 6 MAD, 10 lime-treated, 1 TPAD, 1 COM, and 4 MH biosolid samples from across the United States. Culturable S. aureus was detected only in one of two untreated sewage sludge samples and not in finished biosolid products (41). Our study was able to detect S. aureus genomes in 3/16 class B MAD and 4/8 class A TPAD biosolids with all concentrations at or near detection levels. While our qPCR results and the detection of S. aureus in untreated sludges suggest that there is potential for infectious Staphylococcus aureus to be in biosolids, there is still no evidence of infectious S. aureus in biosolids.
qPCR and pathogen viability.
Studies with pathogens in a broad variety of environmental samples suggest that the concentration of total genomes is typically 2 to 4 orders of magnitude greater than that of culturable or infectious pathogen concentrations (25, 34, 37, 55). An indirect comparison of pathogen genomes to culturable indicators in this study delineates a similar result, even with the incorporation of DNA extraction efficiencies and the assurance of no qPCR inhibition from each biosolid matrix. The qPCR results presented here provide two useful ways for interpreting the pathogen load in biosolids. First, the observed reduction in pathogen genome concentrations in class A biosolids was consistent with the decrease observed in culturable indicators (Fig. 3). This reduction suggests that as pathogens are inactivated, there will be an associated loss in genomic material. Whether that loss in genomic material is less than or equivalent to the loss in culturability or infectivity is not known and will likely be a function of pathogen physiology and the environment which caused the inactivation.
Secondly, in a matrix that has very little information on pathogen load, qPCR results provide a conservative estimate of risk. We note that the degree to which qPCR values are conservative is not certain as the use of culturable values in risk assessment can actually underestimate concentrations by not detecting viable but not culturable bacteria (39). Relevant to biosolids, nonculturable Salmonella spp. have shown reactivation potential in composted biosolids with increasing moisture content and storage (43, 44). Similarly, nonculturable fecal coliforms in TPAD-treated biosolids have demonstrated the ability to recover culturability during dewatering by high-speed centrifugation (26). In cases of possible reactivation, qPCR has been shown to be a more accurate measure of viable E. coli.
Quantitative microbial risk assessments for biosolids report that the highest risk of human infection results from an aerosol exposure in workers at the dewatering belt or entrepreneurs spreading their own biosolids; this infectious risk was based on an inhalation dose of one infectious adenovirus particle (54). Here, we show high adenovirus genome concentrations, but we can only assume the rate of infectivity based on a previous estimation that 0.1% of these genomes are infectious in water samples (25). If we apply this infectious percentage to adenovirus genome concentrations in biosolids analyzed in our study, this would correspond to 100 to 300 infectious viral particles per gram in class B MAD samples and 1 to 5 infectious adenovirus particles per gram in class A TPAD and COM samples. We can translate these infectious adenovirus concentrations in bulk biosolid samples to a downwind aerosol concentration using a previously described and calibrated aerosol transport model for respirable biosolid material at off-site locations (31). Table 5 lists the off-site predicted infectious aerosol values for a biosolid-disking scenario at distances from 0 to 165 m.
TABLE 5.
Predicted adenovirus dose in aerosols downwind of biosolid-disking operations
| Treatment (avg bulk concn) | Distance downwind (m) | Respirable biosolid dose (μg)a | Adenovirus spp. doseb
|
|
|---|---|---|---|---|
| Total no. of genomes | Estimated infectious dose | |||
| Class B MAD (4.3 × 105 GU/dry g) | 0 | 7-19 | 3.0-8.2 | 0.0030-0.0082 |
| 65 | 6-15 | 2.6-6.5 | 0.0026-0.0065 | |
| 165 | 3-8 | 1.3-3.5 | 0.0013-0.0035 | |
| 500 | 0.2-2 | 0.09-0.9 | 0.00009-0.0009 | |
| Class A TPAD/COM (2.5 × 104 GU/dry g) | 0 | 7-19 | 0.18-0.48 | 0.0002-0.0005 |
| 65 | 6-15 | 0.15-0.38 | 0.0002-0.0004 | |
| 165 | 3-8 | 0.08-0.2 | 0.00008-0.0002 | |
| 500 | 0.2-2 | 0.005-0.05 | 0.000005-0.00008 | |
Respirable biosolid dose for 8 h based on a wind speed of 1.5 m/s and 4 to 11% biosolid content in <10 μm aerosol particulate matter.
