Abstract
Although research has increasingly focused on the pathogenesis of avian pathogenic Escherichia coli (APEC) infections and the “APEC pathotype” itself, little is known about the reservoirs of these bacteria. We therefore compared outbreak strains isolated from diseased chickens (n = 121) with nonoutbreak strains, including fecal E. coli strains from clinically healthy chickens (n = 211) and strains from their environment (n = 35) by determining their virulence gene profiles, phylogenetic backgrounds, responses to chicken serum, and in vivo pathogenicities in a chicken infection model. In general, by examining 46 different virulence-associated genes we were able to distinguish the three groups of avian strains, but some specific fecal and environmental isolates had a virulence gene profile that was indistinguishable from that determined for outbreak strains. In addition, a substantial number of phylogenetic EcoR group B2 strains, which are known to include potent human and animal extraintestinal pathogenic E. coli (ExPEC) strains, were identified among the APEC strains (44.5%) as well as among the fecal E. coli strains from clinically healthy chickens (23.2%). Comparably high percentages (79.2 to 89.3%) of serum-resistant strains were identified for all three groups of strains tested, bringing into question the usefulness of this phenotype as a principal marker for extraintestinal virulence. Intratracheal infection of 5-week-old chickens corroborated the pathogenicity of a number of nonoutbreak strains. Multilocus sequence typing data revealed that most strains that were virulent in chicken infection experiments belonged to sequence types that are almost exclusively associated with extraintestinal diseases not only in birds but also in humans, like septicemia, urinary tract infection, and newborn meningitis, supporting the hypothesis that not the ecohabitat but the phylogeny of E. coli strains determines virulence. These data provide strong evidence for an avian intestinal reservoir hypothesis which could be used to develop intestinal intervention strategies. These strains pose a zoonotic risk because either they could be transferred directly from birds to humans or they could serve as a genetic pool for ExPEC strains.
Extraintestinal Escherichia coli infections are among the most significant infectious diseases in production birds and result in severe losses due to mortality, production losses, and condemnations. As a clearly defined genotype of avian pathogenic E. coli (APEC) has still not been determined, the term APEC is commonly ill-defined and used for strains isolated from poultry with clinical signs of different local and systemic E. coli infections, like air sacculitis, cellulites, and septicemia (2, 10, 14). Although research has increasingly focused on the pathogenesis of APEC infections and the “APEC pathotype,” we have little knowledge about the reservoir of these bacteria, considerably hampering disease control. Recent investigations comparing E. coli strains isolated from internal organs of poultry suffering from systemic infections with E. coli strains isolated from the intestines of clinically healthy chickens, designated avian fecal E. coli (Afecal E. coli), concentrated on genotyping, serotyping, and the use of various in vitro assays with a collection of strains (6, 28, 33, 37, 40). Virulence genotyping has been suggested to be the best way to differentiate APEC from Afecal E. coli, while other typing methods, especially serotyping, might not be particularly useful due to the overlap in serogroups not only between APEC and chicken fecal isolates but also between APEC and other extraintestinal pathogenic E. coli (ExPEC) isolates, like uropathogenic E. coli (UPEC) isolates, as well as newborn meningitis E. coli (NMEC) strains (12, 36). Nevertheless, serotyping is still a common method for estimating the pathogenic potential of APEC strains as it has been accepted for a long time that some serotypes, including serotypes O1, O2, O8, O18, and O78, are detected more frequently than other serotypes (11, 12, 28, 36, 37, 41).
Rodriguez-Siek et al. (37) suggested that a typical member of the APEC pathotype is likely to contain several iron transporter-encoding genes, like irp2, fyuA, iutA, iroN, and sitA, and plasmid-associated genes, including cvi/cvaC, tsh, and iss. Moreover, pap genes, coding for P fimbriae, were much more likely to be found in APEC strains than in Afecal strains. Another study suggested that a widespread trait of virulent avian E. coli strains is their resistance to serum complement, which allows distinction between virulent strains and nonvirulent strains (32). In contrast, Vandekerchove et al. (41) did not find a significant difference between serum resistance in APEC and serum resistance in fecal E. coli strains, raising the question of whether this trait is useless as a “pathotype marker” and the question of whether fecal strains serve as a reservoir for extraintestinal infections.
There is increasing evidence that virulence in E. coli extraintestinal infections is more likely linked to the phylogenetic background of a strain than to its ecological background (30, 34). The common phylogenetic origins of APEC and other ExPEC, including UPEC in humans and a wide variety of animals, as well as NMEC in infants, emphasizes the potential zoonotic risk of avian-derived E. coli strains (23, 30, 44). In humans, according to the fecal-vaginal-urethral hypothesis, E. coli strains causing urinary tract infection (UTI) are usually derived from the host's own fecal and perineal flora, which implies that fecal colonization with a urovirulent organism is a potentially modifiable risk factor for subsequent UTI (20).
To assess the reservoir function of the chicken intestine, in vivo experiments using defined strains isolated from clinically healthy chickens or their environment that, based on the criteria mentioned above, fit into the current scheme of the APEC pathotype are necessary. Therefore, the aims of this study were not only to characterize a set of fecal and environmental E. coli strains by using molecular typing but also to select various strains from clinically healthy birds with different virulence gene patterns, serotypes, and phylogenetic backgrounds in order to assess their pathogenicity in a chicken infection model. Our findings prove that similar to UTI in humans, the chicken intestine can serve as a reservoir for E. coli strains capable of causing extraintestinal infections in avian and mammalian hosts, resulting in potential zoonotic risk.
MATERIALS AND METHODS
Bacterial strains.
In an 8-year period (1999 to 2008) 367 E. coli strains were isolated either from poultry farms experiencing disease due to systemic infection caused by APEC (outbreak strains; n = 121) or from flocks without any clinical history of APEC infection (nonoutbreak strains; n = 246). The nonoutbreak strains were avian fecal strains (Afecal strains), which originated from cloacal swabs (n = 211) obtained from clinically healthy laying hens of different ages, and avian environmental strains (Aenviron strains), which were isolated from the environment in chicken coops (n = 35), including the air-handling system and water. The outbreak strains originated from different avian hosts, primarily chickens, and from different sites within these hosts, including the heart, liver, spleen, air sacs, and blood. Outbreak strains did not exhibit any epidemiologic link to Afecal and Aenviron strains in terms of time and geographical origin. Swabs were streaked on blood agar plates and Chromo orient agar (Oxoid, Germany), which were then incubated overnight at 37°C. Red colonies, predicted to be E. coli colonies according to the manufacturer's information, were investigated further using standard biochemical procedures, such as fermentation of glucose and lactose, production of indole, and decarboxylation of lysine, to ensure that the species identification was correct.
