Abstract
We have shown that the plant pathogen Alternaria brassicicola exhibited very high susceptibility to ambruticin VS4 and to a lesser extent to the phenylpyrrole fungicide fludioxonil. These compounds are both derived from natural bacterial metabolites with antifungal properties and are thought to exert their toxicity by interfering with osmoregulation in filamentous fungi. Disruption of the osmosensor group III histidine kinase gene AbNIK1 (for A. brassicola NIK1) resulted in high levels of resistance to ambruticin and fludioxonil, while a mutant isolate characterized by a single-amino-acid substitution in the HAMP domain of the kinase only exhibited moderate resistance. Moreover, the natural resistance of Saccharomyces cerevisiae to these antifungal molecules switched to sensitivity in strains expressing AbNIK1p. We also showed that exposure to fludioxonil and ambruticin resulted in abnormal phosphorylation of a Hog1-like mitogen-activated protein kinase (MAPK) in A. brassicicola. Parallel experiments carried out with wild-type and mutant isolates of Neurospora crassa revealed that, in this species, ambruticin susceptibility was dependent on the OS1-RRG1 branch of the phosphorelay pathway downstream of the OS2 MAPK cascade but independent of the yeast Skn7-like response regulator RRG2. These results show that the ability to synthesize a functional group III histidine kinase is a prerequisite for the expression of ambruticin and phenylpyrrole susceptibility in A. brassicicola and N. crassa and that, at least in the latter species, improper activation of the high-osmolarity glycerol-related pathway could explain their fungicidal properties.
Two-component histidine kinase (HK) phosphorelay signaling systems are involved in several signal transduction pathways in plants, slime molds, fungi, and bacteria (5). They play pivotal roles in responses to environmental stimuli and regulate various processes, including virulence in plant and animal pathogens (30). As HKs have not been identified in animals (5), they may be considered as valuable potential targets for antimicrobial agents. Recent studies have shown that molecules such as the phenylpyrrole fungicide fludioxonil exert their fungicidal activity in filamentous fungi through activation of the high-osmolarity glycerol (HOG) pathway, a mitogen-activated protein kinase (MAPK) cascade involved in the response to high osmotic stress (18). In filamentous fungi, this pathway is regulated by a group III HK (Os1 family of two-component HK) that probably acts upstream as a positive activator of the cascade and may correspond to an osmosensor (31). As the yeast Saccharomyces cerevisiae possesses only one hybrid HK (SNL1p), which differs from the group III HKs (24) and is resistant to phenylpyrroles, the susceptibility of filamentous fungi to this class of fungicide may be dependent on the presence of a group III HK. This hypothesis is strongly supported by recent results indicating that expression of an Os1 orthologue from the rice blast fungus in S. cerevisiae can confer fungicide susceptibility (20, 28).
Naturally occurring molecules indeed represent an important source of antifungal agents that may be used as starting points for the synthesis of new compounds (9). Phenylpyrrole fungicides are derived from the natural bacterial antibiotic pyrrolnitrin produced by Pseudomonas pyrrocinia (2). Strobilurin A, a polyketide produced by the fungus Strobilurus tenacellus, has been used to design a number of commercially important antifungal agents. The ambruticins, another class of natural antifungal polyketides originating from the myxobacterium Sorangium cellulosum (11, 25), were reported to have no significant adverse effect on animals (6, 26) and therefore constitute attractive leads for antifungal drug development. However, while they were shown to be active against Aspergillus species that have a high incidence in chronic respiratory infections in humans, their development as commercial drugs has been hampered by their narrow activity spectrum on animals and human fungal pathogens (17) and by the high cost of total chemical synthesis. The recent characterization of the gene cluster involved in their synthesis in S. cellulosum (16) may pave the way toward future development of a less expensive biological production process for these metabolites. Moreover, they have also been tested successfully in vitro against at least one important crop pathogen, i.e., Botrytis cinerea (17). While the mechanism of action of ambruticins is still unclear, it has been shown that, like phenylpyrroles, these molecules interfere with osmoregulation in filamentous fungi (17), probably by targeting the HOG pathway (28).
Here we have analyzed the effect of ambruticin VS4, one of the N-methylated forms of ambruticin, on the growth of different genotypes of Alternaria brassicicola, an economically important seed-borne fungal pathogen of Brassicaceae species. Parallel experiments were also conducted with the model fungus Neurospora crassa. We demonstrated that ambruticin targets group III HK phosphorelay signaling systems in these two filamentous fungi and, at least in N. crassa, exhibits fungicidal activity through improper activation of the HOG-related pathway.
MATERIALS AND METHODS
Strains and media.
All A. brassicicola strains used in this study and listed in Table 1 have previously been described (3, 12), except the laboratory-resistant strain Abra363H5M, which was obtained after transferring, onto selective medium, mycelial tips from fast-growing sectors observed with some sensitive isolates on iprodione-amended media. All strains were maintained by transferring hyphal plugs to dishes of potato dextrose agar medium amended or not with fungicides (fludioxonil or ambruticin VS4) at the desired concentrations. The fungicides used in this study were added to the media from 1,000-fold-concentrated stock solutions in dimethyl sulfoxide. The effects of fungicides on mycelial growth and spore germination were tested as described by Iacomi-Vasilescu et al. (12).
