Abstract
Plant aerial surfaces comprise a complex habitat for microorganisms, and many plant-associated bacteria, such as the pathogen Pseudomonas syringae, exhibit density-dependent survival on leaves by utilizing quorum sensing (QS). QS is often mediated by diffusible signals called N-acyl-homoserine lactones (AHLs), and P. syringae utilizes N-3-oxo-hexanoyl-dl-homoserine lactone (3OC6HSL) to control traits influencing epiphytic fitness and virulence. The P. syringae pathovar syringae B728a genome sequence revealed two putative AHL acylases, termed HacA (Psyr_1971) and HacB (Psyr_4858), which are N-terminal nucleophile hydrolases that inactivate AHLs by cleaving their amide bonds. HacA is a secreted AHL acylase that degrades only long-chain (C ≥ 8) AHLs, while HacB is not secreted and degrades all tested AHLs. Targeted disruptions of hacA, hacB, and hacA and hacB together do not alter endogenous 3OC6HSL levels under the tested conditions. Surprisingly, targeted disruptions of hacA alone and hacA and hacB together confer complementable phenotypes that are very similar to autoaggregative phenotypes seen in other species. While AHL acylases might enable P. syringae B728a to degrade signals of competing species and block expression of their QS-dependent traits, these enzymes also play fundamental roles in biofilm formation.
Quorum sensing (QS) allows bacterial populations to coordinate gene expression in a cell-density-dependent manner (3, 22). N-Acyl-homoserine lactones (AHLs) are diffusible signal molecules that mediate QS in many species of gram-negative bacteria residing on leaves (10, 18, 19, 50, 68) and are comprised of homoserine lactone rings linked with amide bonds to either long (C ≥ 8) or short (C < 8) acyl chains. Pseudomonas syringae pathovar syringae strain B728a is a gram-negative leaf surface epiphyte and pathogen capable of causing brown spot disease in bean plants (Phaseolus vulgaris) (25). QS in strain B728a is mediated by a single AHL, N-3-oxohexanoyl-dl-homoserine lactone (3OC6HSL) (10, 52) and serves as a model for studying the influence of QS on motility, epiphytic fitness, and virulence to plants (51).
The fate of AHLs once they are released into extracellular environments is relatively unknown, although AHLs have been found to be degraded quickly in soil (69). AHLs can most likely accumulate to physiologically relevant levels in relatively small cell populations (18); on dry leaves, where AHL diffusion is thought to be especially limited, only a few P. syringae pv. syringae strain B728a cells in close proximity can produce enough signals to influence gene expression and subsequent behavior (17). For QS to function, mechanisms of inactivation must exist to modify AHLs so they can no longer bind to their cognate transcription factors. While AHLs can be inactivated both chemically (7, 74) and by eukaryotic hosts (1, 14, 28, 53), bacteria also have the capacity to inactivate AHLs via the production of AHL-degrading enzymes. AHL lactonases (8, 9, 15, 36, 47, 49, 76) have broad AHL substrate specificities and catalyze the opening of the homoserine lactone ring via hydrolysis of the ester bond to produce an inactive open-chain acyl-homoserine. AHL acylases (26, 27, 35, 37, 48, 56, 62, 66, 67) are N-terminal nucleophile (Ntn) hydrolases (6) that inactivate signals by cleaving the acyl chain from the homoserine lactone via nucleophilic attack on the carboxy carbon in the amide linkage. AHL acylases display high substrate specificities based on the lengths of the AHL acyl chains, and like other Ntn hydrolases (6), they are transcribed as large propolypeptides that undergo autoproteolytic cleavage to reach maturation.
While AHL-degrading enzymes are widespread throughout bacteria, there is uncertainty about their endogenous physiological functions (55). AHL-degrading enzymes might modulate QS by recycling AHLs once QS is achieved (76), or they might enable cells to degrade AHLs produced by competing species (48) and even utilize AHL breakdown products as carbon sources (21, 26, 27, 35). The presence of multiple AHL-degrading enzymes in a single strain adds complexity to their potential roles. The gram-negative plant pathogen Agrobacterium tumefaciens employs N-3-oxo-octanoyl-dl-homoserine lactone (3OC8HSL) as a QS signal and harbors two AHL lactonases (8, 9, 11, 76). Pseudomonas aeruginosa PAO1 utilizes N-3-butyryl homoserine lactone (C4HSL) and N-3-oxo-dodecanoyl homoserine lactone (3OC12HSL) for QS, and contains in its genome four Ntn hydrolases, of which two, PvdQ and QuiP, have been experimentally determined to be AHL acylases. PvdQ was originally found to influence accumulation of the siderophore pyoverdine (33, 46). PvdQ expression in E. coli conferred on cells the ability to degrade long-chain (C ≥ 8) AHLs, but PvdQ was not necessary for the ability of P. aeruginosa PAO1 to utilize AHLs for nutrition (26). QuiP, a second AHL acylase, also degrades only long-chain AHLs and enables P. aeruginosa to catabolize AHLs (27).
We searched the strain B728a genome (20) for AHL-degrading enzymes so that we might investigate their influence on QS in a model plant pathogen. No AHL lactonase homologues were present in the strain B728a genome, but we did identify three uncharacterized open reading frames (ORFs; Psyr_1971, Psyr_4858, and Psyr_3871) homologous to characterized AHL acylases. We report here that two of these strain B728a enzymes, termed HacA and HacB for AHL acylase, can indeed inactivate AHLs with differing substrate specificities and that these AHL acylases play potentially important roles in biofilm formation.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
AHLs used in this study were 3OC6HSL (Sigma), 3OC8HSL (Sigma), N-3-hexanoyl-dl-homoserine lactone (C6HSL; Fluka), N-3-octanoyl-dl-homoserine lactone (C8HSL; Fluka), N-3-decanoyl-dl-homoserine lactone (C10HSL; Fluka), and N-3-dodecanoyl-dl-homoserine lactone (C12HSL; Fluka). AHL stock solutions were prepared in ethyl acetate containing 0.01% (vol/vol) glacial acetic acid and stored at −20°C. The bacterial strains and plasmids used in this study are described in Table 1, and the source and characteristics of strain B728a have been previously described (39). Strain B728a was grown in King's B medium (KB) (29) at 28°C, which was buffered at pH 6.5 with 10 mM potassium phosphate when indicated. For the AHL growth assay, strain B728a was incubated for 7 days in M9 minimal medium (58) containing 1 mM C10HSL as a carbon source and buffered at pH 6.5 with 10 mM potassium phosphate. Escherichia coli was grown in Luria-Bertani (LB) medium (58) at 37°C, unless otherwise indicated. The following concentrations of antibiotics were used: ampicillin, 100 μg/ml; gentamicin, 15 μg/ml; kanamycin, 50 μg/ml; rifampin, 100 μg/ml; spectinomycin, 20 μg/ml; and tetracycline, 15 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen, Carlsbad, CA |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen, Madison, WI |
| MT102 | araD139 Δ(ara-leu)7697 Δlac thi hsdR, MC1000 derivative | 73 |
| P. syringae strains | ||
| B728a | Rfr, wild type, bean pathogen | 39 |
| BHSL | Rfr Kmr, ΔahlI::MudI1734, strain B728a derivative | 30 |
| HacA− | Rfr Tetr, ΔhacA::Tet, strain B728a derivative | This study |
| HacB− | Rfr Tetr, ΔhacB::Tet, strain B728a derivative | This study |
| HacA−B− | Rfr Tetr Gmr, ΔhacA::Gm, ΔhacB::Tet, strain B728a derivative | This study |
| Plasmids | ||
| pCR2.1-TOPO | Apr Kmr, cloning vector | Invitrogen |
| pENTR/D-TOPO | Kmr, entry vector for Gateway cloning system | Invitrogen |
| pENTR/D-MCS-Kan | Kmr Gmr, entry vector for Gateway cloning system | T. K. Powell, G. F. J. Dulla |
| pET30 | Kmr, inducible expression vector | Novagen |
| pET30-HacA | Kmr, pET30 containing hacA ORF with C-terminal six-His tag | This study |
| pET30-HacB | Kmr, pET30 containing hacB ORF with C-terminal six-His tag | This study |
| pET30-Psyr_3871 | Kmr, pET30 containing Psyr_3871 ORF with C-terminal six-His tag | This study |
| p519nGFP | Kmr, broad-host-range expression vector | 42 |
| p519-HacA | Kmr, p519 containing hacA ORF with C-terminal six-His tag | This study |
| p519-HacB | Kmr, p519 containing hacB ORF with C-terminal six-His tag | This study |
| P519-Psyr_3871 | Kmr, p519 containing Psyr_3871 ORF with C-terminal six-His tag | This study |
| pLVC/D | Apr Tetr, Gateway destination vector strain B728a suicide plasmid | 40 |
| pBQ9 | Spcr, pPROBE-OT derivative harboring P. syringae ahlI promoter upstream of GFP | 52 |
| JB524 | Tetr, pME6031 derivative harboring V. fischeri luxR,PluxI-gfpmut3* (stable GFP) | 73 |
Ap, ampicillin; Gm, gentamicin; Km, kanamycin; Rf, rifampin; Spc, spectinomycin; Tet, tetracycline.
