Abstract
We resolved the intraspecific diversity of Vibrio fischeri, the bioluminescent symbiont of the Hawaiian sepiolid squid Euprymna scolopes, at two previously unexplored morphological and geographical scales. These scales ranged from submillimeter regions within the host light organ to the several kilometers encompassing two host populations around Oahu. To facilitate this effort, we employed both novel and standard genetic and phenotypic assays of light-organ symbiont populations. A V. fischeri-specific fingerprinting method and five phenotypic assays were used to gauge the genetic richness of V. fischeri populations; these methods confirmed that the symbiont population present in each adult host's light organ is polyclonal. Upon statistical analysis of these genetic and phenotypic population data, we concluded that the characteristics of symbiotic populations were more similar within individual host populations than between the two distinct Oahu populations of E. scolopes, providing evidence that local geographic symbiont population structure exists. Finally, to better understand the genesis of symbiont diversity within host light organs, the process of symbiosis initiation in newly hatched juvenile squid was examined both experimentally and by mathematical modeling. We concluded that, after the juvenile hatches, only one or two cells of V. fischeri enter each of six internal epithelium-lined crypts present in the developing light organ. We hypothesize that the expansion of different, crypt-segregated, clonal populations creates the polyclonal adult light-organ population structure observed in this study. The stability of the luminous-bacterium-sepiolid squid mutualism in the presence of a polyclonal symbiont population structure is discussed in the context of contemporary evolutionary theory.
All multicellular organisms maintain symbiotic associations with microbes (46). These cross-domain relationships fundamentally influence microbial population structure, ecology, and evolutionary biology (57). The search for a clearer understanding of how interspecific interactions affect diversity has a long history (6, 75), yet at its heart lie contemporary issues of population biology and ecology, as well as general evolutionary process (68). Studies of these issues are enriched and expanded by model systems of interspecific biology that may be approached both empirically and theoretically.
The symbiotic relationship between the bioluminescent marine bacterium Vibrio fischeri and the Hawaiian sepiolid squid Euprymna scolopes provides one suitable model in which both ecological scale and evolutionary process may be examined under natural conditions. Soon after hatching, the nascent host establishes and maintains a population of V. fischeri in a specifically adapted structure called the light organ (51, 77). The light organ is composed of two bilaterally symmetrical lobes; each lobe contains three epithelium-lined crypts (Fig. 1) (50, 72). V. fischeri bacteria are harvested from an environment where their concentration (100 to 103 CFU/ml) constitutes less than 0.1% of the total seawater microbial community (42), although higher estimates of their relative abundance have been reported (34). In contrast, the concentration of V. fischeri in the light organ approaches 1010 CFU/ml (4). Bacteria identified as V. fischeri are the only isolates found in the light organ of E. scolopes (4). Thus, this host's light organ serves as a definitive boundary for a distinct and relatively abundant population of symbiotic V. fischeri.
FIG. 1.
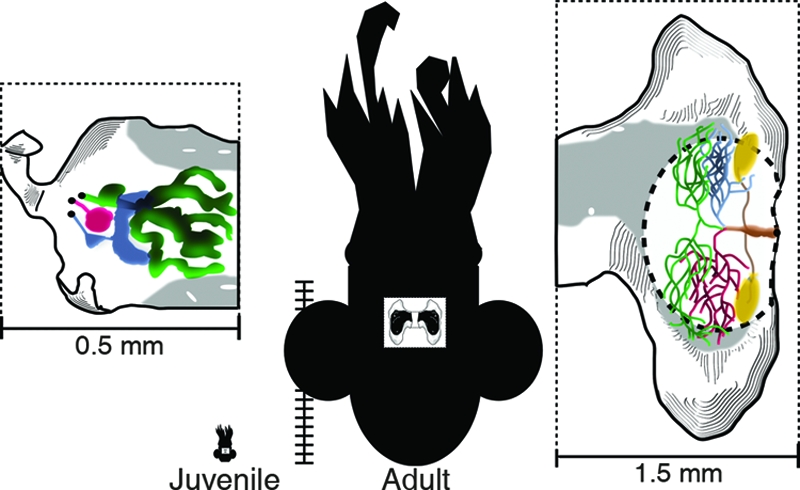
Morphological comparison of juvenile and adult E. scolopes squid and their light organs. The relative body sizes of a juvenile and an adult specimen are depicted by the two central silhouettes (vertical scale bar increments = 1 mm). The white rectangular cutaway within each silhouette frames the position of the entire bilaterally symmetrical light organ. An enlarged half of the light organ, a “lobe,” is depicted to the left (juvenile) or right (adult) of each silhouette. Within each enlarged light-organ lobe, colored areas represent the approximate positions of the three crypts as follows: green, crypt 1; blue, crypt 2; magenta, crypt 3. Other morphological details include the ink sack (light gray area), a yellow tissue or “filter” in adults (51), and juveniles' anterior and posterior appendages (middle left and bottom of the lobe, respectively). The central core region of the adult light-organ lobe is represented by the oval, dash-bordered area. The locations and sizes of both adult and juvenile light-organ crypts are approximations based on composites of confocal micrographs obtained in this study, as well as on previous descriptions (50, 51, 72).
The specific population structure of light-organ-dwelling V. fischeri is not well understood. It is unknown whether the populations in each organ are derived from a single cell (clonal origin) or include several distinct strains (polyclonal origin). Previous work has demonstrated that an individual light organ may contain V. fischeri isolates distinct in both plasmid content and chromosomal restriction sites at certain loci (5, 40). Furthermore, V. fischeri strains differing in bioluminescent output levels have been isolated both from a single light organ and the host's near-shore habitat (41). Finally, other work has demonstrated that different, genetically labeled (but otherwise isogenic) cells of V. fischeri may enter and initially colonize the light organ of individual juvenile hosts (18, 48). Based on these data, we hypothesized that the symbiont population in a single light organ may be composed of several distinct strains of V. fischeri.
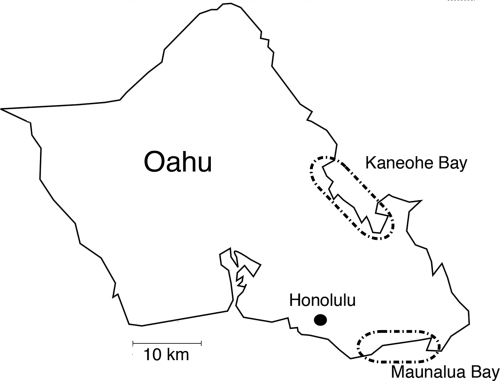
A second scale at which symbiont population structure has been unresolved is related to the host populations surrounding the island of Oahu. Over the years, specimens of E. scolopes have been collected from Maunalua Bay or Kaneohe Bay, Oahu (Fig. 2). Morphological and molecular analyses of individuals from either location support the conclusion that these two E. scolopes populations are distinct and distinguishable (34, 38). Although the issue of host-symbiont coevolution remains in flux (16, 55), evidence of local adaptation of V. fischeri isolates to the host exists at the continental scale (53, 55).
FIG. 2.
Locations of E. scolopes collection sites on Oahu, HI. Specimens were obtained from squid populations in Maunalua Bay (MB; 21°26′3.36"N, 157°47′20.78"W) and Kaneohe Bay (KB; 21°16′51.42"N, 157°43′33.07"W), which are located on either side of the island. Specimens were collected on the nights of 13 (MB2-MB7), 14 (MB11-MB15), and 16 (KB1-KB5) November 2005.
In this work, we had two specific aims: (i) to determine the population structure of symbiotic V. fischeri both within individual host light organs and between host populations on Oahu and (ii) to provide a hypothesis for the creation of this observed population structure by using a simple mathematical model of the process of crypt colonization during symbiosis initiation.