TABLE 1.
Susceptibilities of A. brassicicola and N. crassa strains to DCF
| Strain | Phenotypea | Genotype | Source or reference |
|---|---|---|---|
| A. brassicicola | |||
| Abra18b | DCFs | WT | 3 |
| Abra43 | DCFs | WT | 3 |
| Abra12Ro | DCFs | WT | 3 |
| AbraCM | DCFs | WT | 3 |
| Abra20 | DCFs | WT | 3 |
| Abra363H5 | DCFs | WT | This study |
| Abra363H5M | DCFr | AbNIK1V552A | This study |
| Abra40 | DCFr | AbNIK1E753K | 3 |
| Abra43M | DCFr | AbNIK1Q343NS | 3 |
| Abra3 | DCFr | AbNIK1Q998NS | 3 |
| Abra7407 | DCFr | AbNIK1W634NS | 3 |
| Abra41 | DCFr | AbNIK1ΔCA | 3 |
| AbraCP | DCFr | NDb | 3 |
| nik1Δ1 | DCFr | AbNIK1::hph | This study |
| nik1Δ2 | DCFr | AbNIK1::hph | This study |
| nik1Δ3 | DCFr | AbNIK1::hph | This study |
| N. crassa | |||
| FGSC988 | DCFs | WT | Fungal Genetics Stock Center |
| FGSC824 | DCFlr | Os1Q388S-A578V | Fungal Genetics Stock Center |
| FGSC2432 | DCFr | Os1G580R-L582M | Fungal Genetics Stock Center |
| FGSC4494 | DCFr | Os1Q308NS | Fungal Genetics Stock Center |
| FGSC1509 | DCFr | Os2T97FS | Fungal Genetics Stock Center |
| FGSC4576 | DCFr | Os5K307FS | Fungal Genetics Stock Center |
| FGSC13363 | DCFr | rrg1::hph | Fungal Genetics Stock Center |
| FGSC16195 | DCFs | rrg2::hph | Fungal Genetics Stock Center |
r, resistant; s, susceptible; lr, low resistance.
ND, not determined.
All N. crassa strains were obtained from the Fungal Genetics Stock Center (University of Kansas Medical Center, Kansas City) and grown on agar-solidified Vogel's medium N and 1.2% (wt/vol) sucrose at 25°C (29). S. cerevisiae (strain INVSc1; Invitrogen) was grown at 30°C. YPD (1% yeast extract, 2% peptone, 2% glucose) was used as the complete medium. SD (0.67% yeast nitrogen base without amino acids, 2% glucose, 0.192% dropout supplement without uracil) or SG (0.67% yeast nitrogen base without amino acids, 2% galactose, and 0.192%· dropout supplement without uracil) medium was used as the minimal medium. The media were solidified by the addition of 2% Bacto agar.
Western blot analysis.
To screen the different A. brassicicola strains for their ability to synthesize the HK AbNIK1p (for A. brassicola NIK1p), Western blot analyses with rabbit antibodies raised against a synthetic peptide, whose sequence corresponded to residues 364 to 378 of the AbNIK1p sequence (accession no. AAU10313), were carried out. The phosphorylation of Hog1-related MAPKs in A. brassicicola and N. crassa was examined by the same technique with anti-dually phosphorylated p38 (Cell Signaling Technology, Beverly, MA) and C-terminal anti-Hog1p (Santa Cruz Biotechnology, CA) antibodies. Protein samples were prepared from mycelia obtained by growing a conidial suspension at 25°C for 24 h in potato dextrose broth (A. brassicicola, 200,000 conidia/ml) or Vogel's liquid medium N plus 1% (wt/vol) sucrose (N. crassa, 300,000 conidia/ml). Mycelia were collected by filtration on filter paper and ground with a mortar and pestle to a fine powder under liquid nitrogen. Ice-chilled buffer containing protease and phosphatase inhibitors (50 mM Na phosphate [pH 7.4], 1 mM EDTA, 5% [vol/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 5 mM Na pyrophosphate, 0.1 mM Na vanadate, 10 mM β-glycerophosphate) was added to the powder, and the suspensions were homogeneized. After centrifugation of the suspensions at 10,000 × g for 10 min, the protein concentration in the resulting supernatants was calculated using a bicinchoninic acid protein assay reagent (Pierce, Rockford, IL). Equal quantities (25 μg and 5 μg for AbNIK1p and Hog1-related MAPK detections, respectively) of protein samples were loaded on 10% polyacrylamide gels and blotted onto nitrocellulose membranes (Schleicher and Schuell, Dassel, Germany). Antibody binding was visualized using an ECL Plus enhanced chemiluminescence Western blotting detection reagent (Amersham Biosciences, Buckinghamshire, United Kingdom) after the binding of a horseradish peroxidase-conjugated secondary antibody.
DNA procedure and Southern hybridization.