An infectivity of 0.1% was used to estimate the infective doses from total no. of genomes. Aerosolized adenovirus values were calculated as respirable biosolid concentration × 10−6 g/μg × adenovirus GU/dry g biosolids × 1 infectious dose/103 GU.
Indicator monitoring and predictive indicator-pathogen relationships.
Fecal coliforms are the primary indicator used to represent the level of pathogen inactivation achieved for a particular stabilization method and also as a proxy for the presence of human pathogens. Of the indicators and pathogen genomes considered in this study, fecal coliforms demonstrated the largest decrease in concentration between class B and class A biosolids. Fecal coliforms are either the least resistant to stresses in digesters and compost heaps, or current stabilization procedures have evolved to preferentially remove fecal coliforms. In a cross section of biosolid samples (Fig. 1), male-specific coliphages appear to be a more stringent test of inactivation. When either fecal coliforms or male-specific coliphages are compared to pathogen presence in Fig. 3, predictive relationships emerge for both indicators, irrespective of whether the pathogen is of bacterial or viral origin. Because male-specific coliphage presence was more likely in biosolid samples than fecal coliforms, pathogen concentrations for a given sample were more comparable to male-specific coliphage values; this result suggests that they would be more useful for documenting pathogen presence than fecal coliforms. These observations are further supported by laboratory studies of thermal inactivation in sludge where bacterial indicator resistance was markedly lower than male-specific coliphages, and recommendations were made for using coliphages to indicate E. coli and Salmonella sp. inactivation with a factor of safety (33). Male-specific coliphages are more heat-resistant than fecal coliforms but have also shown the necessary temperature-dependent inactivation required of a good indicator both in batch-scale studies at relevant digester temperatures (<55°C) and in the present study when MAD and TPAD samples are compared (1). These findings support the mounting body of literature that recommends monitoring biosolids with male-specific coliphages. Finally, we note that the linear correlations between indicators and pathogens were enabled in this present study by quantifying pathogen genomes and because of the log-order range of indicator and pathogen concentrations found throughout different biosolid treatments; further studies are warranted to understand how these relationships are relevant according to specific inactivation mechanisms and to other pathogens and environmental matrices.
While no correlations were found between sulfite-reducing clostridia and the pathogens studied, high concentrations of this indicator were found in all biosolid treatments. The low reductions of sulfite-reducing clostridia were expected, since many are thermotolerant and can proliferate in the anaerobic digester environment. Class A COM samples are generated from a MAD product, so even with aerobic conditions during composting, this study and others show the ability of sulfite-reducing clostridia to survive (40). The spores of sulfite-reducing clostridia have been proposed and used as an indicator of the presence of biosolids and have successfully been used in tracking class B biosolid aerosols from land application sites when fecal coliform concentrations were not detected (5, 14). The results here suggest that their use can be extended to tracking class A COM and TPAD biosolids in the environment. Guzman et al. and others also suggest the use of sulfite-reducing clostridial spores as an indicator of parasites, including Cryptosporidium oocysts and helminth ova (22).
Conclusions.
The survey results presented here quantify human pathogen genomes that may survive aerosolization or be transmitted by aerosol routes and relate these pathogens to culturable indicators with a focus on differences in stabilization methods and U.S. EPA class designations for agricultural application. Class A biosolids had less-frequent and lower concentrations of pathogen genomes and indicators than class B biosolids. The reduction from class B to class A was greater for culturable indicators than for qPCR, suggesting that while the qPCR values can reveal inactivation, it may underestimate this inactivation. The addition of qPCR pathogen results to aerosol transport models revealed conservative estimates of aerosol pathogen doses to receptors downwind of land application sites. Finally, the survey results support previous laboratory inactivation studies and environmental observations that promote male-specific coliphages as a more accurate and stringent indicator of pathogen inactivation in class A and class B biosolids.
Acknowledgments
This study was supported by a National Science Foundation career award, BES#0650379.
We extend our gratitude to the 29 treatment facilities involved in this study.
Footnotes
Published ahead of print on 7 November 2008.