MATERIALS AND METHODS
Preparation of bacterial isolates.
Specimens of E. scolopes were collected by dip net at dusk from Kaneohe Bay and Maunalua Bay, Oahu, HI, in November 2005 (Fig. 2). On the night of collection, the squid were anesthetized in 2% ethanol, sexed, measured, and dissected. Crypt-containing central-core tissues (Fig. 1) (51) were removed by the following convention. A ventral dissection, with anterior oriented up, yielded left (A) and right (B) cores from corresponding lobes of the light organ. Cores were placed into sterilized (with a 0.2-μm filter) natural seawater, homogenized, and frozen in seawater-tryptone medium (SWT) (4) containing 17% glycerol. Aliquots of these samples were streaked onto SWT agar, and representative V. fischeri symbionts arising as colonies were isolated and referred to by the following convention: MB11A2 = Maunalua Bay host no. 11, central core A, isolate no. 2; KB1A97 = Kaneohe Bay host no. 1, central core A, isolate no. 97. V. fischeri symbionts collected and identified in 1988 to 1989 have been previously reported (4). For the descriptions of the other bacteria used in this study, see Table S1 in the supplemental material.
Bacterial samples in other squid tissues (accessory nidamental gland, enteric tract, gills, ovaries, and skin) were obtained from two adult female squid that were collected in Maunalua Bay in October 2007 and kept for 6 weeks in artificial-seawater-containing aquaria. Tissues were washed twice, homogenized, and serially diluted in SFIO (filter [0.2 μm]-sterilized instant ocean; Spectrum Brands, Atlanta, GA) and plated on LBS (15) agar. Similarly, several 19-day-old eggs that had been laid in the aquaria were washed twice and homogenized in SFIO and plated on LBS agar. Groups of five juvenile squid were collected, either placed in SFIO immediately upon hatching or left in unfiltered aquarium water for 2 h, and then washed twice in SFIO. The unfiltered aquarium water contained V. fischeri cells that had been naturally expelled by adult hosts (43). The juvenile animals were anesthetized on ice and homogenized and serially diluted in SFIO, and the dilutions were spread on LBS agar.
Bacterial DNA was obtained either directly from bacterial colonies or from SWT cultures diluted (1:20) in TE buffer (10 mM Tris-HCl buffer [pH 8.0], 0.1 mM EDTA) and frozen and thawed at −80°C and room temperature, respectively. Chromosomal DNA was extracted and isolated as previously described (45).
hvnC PCR.
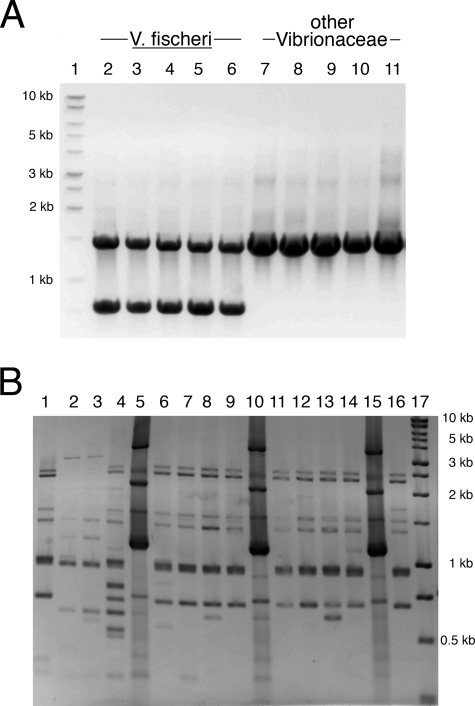
Each PCR mixture contained the following components: 8.3 μl of water, 2.5 μl of 5× GoTaq buffer (Promega Corp., Madison, WI), 0.1 μl of a 25 mM deoxynucleoside triphosphate mixture (dATP, dCTP, dTTP, and dGTP; Promega), 0.075 μl of each of four primers (see below), 0.1 μl of GoTaq DNA polymerase (Promega), and 1.2 μl of DNA. Primer stock concentrations were approximately 150 pmol/μl. Primers 49FVf (5′TNANACATGCAAGTCGNNCG3′) and 1492RVf (5′NGNTACCTTGTTACGACTT3′) amplify an ∼1,500-bp segment of the 16S rRNA gene locus and were modified from previously described universal bacterial 16S rRNA gene primers (39). Primers hvnC1fw (5′GTAACGACACTACTGGCAAG3′) and hvnC1rv (5′CACTGGAATAGCGATTCCTG3′) amplify an ∼750-bp segment of the V. fischeri ES114 hvnC locus (VF_A1105; NC_006841). The PCR was performed with the following program: 95°C for 5 min; 30 repetitions of 94°C for 30 s, 47°C for 30 s, and 72°C for 1 min; and 72°C for 10 min. Samples were run on 1.2 to 1.6% agarose (GenePure LE; ISC BioExpress, Kayesville, UT) gels and photographed with a charge-coupled device (CCD) camera and imaging software (AlphaImager; Alpha Innotech GmbH, Germany). The presence of two fragments (a 1.5-kb 16S rRNA gene product and a 0.75-kb hvnC product) was scored as a positive result; the presence of only the larger fragment was scored as a negative result (Fig. 3A).
FIG. 3.
Genetic analysis of V. fischeri symbionts. (A) Genomic DNA samples from strains of V. fischeri and other members of the family Vibrionaceae were subjected to the hvnC PCR assay. A 1.5-kb band representing the 16S rRNA gene locus was amplified from all strains; the 0.75-kb band is the hvnC product present only upon the amplification of V. fischeri DNA. Lane 1, DNA size standards; lanes 2 to 6, different V. fischeri isolates (lane 2, isolate ES114; lane 3, isolate SA1; lane 4, isolate WH1; lane 5, isolate EM17; lane 6, isolate MB11A1); lanes 7 to 11, representative Vibrionaceae isolates of other species (lane 7, Vibrio logei SA6; lane 8, Vibrio parahaemolyticus KNH1; lane 9, Vibrio harveyi BB392; lane 10, Vibrio salmonicida ATCC 43839; lane 11, Photobacterium leiognathi H90-62). For strain descriptions, see Table S1 in the supplemental material. (B) Genomic DNA samples of 16 isolates from light-organ lobe B of animal KB4 were subjected to the VfRep PCR assay. Three major types of VfRep patterns (as represented in lanes 1, 2, and 5) were observed among the 16 isolates, with some minor banding variation. Lane 17 = DNA size standards.
VfRep PCR.
To measure genetic heterogeneity among light-organ isolates, we developed a V. fischeri-specific rep-PCR (repetitive-sequence-based PCR) (73, 74) fingerprinting approach (VfRep PCR). First, a collection of VfRep PCR primers was developed through analysis of the V. fischeri ES114 genome (63) with the software program Tandem Repeat Finder (2, 13). Next, primer resolution was compared. Of the three primers tested, VfRep1 (5′CCTGAARCCTGAAMCCTGAACCT3′) gave the best balance between informative band number and resolution (data not shown). The VfRep1 primer was designed against a common 7-mer motif (CCTGAAN) on the large chromosome. To confirm that VfREP1 amplified predicted V. fischeri genomic targets, agar gel-resolved products of VfRep PCR on V. fischeri ES114 were cloned and sequenced. These products had sequences matching those predicted by an independent in silico analysis of the V. fischeri ES114 genome (data not shown).