Genomic DNA was extracted from mycelium according to Adachi et al. (1). For Southern analysis, DNA fragments resulting from genomic DNA digestion with NruI and BamHI were separated on 1% agarose gels and vacuum transferred to Hybond N membranes (Amersham Biosciences). Blots were then probed with a PCR fragment, amplified from genomic DNA using the primer set described below for construction of the AbNIK1 disruption cassette, and 32P labeled with a random primer labeling kit (Amersham Biosciences).
Generation of targeted gene disruption construct and fungal transformation.
Based on the previously reported AbNIK1 sequence (3), two primers were designed at the bp 668 position with an exogenous BamHI enzyme site (5′-ggatccTGGCAAACATCAACAGGGTA-3′) and at the bp 995 position with an exogenous HindIII enzyme site (5′-aagcttCAGCCCAAATACCAGCAACT-3′) in relation to the putative start codon. (The respective exogenous enzyme sites are lowercase in the sequences.) These primers were used to amplify a 328-bp partial target gene from the genomic DNA. PCR products were digested with HindIII and BamHI and ligated at the corresponding multiple cloning sites in pCB1636 (27). The ligation was transformed in Escherichia coli strain JM109 (Promega, France). The plasmid was sequenced to check for the presence of an insert and a hygromycin B (Hyg B) cassette and used as template for PCR amplification using M13 forward and M13 reverse primers. The PCR product was purified and used to transform A. brassicicola strain Abra43 based on the protocol described by Cho et al. (7).
Expression of AbNIK1 in S. cerevisiae.
Fungal RNA was extracted, DNase treated, and reverse transcribed as described by Guillemette et al. (8). The cDNA encoding the Os1 family HK gene AbNIK1 of A. brassicicola was amplified by PCR from first-strand cDNAs with the primers STupEco (5′-GAATTCCATAATGGCCGCAGAGACGTACTCGAGCGT-3′) and STdoNot (5′-GCGGCCGCTCAGCTACTGTGACTCCGCAGCAAAAG-3′) with Phusion hot start high-fidelity DNA polymerase (Finnzymes, Espoo, Finland). Unique EcoRI and NotI restriction sites (indicated in boldface) were thus introduced at the upstream start and downstream stop codons (underlined) of the AbNIK1 sequence, respectively. The PCR product was cloned into pGEM-T (Promega) and then transferred into the yeast expression vector pYES2-CT (Invitrogen) after EcoRI-NotI double digestion and ligation to yield the expression clone pYAbNik1. This plasmid can express full-length AbNIK1p without a tag under the control of the GAL1 promoter. The E753K substitution was created by site-directed mutagenesis (Phusion SDM kit; New England Biolabs, Ipswich, MA) on pYAbNik1. S. cerevisiae was transformed using the S.c. EasyComp transformation kit (Invitrogen). Transformants were incubated under GAL1 promoter-repressive conditions (SD without uracil [SD/−Ura]) or under inductive conditions (SG without uracil [SG/−Ura]). Sensitivity to reagents was analyzed on plates. Transformants were precultured overnight in 5 ml of SD/−Ura or SG/−Ura. Serially diluted (1:10 [starting optical density at 600 nm of 0.1]) cell suspensions (5 μl) were spotted onto plates with each of the test reagents, and the plates were incubated for 60 to 240 h.
RESULTS
Alternaria brassicicola is highly susceptible to ambruticin.
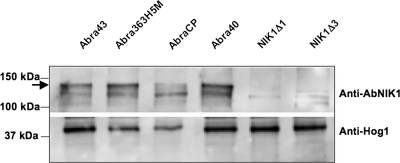
Seven A. brassicicola isolates highly resistant to the dicarboximide fungicide (DCF) iprodione (Table 1) were tested for their susceptibility to the phenylpyrrole fungicide fludioxonil and to ambruticin. All but strain AbraCP were shown to carry mutations at the AbNIK1 locus. Four isolates were previously shown to bear nonsense or frameshift mutations (3) and characterized at the protein level as group III HK-null mutants (13). Immunoblots probed with specific antibodies raised against AbNIK1p (Fig. 1) revealed that strain AbraCP was also unable to synthesize AbNIK1p, suggesting the presence of mutations in the regulatory sequences of the gene. In contrast, a signal at 130 kDa corresponding to the expected size of AbNIK1p was obtained with both strain Abra40, previously reported as a substitution mutant (3), and strain Abra363H5M, an additional laboratory mutant highly resistant to iprodione. Sequence analysis at the AbNIK1 locus revealed that this strain carries a single nucleotide polymorphism at position 1801 relative to the start codon, resulting in the replacement of a valine by an alanine at repeat 4 of the HAMP domain.
FIG. 1.
Immunodetection of AbNIK1p in protein extracts from WT and mutant A. brassicicola isolates. Proteins, extracted from liquid cultures of WT (Abra43), DCF-resistant (AbraCP, Abra40, and Abra363H5M), and ΔAbNIK1 mutant (nik1Δ1 and nik1Δ3) cells, were analyzed by SDS-PAGE and blotting with anti-AbNIK1p N-terminus antibody (upper panel). The arrow indicates the position of immunoreactive polypeptides with a size corresponding to the complete form of the AbNIK1 protein (130 kDa). The fainter signal at ca. 110 kDa probably resulted from nonspecific binding, as was also observed when the specific antibody was replaced with preimmune serum (not shown). A duplicate blot was probed with anti-Hog1 C-terminus antibody (lower panel) to assess loading.