Nucleic acid and protein manipulations.
All DNA and protein manipulations were performed using standard or manufacturer-recommended protocols (58), and general cloning steps were performed in E. coli Top10 cells (Invitrogen). Primers were synthesized by Integrated DNA Technologies (Coralville, IA). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (32) was performed using a Mini-Protean II electrophoresis system (Bio-Rad, Hercules, CA), and proteins were either visualized by Coomassie blue staining or transferred to Immobilon-PSQ polyvinylidene difluoride membranes (Millipore, Bedford, MA) using a Mini-Protean II Electroblot apparatus (Bio-Rad). Western blot analyses were performed with 1:10,000 dilutions of mouse-derived anti-His-tagged monoclonal antibody (Cell Signaling Technology, Danvers, MA) and 1:10,000 dilutions of alkaline phosphatase-coupled anti-mouse secondary antibody (Sigma-Aldrich, St. Louis, MO). Protein concentrations were estimated using a Bio-Rad protein assay kit.
AHL quantification with biosensors.
E. coli MT102-JB524 (73) was grown in LB broth (buffered with 100 mM potassium phosphate, pH 7.0) supplemented with tetracycline. Strain B728a BHSL-pBQ9 was grown in KB broth (buffered with 100 mM potassium phosphate, pH 7.0) supplemented with corresponding antibiotics. Ethyl acetate samples to be assayed for AHLs were dried in 13- by 100-mm borosilicate glass tubes (Fisher Scientific). Biosensor cultures were incubated until early log phase, and 1-ml aliquots were then added to sample tubes and incubated overnight at 28°C with shaking. Five milliliters of sterile water was added to sample tubes, and green fluorescent protein (GFP) was measured using a fluorimeter (Model TD-700; Turner Designs, Sunnyvale, CA) with an excitation wavelength of 390 nm and emission wavelengths of 510 to 700 nm. Raw fluorescence units were normalized for the optical density at 600 nm (OD600) values of the biosensor cultures. AHL concentrations were quantified by comparing normalized fluorescence values with those obtained from standard curves derived from biosensor cells incubated with known concentrations of AHL standards.
Cloning of hacA, hacB, and Psyr_3871 and expression in E. coli.
Genomic DNA was isolated from strain B728a using a DNeasy tissue kit (Qiagen, Valencia, CA). The coding region for hacA, minus the stop codon, was amplified from genomic DNA using primers 5′-AGCTCATATGATTATTTCCAGACCCCTGTG-3′ (NdeI site underlined) and 5′-AGCTCTCGAGGCGATCAGGCTCCT-3′ (XhoI site underlined). The coding region for hacB, minus the stop codon, was amplified from genomic DNA using primers 5′-AGCTCATATGAAACGCAGCCTCTTCA-3′ (NdeI site underlined) and 5′-AGCTGCGGCCGCCGCCCCCGGCAC-3′ (NotI site underlined). The coding region for Psyr_3871, minus the stop codon, was amplified from genomic DNA using primers 5′-AGCTCATATGGCCTCGCCAGCCCTTC-3′ (NdeI site underlined) and 5′-AGCTCTCCGAGCTTGCCCGGCGTCAG-3′ (XhoI site underlined). PCR products were cloned into pCR2.1-TOPO, and inserts were transferred into the appropriately digested expression vector pET30 (Novagen, Madison, WI) to create translational fusions with C-terminal six-His tags. The resulting plasmids pET30-HacACH, pET30-HacBCH, and pET30-Psyr_3871 were transformed into E. coli BL21(DE3) cells (Novagen). Protein expression was induced in liquid cultures by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside at an OD600 of 0.4, followed by overnight incubation at 28°C. Cultures were centrifuged at 7,000 × g, and culture supernatants were passed through 0.45-μm syringe filters. Proteins in culture supernatants were precipitated on ice by the addition of 60% ammonium sulfate or trichloroacetic acid and collected by centrifugation at 10,000 × g. Soluble and insoluble proteins were collected from cell pellets with an EasyLyse kit (Epicentre Biotechnologies, Madison, WI).
AHL degradation assays with intact E. coli cells expressing HacA, HacB, and Psyr_3871.
Overnight, isopropyl-β-d-thiogalactopyranoside-induced liquid cultures of E. coli BL21(DE3) cells harboring plasmids pET30, pET30-HacACH, pET30-HacBCH, and pET30-Psyr_3871 were centrifuged at 7,000 × g. Cells were washed twice with 10 mM potassium phosphate buffer (pH 7.0) and suspended to an OD600 of 1.0 in sterile 13- by 100-mm borosilicate glass tubes containing buffer supplemented with 1 μM AHL. The tubes were incubated at 28°C, and 300-μl samples were collected at various times and centrifuged to pellet the cells. Supernatants (250 μl) were transferred to 1.5-ml Eppendorf tubes and extracted with equal volumes of ethyl acetate. Portions (200 μl) of the ethyl acetate extracts were transferred to fresh 1.5-ml tubes and dried, and each was resuspended in 25 μl ethyl acetate. AHLs were quantified with the E. coli JB524 biosensor.
Constitutive expression of HacA, HacB, and Psyr_3871 in strain B728a.