The controls used for molecular assays were APEC strains IMT5155 (O2:K1:H5) (26) and IMT2470 (O2:K1:H5), UPEC strains IMT7920 (O75:HM) and IMT1200 (O18:H1), and NMEC strain BK658 (O18:K1:H7) (12). For identification of virulence-associated genes (VAGs) typical of enteropathogenic E. coli and Shiga toxin-producing E. coli (STEC), strains IMT8210 (STEC; Ont:H4; positive for stx1 and escV), IMT8215 (STEC; O79:H−; positive for stx2 and escV), and E2348/69 (enteropathogenic E. coli; O127:H6; positive for bfp and escV) were used as PCR controls. The latter controls were chosen after the identities of the stx1, stx2, escV, and bfp genes in these strains were confirmed by sequencing the PCR amplicons and analyzing the DNA sequences using the Basic Local Alignment Search Tool (http://www.ncbi.nlm.nih.gov/BLAST). All strains were stored at −70°C in brain heart infusion broth with 10% glycerol until they were used.
DNA preparation.
Bacterial DNA used for PCR analyses was prepared with a Master Pure genomic DNA purification kit (Biozym Diagnostik GmbH, Hessisch Oldendorf, Germany) used according to the manufacturer's recommendations. The DNA was diluted to obtain a concentration of 20 ng/μl, and 4-μl portions of the dilutions were used for single and multiplex PCR protocols. Alternatively, bacterial DNA was extracted by a boiling procedure. Single colonies were cultured overnight at 37°C in 1.5-ml tubes containing 1 ml LB broth. Each bacterial suspension was centrifuged for 10 min at 14,000 rpm, and the pellet was resuspended in 200 μl deionized distilled water. After 10 min of boiling in a water bath and centrifugation for 10 min at 14,000 rpm, the supernatant was used as template DNA for PCR assays.
Virulence genotyping.
E. coli strains were investigated to determine the presence of various genes by using multiplex protocols described previously (12), as well as single PCRs as described below. The genes targeted were related to (i) bacterial adhesion (afa/draB, bfp, bmaE, fimC, focG, gafD, hra, iha, nfaE, papAH, papC, papEF, papG alleles II and III, sfa/focCD, sfaS, tsh, and mat); (ii) iron acquisition (chuA, fyuA, ireA, iroN, irp2, iucD, iutA, sitD [chromosomal], and sitD ([episomal]); (iii) serum resistance (iss, kpsMTII, neuC, ompA, and traT); (iv) toxins and hemolysins (astA, cnf1/2, sat, stx1, stx2, vat, and hlyA); (v) invasion (ibeA, gimB, and tia); and (vi) the type III secretion system (escV). Miscellaneous genes (cvi/cva, pic, malX, and pks) were also included. Oligonucleotide primers were obtained from Sigma Genosys (Steinheim, Germany). Primer sequences, including sequences that have been described previously (9, 13, 17, 21, 27, 31, 39, 43, 45, 46), and coordinates are shown in Table S1 in the supplemental material.
Primers were sorted into four pools based on primer compatibility and product size by using the previously described protocol. Each primer pool was validated by using control strains containing all the relevant virulence genes. All multiplex PCR procedures were performed with 25-μl reaction mixtures containing 2.5 μl of 10× PCR buffer, 2.0 μl of a 50 mM MgCl2 solution, 1.5 U of Taq DNA polymerase (Rapidozym, Germany), 0.3 μl of a 10 mM solution of each deoxynucleoside triphosphate, 0.1 μl (100 pmol) of a oligonucleotide primer pair (Sigma-Aldrich, Germany), 2 μl (40 ng) of template DNA, and appropriate volumes of double-distilled water. The reaction mixtures were subjected to the following conditions in a thermal cycler (T1; Whatman Biometra, Germany): 3 min at 94°C, 25 cycles of 30 s at 94°C, 30 s at 58°C, and 3 min at 68°C, and a final cycle of 10 min at 72°C, followed by incubation at 10°C.
Single PCRs for amplification of the polyketide synthesis gene pks, the adhesion genes bmaE, focG, gafD, nfaE, papAH, papEF, papG alleles II and III, and sfaS, and the aerobactin receptor gene iutA were performed by using a protocol described previously (11). Briefly, 2 μl (40 ng) of template DNA was added to a reaction mixture (25 ml) containing 0.5 ml (10 pmol) of each primer pair, 0.1 ml of a 10 mM solution of each deoxynucleoside triphosphate, 2.5 ml of 10× PCR buffer, 1.25 ml of a 50 mM MgCl2 solution, and 0.75 U of Taq polymerase. The samples were subjected to 25 cycles of amplification with the thermal cycler mentioned above, and the annealing and elongation times were adjusted for individual primers and the lengths of the products.
Horizontal gel electrophoresis was performed with 1.5% agarose for multiplex PCR products and with 1.0% agarose (Rotigarose; Roth GmbH, Germany) for single PCR products. Amplicons were stained with ethidium bromide and photographed with UV exposure, and their sizes were determined by comparison to a 100-bp DNA marker (Invitrogen, Germany).
EcoR grouping and MLST analysis.
E. coli strains were classified according to the EcoR system (15) by using the rapid phylogenetic grouping PCR technique described by Clermont et al. (5). Multilocus sequence typing (MLST) of E. coli strains used in chicken experiments was carried out by performing a sequence analysis of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) using the typing scheme described recently (44). Sequence analysis and allocation of allele types to sequence types and sequence type complexes were performed with the software program Ridom SeqSphere (Ridom GmbH, Würzburg, Germany). The results were compared with MLST data deposited in the publicly available database (www.mlst.net) using the BioNumerics software (version 4.6; Applied Maths, Belgium).
Serum resistance assay.
The bacterial survival and resistance in serum of 211 Afecal E. coli strains, 35 Aenviron strains, and 121 APEC outbreak isolates were examined by using an assay modified by Dozois et al. (7). Briefly, a 100-fold dilution of an overnight culture was resuspended in 500 μl LB broth and incubated with agitation for 1.75 h at 37°C. This culture was centrifuged for 5 min at 6,000 × g, and the pellet was resuspended in 1 ml phosphate-buffered saline (pH 7.4) to obtain a concentration of approximately 107 CFU ml−1. Fifty microliters of the suspension was added to 450 μl of undiluted chicken serum (PAN Biotech GmbH, Aidenbach, Germany) in a 48-well microtiter plate, resulting in a concentration of 106 CFU ml−1 in 90% chicken serum. After mixing, the serum-bacterium suspension was incubated at 37°C for 3 h. Samples used for determination of viable counts were taken at zero time and after 3 h of incubation. The responses were graded as follows: for grades 1, 2, and 3, the viable counts after 3 h were <0.1, <0.01, and <0.001% of the viable count for the inoculum, respectively; for grade 4, the viable count after 3 h was between 1 log less than and 1 log more than the viable count for the inoculum; and for grades 5, 6, and 7, the viable counts after 3 h were 1, 2, and 3 logs more than the viable counts for the inoculum, respectively. Each strain was tested at least three times, and the results were expressed as the means of the data obtained only for responses in the same direction. The controls included in each experiment were serum-resistant APEC strain MT78 (29), serum-sensitive strain BL21(DE3) (41), and cells incubated with heat-inactivated chicken serum (heat inactivated at 56°C for 25 min).