The VfRep PCR mixture contained 2.5 μl of 5× Gitschier buffer (58) [83 mM (NH4)2SO4, 335 mM Tris-HCl (pH 8.8), 33.5 mM MgCl2, 33.5 μM EDTA, 150 mM ß-mercaptoethanol], 4.92 μl of water, 0.625 μl of a 25 mM deoxynucleoside triphosphate mixture, 0.31 μl of the VfRep1 primer (150 pmol/μl), 0.2 μl of bovine serum albumin (10 mg/ml), 0.2 μl of GoTaq DNA polymerase (Promega), and 3.75 μl of target DNA. The PCR was performed with the following program: 95°C for 2 min; 35 repetitions of 94°C for 3 s, 92°C for 30 s, 50°C for 1 min, and 65°C for 8 min; and 65°C for 8 min.
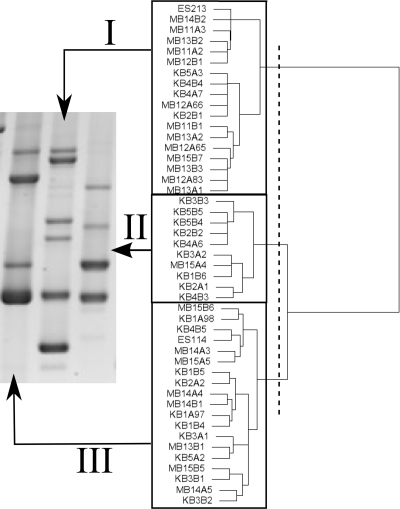
Distinct banding patterns of the VfRep PCR products were identified upon comparison of isolates from a single lobe's central core. A single representative isolate was selected for each VfRep PCR pattern from each lobe, and the resulting 63 isolates were reanalyzed as a group (Fig. 3B) for the assignment of “global” VfRep types as follows. Presence-or-absence data for the 17 observed bands were used to generate a binary code for each isolate. A dissimilarity matrix was created for these data by assuming symmetric binary variables and using a simple matching distance transformation. From these data, dendrograms were created with a variety of agglomerative or divisive hierarchical clustering algorithms and heuristic methods implemented in the R project for statistical computing (32) v2.6.0 and/or the phylogeny inference package PAUP* (71) v4.0b10. The resulting dendrograms were compared, and three clusters were found (Fig. 4). For each isolate, membership in one of these three clusters was recorded as the VfRep type (see Table S2 in the supplemental material).
FIG. 4.
Agglomerative hierarchical cluster analysis, with Ward's algorithm (76), of the VfRep patterns of 47 symbiont strains, 22 isolated from five Maunalua Bay squid light organs (MB11 to MB15) and 23 from five Kaneohe Bay squid light organs (KB1 to KB5), respectively, in 2005, as well as previously described symbiont strains ES114 (obtained from Kaneohe Bay in 1988) and ES213 (obtained from Maunalua Bay in 1989). The vertical dashed line represents the level below which VfRep products exhibited qualitatively similar banding patterns; the three distinguishable patterns are boxed and identified by roman numerals (see Table S2 in the supplemental material). Thus, the qualitative (visible patterns) and quantitative (hierarchical clustering) analyses are congruent. Gel lanes indicated to the left of each cluster represent the typical VfRep pattern for that group. These general groupings were also found by other heuristic (19) and nonheuristic (35, 67) clustering methods.
Phenotypic assays. (i) Bioluminescence.
Dilutions of light-organ homogenates were spread on SWT agar containing 120 nM synthetic 3-oxo hexanoyl homoserine lactone autoinducer (Sigma-Aldrich Corp., St. Louis, MO) (45) and incubated at room temperature for 12 to 24 h. Colonies were then imaged for 10 to 30 min with a CCD camera, and the bioluminescence of “bright” colonies was detectable whereas that of “dim” colonies was not. In the absence of added autoinducer, reproducible variation in bioluminescence was also found among isolates with a TD20/20 photometer (Turner Designs, Sunnyvale, CA) or a CCD camera with a 4- to 6-h exposure (data not shown).
(ii) Colony pigmentation.
After the bacteria were grown for 24 to 28 h at room temperature on SWT, colony pigmentation differences among isolates were observed. Although V. fischeri has been characterized by its yellow-pigmented colony color (26), light-organ lobe isolates reproducibly exhibited either a white or a yellow colony pigmentation.
(iii) Motility.
One microtiter of a mid-log-phase bacterial culture was spotted onto SWT soft agar (0.25%). The diameter of the front of cells swimming through the medium was plotted against time, and the motility rate was calculated from the slope. Empirical analysis of these data showed a bimodal distribution with local maxima at approximately 0.8 and 4 mm/h (data not shown), and the distribution was significantly different from normal (Shapiro-Wilk test for normality, W = 0.836, P < 0.00001; D'Agostino normality test, P < 0.00001). Motility rates were scored either as “fast” (>2 mm/h) or “slow” (≤2 mm/h) for comparative statistical analyses, with the breakpoint identified by k-means clustering (k = 2) and empirical analyses.
(iv) Growth rate.
Isolates were inoculated in triplicate into microtiter plate wells containing SWT and shaken at 25°C. The optical density at 600 nm (OD) of each well was determined periodically for between 8 and 10 h, and the slope between ODs of about 0.1 and 0.5 was used to calculate the linear growth rate during exponential growth. The value of these slopes had a bimodal distribution around local maxima of approximately 0.08 and 0.10 U of OD per h (data not shown); however, this distribution was not significantly different from a normal distribution (Shapiro-Wilk test for normality, W = 0.99, P = 0.94; D'Agostino normality test, P = 0.83). Nevertheless, strains with relatively “high” (≥0.09 U of OD per h) or “low” (<0.09 U of OD per h) growth rates were defined for statistical analyses; this breakpoint was assessed by k-means clustering (k = 2). Growth experiments with a subset of these isolates, performed with 5-ml broth cultures, showed relative growth rate relationships that were qualitatively similar to those determined by the above-described method (data not shown).
(v) Siderophore production.
Agar medium containing the indicator chrome azurol S (Sigma-Aldrich Corp., St. Louis, MO) was used to assay the production of iron-chelating siderophore(s) by V. fischeri (26). The rate of siderophore production was estimated by measuring the diameters of the colony and the orange halo around it; these values were squared, and the latter was divided by the former to calculate the siderophore production index. The distribution of indices was positively skewed (skew = 0.87) with a median of 2.3 and a nonnormal distribution (Shapiro-Wilk test for normality, W = 0.92, P < 0.001; D'Agostino normality test, P = 0.01). Indices above or equal to the median were defined as “high” and those below the median were defined as “low” for further statistical analyses.
Stability of VfRep PCR fingerprints and observed physiological characteristics.
The VfRep PCR banding patterns and physiological characteristics of several strains were determined before and after >200 generations of growth in either solid or liquid medium. Phenotypic traits did not change during the analysis, and only minor banding pattern variations were present among the isolates (data not shown), leading us to conclude that the observed phenotypes and major banding patterns are relatively stable compared to the reported traits of other symbionts (52).
Statistical comparisons.
Statistical comparisons were made with the R project for statistical computing v2.6.0 and its associated graphical user interface, R Commander (21) v1.3-5. Most of the data collected were amenable to nonparametric statistical analyses. Comparisons between continuous and categorical data were made after converting numerical values to categories based on the criteria described above.
Comparisons of phenotypic and genetic characteristics.
The association between any two categorical variables was assessed with Cramér's V (10). A significance test for the value V was accomplished by estimating the significance of the χ2 statistic for the associated contingency table (64). For all comparisons, exact P values were calculated with Fisher's exact test to complement the approximate P value estimates made by use of the χ2 statistic.
Comparisons between Maunalua Bay and Kaneohe Bay isolates.
To determine whether the sampled groups (i.e., Maunalua Bay or Kaneohe Bay isolates) differed with respect to each categorical variable, continuous data from each group were compared by using the appropriate statistical methods. The exact permutation form of the Mann-Whitney-Wilcoxon test (assuming ties) was used to analyze groups without a symmetric distribution, homogeneous variance, or similar skew to determine whether two independent groups were drawn from the same population; a Welch t test was used for all other samples.