ORFs encoding HacA, HacB, and Psyr_3871 with C-terminal histidine tags were amplified from the pET30-based expression constructs generated above with gene-specific sense primers containing NdeI sites and the antisense primer 5′-GAATTCTCAGTGGTGGTGGTGGTGGTG-3′ (EcoRI site underlined) and cloned into the appropriately digested vector p519GFP (42). The resulting constructs, p519-HacACH, p519-HacBCH, and p519-Psyr_3871, were transformed into strain B728a by electroporation, and proteins from the resulting strains were analyzed as described above.
Targeted disruption of hacA and hacB in strain B728a.
The hacA and hacB genes were each disrupted through targeted insertional mutagenesis using the pLVC/D vector system (40). A 796-bp hacA fragment was amplified from strain B728a genomic DNA with the primers 5′-CACCGAATTCAGTTTTGTGTTTGCAGGTCT-3′ and 5′-CTCATCTAGAGTCGAGTTTGCCGGGAATG-3′. A 942-bp hacB fragment was amplified with the primers 5′-CACCGAATTCTCATTCTAGCCATCAT-3′ and 5′-GTCATCTAGAGCTCCAGCCGAAGTCCAT-3′. PCR products were cloned into pENTR/D (Invitrogen) and then transferred using LR Clonase to pLVC/D. The resulting pLVC/D plasmids were introduced into strain B728a by triparental mating, so that gene disruptions occurred via single-crossover events. The resulting strains, HacA− and HacB−, were verified with PCR using primers specific to the pLVC/D vector and to genomic DNA sequences upstream and downstream of the targeted genes.
A hacA hacB double knockout was generated by the insertion of a gentamicin resistance cassette into hacA, followed by the insertion of tetracycline resistance cassette into hacB. Two fragments of hacA were amplified from strain B728a genomic DNA (fragment 1, 5′-AAGCTTAGTTTTGTGTTTGCAGGTCTTTCATTC-3′, HindIII site underlined; 5′-GGTACCGTCGAGTTTGCCGGGAATGGT-3′, KpnI site underlined; fragment 2, 5′-CTCGAGGACCGATTTCGTGCAGCATTCC-3′, XhoI site underlined; 5′-TCTAGACCTCGCTTATCACCTGCACCTCAT-3′, XbaI site underlined) and cloned into the appropriately digested vector pENTR/D-MCS-Gent. The resulting construct, in which the gentamicin resistance cassette was flanked by hacA gene fragments, was transferred into pLVC/D using LR Clonase, and the resulting plasmid was introduced into strain B728a by triparental mating. Resulting colonies were screened for gentamicin resistance and tetracycline susceptibility, indicating that gene disruption had occurred via a double-crossover event. The gene hacB was then disrupted via a single-crossover event, as described above, and colonies were selected with gentamicin and tetracycline. The resulting strain, HacA−B−, was verified using PCR.
3OC6HSL quantification in P. syringae pv. syringae B728a strains.
The strains were grown at 28°C in liquid KB supplemented with appropriate antibiotics. At various time points, culture aliquots (4 ml) were extracted with equal volumes of ethyl acetate, and 3.6 ml of the ethyl acetate extract was dried and resuspended in 300 μl ethyl acetate. Concentrations of 3OC6HSL in the extracts were quantified with the strain B728a BHSL-pBQ9 biosensor.
Purification of HacA and HacB from strain B728a.
Strain B728a cells harboring plasmids p519-HacACH and p519-HacBCH were grown at 28°C in liquid KB with appropriate antibiotics until stationary phase. Cultures were centrifuged at 7,000 × g, and cell pellets were washed twice with 10 mM potassium phosphate buffer before being treated with an EasyLyse kit. Histidine-tagged proteins in soluble fractions were purified with HisTrap-FF Ni-nitrilotriacetic acid (Ni-NTA) affinity columns (Amersham Biosciences, Uppsala, Sweden) according to the manufacturer's instructions, with the exception that imidazole was not included in the wash buffer, and used immediately in AHL degradation assays or analyzed by SDS-PAGE and Western blot analyses. For N-terminal amino acid sequencing, proteins were transferred to polyvinylidene difluoride membranes and Coomassie blue-stained bands were sequenced by Edman degradation at the University of California—Davis Macromolecular Structure Facility.
AHL degradation assays of HacA and HacB with OPA.
The release of homoserine lactone, a product of the acylase reaction, was detected with o-phthaldialdehyde (OPA; Sigma) as described previously (48). Purified HacA or HacB (20 μg), along with boiled negative controls, was mixed with 0.5 mM AHLs or penicillin G in 200 μl 10 mM potassium phosphate (pH 7.0) and incubated at 28°C for 1 h (HacB) or 3 h (HacA). OPA stock solution was added, and the A340 was measured. Pure homoserine lactone (Sigma) was used to generate standard curves in each replicate of the assay.
RESULTS
Strain B728a genome contains three putative AHL acylases.
We initially observed that C10HSL was apparently degraded when added to either wild-type strain B728a cultures in buffered KB media or cell-free culture supernatants (data not shown). BLASTp analyses (2) of the strain B728a genome (20) using amino acid sequences of AHL-degrading enzymes as queries revealed the presence of three putative ORFs with significant similarity to AHL acylases: Psyr_1971 (termed hacA, for AHL acylase), Psyr_4858 (termed hacB), and Psyr_3871. All three ORFs were predicted to contain secretory signal peptides (www.cbs.dtu.dk/services/SignalP/) (5). HacA is predicted to be a 779-amino acid (aa) polypeptide with a molecular mass of 85 kDa and shares 55% aa identity with PA2385 (PvdQ) (26, 33) of P. aeruginosa. HacB is predicted to be a 795-aa polypeptide with a molecular mass of 88 kDa and shares 68% aa identity with PA0305, an uncharacterized protein from P. aeruginosa. The Psyr_3871 ORF is predicted to encode an 824-aa polypeptide with a molecular mass of 90 kDa and shares 63% aa identity with PA1032 (QuiP) (27) of P. aeruginosa. Amino acid alignments of these ORFs with sequences of characterized AHL acylases indicated that HacA and HacB, but not Psyr_3871, contained conserved Gly-Ser-Asn residues reported to be necessary for posttranslational processing in Ntn hydrolases (6, 37).
E. coli cells expressing HacA and HacB, but not Psyr_3871, can inactivate AHLs.
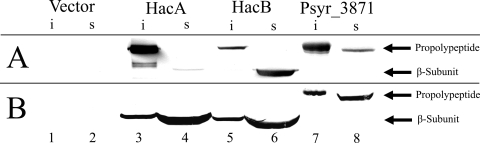
C-terminal histidine tags were added to the putative AHL acylases of strain B728a, as described in other studies of Ntn hydrolases (48, 71), to identify the propolypeptide and the mature β subunit. The insoluble fraction collected from the pET30-HacA-harboring strain contained a prominent reactive band with a molecular mass of ∼85 kDa (Fig. 1A, lane 3), while the soluble fraction contained a reactive band with a molecular mass of ∼60 kDa (Fig. 1A, lane 4). Similar band patterns were detected in insoluble and soluble fractions collected from the pET30-HacB-harboring strain (Fig. 1A, lanes 5 and 6). The molecular masses of the ∼85-kDa and ∼60-kDa bands correspond to predicted molecular masses of the propolypeptides and mature, processed β subunits, respectively, of HacA and HacB. Thus, E. coli-expressed HacA and HacB apparently undergo the posttranslational processing that is characteristic of Ntn hydrolases, but their processing is incomplete. Similar results were reported from attempts to express P. aeruginosa AHL acylases in E. coli (26, 27). The β subunit of HacA was also present in the culture medium (data not shown), indicating that HacA is a secreted enzyme. Psyr_3871 was detected only as an ∼90-kDa propolypeptide in both insoluble and soluble fractions of the pET30-Psyr_3871-harboring strain (Fig. 1A, lanes 7 and 8), indicating that it is not an Ntn hydrolase, perhaps because it does not have the conserved amino acid residues necessary for posttranslational processing (37).