In vivo tests.
Chicken infection experiments were performed as described previously (1). Briefly, groups of six 5-week-old specific-pathogen-free White Leghorn chickens (Lohmann Selected Leghorn; Lohmann Tierzucht GmbH, Cuxhaven, Germany) were infected intratracheally with 0.5 ml of a concentrated log-phase culture (optical density at 600 nm, 1) containing 109 CFU of each strain. Clinical symptoms were monitored hourly, and 24 h after infection clinical scores were assigned to the chickens before they were euthanized and sacrificed. Postmortem examinations were performed immediately after the death of the chickens. Organs were aseptically removed, and the severity of the macroscopic lesions attributed to E. coli was scored by using an organ lesion score, the maximum value of which was 14 (1). Mean lesion scores were determined for all groups, and the significance of differences was calculated by the two-tailed t test; P values of <0.05, <0.02, and < 0.001 indicated moderate, intermediate, and high levels of significance, respectively.
After an organ lesion score was determined, bacteria were reisolated from the lungs, kidneys, liver, spleen, and heart. Tissue samples of these organs were weighed, suspended in phosphate-buffered saline (1 ml/g), and homogenized with an Ultra-Turrax T25 (IKA-Labortechnik, Staufen, Germany). Serial 10-fold dilutions were plated onto LB agar plates that were subsequently incubated at 37°C for 24 h. Colonies were then counted to determine the number of CFU per gram in each organ. Random colonies obtained from various organs and animals were examined to determine their virulence gene patterns by performing multiplex PCR assays as described previously (12) to ensure that the strains reisolated from internal organs were identical to the strains used for infection of animals as far as possible. Animals were treated according to animal welfare norms, and after experimental infection chickens were monitored each hour until the end of the experiment (regulation 0220/06).
Statistical analyses.
Statistical analyses for in vivo animal experiments were carried out using the nonparametric Mann-Whitney U test, which was implemented in the Statistical Package for the Social Sciences (SPSS, version 10.0). Comparisons of proportions for a particular characteristic in different populations were performed by using data from in vitro and molecular assays, and significance was tested with SigmaPlot for Windows (version 10.0) by using the two-tailed t test. Because of the multiple comparisons, the thresholds for statistical significance were the thresholds indicated in the tables, and a P value of <0.0001 indicated strong statistical significance. An analysis of gene combinations for the virulence gene patterns of E. coli strains was performed with Microsoft Office Excel 2003.
RESULTS
Virulence genotyping of outbreak strains (APEC) and nonoutbreak (Afecal and Aenviron) E. coli strains.
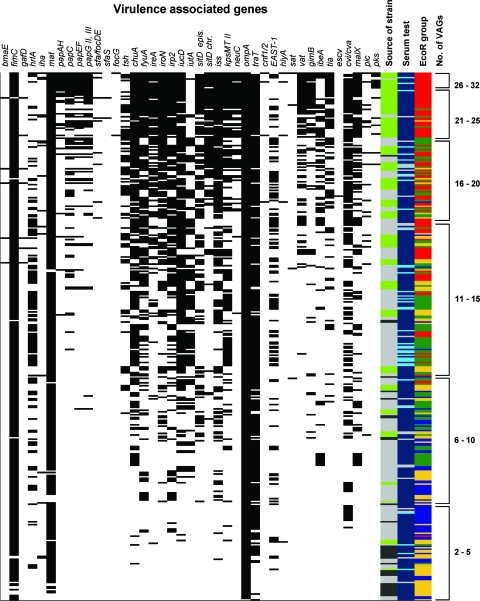
The total number of genes identified among the 46 VAGs tested in 367 E. coli strains ranged from 2 to 32, irrespective of known possible colocalization of some of these genes on plasmids or pathogenicity islands or in operon structures. In general, the three groups of strains could be distinguished by the VAG profiles, as APEC strains contained a mean of 17.4 genes per strain and Afecal and Aenviron strains contained mean numbers of 10.2 and 5.1 genes per strain, respectively. However, single strains, irrespective of the source from which they had been isolated, could not be distinguished, as shown in Fig. 1. For example, 57 (15.5%) of all the strains, including 9 (2.5%) Afecal E. coli strains, harbored at least 20 VAGs.
FIG. 1.
Virulence gene patterns, sources of strains, phylogenetic groups, and results of serum tests for 347 E. coli strains. For VAGs the presence of a gene in a strain is indicated by a black bar. The sources of strains are indicated as follows: light gray, Afecal strains; dark gray, Aenviron strains; and light green, APEC strains. EcoR groups are indicated as follows: yellowish, group A; blue, group B1; red, group B2; and green, group D. The results of the serum test are indicated as follows: dark blue, resistant; light gray, intermediate resistance; cyan, sensitive. Genes not present in any of the strains (afa/dra, bfp, nfaE, stx1, and stx2) are not shown. epis., episomal; chr., chromosomal.
Nearly all isolates, irrespective of the E. coli group to which they belonged, harbored ompA, while the adhesin genes afa/dra, bfp, and nfaE, as well as the toxin genes stx1 and stx2, were not present in any strain. Identical VAG profiles were found only rarely. A substantial number of isolates harbored fimC and mat (at least 71.4% in each group), while the type 1 fimbria-encoding gene fimC was significantly less common in Aenviron strains than in APEC and Afecal E. coli strains. Most of the remaining adhesion genes were found in APEC more frequently than in nonoutbreak E. coli strains. For example, different P fimbria operon genes were present in 22.7 to 40.3% of APEC strains but were not present in any Aenviron strain. Of the strains isolated from diseased birds, 93.8% harbored papG allele II and the remaining three strains harbored papG allele III. In contrast, the heat-resistant agglutinin-encoding gene hra was found significantly more often in Afecal strains (47.4%) than in APEC strains (22.7%) and Aenviron strains (17.1%).