REFERENCES
- 1.Aitken, M. D., M. D. Sobsey, M. Shehee, K. E. Blauth, V. R. Hill, J. Farrell, S. P. Nappier, G. W. Walters, P. L. Crunk, and N. Van Abel. 2005. Laboratory evaluation of thermophilic-anaerobic digestion to produce class A biosolids. 2. Inactivation of pathogens and indicator organisms in a continuous flow reactor followed by batch treatment. Water Environ. Res. 77:3028-3036. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon, B., B. Vicedo, and R. Aznar. 2006. PCR-based procedures for detection and quantification of Staphylococcus aureus and their application in food. J. Appl. Microbiol. 100:352-364. [DOI] [PubMed] [Google Scholar]
- 3.Allard, A., B. Albinsson, and G. Wadell. 2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39:498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthurson, V. 2008. Proper sanitization of sewage sludge: a critical issue for a sustainable society. Appl. Environ. Microbiol. 74:5267-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baertsch, C., T. Paez-Rubio, E. Viau, and J. Peccia. 2007. Aerosol source tracking of anaerobically digested sewage sludge released during land application. Appl. Environ. Microbiol. 73:4522-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastian, R. 1997. The biosolids (sludge) treatment, beneficial use, and disposal situation in the USA. Eur. Water Pollut. Control J. 7:62-72. [Google Scholar]
- 7.Bofill-Mas, S., N. Albinana-Gimenez, P. Clemente-Casares, A. Hundesa, J. Rodriguez-Manzano, A. Allard, M. Calvo, and R. Girones. 2006. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 72:7894-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks, J. P., B. D. Tanner, C. P. Gerba, C. N. Haas, and I. L. Pepper. 2005. Estimation of bioaerosol risk of infection to residents adjacent to a land applied biosolids site using an empirically derived transport model. J. Appl. Microbiol. 98:397-405. [DOI] [PubMed] [Google Scholar]
- 9.Burtscher, C., and S. Wuertz. 2003. Evaluation of the use of PCR and reverse transcriptase PCR for detection of pathogenic bacteria in biosolids from anaerobic digestors and aerobic composters. Appl. Environ. Microbiol. 69:4618-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catalan, V., C. Moreno, M. A. Dasi, C. Munoz, and D. Apraiz. 1994. Nested polymerase chain reaction for detection of Legionella pneumophila in water. Res. Microbiol. 145:603-610. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2000. Legionnaires' disease associated with potting soil—California, Oregon, and Washington, p. 777-778. MMWR Morb. Mortal. Wkly. Rep. 49:777-778. [PubMed] [Google Scholar]
- 12.Chandler, D. 1998. Redefining relativity: quantitative PCR at low template concentrations for industrial and environmental microbiology. J. Ind. Microbiol. Biotechnol. 21:128-140. [Google Scholar]
- 13.Clements, M. O., and S. J. Foster. 1999. Stress resistance in Staphylococcus aureus. Trends Microbiol. 7:458-462. [DOI] [PubMed] [Google Scholar]
- 14.Dowd, S., K. Widmer, and S. D. Pillai. 1997. Thermotolerant Clostridia as an airborne pathogen indicator during land application of biosolids. J. Environ. Qual. 26:194-199. [Google Scholar]
- 15.Eaton, A. E., L. S. Clesceri, E. W. Rice, and A. E. Greenberg (ed.). 2005. Standard methods for the examination of water and wastewater, 21st ed. American Public Health Association, Washington, DC.