Rarefaction analyses.
Two rarefaction analyses were performed with the software EstimateS (8) v8.0. First, the observed richness of VfRep types in the symbiont population within a given light-organ lobe was estimated from approximately 150 different isolates each from MB12A and KB1A and 23 isolates from MB13B. Individual-based rarefaction curves of the observed richness of each subsample were calculated by the Coleman analytical method (9). Second, observed subsample and predicted total richness of VfRep types per light-organ lobe in the Maunalua Bay and Kaneohe Bay host populations was assessed by using data from all lobes sampled in 2005; the three VfRep types determined from hierarchical clustering results described above were used for this analysis. EstimateS was used to calculate sample-based rarefaction curves of observed richness (by both the Mau Tau heuristic and resampling 1,000 times, with replacement) for each population, as well as total richness estimations (MMRuns Mean method) (25).
Construction of green fluorescent protein- and DsRed-expressing V. fischeri derivatives.
Triparental mating was performed as previously described (70), with the following three strains: (i) the conjugative helper strain, (ii) a donor V. fischeri strain carrying either a green fluorescent protein-expressing plasmid (pVSV102) or a red fluorescent protein-expressing plasmid (pVSV208), and (iii) one of three light-organ isolates (MB13B2, MB14A3, or MB15A4), each representing one of the major VfRep type groups determined by hierarchical clustering. As previously reported (18), plasmids were stable in each isolate and did not affect the growth rate of any isolate (data not shown). These three V. fischeri isolates were chosen because the juvenile squid to be colonized were offspring of parents collected in Maunalua Bay.
Juvenile squid colonization experiments.
Juvenile E. scolopes squid were collected immediately after hatching from laboratory-laid eggs. Juveniles were held in SFTW (filter [0.2 μm]-sterilized aquarium water) for at least 30 min prior to inoculation with an equal mixture of isogenic V. fischeri strains containing either the green or the red fluorescent protein-encoding plasmid. Newly hatched juvenile squid were added to SFTW containing this mixed inoculum. After either 3 or 12 h, water samples were taken for bacterial enumeration and the juveniles were moved to individual glass vials filled with 4 ml of SFTW for an additional 40 to 45 h (with a water change at approximately 20 h). Juveniles were anesthetized with 2% ethanol for approximately 15 min and processed for confocal microscopy as previously described (72), with the following modification: fixation in 4% formaldehyde was followed by dissection, light-organ permeabilization, and staining with Alexa Fluor 633-phalloidin (Invitrogen, Carlsbad, CA), an actin-binding agent used to visualize crypt morphology. Light organs were mounted under an LSM510 confocal laser scanning microscope (Carl Zeiss Inc., Thornwood, NY), and their crypts were examined for the presence of green and/or red fluorescent bacteria.
Mathematical modeling of colonization.
The equation p(red)X + p(green)X = p(single) was used to model the probability of observing crypts filled with either red- or green-fluorescing bacteria [p(single)] as a function of the initiating cell number (x) and the sum of the initial concentrations of red- and green-fluorescing bacteria [p(red) + p(green)]. The probability of observing crypts with both red- and green-fluorescing bacteria [p(mixed)] can be determined with the equation p(mixed) = 1 − p(single). For each initiation experiment, a solution to the equation p(red)X + p(green)X = p(single) for x = 1 to 25 was determined, based on empirical values determined by confocal microscopy. The resulting graph of p(single) as a function of x was fitted with an exponential function (in all cases, R2 = >0.99, data not shown); the number of initiating cells (x) was then empirically solved from the equation.
RESULTS
The hvnC PCR assay is a rapid, simple identification method for V. fischeri.
Taxonomic assessments of large numbers of E. scolopes light-organ symbionts have previously relied on the use of large-scale physiological analyses (4). We sought to create a single molecular assay that would both allow the analysis of hundreds of bacterial colonies from symbiotic populations and increase our confidence in the identification of a bacterial DNA sample from dissected light organs or other tissues of E. scolopes. In silico analysis of the V. fischeri ES114 genome yielded two paralogs of the halovibrin A gene (hvnA): hvnB (59, 69) and hvnC. Homologs of the more divergent locus, hvnC (halovibrin C), were found only in the genome of another V. fischeri strain, MJ11, a light-organ symbiont of the Japanese pinecone fish Monocentris japonica (data not shown). With primers designed by alignment of the ES114 and MJ11 loci, a V. fischeri-specific hvnC PCR assay was established.
Two DNA fragments (16S rRNA gene and hvnC loci) were amplified from V. fischeri, while only a single fragment (16S rRNA gene locus) was amplified from all of the other identified bacterial species tested with the hvnC PCR assay (e.g., Fig. 3A). Forty isolates from the light organs of four different E. scolopes squid collected on Oahu in 1988 and 1989 were previously confirmed to be V. fischeri by a series of physiological assays (60); each of these strains tested positive in the hvnC PCR assay. In addition, all of 28 previously identified V. fischeri culture collection strains isolated from a variety of locations and hosts were hvnC PCR positive; most of these isolates have been identified as V. fischeri through sequencing of 16S rRNA loci (54). Finally, the hvnC PCR gave positive results for all 761 light organ isolates from 15 different E. scolopes squid collected on Oahu in 2005 (63 of these are analyzed below; for a list, see Table S1 in the supplemental material). In contrast, other than V. fischeri, all of the tested strains in the family Vibrionaceae (21 strains, 17 species) were hvnC PCR negative (Fig. 3A). All isolates from a variety of other adult E. scolopes tissues (enteric tract, gills, ovaries, and skin), as well as seawater-exposed egg cases and newly hatched juveniles, also were hvnC PCR negative. The culturable bacterial community of the accessory nidamental gland (3) of two female specimens of E. scolopes contained a single hvnC PCR-positive strain among the 24 strains cultured; the nearest-named BLAST and Ribosomal Database Project match to the sequenced 16S rRNA gene loci of this one isolate was V. fischeri (see Fig. S2 in the supplemental material).
The V. fischeri population within a single E. scolopes light organ is physiologically and genetically heterogeneous.
While all of the colonies arising from light-organ homogenates were identified as V. fischeri, physiological differences (e.g., pigmentation and bioluminescence level) were observed among colonies arising from an individual light-organ homogenate (data not shown). Additional physiological assays for traits such as siderophore production, motility rate, and growth rate revealed similar heterogeneity among light-organ isolates. Generally, one to three physiologically distinguishable types were present among isolates from a single lobe (see Table S2 in the supplemental material).
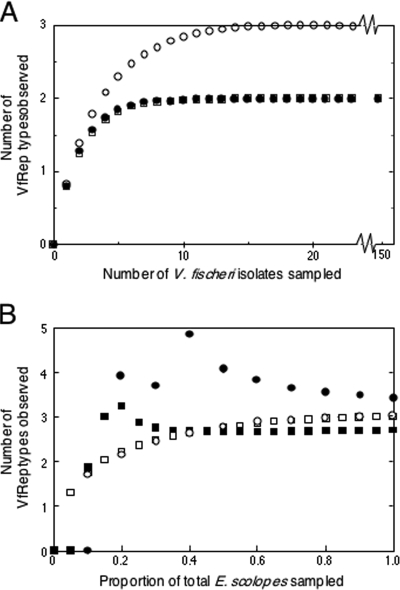
To determine whether physiological types were associated with a shared genetic character set, we designed a repetitive-element PCR (74) (VfRep PCR) primer specific to a common 7-mer repeat on the large chromosome of V. fischeri ES114 (63). To determine a statistically appropriate sample size for a single light-organ lobe, we generated individual-based rarefaction curves (25) by using between 23 and 150 isolates from each of three hosts. From these analyses, we concluded that a sample size of 16 isolates per lobe would predict essentially all of the observed and predicted VfRep type richness in the symbiont population (Fig. 5A).