In general, plasmid-associated genes were found in higher numbers of APEC strains (52.9 [tsh] to 88.2% [iroN]), but they also occurred in a substantial number of Afecal E. coli strains. Among the nonoutbreak strains, Aenviron E. coli strains in general harbored fewer of these episomal genes (5.7% [tsh] to 48.6% [traT]), and traT was significantly less prevalent in these strains than in Afecal strains. Except for chuA, all iron transporter-encoding genes were significantly more prevalent in APEC strains than in Afecal and Aenviron strains.
When protectin genes were examined, the K1 capsule-encoding gene neuC and the serum resistance-conferring gene iss were significantly more prevalent in outbreak strains (41.2 and 88.2%) than in nonoutbreak Afecal strains (4.3 and 32.7%) and Aenviron strains (0 and 25.7%). In addition, the group II capsule antigen-encoding gene kpsMTII was frequently found in APEC strains (51.3%) and in Afecal strains (37.9%), while it was less prevalent in Aenviron strains (8.6%).
The toxin and hemolysin genes cnf1/2, hlyA, and sat were found almost exclusively in outbreak strains, but the numbers of strains were low (1.7 to 4.2%). Detailed results are shown in Table S2 in the supplemental material.
EcoR typing and association with the mean number of VAGs.
The assignments of APEC, Afecal, and Aenviron strains to the four phylogenetic groups are shown in Table 1. The majority of outbreak strains fell into group B2 (44.5%), whereas nonoutbreak strains were significantly associated with groups D (38.9%) and A (65.7%) but were also assigned to EcoR group B2 (2.9% of Aenviron strains and 23.2% of Afecal strains). Substantial numbers of Afecal strains (26.1%) and Aenviron strains (25.7%), but only 2.5% of APEC strains, were assigned to group B1. Strains belonging to phylogenetic group B2 harbored the highest mean number of VAGs (18.7), and the numbers were lower in strains falling into groups D (12.3), A (8.6), and B1 (6.4). Several VAGs, including neuC, ibeA, gimB, malX, vat, and pks, were strongly associated with group B2 strains, irrespective of the E. coli group to which they belonged (P < 0.0001).
TABLE 1.
Assignment of APEC, Afecal, and Aenviron E. coli strains to phylogenetic groups and mean numbers of VAGs in strains belonging to different EcoR groups
| Isolates | n | Mean no. of VAGs per strain
|
% of isolates
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Group A | Group B1 | Group B2 | Group D | Group A | Group B1 | Group B2 | Group D | ||
| APEC | 121 | 12.0 | 13.0 | 22.6 | 15.7 | 34.5 | 2.5 | 44.5a | 18.5 |
| Afecal | 211 | 7.0 | 5.9 | 14.3 | 11.6 | 11.8 | 26.1 | 23.2 | 38.9a |
| Aenviron | 35 | 3.7 | 6.9 | 19.0 | 6.0 | 65.7a | 25.7 | 2.9 | 5.7 |
| Total | 367 | 8.8 | 6.4 | 18.7 | 12.3 | 25.8 | 18.4 | 28.2 | 29.0 |
There was a significant association (P < 0.0001) of strains with phylogenetic groups.
Serum resistance assay.
As serum resistance has previously been shown to be a marker for virulence, we examined the survival of all E. coli strains in chicken serum after 3 h. Strains were classified as highly sensitive (grades 1, 2, and 3), intermediately sensitive (grade 4), or resistant (grades 5, 6, and 7) to serum. As expected, control strain MT78 was serum resistant, whereas BL21 was serum sensitive, a phenotype that was completely abolished when heat-inactivated serum was used. Although the percentage of serum-resistant isolates (grades 5 to 7) was significantly higher among APEC isolates than among Afecal strains and was significantly higher among Aenviron strains than among Afecal isolates, the overall percentages of serum-resistant strains in these three groups of strains were comparable and high; 79.2% of the Afecal strains, 88.6% of the Aenviron strains, and 89.3% of the APEC strains exhibited this phenotype. (Fig. 1; see Table S3 in the supplemental material). Accordingly, the proportion of serum-sensitive strains (grades 1 to 3) among Afecal strains was rather low (13.7%), and only 1 of 35 Aenviron strains had a serum-sensitive phenotype.
While the isolates that were serum resistant and the isolates that had an intermediate level of resistance were nearly equally distributed among phylogenetic groups A, B1, B2, and D, we observed a highly significant association (P < 0.0001) between the serum-sensitive phenotype of E. coli strains and phylogenetic group D. Most noticeably, there was no statistical evidence for an association of serum resistance with the profile of VAGs known to account for this phenotype in APEC strains, such as iss and traT, whereas other genes, including hra, chuA, iucD, iutA, and east-1, exhibited significant associations with the serum resistance phenotype of the strains investigated.
MLST.
Sixteen E. coli strains that were selected for in vivo experiments based on the virulence gene profile, assignment to an EcoR group, and the serum resistance phenotype were characterized further by MLST (Table 2). Prototype APEC strain IMT5155 (O2:K1:H5), which was used as a positive control in chicken infection studies, was identified as a sequence type 140 (ST140) strain which belongs to sequence type complex 95 (STC95). STC95 currently accounts for the majority of phylogenetically typed ExPEC strains and comprises all known ExPEC pathotypes from humans, as well as from animals (www.mlst.net). Nonoutbreak Afecal strain IMT15146 (O1:K1:H7) could also be assigned to this complex (ST95 and STC95). Another Afecal strain, IMT14782 (Ont:NM) was grouped in ST69 of STC69, which to date includes almost exclusively human UTI strains. Other sequence types identified among nonoutbreak Afecal and Aenviron strains were ST58, ST115, ST420, ST428, ST429, and ST920, most of which have not been assigned to a sequence type complex. The remaining strains either exhibited new sequence types (n = 4) or were not typed (n = 1).
TABLE 2.