- 16.Eisenberg, J. N. S., J. A. Soller, J. Scott, D. M. Eisenberg, and J. M. Colford, Jr. 2004. A dynamic model to assess microbial health risks associated with beneficial uses of biosolids. Risk Anal. 24:221-235. [DOI] [PubMed] [Google Scholar]
- 17.Fiume, L., M. A. Bucca Sabattini, and G. Poda. 2005. Detection of Legionella pneumophila in water samples by species-specific real-time and nested PCR assays. Lett. Appl. Microbiol. 41:470-475. [DOI] [PubMed] [Google Scholar]
- 18.Gantzer, C., P. Gaspard, L. Galvez, A. Huyard, N. Dumouthier, and J. Schwartzbrod. 2001. Monitoring of bacterial and parasitological contamination during various treatment of sludge. Water Res. 35:3763-3770. [DOI] [PubMed] [Google Scholar]
- 19.Gerba, C. P., and J. E. Smith, Jr. 2005. Sources of pathogenic microorganisms and their fate during land application of wastes. J. Environ. Qual. 34:42-48. [PubMed] [Google Scholar]
- 20.Godfree, A., and J. Farrell. 2005. Processes for managing pathogens. J. Environ. Qual. 34:105-113. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein, N., and E. Striano. 2003. Summit identifies biosolids research needs. BioCycle 44:67-74. [Google Scholar]
- 22.Guzman, C., J. Jofre, M. Montemayor, and F. Lucena. 2007. Occurrence and levels of indicators and selected pathogens in different sludges and biosolids. J. Appl. Microbiol. 103:2420-2429. [DOI] [PubMed] [Google Scholar]
- 23.Harrison, E., and S. R. Oakes. 2002. Investigation of alleged health incidents associated with land application of sewage sludges. New Solut. 12:387-408. [DOI] [PubMed] [Google Scholar]
- 24.Haugland, R. A., S. C. Siefring, L. J. Wymer, K. P. Brenner, and A. P. Dufour. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559-568. [DOI] [PubMed] [Google Scholar]
- 25.He, J.-W., and S. Jiang. 2005. Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71:2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins, M. J., C. Yen-Chih, S. N. Murthy, D. Hendrickson, J. Farrel, and P. L. Schafer. 2007. Reactivation and growth of non-culturable indicator bacteria in anaerobically digested biosolids after centrifuge dewatering. Water Res. 41:665-673. [DOI] [PubMed] [Google Scholar]
- 27.Jothikumar, N., T. L. Cromeans, V. R. Hill, X. Lu, M. D. Sobsey, and D. D. Erdman. 2005. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 71:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Cann, P., S. Ranarijaona, S. Monpoeho, F. Le Guyader, and V. Ferre. 2004. Quantification of human astroviruses in sewage using real-time RT-PCR. Res. Microbiol. 155:11-15. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S.-A., A. Adhikari, S. A. Grinshpun, R. McKay, R. Shukla, and T. Reponen. 2006. Personal exposure to airborne dust and microorganisms in agricultural environments. J. Occup. Environ. Hyg. 3:118-130. [DOI] [PubMed] [Google Scholar]
- 30.Lewis, D. L., D. K. Gattie, M. E. Novak, S. Sanchez, and C. Pumphrey. 2002. Interactions of pathogens and irritant chemicals in land-applied sewage sludges (biosolids). BMC Public Health 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Low, S. Y., T. Paez-Rubio, C. Baertsch, M. Kucharski, and J. Peccia. 2007. Off-site exposure to respirable aerosols produced during the disk incorporation of class B biosolids. J. Environ. Eng. 133:987-994. [Google Scholar]
- 32.Mandilara, G. D., E. M. Smeti, A. T. Mavridou, M. P. Lambiri, A. Vatopoulos, and F. P. Riga. 2006. Correlation between bacterial indicators and bacteriophages in sewage and sludge. FEMS Microbiol. Lett. 263:119-126. [DOI] [PubMed] [Google Scholar]
- 33.Moce-Llivina, L., M. Muniesa, H. Pimenta-Vale, F. Lucena, and J. Jofre. 2003. Survival of bacterial indicator species and bacteriophages after thermal treatment of sludge and sewage. Appl. Environ. Microbiol. 69:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monpoeho, S., A. Maul, C. Bonnin, L. Patria, S. Ranarijaona, S. Billaudel, and V. Ferre. 2004. Clearance of human-pathogenic viruses from sludge: study of four stabilization processes by real-time reverse transcription-PCR and cell culture. Appl. Environ. Microbiol. 70:5434-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monpoeho, S., A. Maul, B. Mignotte-Cadiergues, L. Schwartzbrod, S. Billaudel, and V. Ferre. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Research Council. 2002. Biosolids applied to land: advancing standards and practices. National Academy Press, Washington, DC.