FIG. 1.
Western blot analyses of putative strain B728a AHL acylases expressed in E. coli and strain B728a. (A) Insoluble (i) and soluble (s) fractions collected from induced E. coli harboring plasmids pET30 (lanes 1 and 2), pET30-HacA (lanes 3 and 4), pET30-HacB (lanes 5 and 6), and pET30-Psyr_3871 (lanes 7 and 8). (B) Insoluble (i) and soluble (s) fractions collected from strain B728a harboring plasmids p519nGFP (lanes 1 and 2), p519-HacA (lanes 3 and 4), p519-HacB (lanes 5 and 6), and p519-Psyr_3871 (lanes 7 and 8).
Two whole-cell biosensors were used to quantify the degradation of a variety of AHLs by the putative P. syringae pv. syringae strain B728a AHL acylases. E. coli MT102-JB524 detects all AHLs (73), while strain B728a BHSL-pBQ9 is highly selective for only 3OC6HSL and 3OC8HSL (52). To ensure that biosensors could be used to measure AHL inactivation via amide bond cleavage, we confirmed that neither responded to the degradation product homoserine lactone (data not shown). Inactivation assays were then performed against a variety of AHLs (Table 2). E. coli cells harboring pET30-HacA inactivated only AHLs with acyl chain lengths of greater than eight carbons, and cells harboring pET30-HacB inactivated all AHLs tested. Cells harboring pET30-Psyr_3871 and pET30 showed no inactivation activities toward any AHL. We note that the specificities of HacA toward long-chain AHLs are reflected in the reported activity of PvdQ, its homologue in P. aeruginosa, while HacB degraded a much-wider range of AHLs than most other characterized AHL acylases.
TABLE 2.
Substrate specificities of putative and characterized AHL acylases from P. syringae pv. syringae strain B728a and other microorganismsa
| Microorganism | Enzyme | 3OC6HSL | C6HSL | C8HSL | C10HSL | 3OC12HSL | C12HSL | Source or reference(s) |
|---|---|---|---|---|---|---|---|---|
| P. syringae B728a | HacA | − | − | + | + | NT | + | This study |
| HacB | + | + | + | + | NT | + | This study | |
| Psyr_3871 | − | − | − | − | NT | − | This study | |
| P. aeruginosa PAO1 | PA2385 (PvdQ) | − | − | + | + | + | + | 26, 62 |
| PA1032 (QuiP) | − | − | + | + | + | + | 27 | |
| Ralstonia sp. strain XJ12B | AiiD | + | NT | NT | NT | + | NT | 37 |
| Streptomyces sp. strain M664 | AhlM | − | − | + | + | + | NT | 48 |
+, degradation ability; −, no degradation ability; NT, not tested.
HacA and HacB were expressed in strain B728a and purified to determine substrate specificities.
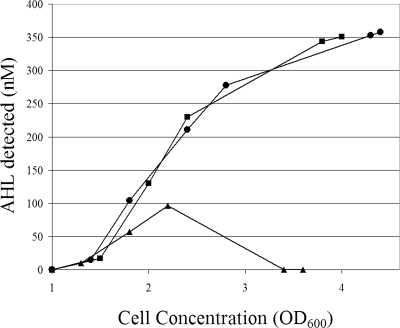
The putative AHL acylases with C-terminal histidine tags were expressed constitutively in strain B728a from plasmids p519-HacA, p519-HacB, and p519-Psyr_3871. Both HacA and HacB underwent complete posttranslational processing and were visible only as ∼60-kDa processed β subunits. The β subunit of HacA was present in the soluble and insoluble cellular fractions (Fig. 1A, lanes 3 and 4), as was the β subunit of HacB (Fig. 1A, lanes 5 and 6). Also, the β subunit of HacA was present in the culture medium (data not shown), again indicating that HacA is a secreted Ntn hydrolase while HacB is not secreted. Psyr_3871 was visible only as an 85-kDa propolypeptide form in both the soluble and insoluble cellular fractions (Fig. 1A, lanes 7 and 8), and we thus conclude that Psyr_3871 is not an Ntn hydrolase because it does not undergo the posttranslational processing necessary for enzyme maturation. Endogenous 3OC6HSL levels in planktonic cultures of strain B728a harboring p519-HacA were similar to levels of the wild-type strain, while strain B728a harboring p519-HacB had decreased 3OC6HSL levels (Fig. 2). These data provide further evidence that HacB can inactivate 3OC6HSL and can function in strain B728a under normal physiological conditions. We note that constitutive expression of HacA or HacB did not confer on strain B728a the ability to grow in media containing C10HSL as a sole carbon source in M9 minimal media (data not shown).
FIG. 2.
Endogenous AHL (3OC6HSL) concentration (nM) in various strains grown in KB medium. Wild-type strain B728a (•), strain B728a harboring p519-HacA (▪), and strain B728a harboring p519-HacB (▴).
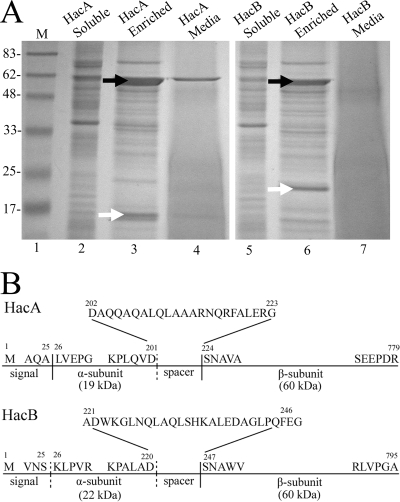
Histidine tag affinity chromatography was used to recover HacA and HacB heterodimers from soluble cellular extracts of strain B728a harboring plasmids p519-HacA and p519-HacB (Fig. 3A). The HacA α subunit has an apparent molecular mass of 19 kDa (Fig. 3A, lane 3), and the HacB α subunit has an apparent molecular mass of 22 kDa (Fig. 3A, lane 6). Filter-sterilized culture supernatants were also passed through histidine tag affinity columns, and only HacA was recovered (Fig. 3A, lane 4), again indicating that HacA is a secreted enzyme while HacB is not secreted. N-terminal amino acid sequences of subunits were also determined (HacA, α subunit LVEPG and β subunit SNAVA; HacB, β subunit SNAWVI), revealing HacA and HacB to be Ntn hydrolases because the residues serine and asparagine form the N termini of their β subunits (Fig. 3B).