Characteristics of E. coli strains used for in vivo experimentsa
| Strain | Serotype | EcoR group | MLST results | Serum testb | Virulence gene profile
|
No. of VAGs | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adhesins
|
Iron acquisition
|
Protectin
|
Invasins, toxins, and others
|
||||||||||||||||||||||||||||||
| hra | iha | mat | papAH | papC | papEF | papG | tsh | chuA | fyuA | ireA | iroN | irp2 | iucD | iutA | sit (chromosomal) | sit (episomal) | iss | kpsMT | neuC | traT | EAST-1 | vat | gimB | ibeA | tia | cvi/cva | malX | ||||||
| Outbreak APEC strain | |||||||||||||||||||||||||||||||||
| IMT5155 | O2:K1:H5 | B2 | ST140/STC95 | R | − | − | + | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | − | + | + | 22 |
| Nonpathogenic control strain | |||||||||||||||||||||||||||||||||
| IMT11327 | Ont:H16 | B1 | ST295/none | R | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 2 |
| Nonoutbreak strains (Afecal/Aenviron) | |||||||||||||||||||||||||||||||||
| IMT6483 | Ont:H4 | B2 | New/none | R | − | − | + | − | − | − | − | − | + | + | − | − | + | − | − | + | − | + | + | − | + | − | + | + | − | − | + | + | 14 |
| IMT6487 | O8:H51 | B2 | New/none | R | + | − | + | − | − | − | − | − | + | + | − | − | + | + | + | − | − | − | − | − | + | + | + | − | + | − | + | + | 15 |
| IMT10666c | O59:H21 | B1 | ST58/STC155 | R | − | − | + | − | − | − | − | + | − | − | − | + | − | + | + | − | + | + | − | − | + | − | − | + | − | − | − | − | 11 |
| IMT10676c | O8:Hnt | B1 | New/none | R | − | − | + | − | − | − | − | − | − | − | − | + | − | + | + | − | + | + | − | − | + | − | − | − | − | − | + | − | 10 |
| IMT10740c | O2:H6 | B2 | New/none | R | − | − | + | − | − | − | − | + | + | + | − | + | + | + | + | + | + | + | + | − | + | − | + | − | + | − | + | + | 19 |
| IMT12205 | O43:H2 | B1 | ST920/none | S | + | − | + | − | − | − | − | − | − | + | − | − | − | + | + | − | − | − | − | − | + | + | + | − | + | − | + | + | 13 |
| IMT12226 | O77:H18 | D | NTd | R | − | − | + | + | + | + | + | + | + | − | + | + | − | + | + | + | − | + | − | − | + | + | − | − | − | + | + | − | 19 |
| IMT13060 | Ont:H6 | B2 | ST115/none | R | − | − | + | − | + | − | - | + | + | − | + | + | − | + | + | + | − | + | − | − | + | + | − | − | − | + | + | + | 17 |
| IMT14782 | Ont:NM | B2 | ST69/STC69 | R | + | + | + | − | + | − | + | − | + | + | + | + | + | + | + | + | − | + | − | − | + | + | − | − | − | + | + | − | 21 |
| IMT15146 | O1:H7 | B2 | ST95/STC95 | R | − | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | − | + | + | + | 26 |
| IMT15147 | O21:H4 | B2 | ST429/none | R | − | + | + | − | − | − | − | − | + | + | − | + | + | + | + | + | + | + | + | + | + | − | + | − | + | + | + | 20 | |
| IMT15151 | Ont:H31 | D | ST420/none | R | + | − | + | − | − | − | − | + | + | + | − | − | + | + | + | + | + | + | + | + | + | + | − | − | + | − | + | − | 19 |
| IMT15152 | Ont:H4 | B2 | ST428/none | R | − | − | + | − | − | − | − | − | + | + | − | + | + | − | − | + | + | + | + | + | + | − | + | − | + | − | + | + | 17 |
| IMT15153 | O21:H4 | B2 | ST429/none | R | − | − | + | − | − | − | − | − | + | − | + | + | + | − | − | + | + | + | + | + | + | + | + | − | + | − | + | + | 18 |
Results for genes for which all strains were either negative (afa/draB, bfp, bmaE, gafD, nfaE, sfa/foc, sfaS, focG, cnf1/2, hlyA, sat, stx1/2, pic, escV, and pks) or positive (fimC and ompA) are not shown, but they were used for calculation of the total number of VAGs per strain. +, gene present; −, gene not present.
R, serum resistant; S, serum sensitive.
Aenviron strain.
NT, not tested.
Chicken experiments.
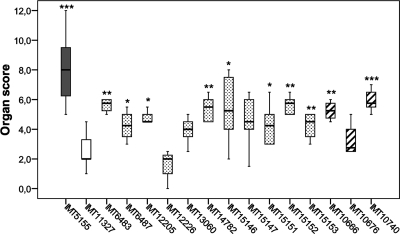
As mentioned above, in addition to the prototype colisepticemia outbreak strain IMT5155 and the negative control strain IMT11327, both of which were used to establish the systemic chicken infection model (1), we selected 14 nonoutbreak Afecal and Aenviron E. coli strains for in vivo experiments (Table 2). Briefly, the viable counts of bacteria in internal organs, as well as the ability to cause pathomorphological changes, scored based on the severity of organ lesions, are shown in Fig. 2 and 3. The most severe pathomorphological changes were observed in chickens infected with outbreak strain IMT5155, and the maximum average score was 7.5 ± 2.5 (Fig. 2). As expected, nonpathogenic control strain IMT11327 induced pathomorphological changes with a maximum average score of only 1.4, which was due mainly to moderate lesions in the lungs and air sacs.
FIG. 2.
Organ lesion scores for groups of chickens 24 h after infection with APEC outbreak strain IMT5155 (gray bar), nonpathogenic control strain IMT11327 (open bar), Afecal E. coli strains (dotted bars), and Aenviron E. coli strains (striped bars). *, P ≤ 0.05; **, P ≤ 0.02; ***, P ≤ 0.001. IMT, Institut für Mikrobiologie und Tierseuchen (Institute of Microbiology and Epizootics).
FIG. 3.
Bacterial loads in (A) lungs, (B) spleens, and (C) kidneys of groups of chickens 24 h after intratracheal infection with APEC outbreak strain IMT5155 (gray bars), nonpathogenic control strain IMT11327 (open bars), Afecal E. coli strains (dotted bars), and Aenviron E. coli strains (striped bars). *, P ≤ 0.05; **, P ≤ 0.02; ***, P ≤ 0.001. IMT, Institut für Mikrobiologie und Tierseuchen (Institute of Microbiology and Epizootics).
However, a considerable number of the nonoutbreak Afecal and Aenviron strains caused macroscopic lesions typical of systemic APEC infection. Among other symptoms, we observed air sacculitis, pneumonia, and pericarditis that resulted in organ lesion scores significantly higher than those obtained for chickens infected with IMT11327. Only Afecal E. coli strain IMT12226 had an organ lesion score similar to the score for the negative control strain, while the remaining scores ranged from 3.3 ± 1.2 for IMT10676 to 6.2 ± 1.9 for IMT10740, both of which have been isolated from the air-handling system of a chicken coop. The latter strain thus caused a highly significant increase in the organ score (P < 0.001) compared to the negative control strain IMT11327. There were four more strains that caused higher organ scores at a significance level of P < 0.02 and another four strains that caused moderately higher scores (P < 0.05).