- 37.Noble, R. T., J. F. Griffith, A. D. Blackwood, J. A. Fuhrman, J. B. Gregory, X. Hernandez, X. Liang, A. A. Bera, and K. Schiff. 2006. Multitiered approach using quantitative PCR to track sources of fecal pollution affecting Santa Monica Bay, California. Appl. Environ. Microbiol. 72:1604-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novak, J. T. 2007. The future of sludge and biosolids research. Water Environ. Res. 79:459-460. [PubMed] [Google Scholar]
- 39.Oliver, J. D. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93-100. [PubMed] [Google Scholar]
- 40.Pourcher, A. M., P. Morand, F. Picard-Bonnaud, S. Billaudel, S. Monpoeho, M. Federighi, V. Ferre, and G. Moguedet. 2005. Decrease of enteric micro-organisms from rural sewage sludge during their composting in straw mixture. J. Appl. Microbiol. 99:528-539. [DOI] [PubMed] [Google Scholar]
- 41.Rusin, P., S. Maxwell, J. P. Brooks, C. P. Gerba, and I. L. Pepper. 2003. Evidence for the absence of Staphylococcus aureus in land applied biosolids. Environ. Sci. Technol. 18:4027-4030. [DOI] [PubMed] [Google Scholar]
- 42.Sartory, D. P., A. M. Pritchard, and P. Holmes. 1993. Enumeration of sulphite-reducing Clostridia from potable water supplies. Water Sci. Technol. 27:279-282. [Google Scholar]
- 43.Sidhu, J., R. A. Gibbs, G. E. Ho, and I. Unkovich. 2001. The role of indigenous microorganisms in suppression of Salmonella regrowth in composted biosolids. Water Res. 35:913-920. [DOI] [PubMed] [Google Scholar]
- 44.Soares, H. M., and B. Cardenas. 1995. Evaluating pathogen regrowth in biosolids compost. BioCycle 36:70-74. [Google Scholar]
- 45.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 46.Tasara, T., and R. Stephan. 2005. Development of an F57 sequence-based real-time PCR assay for detection of Mycobacterium avium subsp. paratuberculosis in milk. Appl. Environ. Microbiol. 71:5957-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Temmerman, R., H. Vervaeren, B. Noseda, N. Boon, and W. Verstraete. 2006. Necrotrophic growth of Legionella pneumophila. Appl. Environ. Microbiol. 72:4323-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.U.S. Environmental Protection Agency. 1999. Environmental regulations and technology: control of pathogens and vector attraction in sewage sludge. EPA/625/R-92-013. Office of Water, U.S. Environmental Protection Agency, Washington, DC.
- 49.U.S. Environmental Protection Agency. 2001. Method 1601: male-specific (F+) and somatic coliphage in water by two-step enrichment procedure. EPA-821-R-01-030. Office of Water, U.S. Environmental Protection Agency, Washington, DC.
- 50.U.S. Environmental Protection Agency. 2006. Method 1680: fecal coliforms in sewage sludge (biosolids) by multiple-tube fermentation using lauryl tryptose broth (LTB) and EC medium. EPA-821-R-06-012. Office of Water, U.S. Environmental Protection Agency, Washington, DC.
- 51.U.S. Environmental Protection Agency. 1989. USEPA manual of methods for virology. EPA/600/4-84/013 (R-7). U.S. Environmental Protection Agency, Washington, DC.
- 52.van den Berg, R. J., N. Vaessen, H. P. Endtz, T. Schulin, E. R. van der Vorm, and E. J. Kuijper. 2007. Evaluation of real-time PCR and conventional diagnostic methods for the detection of Clostridium difficile-associated diarrhoea in a prospective multicentre study. J. Med. Microbiol. 56:36-42. [DOI] [PubMed] [Google Scholar]
- 53.Wery, N., A.-M. Pourcher, V. Stan, J.-P. Delgenes, F. Picard-Bonnaud, and J.-J. Godon. 2006. Survival of Listeria monocytogenes and Enterococcus faecium in sludge evaluated by real-time PCR and culture methods. Lett. Appl. Microbiol. 43:131-136. [DOI] [PubMed] [Google Scholar]
- 54.Westrell, T., C. Schonning, T. A. Stenstrom, and N. J. Ashbolt. 2004. QMRA (quantitative microbial risk assessment) and HACCP (hazard analysis and critical control points) for management of pathogens in wastewater and sewage sludge treatment and reuse. Water Sci. Technol. 50:23-30. [PubMed] [Google Scholar]
- 55.Yaradou, D. F., S. Hallier-Soulier, S. Moreau, F. Poty, Y. Hillion, M. Reyrolle, J. Andre, G. Festoc, K. Delabre, F. Vandenesch, J. Etienne, and S. Jarraud. 2007. Integrated real-time PCR for detection and monitoring of Legionella pneumophila in water systems. Appl. Environ. Microbiol. 73:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]