FIG. 3.
The enrichment and characterization of HacA and HacB in strain B728a. (A) SDS-PAGE analysis of soluble cell extracts (HacA, lane 2; HacB, lane 5), fractions after Ni-NTA affinity purification of soluble extracts (HacA, lane 3; HacB, lane 6), and fractions from media samples after nondenaturing Ni-NTA affinity purification (HacA, lane 4; HacB, lane 7). White arrows denote α subunits. Black arrows denote β subunits. Molecular masses of standards (M) are given in kilodaltons. (B) Schematic representations of HacA and HacB. Solid vertical lines indicate sites of autoproteolytic cleavage determined experimentally by amino acid sequencing. Dashed vertical lines indicate cleavage sites inferred from amino acid alignments with other characterized AHL acylases.
An OPA in vitro assay was used to measure the production of homoserine lactone from AHL by purified HacA and HacB, similar to studies performed with purified AhlM (48). The OPA assay also verifies the degradation of AHLs via cleavage of their amide bonds and the resultant production of primary amines. The substrate specificities of enriched HacA and HacB for AHLs with various acyl chains are shown in Table 3. For both enzymes, AHL degradation activities increased as acyl chain length increased, and HacB had higher overall degradation activity for a given AHL than HacA. Neither HacA nor HacB degraded penicillin G. The specific activities of HacB against AHLs are comparable to those found with AhlM (48), but HacB displayed more activity toward short-chain AHLs than other characterized AHL acylases.
TABLE 3.
Specific and relative activities of HacA and HacB against AHLs and penicillin Ga
| Substrate | HacA specific activityb | HacA relative activity (%) | HacB specific activityb | HacB relative activity (%) |
|---|---|---|---|---|
| C4HSL | 0 | 0 | 1.9 | 4.1 |
| C6HSL | 0 | 0 | 27.6 | 59.6 |
| C8HSL | 3.2 | 84.2 | 46.3 | 100.0 |
| C10HSL | 3.8 | 100.0 | 40.6 | 87.7 |
| C12HSL | +, NT | NT | +, NT | NT |
| 3OC6HSL | 0 | 0 | 4.9 | 10.6 |
| 3OC8HSL | 1.1 | 28.9 | 40.8 | 88.1 |
| Penicillin G | 0 | 0 | 0 | 0 |
+, fluorescence was observed in the OPA assay, indicating that a primary amine was generated, but substrate precipitation interfered with quantification.
Specific activities are given in units of pmol·min−1·μg−1. Values represent the means of three independent experiments. NT, not tested.
Targeted disruptions of AHL acylases do not alter 3OC6HSL levels but do confer complementable phenotypes.
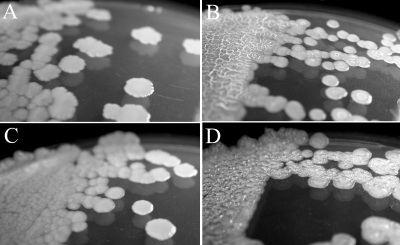
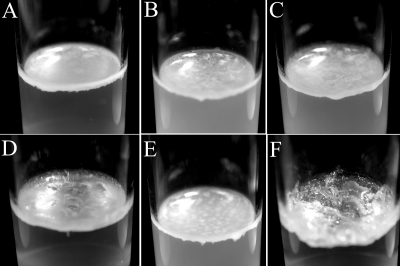
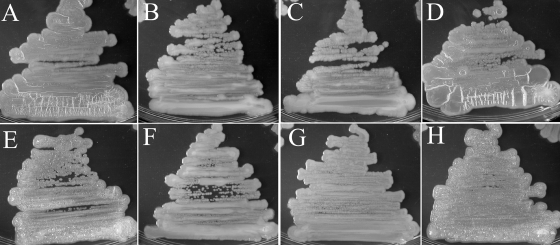
Targeted disruptions of hacA, hacB, and both hacA and hacB together were obtained by the insertion of tetracycline and/or gentamicin expression cassettes, resulting in strains HacA−, HacB−, and HacA−B−. No detectable differences in 3OC6HSL concentrations were found between broth cultures of wild-type strain B728a and strains HacA−, HacB−, and HacA−B−, and we note that both hacA and hacB are expressed in planktonic wild-type cells (data not shown). The colony morphology of strain HacB− was not different than that of the wild-type strain B728a, but strains HacA− and HacA−B exhibited dramatically different morphologies when grown on KB agar (Fig. 4). Individual colonies of strain HacA− appeared to be similar to the wild-type colonies when first visible, but soon developed a dry and wrinkled (rugose) morphology. When plated as a lawn, strain HacA− also was easily distinguishable after ∼2 days due to the formation of ∼1-mm-high, ridgelike structures that spread across the entire surface. Strain HacA−B− was deficient in the production of the fluorescent siderophore pyoverdine and was constitutively moist and bubbly as both individual colonies and lawns. Both strains HacA− and HacA−B− formed pellicles at the air-liquid interfaces when grown for 7 days in static KB cultures (Fig. 5). The phenotypes also persisted when hacA and hacA hacB disruptions were generated in strain BHSL (data not shown), a QS-deficient strain B728a mutant (30), indicating that 3OC6HSL was not necessary for phenotype development. Colony morphologies of strains HacA− and HacA−B− became indistinguishable from wild-type strain B728a by the constitutive expression of either HacA or HacB with plasmids p519-HacA or p519-HacB, but not by the control plasmid p519ngfp (Fig. 6).
FIG. 4.
Colony morphologies of B728a strains on KB agar. Shown are the wild-type B728a (A), HacA− (B), HacB− (C), and HacA−B− (D) strains.
FIG. 5.
Pellicles formed by various B728a strains in static cultures of KB medium. Shown are the wild-type B728a (A), B728a harboring p519-HacA (B), B728a harboring p519-HacB (C), HacA− (D), HacB− (E), and HacA−B− (F) strains.
FIG. 6.
Complementation of the HacA− and HacA−B− phenotypes. Shown are HacA− (A), HacA− plus p519-HacA (B), HacA− plus p519-HacB (C), HacA− plus p519nGFP (D), HacA−B− (E), HacA−B− plus p519-HacA (F), HacA−B− plus p519-HacB (G), and HacA−B− plus p519nGFP (H).
DISCUSSION
The aim of this study was to determine if the model plant pathogen strain B728a had the capacity to degrade AHLs enzymatically and whether this trait was associated with the regulation of QS signals. Here we report that HacA and HacB are two dissimilar AHL acylases which impart on strain B728a the ability to inactivate AHLs via cleavage of amide bonds. HacA degrades only long-chain AHLs (C ≥ 8), while HacB can degrade all AHLs. These enzymes do not apparently influence endogenous 3OC6HSL accumulation under the tested conditions, and their constitutive expression does not confer the ability to grow on C10HSL as a sole carbon source. We are therefore left with the following question—why do such enzymes exist, and what benefit, if any, does their production bestow on strain B728a?