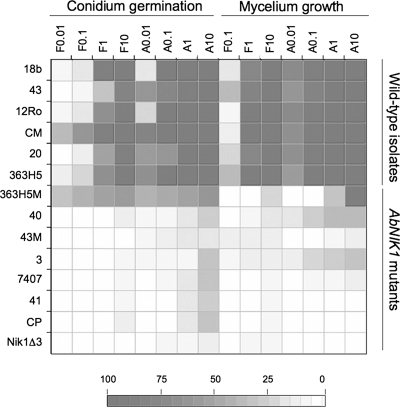
As shown in Fig. 2, based on their susceptibility to fludioxonil and ambruticin, the DCF-resistant strains could be separated into two groups. For most of the strains, i.e., the AbNIK1-null mutants and the Abra40 substitution mutant, the mycelial growth and conidium germination rates were not or only slightly affected in the presence of high concentrations (up to 10 mg/liter) of ambruticin or fludioxonil as compared to control conditions. In contrast, conidium germination and mycelial growth in the Abra363H5M substitution mutant were susceptible to high ambruticin concentrations (10 mg/liter).
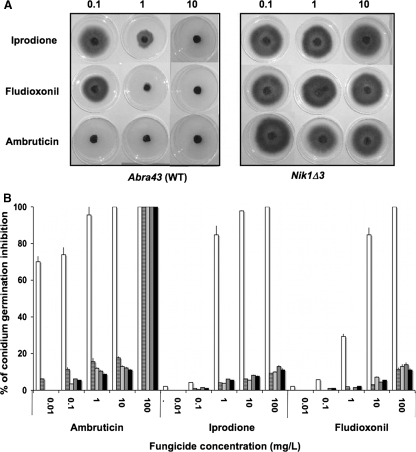
FIG. 2.
Antifungal activities of the phenylpyrrole fludioxonil and the polyketide ambruticin on A. brassicicola isolates. Conidia or mycelium from WT isolates and AbNIK1 mutants of A. brassicicola was exposed to either fludioxonil (F) or ambruticin (A) at various concentrations (0.01 to 10 mg/liter). For each condition, the reduction in germination or radial growth was expressed as a percentage relative to the dimethyl sulfoxide control. In the diagram, the color intensity is proportional to the inhibitory activity. Correspondences between color intensities and the percentages of fungal growth inhibition are given in the scale at the bottom.
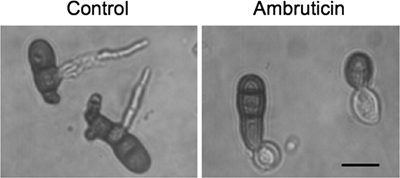
These two growth parameters were strongly affected by ambruticin, with almost complete inhibition at 0.1 mg/liter for all wild-type (WT) strains tested in this study. Irrespective of the growth parameter studied, the toxicity of fludioxonil to A. brassicicola was always lower than that of ambruticin. When conidia from WT strains were germinated in the presence of ambruticin, only short germ tubes with a tendency to swell were observed (Fig. 3), similar to what was previously observed with DCFs (3).
FIG. 3.
Effect of ambruticin treatment on A. brassicicola germinating conidia. Conidia from the WT isolate Abra43 were germinated in water supplemented or not (control) with 0.1 mg/liter of ambruticin. Scale bar, 10 μm.
AbNIK1 is involved in ambruticin susceptibility.
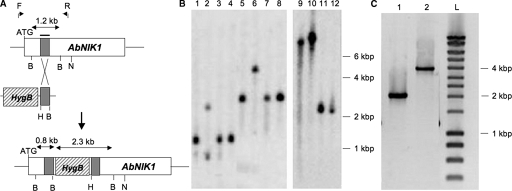
Additional support for the conclusions from the study of the above-mentioned mutants was provided by construction of a loss-of-function allele by homologous integration of a linear minimal element (LME) construct (7) into the AbNIK1 gene. This construct was expected to disrupt the target gene within the second HAMP domain repeat (Fig. 4A). After transformation of protoplasts of the Abra43 WT strain, regeneration, and purification by single spore isolation, three Hyg B-resistant transformants were found to be iprodione (DCF) resistant, thus suggesting that homologous recombination occurred in these strains. Based on Southern blot analysis (Fig. 4B), two of the three transformants, the nik1Δ1 and nik1Δ2 strains, had more than one copy of insertion of the LME construct in the target gene, while the third transformant, the nik1Δ3 strain, was clearly disrupted within the AbNIK1 locus by a single-copy insertion of the LME construct. This was further confirmed by long-range PCR (Fig. 4C) with flanking primers followed by nucleotide sequence analysis. The disruptant strains were confirmed to be null mutants by Western blot analysis with anti-HK antibodies (Fig. 1). As illustrated for the nik1Δ3 strain in Fig. 5A, comparison of the mycelial growth of these strains with that of the WT parental strain Abra43 on medium amended with iprodione, fludioxonil, and ambruticin showed that they were highly resistant to these three molecules at concentrations that completely inhibited growth of the WT strain. Similar differences between the WT strain Abra43 and the three nik1Δ disruptants or the Abra43M nonsense mutant were observed with respect to the effects on conidial germination (Fig. 5B).
