Abstract
The abundances and activities of aerobic methane-oxidizing bacteria (MOB) were compared in depth profiles of littoral and profundal sediments of Lake Constance, Germany. Abundances were determined by quantitative PCR (qPCR) targeting the pmoA gene and by fluorescence in situ hybridization (FISH), and data were compared to methane oxidation rates calculated from high-resolution concentration profiles. qPCR using type I MOB-specific pmoA primers indicated that type I MOB represented a major proportion in both sediments at all depths. FISH indicated that in both sediments, type I MOB outnumbered type II MOB at least fourfold. Results obtained with both techniques indicated that in the littoral sediment, the highest numbers of methanotrophs were found at a depth of 2 to 3 cm, corresponding to the zone of highest methane oxidation activity, although no oxygen could be detected in this zone. In the profundal sediment, highest methane oxidation activities were found at a depth of 1 to 2 cm, while MOB abundance decreased gradually with sediment depth. In both sediments, MOB were also present at high numbers in deeper sediment layers where no methane oxidation activity could be observed.
Methane is formed by natural processes and by anthropogenic processes and is 25 times more effective as a greenhouse gas than carbon dioxide (18). Methane emissions from lakes contribute about 6 to 16% of the total nonanthropogenic global methane emission (4). Microbial methane oxidation is an important process for prevention of the escape of the methane produced in anoxic sediment layers (4, 61) to the atmosphere and ultimately controls global warming.
In mesotrophic or oligotrophic lakes that are oxic down to the sediment surface, aerobic oxidation of methane occurs at the sediment-water interface (35). Methane oxidation in freshwater lakes was considered to be an exclusively aerobic process (24, 34), but recently, anaerobic oxidation of methane in Lake Plußsee has been reported (21). In Lake Constance, a large, deep, oligotrophic lake, methane production and oxidation processes have been studied intensively in the past (7, 58). Recently, a diffusion methane sensor was used to measure methane profiles at millimeter-level resolution both in littoral sediments and in profundal sediments of Lake Constance (14). The sensor consists of a steel cannula with small openings, which are covered by thin silicone tubing. Methane diffuses into the cannula and is flushed directly to a flame ionization detector for quantification.
Aerobic methane-oxidizing bacteria (MOB) have been classified as type I and type II methanotrophs based on their phylogenetic position, carbon assimilation pathways, and the arrangement of intracellular membranes, and they belong to the classes Gammaproteobacteria and Alphaproteobacteria, respectively (9). Very recently, Verrucomicrobium-like bacteria have also been reported to oxidize methane in acidic environments (28, 44). MOB from natural ecosystems such as soil, rice paddies, freshwater marshes, and lakes have been quantified by cultivation methods like the most-probable-number method (MPN) (12, 21) and gradient cultivation (13) and by culture-independent techniques such as Southern hybridization with probes for the pmoA or the mmoX gene (3, 17), phospholipid fatty acid profiles (60), methane oxidation rates (37), fluorescence in-situ hybridization (FISH) (15, 22), and quantitative real-time PCR (qPCR) (32). Using quantitative cultivation of methanotrophs in the littoral sediment of Lake Constance, we determined 104 cells per ml by the MPN method in microtiter plates (12) and up to 105 cells per ml by gradient cultivation (13).
Littoral sediments differ from profundal sediments by their exposure to daily light/dark cycles (26), their higher content of organics, and the frequency of disturbances by either bioturbation or sediment resuspension (62). We therefore expected major differences in community structures (49) and abundances of methanotrophs in these two different compartments of the lake.
In the present study, we determined methanotrophic abundance at high spatial resolution by using two independent molecular methods, qPCR targeting the pmoA gene and FISH targeting 16S rRNA. The abundance of MOB was correlated with the rates of methane oxidation calculated from high-resolution profiles of methane concentrations in the littoral and profundal sediments of Lake Constance.
MATERIALS AND METHODS
Sediment sampling.
Littoral sediment samples were collected by scuba diving from the lower infralittoral zone (“Litoralgarten”) of Lake Constance at a water depth of 2 to 5 m. Profundal sediment was collected with a ship-borne multicorer from a depth of 80 m in the “Überlinger See.” Littoral and profundal sediment cores were collected in late winter (February and April 2007) and were taken to the laboratory within 0.5 to 3 h. To simulate in situ conditions, cores were kept in a water bath at in situ temperature and their surface was flushed continuously with aerated lake water. Water temperatures were 5°C for the profundal core in February 2007 and 8°C for the littoral core in April 2007. The methane and oxygen sensors were calibrated before sediment sampling, and measurements started within 30 min after arrival.
Density and porosity of the sediment samples were determined by drying 0.5-cm slices of sediment for 2 days at 70°C (data determined as wet weight and dry weight), followed by volume determination of the dried sediment in 50-ml volumetric flasks (according to the method described at http://www.ifm-geomar.de/index.php?id = mg_dichtebestimmung). The littoral sediment, with a porosity of 0.54 to 0.62, consisted of fine sand. The profundal sediment, with a porosity of 0.85 to 0.89, consisted of fine-grained material and clay.
Microsensor profiles and methane oxidation activities.
Upon retrieval of the sediment cores, three to five oxygen profiles were measured with a Clark-type microelectrode (Ox-50; Unisense, Denmark). The oxygen sensor was two-point calibrated in air-saturated water and in anoxic sediment. The detection limit was 0.3 μmol liter−1. Molar concentrations of oxygen were calculated according to the methods used in reference 25. The sensor was mounted on a micromanipulator and was moved into the sediment at 0.5-mm steps.
Methane profiles of high spatial resolution were determined with a diffusion-based microsensor (14). For calibration, three methane standard solutions were prepared with glass beads to mimic sediment diffusivity (14). Standard solutions were incubated in a water bath at the respective in situ temperatures. The aerated water bath was taken as a zero-methane standard. Additional standards contained 3, 44, and 93 μM of methane. The detection limit was 2 μmol liter−1. The relative accuracy of the sensor was ±15% with a precision of ±7.5%. Methane in sediment cores was measured in three parallel profiles at 2-mm intervals to a depth of 6 cm.