The phyllosphere harbors many diverse types of bacteria that utilize AHL-mediated QS, and AHLs are believed to be common components of the leaf surface milieu (10, 18, 19, 50, 68). While AHLs can be inactivated by lactonolysis in alkaline conditions (7, 74), abundant 3OC6HSL and 3OC12HSL have been recovered from leaf surfaces of greenhouse-grown Nicotiana tabacum plants constitutively expressing bacterial AHL synthases (61), and exogenous AHLs applied to plants are also stable enough to influence plant gene expression (4, 41, 60). AHLs have been shown to diffuse away from producing bacterial cells inhabiting the rhizosphere (23), so in the microbial communities inhabiting leaf surfaces, strain B728a cells most likely would encounter AHLs produced by neighboring cells that could disrupt their normal patterns of gene expression. Antibiotic production is controlled by QS in Erwinia spp. (44), Serratia spp. (64), and numerous pseudomonads (43, 72), and rhizosphere strains have been isolated that can influence antibiotic production of other strains via QS interference (45). The production of an AHL acylase, and particularly a secreted AHL acylase such as HacA, could therefore be an adaptation that allows strain B728a to degrade AHLs of neighboring bacterial competitors and suppress expression of deleterious traits. Many plants also secrete QS-disruptive biochemicals (1, 14, 28, 53), and while the specific compounds are as-yet uncharacterized, AHL acylases of strain B728a might block inappropriate plant-mediated QS.
While both HacA and HacB have the capacity to degrade AHLs, it is very likely that they have other biologically significant substrates. AhlM, the AHL acylase of Streptomyces spp., can degrade the antibiotic penicillin G (48), suggesting that this enzyme might function to degrade extracellular inhibitors produced by other organisms. HacA and HacB do not have the ability to degrade penicillin G, but they may act against other antibiotics produced by plant-associated bacteria, and strain B728a itself may even produce an endogenous substrate. An AHL synthase from Pseudomonas fluorescens F113, termed HdtS, has been reported to make long-chain, AHL-like molecules (34). While a further report suggests that HdtS functions mainly to produce lipopolysaccharides (13), an orthologue of HdtS in Acidithiobacillus ferrooxidans termed Act has also been reported to produce long-chain AHLs (54). Psyr_0009, an uncharacterized gene in the P. syringae pv. syringae strain B728a genome, is homologous to both HdtS and Act and may produce an AHL-like substrate for AHL acylases. Additionally, Rhodopseudomonas palustris and other strains have recently been shown to produce and respond to p-coumaryl homoserine lactone (59). This new QS signal is synthesized from environmental p-coumaric acid rather than from cellular pools of fatty acids and raises the possibility that other signaling molecules containing amide bonds might exist that could be substrates for AHL acylases.
The dramatic phenotypes exhibited by strains HacA− and HacA−B− strongly indicate that HacA and HacB act against an as-yet unidentified endogenous substrate, especially since the phenotypes occur in both the wild-type and 3OC6HSL-deficient P. syringae pv. syringae strain B728a backgrounds. Constitutive expression of either HacA or HacB also complements the phenotypes of strains HacA− and HacA−B−, suggesting that a shared substrate is perhaps involved in a common regulatory pathway. In P. aeruginosa, the disruption of PvdQ influenced pyoverdine accumulation but was not reported to influence colony morphology (27), and no alterations in colony morphology were observed in pyoverdine-deficient strain B728a mutants (39). Since only strain HacA−B− is pyoverdine deficient, HacA and HacB are apparently involved in pyoverdine accumulation, perhaps through the editing of a precursor or modulation of a regulator molecule, but it is puzzling that other dramatic changes in colony morphology could be influenced by this process. The phenotypes of strains HacA− and HacA−B− appear very similar to autoaggregative phenotypes associated with the increased production of extracellular polymeric substances. These include the wrinkly spreader phenotype of P. fluorescens SBW25 (63, 65), the rdar (rough, dry, and red) phenotype of Salmonella enterica serovar Typhimurium (57), the rugose phenotype of Vibrio cholerae (75), and the phenotype exhibited by P. aeruginosa cells growing on medium containing SDS (31). These phenotypes are believed to increase cell survival in harsh environments and are modulated by 3′,5′-cyclic diguanylic acid, an intracellular second messenger that fluctuates in response to environmental conditions. AHL-mediated QS in V. cholerae controls biofilm formation by influencing levels of 3′,5′-cyclic diguanylic acid (70), but this link has not been established in strain B728a, and further study is required to elucidate the phenotypes exhibited by strains HacA− and HacA−B−.
Despite the uncertainties regarding the alternative physiological roles of HacA and HacB, they are of great interest because they might be utilized to inhibit the onset of bacterial diseases through so-called quorum quenching (16, 24, 78). As QS controls the expression of many traits important for a bacterium's lifestyle, including the expression of virulence factors, AHL-degrading enzymes have been utilized to confuse potential pathogens and disrupt the infection process. The potential utility of an engineered AHL-degrading bacterial strain as a biocontrol agent was illustrated when the AHL lactonase AiiA was introduced into Burkholderia sp. strain KJ006, a nonpathogenic bacterial endophyte of rice (12). The engineered Burkholderia sp. strain degraded the QS signal of pathogenic Burkholderia glumae and reduced the incidence of disease when coinoculated (12). Additionally, secretion of AiiA from the biocontrol strain Bacillus thuringiensis increased the strain's effectiveness at controlling plant disease by Erwinia carotovora (77). Expression of the secreted HacA of strain B728a might prove particularly useful as a trait to be added to biocontrol strains since it might enable quorum quenching over a longer distance than a nonsecreted AHL-degrading enzyme.
AHLs, antibiotics, and other small diffusible molecules produced by bacteria are widespread components of the mixed microbial communities inhabiting phyllosphere environments (38). While AHL acylases of strain B728a do have the capacity to degrade AHLs, the phenotypes associated with their targeted disruptions strongly indicate that they act against an endogenous substrate to influence colony and biofilm morphology and will thus most likely influence epiphytic fitness and disease development. The elucidation of other physiological functions of these dissimilar AHL acylases as well as their potential to interfere with QS of other species should prove very rewarding.
Acknowledgments
We thank L. Eberl for providing the E. coli MT102-JB524 biosensor. We also thank Tracy K. Powell and Glenn F. J. Dulla of the University of California—Berkeley for providing the plasmid pENTR/D-MCS-Gent.
This research was supported in part by United States Department of Agriculture National Research Initiative grant 2004-35319-14145 and a postdoctoral fellowship from the National Science Foundation awarded to R.W.S.
Footnotes
Published ahead of print on 7 November 2008.