FIG. 5.
Rarefaction analyses of VfRep types. (A) Individual-based rarefaction analysis of VfRep PCR patterns of V. fischeri isolates (23, 150, and 150, respectively) from a single light-organ lobe of three different adult specimens of E. scolopes: MB13B (open circles), KB1A (closed circles), and MB12A (open squares), respectively. In each case, richness had saturated after the sampling of 16 isolates. (B) Sample-based rarefaction (open shapes) and richness estimate (closed shapes) analyses of major VfRep types of each of the two squid populations sampled: 10 Maunalua Bay squids (squares) and 5 Kaneohe Bay squids (circles). The software program EstimateS v8.0 was applied in all of the analyses (8) with the analytic Coleman method for determining individual-based rarefaction (9) and the Mao Tau heuristic and MMRuns Mean methods (25) for sample-based rarefaction determination and richness estimation.
VfRep PCR was used to analyze an additional 438 V. fischeri isolates from 15 hosts; 63 of these were further analyzed as described below (see Table S2 in the supplemental material). Multiple VfRep patterns per lobe (X̄ = 1.5 ± 0.5), per animal (X̄ = 1.9 ± 0.8), and per sampling site were found. Because each lobe was composed of one to three VfRep types with similarly grouped physiological characteristics (see below), representative strains were chosen from each of the 30 lobes for further analyses. The VfRep PCR banding of these strains was compared on agarose gels, and 17 different bands were recognized. Binary (presence or absence) band data were compiled for each representative strain, and these data were subjected to hierarchical cluster and heuristic analyses, which separated all of the isolate patterns into three major VfRep types (Fig. 4). Sample-based rarefaction analyses and richness estimation of the VfRep types present at each sampling site demonstrated that analyses of between three and eight hosts yielded approximately 100% of the predicted VfRep type richness characteristic of both sampled host populations (Fig. 5B).
Certain genetic and physiological characteristics of V. fischeri strains are associated and exhibit different frequencies in each host population sampled.
For all hosts, similar VfRep PCR-patterned isolates were grouped together and tested for a statistically significant association with each physiological trait. Relatively large significant associations with VfRep type were found for three traits: bioluminescence level, motility rate, and colony pigmentation (χ2, >30 [df = 2], P < 0.01; Cramér's V, >0.7). In contrast, growth rate and siderophore production both had only a low to moderate, and not statistically significant, association with VfRep type (Table 1).
TABLE 1.
Statistical analyses of associations between genetic and phenotypic traits of V. fischeri isolates collected in 2005
| Trait | χ2 | df | % of cells <5b | Cramér's V | P valuea | Fisher's exact test P valuea |
|---|---|---|---|---|---|---|
| Bioluminescence level | 32.9 | 2 | 17 | 0.71 | **** (****) | **** (****) |
| Colony pigmentation | 33.1 | 2 | 17 | 0.71 | **** (****) | **** (****) |
| Motility rate | 57.7 | 2 | 17 | 0.94 | **** (****) | **** (****) |
| Growth rate | 6.8 | 2 | 17 | 0.32 | *** (**) | *** (*) |
| Siderophore production | 4.0 | 2 | 17 | 0.25 | * (*) | * (*) |
The correction of P values for multiple testings was performed by considering all of the data as a family of tests. The P values were analyzed and adjusted according to the method of Hommel (31); corrected values are in parentheses. Ranges of calculated P values: *, P > 0.1; **, 0.1 > P > 0.05; ***, 0.01 < P < 0.05; ****, P < 0.01.
The percentage of cells <5 refers to a common benchmark for the validity of χ2 analysis.
We tested whether there were statistically significant differences in the frequencies of both genetic and physiological characteristics of symbionts sampled from the two E. scolopes collection sites. Statistically significantly different frequencies between the two collection sites were found for VfRep type, bioluminescence level, motility rate, colony pigmentation, and growth rate; however, the siderophore production frequencies were not statistically significantly different (Table 2). The frequencies of all characteristics were not found to be statistically significantly different when bacteria collected from the same site but on different nights were similarly compared (see Table S3 in the supplemental material), suggesting that the variation in the frequencies of phenotypic traits and the genetic fingerprints at either collection site was less than the variation between the two collection sites. The presence of statistically significant differences in phenotypes between the two collection sites (i.e., growth rate), differences that were not associated with other phenotypes, indicates that there is some degree of genetic variation in the population that is not detected by the VfRep assay.
TABLE 2.
Statistical analyses of frequency differences of various genetic and phenotypic traits of V. fischeri isolates collected from different host populations on Oahu in 2005
| Test and trait | χ2 | t | df | U | % of cells <5b | P valuea | Fisher's exact test P valuea |
|---|---|---|---|---|---|---|---|
| Chi-square test | |||||||
| VfRep type | 20.4 | 2 | 17 | **** (****) | **** (****) | ||
| Bioluminescence level | 5.5 | 1 | 0 | *** (***) | *** (***) | ||
| Colony pigmentation | 6.7 | 1 | 0 | **** (***) | *** (***) | ||
| Mann-Whitney-Wilcoxon test | |||||||
| Motility rate | 776 | **** (****) | |||||
| Siderophore production | 451 | * (*) | |||||
| Welch t test, growth rate | 2.46 | 50.75 | *** (***) |
The correction of P values for multiple testings was performed by considering each test as a family of tests. The P values were analyzed and adjusted according to the method of Hommel (31); corrected values are in parentheses. Ranges of calculated P values: *, P > 0.1; **, 0.1 > P > 0.05; ***, 0.01 < P < 0.05; ****, P < 0.01.
The percentage of cells <5 refers to a common benchmark for the validity of χ2 analysis.
One or two V. fischeri cells initially enter each light-organ crypt during the colonization of juvenile E. scolopes squid.
The intrinsic morphology of newly hatched juveniles, as well as the ecology of V. fischeri in Hawaiian seawater, led us to hypothesize that the initiation of crypt colonization by only a few cells could give rise to homogenous symbiont populations. Multiple clones within a light organ lobe were detected (Fig. 3B). However, each lobe consists of three crypts (Fig. 1), which, if occupied by different homogenous populations, could create the overall heterogenous population structure observed in many lobes (Fig. 5A).
To determine the number of bacterial cells that typically initiate symbiosis in individual crypts, we created a simple, deterministic model predicting the probability of observing mixed or homogenous populations of symbionts in a crypt sample based on the initial proportions of two isogenic strains and the number of initiating cells. A 12-h exposure to an inoculum consisting of cells of a single strain, labeled with either a red or a green fluorescent protein-expressing plasmid, was used to initiate the colonization of juvenile squids. After 48 h, the percentages of mixed- and single-color crypts were observed by confocal microscopy. As has been previously noted (72), due to the order in which the crypts mature, symbionts were more frequently observed in crypts 1 and 2 than in crypt 3. In 40 separate experiments, with over 900 crypts analyzed, approximately 21% (±17%) of the crypts contained a bacterial population expressing both red and green fluorescence whereas 79% (±17%) of the crypts contained a bacterial population expressing a single color (Fig. 6). A comparison of empirical and modeling results predicted that fewer than two cells were responsible for initiating symbiosis in each crypt (see Table S4 in the supplemental material). When, instead, a 3-h inoculation or a 10-fold range of bacterial inoculum concentrations was used, no significant difference in the mean number of initiating cells was found (Welch t test, df = 36.5, P value = 0.62) and no correlation between the inoculation concentration (up to 100,000 CFU/ml) and the initiating cell number could be demonstrated (see Fig. S1 in the supplemental material) (data not shown). We conclude that the number of bacterial cells colonizing a crypt is not significantly influenced by the duration of exposure or the concentration of V. fischeri in the ambient environment.