FIG. 4.
AbNIK1 gene inactivation by homologous recombination. (A) Schematic representation of the AbNIK1 locus, the LME disruption construct with the Hyg B resistance gene cassette (hatched box) and the 328-bp partial gene fragment (gray box), and the typical gene disruption mutant locus. B, BamHI; H, HindIII; N, NruI. Arrows above the AbNIK1 locus indicate the positions of the forward (F) and reverse (R) primers used for long-range PCR. (B) Southern hybridization of genomic DNA from WT isolates (lanes 1, 3, 4, 5, 7, 8, 11, and 12) and three transformants (nik1Δ1, lane 9; nik1Δ2, lane 10, and nik1Δ3, lanes 2 and 6). Each DNA set was digested with BamHI (lanes 1 to 4) or NruI (lanes 5 to 12), and the blot was probed with the 32P-labeled partial gene fragment (shown by the gray boxes in panel A). (C) Gel electrophoresis of long-range PCR products obtained from DNA of the Abra43 WT isolate (lane 1) or of the nik1Δ3 transformant (lane 2) using the flanking primers F and R. The size of DNA fragments was estimated using the DNA Smart ladder (lane L [Eurogentec, Seraing, Belgium]) and is indicated on right of panels B and C.
FIG. 5.
Effects of fungicides on the mycelium radial growth and conidia germination rate of the A. brassicicola wild-type isolate and AbNIK1-null mutants. (A) In vitro growth tests were carried out on potato dextrose agar medium supplemented with iprodione, fludioxonil, or ambruticin at 0.1, 1, and 10 mg/liter. Growth of the Abra43 and nik1Δ3 strains was scored after 4 days of incubation at 25°C. (B) Germination rates were estimated by counting, under a microscope, the number of germinated conidia after incubation for 24 h at 25°C in the presence of iprodione, fludioxonil, and ambruticin at 0.01, 0.1, 1, 10, and 100 mg/liter. The results are expressed as the percentage of inhibition in treated samples compared to the control without fungicide. Values are the means of three replicates; error bars indicate standard deviations. White bars, Abra43; striped bars, Abra43M; light gray bars, nik1Δ1; medium gray bars, nik1Δ2; black bars, nik1Δ3.
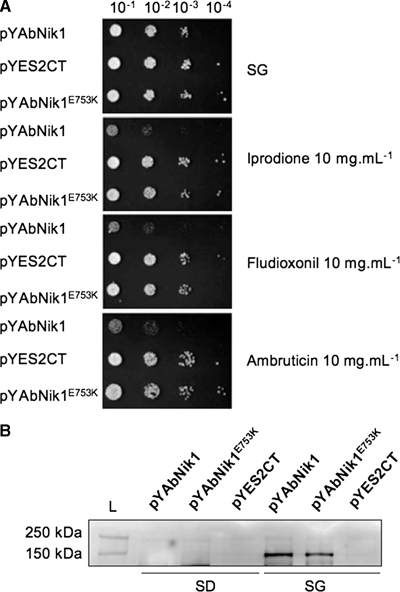
To further demonstrate the involvement of group III HK in the susceptibility of A. brassicicola to ambruticin, the AbNIK1 gene was expressed in S. cerevisiae, which is naturally resistant to this compound. Under inducing conditions, in the absence of fungicide, pYAbNik1 and pYES2-CT (empty vector) transformants showed similar growth, while the pYAbNik1-containing strain showed a growth defect in the presence of fludioxonil or ambruticin (Fig. 6A). Furthermore, yeast transformants expressing a mutated form of AbNIK1p harboring the same substitution as strain Abra40(pYAbNik1E753K) showed resistant phenotypes similar to that of the S. cerevisiae control strain. The successful expression of AbNIK1p by S. cerevisiae cells carrying recombinant plasmids under the inducing condition was demonstrated by Western blot analysis with anti-HK antibodies (Fig. 6B).
FIG. 6.
Fungicide susceptibility of S. cerevisiae cells expressing AbNIK1p. (A) Identical volumes of 10-fold serial dilutions (10−1 to 10−4) of exponentially growing S. cerevisiae cells transformed with empty vector (pYES2-CT) or recombinant vectors carrying the WT (pYAbNik1) or mutated (pYAbNik1E753K) AbNIK1 coding sequences were spotted onto SG plates containing or not containing iprodione, fludioxonil, or ambruticin (final concentration, 10 mg/liter). Colony growth was observed after 48 h of incubation at 30°C. (B) Immunodetection of recombinant AbNIK1p in protein extracts from S. cerevisiae cells transformed with empty vector or recombinant vectors carrying WT or mutated AbNIK1 coding sequences and cultivated in noninducing (SD) and inducing (SG) liquid medium.