REFERENCES
- 1.Adonizio, A., K.-F. Kong, and K. Mathee. 2008. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by south Florida plant extracts. Antimicrob. Agents Chemother. 52:198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, C. W. Myers, and D. L. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., and R. Losick. 2006. Bacterially speaking. Cell 125:237-246. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, W. D., and U. Mathesius. 2004. Plant responses to bacterial quorum sensing signals. Curr. Opin. Plant Biol. 7:429-433. [DOI] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Brannigan, J. A., G. Dodson, H. J. Duggleby, P. C. Moody, J. L. Smith, D. R. Tomchick, and A. G. Murzin. 1995. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378:416-419. [DOI] [PubMed] [Google Scholar]
- 7.Byers, J. T., C. Lucas, G. P. Salmond, and M. Welch. 2002. Nonenzymatic turnover of an Erwinia carotovora quorum-sensing signaling molecule. J. Bacteriol. 184:1163-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlier, A., R. Chevrot, Y. Dessaux, and D. Faure. 2004. The assimilation of γ-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol. Plant-Microbe Interact. 17:951-957. [DOI] [PubMed] [Google Scholar]
- 9.Carlier, A., S. Uroz, B. Smadja, R. Fray, X. Latour, Y. Dessaux, and D. Faure. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 69:4989-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 11.Chai, Y., C. S. Tsai, H. Cho, and S. C. Winans. 2007. Reconstitution of the biochemical activities of the AttJ repressor and the AttK, AttL, and AttM catabolic enzymes of Agrobacterium tumefaciens. J. Bacteriol. 189:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, H. S., S. Y. Park, C. M. Ryu, J. F. Kim, J. G. Kim, and S. H. Park. 2007. Interference of quorum sensing and virulence of the rice pathogen Burkholderia glumae by an engineered endophytic bacterium. FEMS Microbiol. Ecol. 60:14-23. [DOI] [PubMed] [Google Scholar]
- 13.Cullinane, M., C. Baysse, J. P. Morrissey, and F. O'Gara. 2005. Identification of two lysophosphatidic acid acyltransferase genes with overlapping function in Pseudomonas fluorescens. Microbiology 151:3071-3080. [DOI] [PubMed] [Google Scholar]
- 14.Degrassi, G., G. Devescovi, R. Solis, L. Steindler, and V. Venturi. 2007. Oryza sativa rice plants contain molecules that activate different quorum-sensing N-acyl homoserine lactone biosensors and are sensitive to the specific AiiA lactonase. FEMS Microbiol. Lett. 269:213-220. [DOI] [PubMed] [Google Scholar]
- 15.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. Aiia, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong, Y. H., and L. H. Zhang. 2005. Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43:101-109. [PubMed] [Google Scholar]
- 17.Dulla, G., and S. E. Lindow. 2008. Quorum size of Pseudomonas syringae is small and dictated by water availability on the leaf surface. Proc. Natl. Acad. Sci. USA 105:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dulla, G., M. L. Marco, B. Quinones, and S. E. Lindow. 2005. A closer look at Pseudomonas syringae as a leaf colonist. ASM News 71:469-475. [Google Scholar]
- 19.Elasri, M., S. Delorme, P. Lemanceau, G. Stewart, B. Laue, E. Glickmann, P. M. Oger, and Y. Dessaux. 2001. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 67:1198-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feil, H., W. S. Feil, P. Chain, F. Larimer, G. DiBartolo, A. Copeland, A. Lykidis, S. Trong, M. Nolan, E. Goltsman, J. Thiel, S. Malfatti, J. E. Loper, A. Lapidus, J. C. Detter, M. Land, P. M. Richardson, N. C. Kyrpides, N. Ivanova, and S. E. Lindow. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 102:11064-11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flagan, S., W. K. Ching, and J. R. Leadbetter. 2003. Arthrobacter strain VAI-A utilizes acyl-homoserine lactone inactivation products and stimulates quorum signal biodegradation by Variovorax paradoxus. Appl. Environ. Microbiol. 69:909-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Gen. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 23.Gantner, S., M. Schmid, C. Durr, R. Schuhegger, A. Steidle, P. Hutzler, C. Langebartels, L. Eberl, A. Hartmann, and F. B. Dazzo. 2006. In situ quantitation of the spatial scale of calling distances and population density-dependent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 56:188-194. [DOI] [PubMed] [Google Scholar]
- 24.González, J. E., and N. D. Keshavan. 2006. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 70:859-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, J. J., J. I. Han, L. H. Zhang, and J. R. Leadbetter. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PA01. Appl. Environ. Microbiol. 69:5941-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, J. J., A. Peterson, M. Whiteley, and J. R. Leadbetter. 2006. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PA01. Appl. Environ. Microbiol. 72:1190-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karamanoli, K., and S. E. Lindow. 2006. Disruption of N-acyl homoserine lactone-mediated cell signaling and iron acquisition in epiphytic bacteria by leaf surface compounds. Appl. Environ. Microbiol. 72:7678-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 30.Kinscherf, T. G., and D. K. Willis. 1999. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J. Bacteriol. 181:4133-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klebensberger, J., K. Lautenschlager, D. Bressler, J. Wingender, and B. Philipp. 2007. Detergent-induced cell aggregation in subpopulations of Pseudomonas aeruginosa as a preadaptive survival strategy. Environ. Microbiol. 9:2247-2259. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lamont, I. L., and L. W. Martin. 2003. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 149:833-842. [DOI] [PubMed] [Google Scholar]
- 34.Laue, B. E., Y. Jiang, S. R. Chhabra, S. Jacob, G. S. Stewart, A. Hardman, J. A. Downie, F. O'Gara, and P. Williams. 2000. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology 146:2469-2480. [DOI] [PubMed] [Google Scholar]
- 35.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, S. J., S. Y. Park, J. J. Lee, D. Y. Yum, B. T. Koo, and J. K. Lee. 2002. Genes encoding the N-acyl homoserine lactone-degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Appl. Environ. Microbiol. 68:3919-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, Y. H., J. L. Xu, J. Hu, L. H. Wang, S. L. Ong, J. R. Leadbetter, and L. H. Zhang. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47:849-860. [DOI] [PubMed] [Google Scholar]
- 38.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loper, J. E., and S. E. Lindow. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449-1454. [Google Scholar]
- 40.Marco, M. L., J. Legac, and S. E. Lindow. 2005. Pseudomonas syringae genes induced during colonization of leaf surfaces. Environ. Microbiol. 7:1379-1391. [DOI] [PubMed] [Google Scholar]
- 41.Mathesius, U., S. Mulders, M. Gao, M. Teplitski, G. Caetano-Anolles, B. G. Rolfe, and W. D. Bauer. 2003. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. USA 100:1444-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matthysse, A. G., S. Stretton, C. Dandie, N. C. McClure, and A. E. Goodman. 1996. Construction of GFP vectors for use in gram-negative bacteria other than Escherichia coli. FEMS Microbiol. Lett. 145:87-94. [DOI] [PubMed] [Google Scholar]