Depth profiles of methane oxidation and production were calculated by two methods. One method used was a computer-implemented diffusion-reaction model (5). In a first step, the best-fitting concentration profile was calculated. In the next step, the simplest production-consumption profile that reproduces the concentration profile was chosen. The other method was an application of Fick's second law of diffusion to the best-fitting concentration profile. The calculated activities were then smoothed by a running average. Sediment diffusivity (Ds) was determined by the equation Ds = Φ2 × D, where Φ is porosity and D is the diffusivity of methane in free water. The methane diffusion coefficient ranged from 1.13 × 10−5 cm2 s−1 at 5°C to 1.25 10−5 cm2 s−1 at 8°C, for profundal and littoral sediments, respectively (gas tables from Unisense, Denmark). The in situ biodiffusivities for profundal and littoral sediments were calculated by multiplying Db (biodiffusion coefficient for each organism group) by the average in situ density of the respective group (39).
Sample preservation and DNA isolation.
After the methane and oxygen profiles were measured, the uppermost 5-cm parts of the sediment cores were cut into 0.5-cm slices and stored at −20°C. DNA was extracted from 300 to 400 mg wet sediment by using the Fast DNA spin kit for soil (MP Biomedicals Germany). The final concentration of the diluted DNA was determined by the Sybr green quantification method (with Sybr green I; Cambrex Bioscience, Maine) (63).
PCR and qPCR.
Trial qPCR assays, namely, MTOT, MBAC and TYPEII, as described in reference 32, were run using DNA from littoral and profundal sediments as templates. The MTOT assay (with the A189f-mb661r primer set [2]) has been designed to quantify the pmoA gene as a target for all methanotrophs (32). For this assay, a plasmid carrying the pmoA clone (littoral site 1, clone 12, NCBI accession number DQ235460) from the pmoA clone library of DNA found from the Lake Constance littoral sediment was used as a standard (49). For exact quantification, the concentration of the plasmid was determined by the Sybr green quantification method. pmoA target molecules per ng of DNA were calculated assuming a molecular mass of 660 Da per DNA base pair (23). A dilution series with 10-fold dilution steps resulting in 101 to 107 target molecules of DNA μl−1 was used as standards.
Usually a small amount (i.e., 1 to 2 ng) of sediment DNA was used in a 20-μl PCR mixture, to avoid any possible effects of PCR inhibitors in the sediment DNA. Power Sybr green qPCR kit (Applied Biosystems) was used, and the qPCR was performed in an ABI-7500 instrument (Applied Biosystems). The reaction mixture consisted of 10 μl of the master mix and 10 pmol of each primer in a final 20-μl reaction mixture. Melting curve analyses were performed with samples and standard assays from which the data acquisition temperature was calculated. The data acquisition temperature is the temperature above the melting temperature of the primer dimers and was determined to be 77.5°C for the MTOT assay. The qPCR program for the MTOT assay was modified as follows: 94°C for 15 s, 56°C for 30 s, and 60°C for 30 s, and data acquisition at 77.5°C for 34 s for 40 cycles followed by denaturation. All standards and samples were used in triplicate. Standard graphs of threshold cycle were plotted against the logarithm of the copy number. The copy numbers of the samples were calculated with the help of 7500 system SDS software (Applied Biosystems) or with Microsoft Excel.
The MBAC assay targeting the Methylobacter/Methylosarcina group was found not to amplify all the pmoA sequences retrieved from Lake Constance (13). Therefore, a new qPCR assay was developed to detect the abundance of type I MOB in Lake Constance (LC type I assay) by designing a reverse degenerate primer, LC Type I r (5′TTCTDACRTAGTGGTAACC3′), to cover the detected pmoA diversity of type I MOB from Lake Constance (13). The specificity of the reverse primers was checked by performing a BLAST search at the NCBI site (http://www.ncbi.nlm.nih.gov/) (1), using the MegAlign program in the DNASTAR software and using the ARB software package (version 2.5b; http://www.arb-home.de) (38). Annealing temperatures were determined by amplifying this particular region of the pmoA gene from a pmoA clone (littoral site 1, clone 12, accession number DQ235460) (49), for which only a single band of correct size was obtained and annealing and data acquisition temperatures of 54°C and 78°C, respectively, were determined. The LC type I assay was additionally validated by creating clone libraries from the sediment, using this primer set, and it was found that all clones belonged to type I methanotrophs.
With qPCR, we detected positive products of correct size with the MTOT assay (total methanotrophs), the MBAC assay (for Methylobacter/Methylosarcina type I methanotrophs) (32), and the LC type I assay (this study) but did not get any amplification for the type II MOB-specific assay at the annealing temperature mentioned in reference 32. Thus, only the MTOT assay and the LC type I assay were used further for quantifying the total pmoA genes and pmoA genes of type I methanotrophs, respectively.
For estimation of the bacterial 16S rRNA gene copy numbers, bacterial primers Eub338 and Eub518r (23) were used. The same program as described for the two PCRs described above was used except that the annealing temperature was 53°C, and a plasmid containing a 16S rRNA gene fragment was used as a standard after appropriate dilutions.
Finally, to compare the cell numbers obtained by FISH with those obtained by qPCR, pmoA copy numbers were divided by 2 (average copy number of pmoA in methanotrophs) (32), and for total bacteria, the copy numbers were divided by 4 (16).
Standard errors for qPCR were on average 0.1 × 107 bacteria per g of sediment (wet weight) for the MOB from the littoral sediment (in both the MTOT assay and the LC type I assay), and on average 0.7 × 107 bacteria per g of sediment (wet weight) for the MOB from the profundal sediment (in both the MTOT assay and the LC type I assay). The standard errors for the total bacterial assay for the littoral and profundal sediments were 0.1 × 109 and 0.2 × 109 per g of sediment (wet weight), respectively.
Extraction of cells from the sediment and FISH.
FISH was performed on samples collected in February and April 2007. Immediately after the sediment was sliced, samples of 240 to 820 mg (fresh weight) were fixed by the addition of formalin to a 4% final formaldehyde concentration and incubation at room temperature for at least 1 h or overnight at 4°C. Formaldehyde was removed by centrifugation at 10,000 × g for 2 min. The supernatant was removed, and 1 ml of 1× phosphate-buffered saline (PBS), 160 μl of Na-pyrophosphate (0.1 M), and 1 drop of Tween 80 was added to the pellet. The suspension was mixed vigorously for one minute and then incubated at room temperature for 30 min. After a further brief mixing, the samples were centrifuged for 2 min at 720 × g and the pellet was washed twice with 1 ml PBS in a similar way. All three supernatants were pooled and centrifuged for 10 min at 14,000 × g. The obtained cell pellet was resuspended in 100 μl of PBS-ethanol mix (1:1). After sonication for eight short intervals (in total, 10 s) at cycle 0.5/amplitude 50 (instrument settings), samples were stored at −20°C. Extraction efficiency was checked with three littoral sediment samples, three profundal sediment samples, and three stored fixed sediment samples. For an estimate of the extraction efficiency of our protocol, we counted the bacterial numbers after each extraction step. The third extraction step yielded less than 20% of the total number of extracted cells. Therefore a fourth extraction step was omitted. Sonication of the samples (15 intervals at cycle 0.5/amplitude 50) increased the total bacterial count by 20%, but the background fluorescence of the sediment increased dramatically to render counting, especially of profundal sediment samples, barely possible.