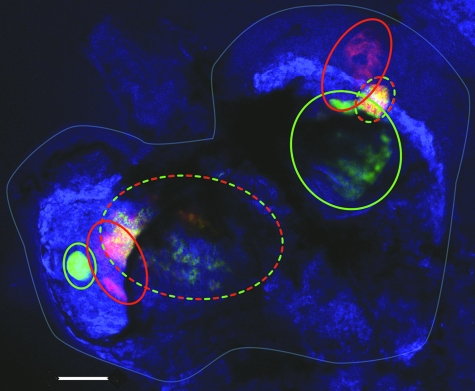
FIG. 6.
Confocal micrograph of fluorescently labeled V. fischeri cells colonizing the light organ of a juvenile E. scolopes squid. A 1:1 ratio of red and green fluorescent protein-expressing cells of V. fischeri strain MB13B2 was incubated with freshly hatched juvenile E. scolopes for 12 h at a total concentration of 4,500 CFU/ml. After an additional 36 h of symbiotic development, light organs were fixed and the animal tissue was stained (blue) and observed for the presence of mixed or single-color bacterial populations in each morphologically distinguishable crypt. The entire light organ (light blue outline) contains six crypts outlined with colored ovals. Two (33%) of the six crypts were cocolonized (yellow with a red and green dashed outline), while four (67%) were colonized by a single strain (either red or green and outlined with the corresponding solid color). Bar = 100 μm.
DISCUSSION
Naturally occurring populations of symbiotic bioluminescent bacteria provide at least two complementary opportunities for studying the principles governing host-microbe evolution and ecology. First, binary symbioses are a good model of the process(es) of metazoan-bacterial coevolution. Bioluminescent symbioses have provided evidence of species specificity (29), with a documented exception in one (20), and possibly another (27), host genus; species specificity and cocladogenesis continue to be subjects of vigorous investigation (16, 33).
Such symbioses also provide an opportunity to study microbial populations in environments that are both biologically natural and experimentally tractable. Specifically, bioluminescent microbial populations composed of culturable bacteria existing within defined boundaries for a known duration are well suited for studies that combine an analysis of ecological scale (e.g., temporal, morphological, and geographical) with a description of evolutionary processes (e.g., selection and recombination) that can create intraspecific differences within the population. In this study, we focused on two goals: (i) to define symbiont population structure at unstudied morphological and geographical scales and (ii) to connect the observed intraspecific diversity of symbiont populations of adult hosts with the process of initial colonization.
V. fischeri population diversity in adult host light organs and populations.
Although there is good evidence that V. fischeri is the only symbiont in the E. scolopes light organ (4), we sought to confirm the identity of a larger sample of light-organ isolates. To this end, we created a fast and simple PCR-based molecular assay to determine whether a DNA sample reliably identifies an isolate as V. fischeri. This idea is similar to previously reported species-specific PCR identification techniques (44), but we used a V. fischeri-specific hvnC PCR assay that discriminates this species from a number of other members of the family Vibrionaceae. Our results confirmed that all of the culturable isolates from light-organ populations of E. scolopes are V. fischeri (4) and that this species is found at a high concentration only in the host light organ.
Next, we asked whether the genetic and phenotypic characteristics of individual V. fischeri cells present in light-organ populations provide evidence of a clonal origin of these populations. In fact, adult light-organ populations are often polyclonal, dominated by one to three genetically distinct strains of V. fischeri. Interestingly, these results are similar to those reported for symbiont populations of leiognathid fishes (17, 61). Each squid light organ contained phenotypically distinct isolates that exhibited differences in bioluminescence level, motility rate, colony pigmentation, and other traits. Upon further analysis, distinct phenotypic combinations were found to be significantly associated with particular genetic signatures (Table 1). This association between genetic and phenotypic characteristics supports the existence of intraspecific differences among V. fischeri strains within the same light organ.
A novel extension of our work is the demonstration that the intraspecific variation within light-organ populations is intimately related to host population biology. Specifically, the frequencies of specific phenotypically and genetically distinct isolates were found to be statistically significantly different when strains from Kaneohe Bay hosts were compared to those from Maunalua Bay hosts (Table 1). We conclude that the intraisland geographic scale is relevant to observed intraspecific differences among symbiotic V. fischeri populations.
Our results also revealed that intraspecific differences within a given symbiont population may reflect the morphological organization of the light organ. Taken as a whole, the adult light organ population is polyclonal, harboring at least two or three different V. fischeri strains, yet each light organ is composed of two bilaterally symmetrical lobes and each lobe contains three crypts (Fig. 1). In the newly hatched juvenile, each crypt is a blind-ended sack connected to the environment through a single pore and each pore is the site of potentially independent colonization by ambient V. fischeri cells (56).
Development of population diversity in the light organ.
We hypothesized that the polyclonal population of cells in the light organ is composed of several clonal populations of V. fischeri that arose from a single cell or a small number of cells entering each crypt during symbiosis initiation (18). To indirectly test this hypothesis, we constructed a simple deterministic model of symbiosis initiation and empirically determined the number of cells colonizing the different crypts of a juvenile host. We concluded that individual crypts are initially colonized by, at most, two individual V. fischeri cells. This result is robust over more than an order of magnitude of V. fischeri abundance, as well as over short and long periods of inoculation (see Fig. S1 and Table S4 in the supplemental material) and is consistent with a prior experimental study that used a simple culturing approach to detect between one and three different strains colonizing an individual juvenile light organ (48). To further test the hypothesis that the observed low number of cells initiating crypt colonization leads to generally clonal crypt populations in the E. scolopes-V. fischeri symbiosis, we have begun extending the studies in this work to observations of the development of nonisogenic infections over the full lifetime of the host.
The theme of clonal bacterial symbiont populations at highly resolved morphological scales but polyclonal symbiont population structure at the scale of the individual host is not unique. Both the rhizobium-legume (23, 66) and Xenorhabdus nematophila-Steinernema carpocapsae nematode (47) symbioses can exhibit clonal symbiont population structure at the highly resolved scales of individual nodules and intestinal vesicles, respectively. In the former associations, an individual plant may host many different symbiotic strains although its individual root nodules can be clonal (28, 49). In contrast, the nematode hosts have only a single intestinal vesicle (24).
Stability of polyclonal population structure in symbioses.
This work provides evidence that a sepiolid squid light organ, like the roots of a legume, can be colonized by polyclonal symbiont populations. Such a population structure in symbiosis presents a challenge to preexisting evolutionary theory. Briefly, host-microbe associations in which the symbionts are horizontally passed and develop as polyclonal populations are predicted and observed to be amenable to intraspecific competition. This competition may occur at the expense of host fitness and symbiotic stability, leading to increased virulence in the symbiont and/or increased parasitism and decreased mutualism in the relationship (22, 30). Competition among clonal lineages may exist at different levels of host organization. Polyclonal V. fischeri populations may be in conflict within each crypt, among different crypts in a single lobe, or between lobes in a single squid since all of the light-organ symbionts of a single squid compete for the same shared resource. The observation of polyclonal population structure in both the rhizobium-legume and the luminous-bacterium-sepiolid squid symbioses begs the question: what maintains evolutionarily successful, mutualistic relationships in the face of a supposedly destabilizing polyclonal symbiont population structure and transmission mode (12, 65)?