- 43.Mavrodi, D. M., W. Blankenfeldt, and L. S. Thomashow. 2006. Phenazine compounds in fluorescent Pseudomonas spp. Biosynthesis and regulation. Annu. Rev. Phytopathol. 44:417-445. [DOI] [PubMed] [Google Scholar]
- 44.McGowan, S. J., A. M. L. Barnard, G. Bosgelmez, M. Sebaihia, N. J. L. Simpson, N. R. Thomson, D. E. Todd, M. Welch, N. A. Whitehead, and G. P. C. Salmond. 2005. Carbapenem antibiotic biosynthesis in Erwinia carotovora is regulated by physiological and genetic factors modulating the quorum sensing-dependent control pathway. Mol. Microbiol. 55:526-545. [DOI] [PubMed] [Google Scholar]
- 45.Morello, J. E., E. A. Pierson, and L. S. Pierson III. 2004. Negative cross-communication among wheat rhizosphere bacteria: effect on antibiotic production by the biological control bacterium Pseudomonas aureofaciens 30-84. Appl. Environ. Microbiol. 70:3103-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. Genechip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 47.Park, S. Y., B. J. Hwang, M. H. Shin, J. A. Kim, H. K. Kim, and J. K. Lee. 2006. N-Acylhomoserine lactonase producing Rhodococcus spp. with different AHL-degrading activities. FEMS Microbiol. Lett. 261:102-108. [DOI] [PubMed] [Google Scholar]
- 48.Park, S. Y., H. O. Kang, H. S. Jang, J. K. Lee, B. T. Koo, and D. Y. Yum. 2005. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 71:2632-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park, S. Y., S. J. Lee, T. K. Oh, J. W. Oh, B. T. Koo, D. Y. Yum, and J. K. Lee. 2003. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149:1541-1550. [DOI] [PubMed] [Google Scholar]
- 50.Pierson, L. S., III, and E. A. Pierson. 2007. Roles of diffusible signals in communication among plant-associated bacteria. Phytopathology 97:227-232. [DOI] [PubMed] [Google Scholar]
- 51.Quiñones, B., G. F. J. Dulla, and S. E. Lindow. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:682-693. [DOI] [PubMed] [Google Scholar]
- 52.Quiñones, B., C. J. Pujol, and S. E. Lindow. 2004. Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant-Microbe Interact. 17:521-531. [DOI] [PubMed] [Google Scholar]
- 53.Rasmussen, T. B., T. Bjarnsholt, M. E. Skindersoe, M. Hentzer, P. Kristoffersen, M. Köte, J. Nielsen, L. Eberl, and M. Givskov. 2005. Screening for quorum-sensing inhibitors (QSI) by use of novel genetic system, the QSI selector. J. Bacteriol. 187:1799-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rivas, M., M. Seeger, E. Jedlicki, and D. S. Holmes. 2007. Second acyl homoserine lactone production system in the extreme acidophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 73:3225-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roche, D. M., J. T. Byers, D. S. Smith, F. G. Glansdorp, D. R. Spring, and M. Welch. 2004. Communications blackout? Do N-acylhomoserine-lactone-degrading enzymes have any role in quorum sensing? Microbiology 150:2023-2028. [DOI] [PubMed] [Google Scholar]
- 56.Romero, M., S. P. Diggle, S. Heeb, M. Cámara, and A. Otero. 2008. Quorum quenching activity in Anabaena sp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol. Lett. 280:73-80. [DOI] [PubMed] [Google Scholar]
- 57.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutation in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Schaefer, A. L., E. P. Greenberg, C. M. Oliver, Y. Oda, J. J. Huang, G. Bittan-Banin, C. M. Peres, S. Schmidt, K. Juhaszova, J. R. Sufrin, and C. S. Harwood. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454:595-599. [DOI] [PubMed] [Google Scholar]
- 60.Schuhegger, R., A. Ihring, S. Gantner, G. Bahnweg, C. Knappe, F. Vogg, P. Hutzler, M. Schmid, F. Van Breusegem, L. Eberl, A. Hartmann, and C. Langebartels. 2006. Induction of systemic resistance in tomato by N-acyl-l-homoserine lactone-producing rhizosphere bacteria. Plant. Cell. Environ. 29:909-918. [DOI] [PubMed] [Google Scholar]
- 61.Scott, R. A., J. Weil, P. T. Le, P. Williams, R. G. Fray, S. B. von Bodman, and M. A. Savka. 2006. Long- and short-chain plant-produced bacterial N-acyl-homoserine lactones become components of phyllosphere, rhizosphere, and soil. Mol. Plant-Microbe Interact. 19:227-239. [DOI] [PubMed] [Google Scholar]
- 62.Sio, C. F., L. G. Otten, R. H. Cool, S. P. Diggle, P. G. Braun, R. Bos, M. Daykin, M. Cámara, P. Williams, and W. J. Quax. 2006. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PA01. Infect. Immun. 74:1673-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spiers, A. J., J. Bohannon, S. M. Gehrig, and P. B. Rainey. 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50:15-27. [DOI] [PubMed] [Google Scholar]
- 64.Thomson, N. R., M. A. Crow, S. J. McGowan, A. Cox, and G. P. C. Salmond. 2000. Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 36:539-556. [DOI] [PubMed] [Google Scholar]
- 65.Ude, S., D. L. Arnold, C. D. Moon, T. Timms-Wilson, and A. J. Spiers. 2006. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ. Microbiol. 8:1997-2011. [DOI] [PubMed] [Google Scholar]
- 66.Uroz, S., S. R. Chhabra, M. Cámara, P. Williams, P. M. Oger, and Y. Dessaux. 2005. N-Acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology 151:3313-3322. [DOI] [PubMed] [Google Scholar]
- 67.Uroz, S., P. Oger, S. R. Chhabra, M. Cámara, P. Williams, and Y. Dessaux. 2007. N-Acyl homoserine lactones are degraded via an amidolytic activity in Comamonas sp. strain D1. Arch. Microbiol. 187:249-256. [DOI] [PubMed] [Google Scholar]
- 68.Von Bodman, S. B., W. D. Bauer, and D. L. Coplin. 2003. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41:455-482. [DOI] [PubMed] [Google Scholar]
- 69.Wang, Y. J., and J. R. Leadbetter. 2005. Rapid acyl-homoserine lactone quorum signal biodegradation in diverse soils. Appl. Environ. Microbiol. 71:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waters, C. M., W. Lu, J. D. Rabinowitz, and B. L. Bassler. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wen, Y., X. Shi, Z. Yuan, and P. Zhou. 2004. Expression, purification, and characterization of his-tagged penicillin G acylase from Kluyvera citrophila in Escherichia coli. Protein Exp. Purif. 38:24-28. [DOI] [PubMed] [Google Scholar]
- 72.Wood, D. W., and L. S. Pierson III. 1996. The phzI gene of Pseudomonas aureofaciens 30-84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene 168:49-53. [DOI] [PubMed] [Google Scholar]
- 73.Wu, H., Z. Song, M. Hentzer, J. B. Andersen, A. Heydorn, K. Mathee, C. Moser, L. Eberl, S. Molin, N. Hoiby, and M. Givskov. 2000. Detection of N-acylhomoserine lactones in lung tissues of mice infected with Pseudomonas aeruginosa. Microbiology 146:2481-2493. [DOI] [PubMed] [Google Scholar]
- 74.Yates, E. A., B. Philipp, C. Buckley, S. Atkinson, S. R. Chhabra, R. E. Sockett, M. Goldner, Y. Dessaux, M. Cámara, H. Smith, and P. Williams. 2002. N-Acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70:5635-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, H. B., L. H. Wang, and L. H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, L., L. Ruan, C. Hu, H. Wu, S. Chen, Z. Yu, and M. Sun. 2007. Fusion of the genes for AHL-lactonase and S-layer protein in Bacillus thuringiensis increases its ability to inhibit soft rot caused by Erwinia carotovora. Appl. Microbiol. Biotechnol. 74:667-675. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, L. H. 2003. Quorum quenching and proactive host defense. Trends Plant Sci. 8:238-244. [DOI] [PubMed] [Google Scholar]