FISH was performed in 10-well microscopic slides (Roth, Germany) with 10 μl of sample that had been sonicated briefly once more, as described in reference 22, and stained with DAPI (4′,6-diamidino-2-phenylindole; 1 μg/ml). Hybridizations with probes for type I and type II methanotrophs were performed separately but combined with the Eub338 probe linked to fluorescein. Probes used for type I methanotrophs were Cy3-linked Mγ84 and Mγ705 (22) and Cy3-linked Mα450 for type II methanotrophs (22). All probes were purchased from ThermoHybaid (Germany).
Slides were dried, Citifluor antifading agent (Citifluor Ltd., United Kingdom) was added, and the slides were stored at −20°C until counting. Slides were observed with an Axiophot fluorescence microscope (Zeiss) with the filter sets suitable for observing DAPI, Cy3, and fluorescein and photographed with a cooled charge-coupled-device camera (Magnafire; INTAS). Only cells showing clear signals with all three excitation filters and fluorescing in the proper colors were counted as methanotrophs. For calculation of final numbers, we used a calculation similar to that used in reference 19 except that at minimum 35 fields of view were counted, and this number would correspond to 11,000 to 140,000 DAPI counts.
For DAPI, only five squares were counted. Cells were found evenly distributed on the slides. Because of the high background fluorescence of the sediment, only brightly fluorescing cells were counted. Bacterial cells hybridized with the Eub338 probe were not counted, because background fluorescence of sediment particles was too high for reliable counts with the corresponding filter to be obtained. However, differentiation between bacteria and inorganic particles was facilitated by comparison of three pictures with different filters to determine the MOB-specific counts.
RESULTS
Microsensor profiles and activities.
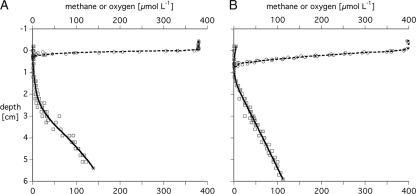
The distributions of oxygen and methane in littoral and profundal sediments were analyzed with microsensors at high vertical and horizontal resolutions. In the littoral sediment, the oxic zone was narrow, with oxygen penetrating down to a depth of 0.35 cm (Fig. 1A). In the profundal sediment, oxygen penetrated to a depth of 0.65 cm (Fig. 1B). The highest methane concentrations in the littoral sediment (140 μM) were measured at a depth of 5.5 cm. A steep decrease of methane was found at a depth of 2 to 3 cm, and concentrations were close to zero in the uppermost 1 cm. Methane determinations in different months during 2005 and 2006 showed similar profiles (data not shown). In the profundal sediment, a maximum of 113 μM of methane was measured at a depth of 6 cm (Fig. 1B), and there was a linear decrease toward zero in the top 1 cm. By performing additional measurements in the profundal sediment during 2005 and 2006, we observed comparable profiles as well (data not shown).
FIG. 1.
Concentration profiles of oxygen (circles) and methane (squares) in littoral (A) and profundal (B) sediments. Lines indicate the means of three oxygen measurements and the calculated best-fitting profile of methane concentration.
In order to localize and quantify the zones of methane consumption or methane production, we assumed steady-state conditions. We calculated the activities directly from the concentration gradients and additionally applied a model to the methane profiles (5).
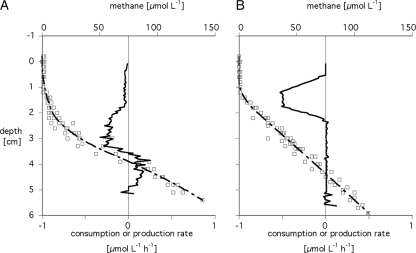
In the littoral sediment, methane was oxidized in the top 3.5 cm, with a zone of low activity (average, 0.04 μmol liter−1 h−1) in the top 1.6 cm (Fig. 2A) followed by a zone of higher activity (average, 0.18 μmol liter−1 h−1) from 1.6 to 3.5 cm. Below 3.5-cm sediment depth, methane production started. In total, 4.01 μmol methane m−2 h−1 was oxidized. Use of the model of Berg et al. (5) revealed the same depth zonation and similar activities (total oxidation rate, 3.4 μmol m−2 h−1).
FIG. 2.
Methane consumption and production rates (bold continuous line) calculated via Fick's second law of diffusion from the concentration profiles (dashed line) measured for littoral (A) and profundal (B) sediments. The individual data are plotted as squares.
In the profundal sediment, methane oxidation was restricted to the top 0.5 to 2.3 cm, with an average rate of 0.28 μmol liter−1 h−1 (Fig. 2B). The total methane oxidation rate was 5.74 μmol m−2 h−1. Within a depth of 2.3 to 5 cm, no notable activity of methane oxidation or production was observed. The methane production zone presumably started below the investigated depth. Use of the model of Berg et al. (5) revealed a much broader zone of methane oxidation, reaching from 0 cm to almost 3 cm. However, the overall methane oxidation rate was almost the same (5.76 μmol m−2 h−1).
For both the profundal sediment and the littoral sediment, we found the zone of methane oxidation extending 2 to 3.5 cm into the sediment, which is significantly deeper than the respective oxygen penetration depths (0.35 and 0.65 cm, respectively).
Abundance of methanotrophs.