Although the potential to induce lesions in internal organs is an important criterion for the virulence grade of a strain, a bacterium's ability to persist systemically is crucial and was therefore assessed in our study. At 24 h postinfection, viable bacterial counts comparable to those observed for outbreak strain IMT5155 were detected in the lungs of four groups of chickens infected with nonoutbreak strains IMT6487, IMT10666, IMT10676, and IMT10740, while approximately 10-fold reductions in the bacterial loads were observed for the lungs of the remaining chicken groups (Fig. 3). Conversely, it was observed that on average bacterial spread in internal organs was reduced in chickens infected with negative control strain IMT11327, and the decreases in the viable counts were almost 100- to 1,000-fold in the spleen, kidneys, and liver (examples are shown for the kidneys and spleen in Fig. 3). Comparable decreases in the bacterial counts were observed for organs of chickens infected with Afecal strains IMT12205, IMT12226, and IMT15153. Although none of the other nonoutbreak strains had viable counts in internal organs that were as high as those observed for IMT5155, we found 10- to 1,000 fold increases in the spleen, kidneys, and liver. On average, higher numbers of Aenviron strain IMT10740 and Afecal strain IMT15146 than of the negative control strain and the remaining nonoutbreak strains were reisolated from all internal organs, as shown for the lung, spleen, and kidneys in Fig. 3.
DISCUSSION
In the present study we analyzed the virulence traits, phylogenetic background, and serum resistance of E. coli nonoutbreak isolates obtained from clinically healthy chickens and their environment (Afecal and Aenviron strains) and compared these strains with APEC outbreak strains. We found that a number of Afecal E. coli strains have characteristics typical of APEC and basically of human and animal ExPEC and that some nonoutbreak strains are capable of causing systemic disease in immunocompetent 5-week-old chickens. Thus, our findings corroborate the avian intestine reservoir hypothesis. Most interestingly, some Afecal strains belonged to multilocus sequence types that are regularly observed not only among avian strains but also among human ExPEC strains, including strains causing UTI, septicemia, and newborn meningitis. Our data therefore strongly suggest that the virulence of APEC strains is determined not by their extraintestinal habitat but rather by their phylogeny. Another important finding was that the serum resistance of avian E. coli strains is not a useful marker for in vivo virulence, as this feature is widely distributed among all three groups of strains tested irrespective of their virulence for adult chickens.
The virulence genotyping data were basically compatible with previous studies that established the fact that a few VAGs which are probably essential in this host environment, like fimC, mat, ompA, and traT, were regularly detected in Afecal E. coli strains. For the other VAGs, a high level of diversity was observed, which is consistent with the situation in APEC, demonstrating that all stages of infection could putatively be mediated by several alternative virulence factors (11, 12, 16, 37). In general, nearly all VAGs tested in our study were present significantly more often in the outbreak strains, a finding in agreement with the findings of previous studies (6, 24, 28, 41).
However, this reflects a narrow view of the virulence features of the E. coli groups analyzed as a whole, neglecting the possibility that one or more exceptional supposedly nonpathogenic strains are potential risks as infectious agents. Taking a closer look at the virulence gene patterns of single nonoutbreak avian E. coli strains in particular, we recognized a considerable number of strains that were absolutely indistinguishable from outbreak strains because they harbored genes like fimC, papC, papG, hra, tsh, fyuA, chuA, iroN, iucD, iutA, sit, cvi/cva, iss, vat, ibeA, gimB, and neuC in various combinations, some of which have recently been directly linked with the “APEC pathotype” (37, 41), in addition to the similarity to other ExPEC pathovars (12, 20, 36). We also found that, although significantly higher numbers of Afecal strains were sensitive to chicken serum, nearly 80% of these strains exhibited a serum-resistant phenotype, which currently is a commonly accepted virulence feature of APEC (32). A similar proportion (4%) of serum-sensitive isolates among APEC strains has been reported by Vandemaele et al.; however, these workers did not compare the outbreak isolates with Afecal strains (42). Also similar to our data, Vandekerchove et al. did not find a significant difference between serum resistance in outbreak isolates and serum resistance in control isolates, supporting our hypothesis that the chicken intestine may accommodate virulent ExPEC strains (41). In the study of Vandekerchove et al. cecal (95%) and extraintestinal (100%) outbreak isolates showed negligibly higher percentages of serum resistance than cecal (88%) and extraintestinal (87%) control isolates. Thus, these authors concluded that either this trait might not be useful as a marker for the APEC pathotype or fecal strains could possibly serve as a reservoir for extraintestinal infections. In our study some serum-resistant strains were indeed highly pathogenic, while an association of this phenotype with a strain's ability to cause severe systemic disease is questionable considering the different degrees of virulence of the remaining serum-resistant strains. A further point supporting the hypothesis that serum resistance may not be useful as a “pathotype marker” per se is that nearly all Aenviron E. coli strains isolated from different sources in chicken coops appeared to be as serum resistant as APEC, irrespective of their virulence genes. Thus, in general, this phenotype might instead confer tenacity to the bacteria, enabling them to survive even outside a host, which would in a way contribute to their transmission, a suggestion that clearly needs to be explored further in the future.
Pyelonephritis-associated fimbriae are known to play a crucial role in the pathogenesis of APEC and UPEC infections by means of their PapG adhesin, which occurs as different molecular variants encoded by the three alleles of the corresponding gene, papG (8, 20, 35). In our study, the papG-positive E. coli strains included not only APEC strains (40.3%) but also Afecal strains (9.0%), whereas Aenviron strains were negative for this adhesin gene. Consistent with the findings of previous studies of the prevalence of papG alleles in APEC, our analysis showed that papG allele II was by far the most predominant allele in both outbreak strains (93.8%) and Afecal nonoutbreak strains (94.7%), while papG allele III was detected in only one strain of these two groups (41, 42). It has been shown that strains which cause pyelonephritis and bacteremia in humans more commonly contain papG allele II (22), whereas papG allele III has been associated with human cystitis (19). Johnson et al. showed that PapGIII sequences from humans and dogs are largely indistinguishable and suggested that if humans do acquire ExPEC from dogs, the acquisition is probably primarily related to papG allele III-containing strains (19, 22). Similarly, Vandemaele et al. (42) found that the PapGII sequences of two avian strains were highly homologous (99%) or identical to the PapGII sequences of human strains. Although we did not sequence the papG allele sequences, we hypothesize that both avian pathogenic and Afecal strains from clinically healthy chickens probably have zoonotic potential, as numerous scenarios of transmission between birds and humans via physical contact, contaminated dust, eggs, etc. can be envisioned.