Ambruticin activates the HOG pathway.
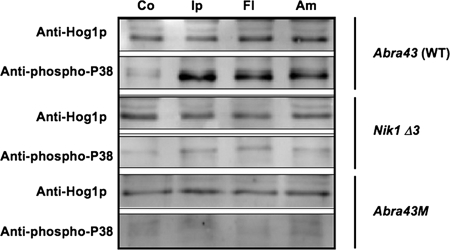
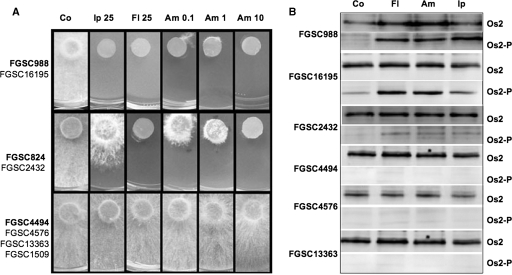
To investigate the phosphorylation of Hog1-related MAPK in A. brassicicola when exposed to fungicides, a Western blot approach using an antibody that specifically recognizes the phosphorylated form of p38-type kinase (18) was used. The nik1Δ3 disruptant strain, the Abra43M mutant strain, and their parental Abra43 WT strain were grown for 24 h in liquid medium; exposed to either iprodione (DCF), fludioxonil (phenylpyrrole), or ambruticin for 10 min; and then harvested for total protein extraction. Duplicate blots were prepared after protein separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The control blot, which had been incubated with an antibody raised against the C terminus of Hog1p, revealed a single band at ca. 41 kDa in all samples, with almost equal intensity (Fig. 7). In contrast, detection with the anti-phospho-p38 antibody on the second blot generated a strong signal at the expected size in protein samples from the WT strain treated with iprodione, fludioxonil, and ambruticin, while only a faint signal was observed in the untreated sample from the WT strain or in all samples from the nik1Δ3 and Abra43M mutant strains, irrespective of the treatment (Fig. 7). To further study the link between ambruticin susceptibility and activation of the HOG/Os pathway, we studied the effects of exposure to this compound on the growth of different N. crassa mutants. As shown in Fig. 8A, three different patterns of sensitivity toward iprodione, fludioxonil, and ambruticin were obtained. Growth of the WT N. crassa strain FGSC988 and the rrg2 knockout mutant FGSC16195 were completely inhibited by ambruticin at concentrations as low as 0.1 μg/ml, while the os1-null mutant FGSC4494, the os5-null mutant FGSC4576, the os2-null mutant FGSC1509, and the rrg1-knockout mutant FGSC13363 had normal mycelial growth on media amended with up to 10 μg/ml ambruticin. As expected from previously published results (15), these four strains were also found to be highly resistant to iprodione and fludioxonil. The os1 substitution mutant strains FGSC2432 and FGSC824 were found to express high and moderate levels, respectively, of resistance to iprodione but only low levels of resistance to ambruticin. Phosphorylation of the Hog1-related protein Os2p was observed in the WT N. crassa strain FGSC988 and the rrg2 knockout mutant FGSC16195 after exposure to the three molecules, while only the nonphosphorylated form was present in the os1-, os5-, and rrg1-null mutants (Fig. 8B). In the os1 substitution mutant strain FGSC2432, a weak signal was observed with anti-phospho-p38 antibodies after exposure to iprodione, fludioxonil, and ambruticin, thus indicating residual activation of the HOG1-related pathway in these mutants.
FIG. 7.
Phosphorylation of the Hog1-like MAPK in A. brassicicola after exposure to fungicides. Germinated conidia from the WT strain (Abra43), the derived spontaneous mutant (Abra43M), and the disrupted mutant (nik1Δ3) were grown for 24 h in PDB and exposed to 100 mg/liter iprodione (Ip) or fludioxonil (Fl) or to 1 mg/liter ambruticin (Am) for 10 min. Control (Co) cultures were not supplemented with fungicides. Total protein extracts, prepared from the harvested mycelia, were analyzed by SDS-PAGE and blotting with either anti-Hog1 C-terminus antibody or anti-dually phosphorylated p38 antibody.
FIG. 8.
Effects of fungicides on the growth and phosphorylation of Os2 MAPK of N. crassa isolates. (A) The growth of WT and mutant N. crassa isolates was monitored after 24 h of incubation at 25°C on agar-solidified Vogel's medium supplemented with 25 mg/liter iprodione (Ip25) or fludioxonil (Fl25) or 0.1, 1, or 10 mg/liter ambruticin (Am 0.1, Am 1, or Am 10, respectively) or not supplemented (control [Co]). Three typical growth inhibition patterns were obtained (upper panel, isolates FGSC988 and FGSC16195; middle panel, isolates FGSC2432 and FGSC824; lower panel, isolates FGSC4494, FGSC4576, FGSC13363, and FGSC1509). Names of isolates that were selected for this picture are indicated in bold letters. (B) Phosphorylation of Os2 MAPK in the different N. crassa isolates exposed to fludioxonil (Fl), ambruticin (Am), or iprodione (Ip) or not exposed (Co) was assessed as described in the legend to Fig. 7. Three typical phosphorylation patterns were obtained: one with isolates FGSC988 and FGSC16195; one with isolate FGSC2432; and one with isolates FGSC4494, FGSC4576, and FGSC13363. The phosphorylation pattern of isolate FGSC824 was not studied, and no signal was obtained with proteins from the Os2-null mutant FGSC1509.