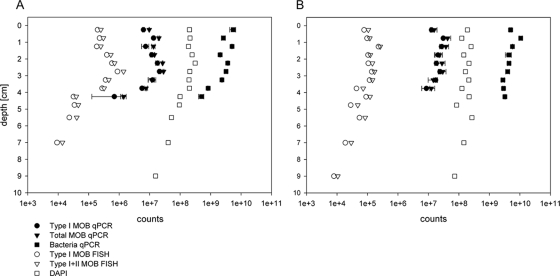
Quantification of MOB by qPCR of pmoA genes (MTOT assay) revealed that in the littoral sediment (Fig. 3A), the numbers of methanotrophs as determined by quantification of the pmoA copies in the littoral sediment were slightly lower in the top 0.5 cm (9.6 × 106 MOB per g of sediment [wet weight]) than in the sediment below. Their numbers increased with depth and were almost constant down to a depth of 3.5 cm. The highest numbers of MOB were found at a depth of 2 to 3 cm (average of 2.7 × 107 MOB per g of sediment [wet weight]). Below 3.5 cm, the pmoA copy number decreased. Type I MOB (in the LC type I assay) constituted a large proportion of total MOB in all layers. They contributed to 60 to 100%, and on average 70%, of the total methanotrophs in terms of copy numbers. In the zone of highest abundance, i.e., 2 to 3 cm, they contributed 70% of the total pmoA copies.
FIG. 3.
Abundance of methanotrophs in littoral (A) and profundal (B) sediments. MOB as quantified by FISH (cells per g of sediment [wet weight]) and by qPCR (cells per g of sediment [wet weight]). Type I MOB (determined with Mγ84 and Mγ705 for FISH and the LC type I assay for qPCR), total numbers of MOB (determined with Mγ84, Mγ705, and Mα450 for FISH and the MTOT assay for qPCR), and total bacterial numbers (determined by DAPI counts and 16S rRNA gene qPCR) are shown.
The detection of MOB in the littoral sediment with FISH also showed a clear maximum of MOB abundance at a depth of 2 to 3.5 cm (6 × 105 cells per g of sediment [wet weight]). Type II MOB were found in lower numbers (2 × 105 per g of sediment [wet weight]). In the upper 4 cm of the sediment, type I MOB were about four times more abundant than type II MOB. Both types were detected down to a depth of 8 cm. Type I methanotrophs were sometimes visible as pairs of cylindrical or elliptical cells. These cells were quite large (2 to 4 μm) compared to the other bacteria. The type II methanotrophs were mainly single coccoid cells.
According to qPCR analysis, the total number of bacteria in the littoral sediment was maximal in the top 0.5 to 1.5 cm, with 5 × 109 bacteria per g of sediment (wet weight) (Fig. 3A) and on an average 2.8 × 109 bacteria per g of sediment (wet weight) were counted at a depth of 4.5 cm. Counting the cells after DAPI staining revealed on average a similar distribution of cells in the top 4.5 cm but numbers lower by 1 order of magnitude (2.1 × 108 cells per g of sediment [wet weight]) (Fig. 3A). With both methods, the abundance of total bacteria decreased by 1 order of magnitude toward deeper sediment layers.
In the profundal sediment, pmoA qPCR revealed comparable, high MOB numbers at a depth of 0.5 to 1.5 cm, which were around 4 × 107 per g (wet weight) (Fig. 3B). Numbers decreased only slightly below a depth of 1.5 cm. Average numbers of 2 × 107 copies per g fresh sediment were observed at a sediment depth of 2 to 4 cm. Type I MOB (LC type I assay) represented a large proportion, i.e., 65 to 100%, and on average 81%, of the total methanotrophs.
Quantification of MOB with FISH probes revealed in profundal sediment maximal numbers between 1- and 1.5-cm depth. Both type I and type II MOB were detectable down to a depth of 10 cm, which was the deepest layer investigated. The ratio of type I MOB to type II MOB was similar to that observed in the littoral sediment. Total bacteria as detected by qPCR were almost constant from a depth of 0 to 4.0 cm, and on average, 4.7 × 109 cells per g of sediment (wet weight) were detected. Total MOB abundance in the uppermost 4 cm in the profundal sediment ([1.4 ± 0.6] × 105 cells per g of sediment [wet weight]) was lower by a factor of 4 to 5 than that in the littoral sediment ([5.3 ± 3.9] × 105 cells per g of sediment [wet weight]).
The total numbers of bacteria in the littoral and profundal sediments by qPCR were approximately (4.4 ± 1.1) × 109 and (5.0 ± 1.8) × 109 bacteria per g of sediment (wet weight), respectively. With DAPI staining, one-tenth as many bacteria were found in each of the two sediments, i.e., 2 × 108 cells per g of sediment (wet weight). For both sediments, plots of the ratios of total MOB (sum of type I and type II MOB) to DAPI counts (details not shown) corresponded well depthwise with the zones of methane oxidation. The highest ratios were found in the littoral sediment at a depth between 1.5 and 3.5 cm (zone of high methane oxidation activity) and a depth between 0.5 cm and 2.0 cm in the profundal sediment. According to qPCR results, MOB represented around 0.2 to 0.9% of the total bacteria in the littoral sediment and 0.3 to 0.7% in the profundal sediment. However, the ratios of total MOB to total bacteria when plotted against sediment depths showed a more or less unequal distribution (i.e., a zigzag line).
DISCUSSION
Concentration and activity profiles.
Methane consumption and production rates were calculated from high-resolution determinations of methane concentrations in sediment samples. In contrast to the case for previously used methods, the sediment cores were not destroyed and could be used for microbiological investigations afterwards. Additionally, the profile measurements lasted only approximately 3 h, and thus, incubation artifacts could be minimized.
We used two approaches to calculate the methane oxidation rate: direct application of Fick's second law of diffusion and model calculations (5). For littoral sediment, our calculated flux data (0.46 μmol m−2 h−1) compared well with modeled flux data (0.56 μmol m−2 h−1) and flux data calculated earlier on the basis of sediment core incubations (0.61 μmol m−2 h−1) (11). The results of the two approaches for the profundal sediment did not agree as well as the results for the littoral sediment. Although the overall methane oxidation rates were similar, the zonations were different. Direct calculation of the activity revealed a zone of methane oxidation activity which was much more consistent with the concentration profiles. From core incubations of profundal sediment of Lake Constance, a methane oxidation rate of 18.7 μmol m−2 h−1 has been calculated (24), and this rate is twice as high as the rate we calculated from our data (8.8 μmol m−2 h−1). However, these data were obtained later in the year and at a time when the phosphate content of Lake Constance water was still about six times higher than it is presently. Thus, our estimates of methane oxidation activities appear to represent realistic values for the in situ activity.