In the rhizobium-legume system, this question has been approached under a group of evolutionary models generally called “partner choice” (7). Specifically, it has been hypothesized that uncooperative strains of rhizobia are sanctioned by the host while cooperative strains are favored (11). Both experimental and empirical evidence supports this hypothesis: Bradyrhizobium japonicum isolates from nodules of dwarf soybean containing experimentally created “noncooperative” strains have lower reproductive success (36, 37), and small nodule size is associated with the presence of less cooperative strains of rhizobia in the wild legume Lupinus arboreus (66).
The role of partner choice in the luminous-bacterium-sepiolid squid symbiosis remains an open question. Preliminary evidence of different initiation efficacies and population sizes in juvenile hosts among sympatric symbiont strains exists (62). Furthermore, one natural strain may outcompete another, sympatric, natural strain in the juvenile host light organ (41). To date, however, there has been no work explicitly defining cooperation in this system and addressing the host's response to cooperating or noncooperating symbionts.
Another common class of models explaining the persistence of mutualism in the presence of horizontal transmission, termed “partner fidelity,” remains unstudied in the luminous-bacterium-sepiolid squid symbioses. Generally, partner fidelity refers to the theory that repeated interaction of symbiotic partners and adjustment of contemporary interactions based on the outcome of previous interactions promote a mutualistic relationship (1). Under models of partner fidelity, spatial structuring of populations has been found to stabilize mutualistic associations (14). In this study, we have found significant differences between host-associated populations of V. fischeri at two locations on Oahu, which might indicate local adaptation to the host and, through association, localized population structuring of the symbiosis on Oahu. Another study has presented evidence that V. fischeri symbionts of sepiolid squids are locally adapted to their hosts at the continental scale (53). Both of these studies will preface future research defining what, if any, role spatial structuring plays in stabilizing this mutualism. Evolutionary models such as partner choice or partner fidelity will provide a valuable conceptual framework under which the stability of the polyclonal structure of the luminous-bacterium-sepiolid squid mutualism may be studied. In addition, this binary symbiosis, delicately balanced between natural system and experimental model, will help us understand the specific population biology of a symbiotic bioluminescent marine bacterium, as well as continue to provide insight into the evolutionary ecology of microbe-metazoan symbioses in general.
Supplementary Material
Acknowledgments
We thank C. Brennan, K. Budsberg, A. Charkowski, H. Goodrich-Blair, J. Graber, A. Johnson, M. Mandel, M. McFall-Ngai, T. McMahon, N. Perna, A. Pollack, A. Schaefer, S. Studer, and Y. Wang for discussing this research and the resulting manuscript. J. Troll and M. Altura provided helpful advice with confocal microscopy. R. Gates provided support and laboratory resources at the Hawaiian Institute for Marine Biology during collection. C. Wimpee and E. O'Grady provided advice and suggestions for molecular fingerprinting methodology. E. Caldera suggested binning the fingerprinting data, to the improvement of the manuscript.
This work was supported by two awards to M.S.W., an NSF Graduate Research Fellowship and an NIH Molecular Biosciences Training Grant through UW—Madison. Additional research support was provided by NIH RR12294 to E.G.R. and M. McFall-Ngai and NSF IOB 0517007 to M. McFall-Ngai and E.G.R.
Footnotes
Published ahead of print on 7 November 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Axelrod, R., and W. D. Hamilton. 1981. The evolution of cooperation. Science 211:1390-1396. [DOI] [PubMed] [Google Scholar]
- 2.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloodgood, R. A. 1977. The squid accessory nidamental gland: ultrastructure and association with bacteria. Tissue Cell 9:197-208. [DOI] [PubMed] [Google Scholar]
- 4.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boettcher, K. J., and E. G. Ruby. 1994. Occurrence of plasmid DNA in the sepiolid squid symbiont Vibrio fischeri. Curr. Microbiol. 29:279-286. [Google Scholar]
- 6.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York, NY.
- 7.Bull, J. J., and W. R. Rice. 1991. Distinguishing mechanisms for the evolution of cooperation. J. Theor. Biol. 149:63-74. [DOI] [PubMed] [Google Scholar]
- 8.Colwell, R. K. 2007. EstimateS: statistical estimation of species richness and shared species from samples. http://viceroy.eeb.uconn.edu/estimates.
- 9.Colwell, R. K., C. X. Mao, and J. Chang. 2004. Interpolating, extrapolating, and comparing incidence-based species accumulation curves. Ecology 85:2717-2727. [Google Scholar]
- 10.Cramér, H. 1946. Mathematical methods of statistics. Princeton University Press, Princeton, NJ.
- 11.Denison, R. F. 2000. Legume sanctions and the evolution of symbiotic cooperation by rhizobia. Am. Nat. 156:567-576. [DOI] [PubMed] [Google Scholar]
- 12.Denison, R. F., and E. T. Kiers. 2004. Why are most rhizobia beneficial to their plant hosts, rather than parasitic? Microbes Infect. 6:1235-1239. [DOI] [PubMed] [Google Scholar]
- 13.Denoeud, F., and G. Vergnaud. 2004. Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a web-based resource. BMC Bioinformatics 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doebeli, M., and N. Knowlton. 1998. The evolution of interspecific mutualisms. Proc. Natl. Acad. Sci. USA 95:8676-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunlap, P. V. 1989. Regulation of luminescence by cyclic AMP in cya-like and crp-like mutants of Vibrio fischeri. J. Bacteriol. 171:1199-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunlap, P. V., J. C. Ast, S. Kimura, A. Fukui, T. Yoshino, and H. Endo. 2007. Phylogenetic analysis of host-symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 23:507-532. [Google Scholar]
- 17.Dunlap, P. V., A. Jiemjit, J. C. Ast, M. M. Pearce, R. R. Marques, and C. R. Lavilla-Pitogo. 2004. Genomic polymorphism in symbiotic populations of Photobacterium leiognathi. Environ. Microbiol. 6:145-158. [DOI] [PubMed] [Google Scholar]
- 18.Dunn, A. K., D. S. Millikan, D. M. Adin, J. L. Bose, and E. V. Stabb. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein, J. 2007. PHYLIP (Phylogeny Inference Package) v3.66. http://evolution.genetics.washington.edu/phylip.html.
- 20.Fidopiastis, P. M., S. von Boletzky, and E. G. Ruby. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox, J. 2005. The R Commander: a basic-statistics graphical user interface to R. J. Stat. Software 14:1-42. [Google Scholar]
- 22.Frank, S. A. 1996. Host-symbiont conflict over the mixing of symbiotic lineages. Proc. Biol. Sci. 263:339-344. [DOI] [PubMed] [Google Scholar]
- 23.Gage, D. J. 2002. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 184:7042-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodrich-Blair, H., and D. J. Clarke. 2007. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol. Microbiol. 64:260-268. [DOI] [PubMed] [Google Scholar]
- 25.Gotelli, N., and R. K. Colwell. 2001. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4:379-391. [Google Scholar]
- 26.Graf, J., and E. G. Ruby. 2000. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol. Microbiol. 37:168-179. [DOI] [PubMed] [Google Scholar]
- 27.Guerrero-Ferreira, R. C., and M. K. Nishiguchi. 2007. Biodiversity among luminescent symbionts from squid of the genera Uroteuthis, Loliolus and Euprymna (Mollusca: Cephalopoda). Cladistics 23:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagen, M. J., and J. L. Hamrick. 1996. Population level processes in Rhizobium leguminosarum bv. trifolii: the role of founder effects. Mol. Ecol. 5:707-714. [DOI] [PubMed] [Google Scholar]
- 29.Hastings, J. W., and K. H. Nealson. 1981. The symbiotic luminous bacteria, p. 1332-1345. In M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes, vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 30.Herre, E. A. 1993. Population structure and the evolution of virulence in nematode parasites of fig wasps. Science 259:1442-1445. [DOI] [PubMed] [Google Scholar]
- 31.Hommel, G. 1988. A stagewise rejective multiple test procedure based on a modified Bonferroni test. Biometrika 75:383-386. [Google Scholar]
- 32.Hulton, C. S., C. F. Higgins, and P. M. Sharp. 1991. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol. Microbiol. 5:825-834. [DOI] [PubMed] [Google Scholar]
- 33.Jones, B. W., J. E. Lopez, J. Huttenburg, and M. K. Nishiguchi. 2006. Population structure between environmentally transmitted vibrios and bobtail squids using nested clade analysis. Mol. Ecol. 15:4317-4329. [DOI] [PubMed] [Google Scholar]
- 34.Jones, B. W., A. Maruyama, C. C. Ouverney, and M. K. Nishiguchi. 2007. Spatial and temporal distribution of the Vibrionaceae in coastal waters of Hawaii, Australia, and France. Microb. Ecol. 54:314-323. [DOI] [PubMed] [Google Scholar]
- 35.Kaufman, L., and P. J. Rousseeuw. 1990. Finding groups in data: an introduction to cluster analysis. J. Wiley, Inc., New York, NY.