The supposed zoonotic potential of APEC strains, particularly Afecal strains from healthy birds, is further supported by the phylogenetic background of the strains and their in vivo pathogenicity. In a rigorous study comparing human ExPEC and commensal strains using a mouse model, Picard et al. (34) hypothesized that the virulence or nonvirulence of a strain cannot be systematically inferred from its “commensal” or “pathogenic” origin but can be inferred from its phylogenetic background (34). They observed that phylogenetic group B2 strains were the strains most virulent for mice, which was in agreement with the presence of a large number of VAGs, and this is why virulence was suggested to be an ancestral characteristic in the B2 phylogenetic group (25). Le Gall et al. (25) performed association studies which revealed that the extraintestinal virulence of group B2 strains involves a multigenic process with a common set of virulence determinants which could also be viewed as intestinal colonization and survival factors linked to commensalism, as they could increase the fitness of the strains in the normal gut environment. Indeed, it has recently been demonstrated that the presence of intestinal EcoR group B2 E. coli strains harboring typical ExPEC VAGs correlates with successful colonization in piglets (38). Other workers have also shown that extraintestinal pathogenic human and animal group B2 and D strains have a greater frequency and diversity of virulence traits than group A and B1 strains (3, 4, 12, 18, 25), which is compatible with our results. Moreover, MLST analysis revealed that a considerable number of nonoutbreak strains could be assigned to sequence types that are quite common in ExPEC strains not only of avian origin but also of human origin. Afecal strain IMT15146 (O1:K1:H7) was allocated to ST95 of STC95, which according to the data available to date (www.mlst.net) comprises 150 K1-positive human and animal strains capable of causing extraintestinal infections like septicemia, UTI, and newborn meningitis, as well as human commensal E. coli strains obtained from the guts of clinically healthy patients, indicating that there may be similar intestinal reservoirs in birds and humans. Another Afecal strain, IMT14782 (Ont:NM), belonged to ST69 of STC69, which to date includes almost exclusively human UTI strains (n = 39) and contains only four strains from animals with unknown clinical histories. Other sequence types (ST58, ST115, ST420, ST428, and ST429) identified among nonoutbreak strains in our study are quite small groups in the database but nevertheless contain K1-positive ExPEC strains from avian and human sources. Capsular serotype K1 E. coli strains are highly associated with invasive diseases, and interestingly, the Afecal strains that have been grouped in the sequence types mentioned above also possess the K1 biosynthesis gene neuC, supporting the hypothesis that they have pathogenic potential. The fact that these strains and the strains whose sequence types so far are unknown were potent colonizers of internal organs of 5-week-old intratracheally infected chickens and the fact that most of them in addition produced organ lesions that were significantly larger than those produced by negative control strain IMT11327 support our hypothesis that an E. coli subpopulation within the avian intestine serves as a reservoir for extraintestinal pathogens that could emerge as virulent strains if there were a change in the environment or host immunity.
In conclusion, virulence genotyping and phylogenetic data, including EcoR analysis and MLST data, show that certain nonoutbreak strains, particularly Afecal strains originating from the intestine of clinically healthy poultry, have zoonotic potential because they either are directly transferred from birds to humans or serve as a genetic pool for extraintestinal pathogenic strains. A rigorous epidemiological study to determine the diversity of the E. coli population in the chicken intestine is currently being performed in our lab in order to analyze the frequency of such strains in the chicken gut, with the goal of developing intestinal intervention strategies in the future.
Supplementary Material
Acknowledgments
We thank Uwe Boettcher, Timo Homeier, and Astrid Bethe for their indispensable assistance during animal experiments, Bert-Andrée Zucker for providing environmental E. coli strains, and John M. Fairbrother for providing APEC strain MT78. We are also indebted to Rudolf Preisinger of Lohmann Tierzucht GmbH, Cuxhaven, Germany, for providing specific-pathogen-free chickens.
This work was supported by the National Genome Research Network within the framework of the Funktionelle Genomanalyse Tierischer Organismen (FUGATO) “E. coli chick” project funded by the Federal Ministry of Education. The German Research Foundation (DFG) supported this work through project Wi-1436/5-1 and by providing a fellowship from the Graduate School to Esther-Maria Antão (GRK 1121 project A2).
Footnotes
Published ahead of print on 7 November 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Antão, E.-M., S. Glodde, G. Li, T. Homeier, C. Laturnus, I. Diehl, A. Bethe, H.-C. Philipp, R. Preisinger, L. H. Wieler, and C. Ewers. 20 September 2008, posting date. The chicken as a natural model for extraintestinal infections caused by avian pathogenic Escherichia coli (APEC). Microb. Pathog. doi: 10.1016/j.micpath.2008.005. [DOI] [PubMed]
- 2.Barnes, H. J., and W. B. Gross. 1999. Colibacillosis, p. 131-141. In W. B. Gross (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames.
- 3.Bingen, E., B. Picard, N. Brahimi, S. Mathy, P. Desjardins, J. Elion, and E. Denamur. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642-650. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E. F., and D. L. Hartl. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delicato, E. R., B. G. de Brito, L. C. Gaziri, and M. C. Vidotto. 2003. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet. Microbiol. 94:97-103. [DOI] [PubMed] [Google Scholar]
- 7.Dozois, C. M., J. M. Fairbrother, J. Harel, and M. Bosse. 1992. pap- and pil-related DNA sequences and other virulence determinants associated with Escherichia coli isolated from septicemic chickens and turkeys. Infect. Immun. 60:2648-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dozois, C. M., S. A. Pourbakhsh, and J. M. Fairbrother. 1995. Expression of P and type 1 (F1) fimbriae in pathogenic Escherichia coli from poultry. Vet. Microbiol. 45:297-309. [DOI] [PubMed] [Google Scholar]
- 9.Ewers, C., T. Janssen, S. Kiessling, H. C. Philipp, and L. H. Wieler. 2005. Rapid detection of virulence-associated genes in avian pathogenic Escherichia coli by multiplex polymerase chain reaction. Avian Dis. 49:269-273. [DOI] [PubMed] [Google Scholar]
- 10.Ewers, C., T. Janssen, and L. H. Wieler. 2003. Avian pathogenic Escherichia coli (APEC). Berl. Munch. Tierarztl. Wochenschr. 116:381-395. (In German.) [PubMed] [Google Scholar]