DISCUSSION
In this study, we demonstrated that the polyketide drug ambruticin exerted antifungal activity against the ascomycetous plant fungal pathogen A. brassicicola. The high toxicity of this bacterial metabolite for this fungus was well illustrated by the very low concentrations that were found to significantly inhibit in vitro the mycelial growth and germination of conidia of several WT isolates. A previous study already pointed out the high efficacy of ambruticin to kill growing yeast cells of Hansenula anomala, probably as a consequence of interference with osmoregulation (17). The fungicidal activity of phenylpyrroles, which are commercially used to control fungal crop diseases, is also linked to improper activation of the osmosensing pathway, and Knauth and Reichenbach (17) observed cross-resistance of H. anomala ambruticin-resistant mutants to pyrolnitrin. Similarly, we showed here that previously characterized A. brassicicola AbNIK1-null mutants expressing high resistance to the DCF iprodione (13) were also highly resistant to the phenylpyrrole fludioxonil as well as to ambruticin. Such cross-resistance was also observed for the N. crassa os1-null mutant FGSC4494. Two additional mutants carrying a single-amino-acid substitution in the AbNIK1 gene were also tested and showed differences in their susceptibilities to fungicides. While both were found to be highly resistant to iprodione, only the Abra40 mutant, which probably synthesizes a nonfunctional HK due to an E-K substitution in its kinase domain (3), displayed a phenotype similar to that of null mutants. In contrast, growth of strain Abra363H5M, characterized by a V-A substitution in the fourth repeat of the HAMP domain, was affected by high concentrations of fludioxonil and ambruticin. Similar observations (i.e., amino acid changes located on the repeat domain of group III HKs and fludioxonil susceptibility) were obtained for Botrytis cinerea field mutants (22, 23) and for N. crassa laboratory mutants (19) resistant to iprodione. Here we show that such N. crassa mutants producing abnormal forms of Os1p with substitutions within the third and fifth repeats were also susceptible to ambruticin.
In fungi, the ability to synthesize a functional group III HK therefore seems to be a prerequisite for the expression of ambruticin susceptibility. This was further demonstrated for A. brassicicola by two complementary approaches, i.e., phenotypic characterization of AbNIK1 knockout mutants and of S. cerevisiae strains expressing AbNIK1p. As expected, considering the above-mentioned hypothesis, the natural ambruticin suceptibility expressed by A. brassicicola WT isolates was abolished in ΔNIK1 mutants and the natural ambruticin resistance of S. cerevisiae cells switched to susceptibility in pYAbNik1 transformants under growth-inducing conditions. S. cerevisiae strains expressing HIK1p, an Os1p homologue from Magnaporthe grisea, have already been characterized as susceptible to fludioxonil (20) and to ambruticin (27), but the resistance of M. grisea HIK1-deficient strains to the latter compound has not yet been reported. These observations strongly suggest that, considering their essential role in the response of fungi to environmental stress, group III HK phosphorelay signaling systems from filamentous fungi probably constitute strategic targets for some fungicidal metabolites produced in the environment by bacteria.
Indeed, group III HKs are key proteins in the high-osmolarity stress response of filamentous fungi that regulate the phosphorylation of downstream components of a signal transduction pathway, including Hog1-type MAPK (15, 18, 21, 31). We therefore explored the possibility of abnormal phosphorylation of such MAPKs in fungi exposed to ambruticin. We report here that, like iprodione and fludioxonil, ambruticin induces phosphorylation of the MAPKs Os2p and AbHog1p in WT strains of N. crassa and A. brassicicola, respectively. These observations supplement recently published data of Vetcher et al. (28), who detected Hog1 in the phosphorylated state in HIK1-expressing S. cerevisiae strains exposed to ambruticin. As expected, no or only weak phosphorylation of Hog1-type MAPK was observed in AbNIK1-, os1-, rrg1- and os5-deficient strains when treated with the different antifungal agents tested. The fact that improper activation of the Hog-related pathway may be sufficient to explain the fungicidal effect of ambruticin on N. crassa is supported by the highly resistant phenotype of rrg1-, os2-, and os5-null mutants and the susceptible phenotype of the rrg2-null mutant. Similar observations have already been reported for dicarboximides and phenylpyrroles on N. crassa (15). With regard to A. brassicicola, due to the lack of mutants deficient in equivalent genes from the Hog-related pathway, a similar conclusion cannot yet be drawn. Indeed, there is accumulating evidence that two pathways downstream of group III HKs additively contribute to the susceptibility of some fungi toward antifungal agents targeting these HKs (4, 10, 14).
Acknowledgments
This work was supported by the Contrat de Plan Etat-Région Pays de la Loire (2000 to 2006) and by a postdoctoral fellowship (A. Dongo) provided by the CADRES.