In the littoral sediment of Lake Constance, the group of Chironomidae is the most abundant macrofaunal group (41). In profundal sediments, the Tubificidae are the dominant infauna (54). Including biodiffusivity in the model did not change the zonation of methane oxidation rates in littoral and profundal sediments. Chironomids dwelling in the top 1 cm of littoral sediment (59) had only a minor influence on the overall methane oxidation rate. Tubificidae with deeper-reaching burrows in profundal sediment increased the overall methane oxidation rate by a factor of 1.7. However, more data on the distribution and activity pattern of the respective infauna would be necessary to assess their influence more precisely.
Methane production rates were calculated based on methane fluxes and oxidation rates. It turned out that in the profundal sediment, about 98% of the produced methane was oxidized, and in the littoral sediment, this value was about 90%. Earlier studies reported 93% methane oxidation in the profundal sediment and 79% in littoral sediment of Lake Constance (7, 24). It should be noted that these data (7, 24) were obtained when Lake Constance was still rather eutrophic. The overall oxygen consumption rates in littoral and profundal sediment were 250 and 228 μmol m−2 h−1. Given a stoichiometry of 2 mol O2 per mol CH4 oxidized, methane oxidation contributed to about 3% and 5% of the total oxygen consumption in littoral and profundal sediments, respectively.
Quantification and abundance of methanotrophs.
For quantification of methanotrophs, we used two independent molecular methods, namely, qPCR based on the abundance of a functional gene (pmoA) amplified from the DNA obtained from the sediment and FISH based on the hybridization of 16S rRNA with specific probes for type I and type II MOB, to estimate the abundance of total methanotrophs relative to the active fraction. Although FISH and qPCR showed very similar profiles of distribution of MOB in the sediment, there was a considerable difference in the absolute numbers obtained. Under the assumption that every cell contains two copies of the pmoA gene (32), we detected an average of 1.7 × 107 MOB cells per g fresh weight in the littoral sediment and 2.5 × 107 MOB cells per g fresh weight in the profundal sediment in the upper 4 cm. With FISH, we obtained much lower numbers, i.e., a total of 5.3 × 105 MOB per g of sediment (wet weight) in the littoral sediment and 1.4 × 105 MOB per g of sediment (wet weight) in the profundal sediment. To explain this discrepancy, we compared clone library data for type I and type II methanotrophs from the study site (49) with the oligonucleotide sequence of the FISH probe and assumed that one mismatch resulted in no detectable signal (20). Thus, with the FISH probe set we employed (mg705 and mg84), we missed about 32% of the clones present in littoral sediment and about 63% of the profundal clones. When the FISH numbers were corrected for these mismatches, we obtained 7.1 × 105 and 3.3 × 105 cells per g of sediment (wet weight) for littoral and profundal sediments, respectively. In a comparison of these numbers with the qPCR data, the FISH numbers for the littoral and profundal sediments were still 23 and 75 times lower, respectively.
The high numbers determined by qPCR could be due to the fact that growing cells contain more than one genome copy per cell, and this could result in copy numbers being higher than the number of cells (32). Another reason for this discrepancy could be that binding of the FISH probe to cells depends on the number of rRNA molecules, which in turn depends on the activity status of the cells (8). Thus, inactive or slowly growing cells would not be detected with the FISH method, thus resulting in lower counts. In contrast, qPCR is based on DNA, and therefore, all cells would be counted, even if they are not active. In addition, the extraction of cells from the sediment for FISH could cause considerable losses. However, additional sonication used to estimate losses due to our extraction protocol yielded only 21% more cells. The true extraction efficiency is still unknown. Nevertheless, DAPI counts in littoral and profundal sediments of Lake Constance in the present study were comparable with those of previous studies of the littoral sediment; in these studies, 4 × 108 to 8 × 108 bacteria per g of sediment (wet weight) were detected, whereas in the profundal sediment, 1 × 109 to 4 × 109 bacteria per g of sediment (wet weight) were detected (53). Bacterial numbers detected by qPCR were a bit higher than those detected earlier in the case of the littoral sediment and in the same range as that of the profundal sediment (53).
qPCR designed to amplify type II MOB was not successful, suggesting that type II MOB were present in low abundance. The type I MOB qPCR assay also showed that type I MOB represented a major proportion of total MOB in terms of copy numbers. Various molecular approaches used so far, i.e., terminal restriction fragment length polymorphism using the pmoA gene (43, 50), clone libraries based on pmoA, and the 16S rRNA gene, all have suggested low abundance of type II MOB in the littoral and profundal sediments (50) compared to that of type I MOB. Although all the type II MOB sequences deposited thus far from Lake Constance sediment were covered by the primers (32), there could be yet-undescribed type II MOB present that were not covered by the existing primer sets. By using another molecular method, FISH, which is not based on PCR, we were able to document the presence of type II MOB which have also been cultivated from this site (12).
Few studies have focused on determining the abundances of methanotrophs in environments such as sediments and soils (17, 32, 30, 31). In the profundal sediment of Lake Washington, 108 to 109 total MOB cells per g (dry weight) have been detected by phospholipid fatty acid analysis and by quantitative slot-blot hybridization (17). This is within the same range as our qPCR number for profundal sediment (1 × 108 cells per g of sediment [dry weight]). FISH and qPCR are the most recent techniques used for the quantification of methanotrophs (40). Very few quantitative data for methanotrophs based on FISH are available, e.g., data for methanotrophs in rhizoplane soil (22) and Sphagnum peat bogs (19). In Siberian permafrost regions of the Lena Delta, similar numbers of methanotrophs were counted by FISH (36). Many freshwater habitats have been shown to be dominated by type I MOB, as observed in our study (6, 17, 21).
Relating activity to cell numbers.
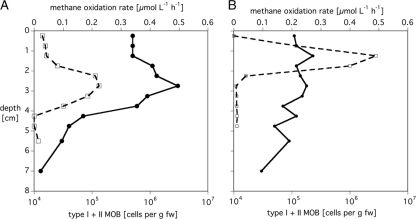
We found low methane oxidation activity in the top 2 cm of the littoral sediment, followed by a zone of higher methane oxidation activity at approximately 2 to 4 cm. This is also the zone where the MOB abundance increased. Thus, we found a good correlation between increased cell numbers and increased methane oxidation activity (Fig. 4A). In profundal sediment, we observed a slight decrease in abundance of MOB with sediment depth, and a zone of high methane oxidation activity at the sediment subsurface (0.5 to 2 cm) (Fig. 4B).
FIG. 4.