- 36.Kiers, E. T., R. A. Rosseau, and R. F. Denison. 2006. Measured sanctions: legume hosts detect quantitative variation in rhizobium cooperation and punish accordingly. Evol. Ecol. Res. 8:1077-1086. [Google Scholar]
- 37.Kiers, E. T., R. A. Rousseau, S. A. West, and R. F. Denison. 2003. Host sanctions and the legume-rhizobium mutualism. Nature 425:78-81. [DOI] [PubMed] [Google Scholar]
- 38.Kimbell, J. R., M. J. McFall-Ngai, and G. K. Roderick. 2002. Two genetically distinct populations of bobtail squid, Euprymna scolopes, exist on the island of Oahu. Pacific Sci. 56:347-355. [Google Scholar]
- 39.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. Wiley, Chichester, United Kingdom.
- 40.Lee, K. H. 1994. Ecology of Vibrio fischeri, the light organ symbiont of the Hawaiian sepiolid squid Euprymna scolopes. Ph.D. dissertation. University of Southern California, Los Angeles.
- 41.Lee, K. H., and E. G. Ruby. 1994. Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J. Bacteriol. 176:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, K.-H., and E. G. Ruby. 1992. Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl. Environ. Microbiol. 58:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, K. H., and E. G. Ruby. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, S. E., S. Y. Kim, S. J. Kim, H. S. Kim, J. H. Shin, S. H. Choi, S. S. Chung, and J. H. Rhee. 1998. Direct identification of Vibrio vulnificus in clinical specimens by nested PCR. J. Clin. Microbiol. 36:2887-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319-331. [DOI] [PubMed] [Google Scholar]
- 46.Margulis, L., and R. Fester. 1991. Symbiosis as a source of evolutionary innovation. MIT Press, Cambridge, MA. [PubMed]
- 47.Martens, E. C., K. Heungens, and H. Goodrich-Blair. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J. Bacteriol. 185:3147-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCann, J., E. V. Stabb, D. S. Millikan, and E. G. Ruby. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 69:5928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McInnes, A., J. E. Thiesb, L. K. Abbotta, and J. G. Howieson. 2004. Structure and diversity among rhizobial strains, populations and communities—a review. Soil Biol. Biochem. 36:1295-1308. [Google Scholar]
- 50.Montgomery, M. K., and M. J. McFall-Ngai. 1990. The anatomy and morphology of the adult bacterial light organ of Euprymna scolopes Berry (Cephalopoda: Sepiolidae). Biol. Bull. 179:332-339. [DOI] [PubMed] [Google Scholar]
- 51.Montgomery, M. K., and M. J. McFall-Ngai. 1998. Late postembryonic development of the symbiotic light organ of Euprymna scolopes (Cephalopoda: Sepiolidae). Biol. Bull. 195:326-336. [DOI] [PubMed] [Google Scholar]
- 52.Nealson, K. H., and J. W. Hastings. 1991. Chapter 25. The luminous bacteria, p. 625-639. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, vol. 1, 2nd ed. Springer-Verlag, New York, NY.
- 53.Nishiguchi, M. K. 2002. Host-symbiont recognition in the environmentally transmitted sepiolid squid-Vibrio mutualism. Microb. Ecol. 44:10-18. [DOI] [PubMed] [Google Scholar]
- 54.Nishiguchi, M. K., and V. S. Nair. 2003. Evolution of symbiosis in the Vibronaceae: a combined approach using molecules and physiology. Int. J. Syst. Evol. Microbiol. 53:2019-2026. [DOI] [PubMed] [Google Scholar]
- 55.Nishiguchi, M. K., E. G. Ruby, and M. J. McFall-Ngai. 1998. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-Vibrio symbioses. Appl. Environ. Microbiol. 64:3209-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nyholm, S. V., and M. J. McFall-Ngai. 2004. The winnowing: establishing the squid-Vibrio symbiosis. Nat. Rev. Microbiol. 2:632-642. [DOI] [PubMed] [Google Scholar]
- 57.Ogunseitan, O. 2005. Microbial diversity. Blackwell Publishing, Oxford, United Kingdom.
- 58.Rademaker, J. L. W., F. J. Louws, and F. J. de Bruijn. 1998. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, v3.4.3. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 59.Reich, K. A., and G. K. Schoolnik. 1996. Halovibrin, secreted from the light organ symbiont Vibrio fischeri, is a member of a new class of ADP-ribosyltransferases. J. Bacteriol. 178:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichelt, J. L., and P. Baumann. 1973. Taxonomy of the marine, luminous bacteria. Arch. Mikrobiol. 94:283-330. [DOI] [PubMed] [Google Scholar]
- 61.Reichelt, J. L., K. H. Nealson, and J. W. Hastings. 1977. The specificity of symbiosis: pony fish and luminescent bacteria. Arch. Microbiol. 112:157-161. [Google Scholar]
- 62.Ruby, E. G., and K. H. Lee. 1998. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl. Environ. Microbiol. 64:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siegel, S., and N. J. Castellan, Jr. 1988. Nonparametric statistics for the behavioral sciences, 2nd ed. McGraw-Hill Book Co., New York, NY.
- 65.Simms, E. L., and D. L. Taylor. 2002. Partner choice in nitrogen-fixation mutualisms of legumes and rhizobia. Int. Comp. Biol. 42:369-380. [DOI] [PubMed] [Google Scholar]
- 66.Simms, E. L., D. L. Taylor, J. Povich, R. P. Shefferson, J. L. Sachs, M. Urbina, and Y. Tausczik. 2006. An empirical test of partner choice mechanisms in a wild legume-rhizobium interaction. Proc. Biol. Sci. 273:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sneath, P. K. H., and R. R. Sokal. 1973. Numerical taxonomy: the principles and practice of numerical classification. Freeman Press, San Francisco, CA.
- 68.Spratt, B. G., J. T. Staley, and M. C. Fisher. 2006. Introduction: species and speciation in microorganisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1897-1898. [Google Scholar]
- 69.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 71.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Inc., Sunderland, MA.
- 72.Sycuro, L. K., E. G. Ruby, and M. McFall-Ngai. 2006. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J. Morphol. 267:555-568. [DOI] [PubMed] [Google Scholar]
- 73.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Versalovic, J., M. Schneider, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based PCR (rep-PCR). Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 75.Wallick, H., and C. A. Stuart. 1943. Antigenic relationships of Escherichia coli isolated from one individual. J. Bacteriol. 45:121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ward, J. H. 1963. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58:236-244. [Google Scholar]
- 77.Wei, S. L., and R. E. Young. 1989. Development of symbiotic bioluminescence in a nearshore cephalopod, Euprymna scolopes. Mar. Biol. 103:541-546. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.