- 11.Ewers, C., S. Kießling, L. H. Wieler, T. Janßen, and H.-C. Philipp. 2004. Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Vet. Microbiol. 104:91-101. [DOI] [PubMed] [Google Scholar]
- 12.Ewers, C., G. Li, H. Wilking, S. Kiessling, K. Alt, E.-M. Antão, C. Laturnus, I. Diehl, S. Glodde, T. Homeier, U. Boehnke, H. Steinrueck, H.-C. Philipp, and L. H. Wieler. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297:163-176. [DOI] [PubMed] [Google Scholar]
- 13.Ewers, C., C. Schueffner, R. Weiss, G. Baljer, and L. H. Wieler. 2004. Molecular characteristics of Escherichia coli serogroup O78 strains isolated from diarrheal cases in bovines urge further investigations on their zoonotic potential. Mol. Nutr. Food Res. 48:504-514. [DOI] [PubMed] [Google Scholar]
- 14.Gross, W. B. 1994. Diseases due to Escherichia coli in poultry, p. 237-259. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 15.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janßen, T., C. Schwarz, P. Preikschat, M. Voss, H. C. Philipp, and L. H. Wieler. 2001. Virulence-associated genes in avian pathogenic Escherichia coli (APEC) isolated from internal organs of poultry having died from colibacillosis. Int. J. Med. Microbiol. 291:371-378. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. R., J. J. Brown, U. B. Carlino, and T. A. Russo. 1998. Colonization with and acquisition of uropathogenic Escherichia coli as revealed by polymerase chain reaction-based detection. J. Infect. Dis. 177:1120-1124. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. R., T. T. O'Bryan, D. A. Low, G. Ling, P. Delavari, C. Fasching, T. A. Russo, U. Carlino, and A. L. Stell. 2000. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG allele III. Infect. Immun. 68:3327-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, J. R., and T. A. Russo. 2005. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int. J. Med. Microbiol. 295:383-404. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, J. R., A. L. Stell, and P. Delavari. 2001. Canine feces as a reservoir of extraintestinal pathogenic Escherichia coli. Infect. Immun. 69:1306-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, T. J., K. E. Siek, S. J. Johnson, and L. K. Nolan. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188:745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Gall, T., O. Clermont, S. Gouriou, B. Picard, X. Nassif, E. Denamur, and O. Tenaillon. 2007. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol. Biol. Evol. 24:2373-2384. [DOI] [PubMed] [Google Scholar]
- 26.Li, G., C. Ewers, C. Laturnus, I. Diehl, K. Alt, J. Dai, E.-M. Antão, K. Schnetz, and L. H. Wieler. 2008. Characterization of a yjjQ mutant of avian pathogenic Escherichia coli (APEC). Microbiology 154:1082-1093. [DOI] [PubMed] [Google Scholar]
- 27.Marklund, B. I., J. M. Tennent, E. Garcia, A. Hamers, M. Baga, F. Lindberg, W. Gaastra, and S. Normark. 1992. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol. Microbiol. 6:2225-2242. [DOI] [PubMed] [Google Scholar]
- 28.McPeake, S. J., J. A. Smyth, and H. J. Ball. 2005. Characterisation of avian pathogenic Escherichia coli (APEC) associated with colisepticaemia compared to faecal isolates from healthy birds. Vet. Microbiol. 110:245-253. [DOI] [PubMed] [Google Scholar]
- 29.Mellata, M., M. Dho-Moulin, C. M. Dozois, R. Curtiss III, P. K. Brown, P. Arne, A. Bree, C. Desautels, and J. M. Fairbrother. 2003. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect. Immun. 71:536-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moulin-Schouleur, M., M. Reperant, S. Laurent, A. Bree, S. Mignon-Grasteau, P. Germon, D. Rasschaert, and C. Schouler. 2007. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J. Clin. Microbiol. 45:3366-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller, D., P. Hagedorn, S. Brast, G. Heusipp, M. Bielaszewska, A. W. Friedrich, H. Karch, and M. A. Schmidt. 2006. Rapid identification and differentiation of clinical isolates of enteropathogenic Escherichia coli (EPEC), atypical EPEC, and Shiga toxin-producing Escherichia coli by a one-step multiplex PCR method. J. Clin. Microbiol. 44:2626-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nolan, L. K., S. M. Horne, C. W. Giddings, S. L. Foley, T. J. Johnson, A. M. Lynne, and J. Skyberg. 2003. Resistance to serum complement, iss, and virulence of avian Escherichia coli. Vet. Res. Commun. 27:101-110. [DOI] [PubMed] [Google Scholar]
- 33.Pfaff-McDonough, S. J., S. M. Horne, C. W. Giddings, J. O. Ebert, C. Doetkott, M. H. Smith, and L. K. Nolan. 2000. Complement resistance-related traits among Escherichia coli isolates from apparently healthy birds and birds with colibacillosis. Avian Dis. 44:23-33. [PubMed] [Google Scholar]
- 34.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pourbakhsh, S. A., M. Dho-Moulin, A. Bree, C. Desautels, B. Martineau-Doize, and J. M. Fairbrother. 1997. Localization of the in vivo expression of P and F1 fimbriae in chickens experimentally inoculated with pathogenic Escherichia coli. Microb. Pathog. 22:331-341. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151:2097-2110. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, and L. K. Nolan. 2005. Characterizing the APEC pathotype. Vet. Res. 36:241-256. [DOI] [PubMed] [Google Scholar]
- 38.Schierack, P., N. Walk, C. Ewers, H. Wilking, H. Steinrueck, M. Filter, and L. H. Wieler. 2008. ExPEC-typical virulence-associated genes correlate with successful colonization by intestinal E. coli in a small piglet group. Environ. Microbiol. 10:1742-1751. [DOI] [PubMed] [Google Scholar]
- 39.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skyberg, J. A., K. E. Siek, C. Doetkott, and L. K. Nolan. 2007. Biofilm formation by avian Escherichia coli in relation to media, source and phylogeny. J. Appl. Microbiol. 102:548-554. [DOI] [PubMed] [Google Scholar]
- 41.Vandekerchove, D., F. Vandemaele, C. Adriaensen, M. Zaleska, J. P. Hernalsteens, L. De Baets, P. Butaye, F. Van Immerseel, P. Wattiau, H. Laevens, J. Mast, B. Goddeeris, and F. Pasmans. 2005. Virulence-associated traits in avian Escherichia coli: comparison between isolates from colibacillosis-affected and clinically healthy layer flocks. Vet. Microbiol. 108:75-87. [DOI] [PubMed] [Google Scholar]
- 42.Vandemaele, F. J., J. P. Mugasa, D. Vandekerchove, and B. M. Goddeeris. 2003. Predominance of the papGII allele with high sequence homology to that of human isolates among avian pathogenic Escherichia coli (APEC). Vet. Microbiol. 97:245-257. [DOI] [PubMed] [Google Scholar]
- 43.Watt, S., P. Lanotte, L. Mereghetti, M. Moulin-Schouleur, B. Picard, and R. Quentin. 2003. Escherichia coli strains from pregnant women and neonates: intraspecies genetic distribution and prevalence of virulence factors. J. Clin. Microbiol. 41:1929-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto, S., A. Terai, K. Yuri, H. Kurazono, Y. Takeda, and O. Yoshida. 1995. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol. Med. Microbiol. 12:85-90. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto, T., and P. Echeverria. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.