Abundances of type I and II methanotrophs as determined by FISH (circles) together with the corresponding average methane oxidation rates (squares) for littoral (A) and profundal (B) sediments. fw, fresh weight.
The activity of a population is the product of its cell number (population size) and the activity per cell (50). We applied this concept to our data set. The average methane oxidation rate was estimated in 0.5-cm steps, similar to the sampling intervals in the determination of cell numbers by FISH. Analysis by cell numbers based on qPCR showed similar results. In the littoral sediment, the slope of the double logarithmic plot (log methane consumption rate versus log MOB cell number) was significantly different from zero, but it was not near 1 (0.44 ± 0.10, P < 0.005, n = 10). Thus, in the littoral sediment, the methane oxidation activity was controlled by changes in population size and by a changed activity per cell. In the profundal sediment, the slope of the double logarithmic plot did not differ significantly from zero (0.13 ± 0.07, P = 0.08, n = 10), indicating that in the profundal sediment, the methane oxidation rate was controlled mainly by a changed activity per cell and not by the number of MOB. The uncoupling of activity and cell number in the profundal sediment is probably due to lower growth efficiency. Growth efficiency of bacteria is positively influenced by grazing and nutrient addition (N and especially P) (29, 45). In the littoral zone, with frequent sediment resuspension, nutrients can be released from deeper sediment layers (27), which could lead to a higher growth efficiency. Protozoa graze on MOB (42); however, no information on grazing or protozoan abundance in littoral sediments in comparison to that in profundal sediments of Lake Constance is available, and it remains unclear if lower grazing activity in profundal sediment could explain the low growth efficiency.
Additionally, in the littoral sediment, maximum ratios of MOB over bacterial numbers (FISH/DAPI) coincided with the zone of high methane oxidation, whereas in the profundal sediment, the ratios decreased with depth, not influenced by the depth distribution of the methane oxidation rate. Ratios of MOB over bacterial numbers determined by qPCR did not show any conclusive pattern, which could be due the fact that these ratios were very low and the method was not sensitive enough to detect the small differences in this range accurately.
Depth zonation of methanotrophs.
The high spatial resolution of MOB abundance together with the corresponding methane oxidation activity permitted definition of three zones in the sediment. In the surface zone, both oxygen and methane are present, and methane can be oxidized aerobically by MOB. In the second zone, most obvious in the littoral sediment, most of the methane was actually consumed where oxygen was not detectable. This was also where MOB were most abundant. In the deepest zone (deeper than 3 to 4 cm), we found MOB still in large numbers even though methane oxidation activity was not detectable.
In the littoral sediment, the noticeable disappearance of methane in the absence of oxygen in the second zone is hard to explain; sulfate-dependent oxidation of methane is energetically difficult under the conditions of low sulfate availability and low methane pressure prevailing there (55, 56). One can argue that the described oxygen profiles were shifted artificially by transport and by the incubation conditions that prevented water flow and wave action at the sediment surface. Thus, in situ, the oxygen would penetrate deeper into the sediment, as has been described repeatedly in studies on sandy marine sediments (46). However, even with strong illumination of littoral cores, oxygen did not penetrate deeper than 6 mm into the sediment (26), and in sediment cores incubated in a flume tank, oxygen penetrated to 4-mm depth at most (I. Bussmann, unpublished data). Bioturbation by Tubificidae or chironomid larvae can transport oxygen deeper into the sediment (10), but at least for chironomids, this occurs no deeper than 6 mm (59). Comparisons of microelectrode measurements of oxygen in sediment cores with in situ measurements revealed no significant difference of the oxygen penetration depth for profundal cores; in littoral cores, oxygen penetrated twice as deep in the in situ measurement (33). Thus, even after correcting for laboratory artifacts and bioturbation, oxygen could be available only in traces at 20- to 30-mm sediment depth, at which we found maximal methane oxidation and maximal numbers of aerobic MOB (Fig. 4A). By analogy, aerobic methanotrophs in the Black Sea water column are thought to be responsible for methane oxidation at the chemocline, although no free oxygen could be detected at these water depths (57). Thus, further studies are necessary to check for traces of oxygen in these respective depths or to check if alternatively nitrate could be involved as terminal electron acceptor (47). In the profundal sediment, the profiles of oxygen and methane distribution showed maximal methane oxidation activity in a sediment layer at a depth of 0.8 to 1 cm, which is only a few mm below the measured oxygen penetration maximum. Again, we have to assume that in situ oxygen penetrated deeper into the sediment than our measurements indicate.
In the deepest zone, MOB were present at high numbers, and they obviously did not oxidize methane at this depth. The presence of aerobic methanotrophs in anoxic zones has been documented several times by MPN counts (21, 52) and by cultivation-independent analyses (15). These bacteria may be in a dormant state, possibly thriving on endogenous storage material (48). Nonetheless, they are able to respond quickly to environmental changes (51) when oxygen or other electron acceptors become available again by bioturbation or sediment resuspension.
In sediments of Lake Constance, littoral and profundal type I MOB dominated over type II MOB. In the littoral sediment, the methane oxidation rate was controlled by cell number and activity per cell, while in the profundal sediment, the methane oxidation rate was controlled mainly by changing the activity per cell. For both sediments, we found a depth zonation of MOB, with maximal activities and highest MOB abundance at depth layers where oxygen was not detectable.
Acknowledgments
This study was supported by Sonderforschungsbereich 454 “Bodenseelitoral” of the Deutsche Forschungsgemeinschaft, Bonn, Germany, and research funds of the Universität Konstanz.
Special thanks are due to Alfred Sulger and colleagues for help in sample collection. We also thank Rahul Bahulikar for help during standardization of real-time PCR.
Footnotes
Published ahead of print on 7 November 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Auman, A., S. Stolyar, and M. E. Lidstrom. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66:5259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auman, A. J., and M. E. Lidstrom. 2002. Analysis of sMMO-containing type I methanotrophs in Lake Washington sediment. Environ. Microbiol. 4:517-524. [DOI] [PubMed] [Google Scholar]
- 4.Bastviken, D., J. Cole, M. Pace, and L. Tranvik. 2004. Methane emissions from lakes: dependence of lake characteristics, two regional assessments, and a global estimate. Global Biogeochem. Cycles 18:GB4009. [Google Scholar]
- 5.Berg, P., N. Risgaard-Petersen, and S. Rysgaard. 1998. Interpretation of measured concentration profiles in sediment pore water. Limnol. Oceanogr. 43:1500-1510. [Google Scholar]
- 6.Bodelier, P. L. E., M. Meima-Franke, G. Zwart, and H. J. Laanbroek. 2005. New DGGE strategies for the analyses of methanotrophic microbial communities using different combinations of existing 16S rRNA-based primers. FEMS Microbiol. Ecol. 52:163-174. [DOI] [PubMed] [Google Scholar]
- 7.Bosse, U., P. Frenzel, and R. Conrad. 1993. Inhibition of methane oxidation by ammonium in the surface layer of a littoral sediment. FEMS Microbiol. Ecol. 13:123-134. [Google Scholar]
- 8.Bouvier, T., and P. A. del Giorgio. 2003. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): a quantitative review of published reports. FEMS Microbiol. Ecol. 44:3-18. [DOI] [PubMed] [Google Scholar]
- 9.Bowman, J. 2000. The methanotrophs—the families Methylococcaceae and Methylocystaceae, p. 266-289. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, part 1, section 3.1. Springer Verlag, New York, NY.
- 10.Brune, A., P. Frenzel, and H. Cypionka. 2000. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol. Rev. 24:691-710. [DOI] [PubMed] [Google Scholar]
- 11.Bussmann, I. 2005. Methane release through suspension of littoral sediment. Biogeochemistry 74:283-302. [Google Scholar]
- 12.Bussmann, I., M. Pester, A. Brune, and B. Schink. 2004. Preferential cultivation of type II methanotrophic bacteria from littoral sediments (Lake Constance). FEMS Microbiol. Ecol. 47:179-189. [DOI] [PubMed] [Google Scholar]
- 13.Bussmann, I., M. Rahalkar, and B. Schink. 2006. Cultivation of methanotrophic bacteria in opposing gradients of methane and oxygen. FEMS Microbiol. Ecol. 56:331-344. [DOI] [PubMed] [Google Scholar]
- 14.Bussmann, I., and B. Schink. 2006. A modified diffusion-based methane sensor and its application in freshwater sediment. Limnol. Oceanogr. Methods 4:275-283. [Google Scholar]
- 15.Carini, S., N. Bano, G. LeCleir, and S. B. Joye. 2005. Aerobic methane oxidation and methanotroph community composition during seasonal stratification in Mono Lake, California (USA). Environ. Microbiol. 7:1127-1138. [DOI] [PubMed] [Google Scholar]
- 16.Case, R. J., Y. Boucher, I. Dahlloef, C. Holmstroem, W. F. Doolittle, and S. Kjelleberg. 2007. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 73:278-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costello, A. M., A. J. Auman, J. L. Macalady, K. M. Scow, and M. E. Lidstrom. 2002. Estimation of methanotroph abundance in a freshwater lake sediment. Environ. Microbiol. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 18.Dalton, H. 2005. The Leeuwenhoek Lecture 2000: the natural and unnatural history of methane-oxidizing bacteria. Philos. Trans. R. Soc. Lond. B 360:1207-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dedysh, S. N., M. Derakshani, and W. Liesack. 2001. Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of a newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67:4850-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eller, G. 2001. Aktivität, Populationsdynamik und Diversität Methan oxidierender Bakterien im Reisfeld. Ph.D. thesis. Philipps-University Marburg, Marburg, Germany.
- 21.Eller, G., P. Deines, J. Grey, H.-H. Richnow, and M. Krüger. 2005. Methane cycling in lake sediments and its influence on chironomid larval d13C. FEMS Microbiol. Ecol. 54:339-350. [DOI] [PubMed] [Google Scholar]
- 22.Eller, G., S. Stubner, and P. Frenzel. 2001. Group-specific 16S rRNA targeted probes for the detection of type I and type II methanotrophs by fluorescence in situ hybridisation. FEMS Microbiol. Lett. 198:91-97. [DOI] [PubMed] [Google Scholar]
- 23.Fierer, N., J. A. Jackson, R. Vilgalys, and R. B. Jackson. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frenzel, P., B. Thebrath, and R. Conrad. 1990. Oxidation of methane in the oxic surface layer of a deep lake sediment (Lake Constance). FEMS Microbiol. Ecol. 73:149-158. [Google Scholar]
- 25.Garcia, H. E., and L. I. Gordon. 1992. Oxygen solubility in seawater: better fitting equations. Limnol. Oceanogr. 37:1307-1312. [Google Scholar]
- 26.Gerhardt, S., A. Brune, and B. Schink. 2005. Dynamics of redox changes of iron caused by light-dark variations in littoral sediment of a freshwater lake. Biogeochemistry 74:323-339. [Google Scholar]
- 27.Güde, H., M. Seidel, P. Teiber, and M. Weyhmueller. 2000. P-release from littoral sediments in Lake Constance. Verh. Int. Verein. Limnol. 27:2624-2627. [Google Scholar]
- 28.Islam, T., S. Jensen, L. J. Reigstad, Ø. Larsen, and N.-K. Birkeland. 2008. Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. USA 105:300-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansson, M., A. K. Bergström, D. Lymer, K. Vrede, and J. Karlsson. 2006. Bacterioplankton growth and nutrient use efficiencies under variable organic carbon and inorganic phosphorus ratios. Microb. Ecol. 52:358-364. [DOI] [PubMed] [Google Scholar]
- 30.Knief, C., S. Kolb, P. L. E. Bodelier, A. Lipski, and P. F. Dunfield. 2006. The active methanotrophic community in hydromorphic soils changes in response to changing methane concentration. Environ. Microbiol. 8:321-333. [DOI] [PubMed] [Google Scholar]
- 31.Kolb, S., C. Knief, P. F. Dunfield, and R. Conrad. 2005. Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ. Microbiol. 7:1150-1161. [DOI] [PubMed] [Google Scholar]
- 32.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koschorreck, M., I. Brookland, and A. Matthias. 2003. Biogeochemistry of the sediment-water interface in the littoral of an acidic mining lake studied with microsensors and gel-probes. J. Exp. Mar. Biol. Ecol. 285-286:71-84. [Google Scholar]
- 34.Kuivila, K. M., J. W. Murray, A. H. Devol, M. E. Lidstrom, and C. E. Reimers. 1988. Methane cycling in the sediments of Lake Washington. Limnol. Oceanogr. 33:571-581. [Google Scholar]
- 35.Lidstrom, M. E., and L. Somers. 1984. Seasonal study of methane oxidation in Lake Washington. Appl. Environ. Microbiol. 47:1255-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebner, S., and D. Wagner. 2007. Abundance, distribution and potential activity of methane oxidizing bacteria in permafrost soils from the Lena Delta, Siberia. Environ. Microbiol. 9:107-117. [DOI] [PubMed] [Google Scholar]
- 37.Lin, J.-L., S. B. Joye, J. C. M. Scholten, H. Schafer, I. R. McDonald, and J. C. Murrell. 2005. Analysis of methane monooxygenase genes in Mono Lake suggests that increased methane oxidation activity may correlate with a change in methanotroph community structure. Appl. Environ. Microbiol. 71:6458-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matisoff, G., and X. Wang. 2000. Particle mixing by freshwater infaunal bioirrigators: midges (Chironomidae: Diptera) and mayflies (Ephemeridae: Ephemeroptera). J. Great Lakes Res. 26:174-182. [Google Scholar]
- 40.McDonald, I. R., L. Bodrossy, Y. Chen, and C. J. Murell. 2008. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 74:1305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mörtl, M. 2003. Biotic interactions in the infralittoral of Lake Constance. Ph.D. thesis. Universität Konstanz, Constance, Germany.
- 42.Murase, J., and P. Frenzel. 2007. A methane-driven microbial food web in a wetland rice soil. Environ. Microbiol. 9:3025-3034. [DOI] [PubMed] [Google Scholar]
- 43.Pester, M., M. W. Friedrich, B. Schink, and A. Brune. 2004. pmoA-based analysis of methanotrophs in a littoral lake sediment reveals a diverse and stable community. Appl. Environ. Microbiol. 70:3138-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pol, A., K. Heijmans, H. R. Harhangi, D. Tedesco, M. S. M. Jetten, and H. J. M. Op den Camp. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450:874-878. [DOI] [PubMed] [Google Scholar]
- 45.Pradeep Ram, A. S., and T. Sime-Ngando. 2008. Functional responses of prokaryotes and viruses to grazer effects and nutrient additions in freshwater microcosms. ISME J. 2:498-509. [DOI] [PubMed] [Google Scholar]
- 46.Precht, E., and M. Huettel. 2003. Advective pore-water exchange driven by surface gravity waves and its ecological implications. Limnol. Oceanogr. 48:1674-1684. [Google Scholar]
- 47.Raghoebarsing, A. A., A. Pol, K. T. van de Pas-Schoonen, A. J. P. Smolders, K. F. Ettwig, W. I. C. Rijpstra, S. Schouten, J. S. S. Damste, H. J. M. Op den Camp, M. S. M. Jetten, and M. Strous. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918-921. [DOI] [PubMed] [Google Scholar]
- 48.Rahalkar, M., I. Bussmann, and B. Schink. 2007. Methylosoma difficile gen. nov., sp. nov., a novel methanotroph enriched by gradient cultivation from littoral sediment of Lake Constance. Int. J. Syst. Evol. Microbiol. 57:1073-1080. [DOI] [PubMed] [Google Scholar]
- 49.Rahalkar, M., and B. Schink. 2007. Comparison of aerobic methanotrophic communities in littoral and profundal sediments of Lake Constance by a molecular approach. Appl. Environ. Microbiol. 73:4389-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Röling, W. F. M. 2007. Do microbial numbers count? Quantifying the regulation of biogeochemical fluxes by population size and cellular activity. FEMS Microbiol. Ecol. 62:202-210. [DOI] [PubMed] [Google Scholar]
- 51.Roslev, P., and G. M. King. 1995. Aerobic and anaerobic starvation metabolism in methanotrophic bacteria. Appl. Environ. Microbiol. 61:1563-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy, R., R. Knowles, and M. N. Charlton. 1996. Nitrification and methane oxidation at the sediment surface in Hamilton Harbour (Lake Ontario). Can. J. Fish. Aquat. Sci. 53:2466-2472. [Google Scholar]
- 53.Sala, M. M., and H. Güde. 2006. Seasonal dynamics of pelagic and benthic (littoral and profundal) bacterial abundances and activities in a deep prealpine lake (L. Constance). Arch. Hydrobiol. 167:351-369. [Google Scholar]
- 54.Sauter, G., and H. Güde. 1996. Influence of grain size on the distribution of tubificid oligochaete species. Hydrobiologia 334:97-101. [Google Scholar]
- 55.Schink, B. 1997. Energetics of syntrophic cooperations in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61:262-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schink, B., and A. J. M. Stams. 2001. Syntrophism among prokaryotes, p. 25. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, NY.
- 57.Schubert, C. J., M. J. L. Coolen, L. N. Neretin, A. Schippers, B. Abbas, E. Durisch-Kaiser, B. Wehrli, E. C. Hopmans, J. S. S. Damste, S. Wakeham, and M. M. M. Kuypers. 2006. Aerobic and anaerobic methanotrophs in the Black Sea water column. Environ. Microbiol. 8:1844-1856. [DOI] [PubMed] [Google Scholar]
- 58.Schulz, S., and R. Conrad. 1995. Effect of algal deposition on acetate and methane concentrations in the profundal sediment of a deep lake (Lake Constance). FEMS Microbiol. Ecol. 16:251-260. [Google Scholar]
- 59.Stief, P., and D. de Beer. 2006. Probing the microenvironment of freshwater sediment macrofauna: implications of deposit-feeding and bioirrigation for nitrogen cycling. Limnol. Oceanogr. 51:2538-2548. [Google Scholar]
- 60.Sundh, I., D. Bastviken, and L. J. Tranvik. 2005. Abundance, activity, and community structure of pelagic methane-oxidizing bacteria in temperate lakes. Appl. Environ. Microbiol. 71:6746-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thebrath, B., F. Rothfuss, M. J. Whiticar, and R. Conrad. 1993. Methane production in littoral sediment of Lake Constance. FEMS Microbiol. Ecol. 102:279-289. [Google Scholar]
- 62.Wetzel, R. 2001. Limnology, lake and river ecosystems, 3rd ed. Academic Press, San Diego, CA.
- 63.Zipper, H., C. Buta, K. Lämmle, H. Brunner, J. Bernhagen, and F. Vitzthum. 2003. Mechanisms underlying the impact of humic acids on DNA quantification by SYBR green I and consequences for the analysis of soils and aquatic sediments. Nucleic Acids Res. 31:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]