We thank David Manley for correcting the English version of the manuscript.
Footnotes
Published ahead of print on 14 November 2008.
REFERENCES
- 1.Adachi, Y., H. Watanabe, K. Tanabe, N. Doke, S. Nishimura, and T. Tsuge. 1993. Nuclear ribosomal DNA as a probe for genetic variability in the Japanese pear pathotype of Alternaria alternata. Appl. Environ. Microbiol. 59:3197-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arima, K., H. Imanaka, M. Kousaka, A. Fukuda, and G. Tamura. 1965. Studies on pyrrolnitrin, a new antibiotic. I. Isolation and properties of pyrrolnitrin. J. Antibiot. (Tokyo) 18:201-204. [PubMed] [Google Scholar]
- 3.Avenot, H., P. Simoneau, B. Iacomi-Vasilescu, and N. Bataillé-Simoneau. 2005. Characterization of mutations in the two-component histidine kinase gene AbNIK1 from Alternaria brassicicola that confer high dicarboximide and phenylpyrrole resistance. Curr. Genet. 47:234-243. [DOI] [PubMed] [Google Scholar]
- 4.Bahn, Y. S., K. Kojima, G.M. Cox, and J. Heitman. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17:3122-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catlett, N. L., O. C. Yoder, and B. G. Turgeon. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang, L. Y., D. E. Ejzykowicz, Z.-Q. Tian, L. Katz, and S. G. Filler. 2006. Efficacy of ambruticin analogs in a murine model of invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 50:3464-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, Y., J. W. Davis, K. H. Kim, J. Wang, Q. H. Sun, R. A. Cramer, and C. B. Lawrence. 2006. A high throughput targeted gene disruption method for Alternaria brassicicola functional genomics using linear minimal element (LME) constructs. Mol. Plant-Microbe Interact. 19:7-15. [DOI] [PubMed] [Google Scholar]
- 8.Guillemette, T., A. Sellam, and P. Simoneau. 2004. Analysis of a nonribosomal peptide synthetase gene from Alternaria brassicae and flanking genomic sequences. Curr. Genet. 45:214-224. [DOI] [PubMed] [Google Scholar]
- 9.Gullino, M. L., P. Leroux, and C. M. Smith. 2000. Uses and challenges of novel compounds for plant disease control. Crop Prot. 19:1-11. [Google Scholar]
- 10.Hagiwara, D., Y. Matsubayashi, J. Marui, K. Furukawa, T. Yamashino, K. Kanamaru, M. Kato, K. Abo, T. Kobayashi, and T. Mizuno. 2007. Characterization of the NikA histidine kinase implicated in the phosphorelay signal transduction of Aspergillus nidulans, with special reference to fungicide responses. Biosci. Biotechnol. Biochem. 71:844-847. [DOI] [PubMed] [Google Scholar]
- 11.Höfle, G., H. Steinmetz, K. Gerth, and H. Reichenbach. 1991. Ambruticin VS: new members of the antifungal ambruticin family from Sorangium cellulosum. Liebigs Ann. Chem. 1991:941-945. [Google Scholar]
- 12.Iacomi-Vasilescu, B., H. Avenot, N. Bataillé-Simoneau, E. Laurent, M. Guénard, and P. Simoneau. 2004. In vitro fungicide sensitivity of Alternaria species pathogenic to crucifers and identification of Alternaria brassicicola field isolates highly resistant to both dicarboximides and phenylpyrroles. Crop Prot. 23:481-488. [Google Scholar]
- 13.Iacomi-Vasilescu, B., N. Bataillé-Simoneau, C. Campion, A. Dongo, E. Laurent, I. Serandat, B. Hamon, and P. Simoneau. 2008. Effect of null mutations in the AbNIK1 gene on saprophytic and parasitic fitness of Alternaria brassicicola isolates highly resistant to dicarboximide fungicides. Plant Pathol. 57:937-947. [Google Scholar]
- 14.Izumitsu, K., A. Yoshimi, and C. Tanaka. 2007. Two-component response regulators Ssk1p and Skn7p additively regulate high-osmolarity adaptation and fungicide sensitivity in Cochliobolus heterostrophus. Eukaryot. Cell 6:171-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, C. A., S. E. Greer-Phillips, and K. A. Borkovich. 2007. The response regulator RRG-1 functions upstream of a mitogen-activated protein kinase pathway impacting asexual development, female sterility, osmotic stress, and fungicide resistance in Neurospora crassa. Mol. Biol. Cell 18:2123-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julien, B., Z. Q. Tian, R. Reid, and C. D. Reeves. 2006. Analysis of the ambruticin and jerangolid gene clusters of Sorangium cellulosum reveals unusual mechanisms of polyketide biosynthesis. Chem. Biol. 13:1277-1286. [DOI] [PubMed] [Google Scholar]
- 17.Knauth, P., and H. Reichenbach. 2000. On the mechanism of action of the myxobacterial fungicide ambruticin. J. Antibiot. (Tokyo) 53:1182-1190. [DOI] [PubMed] [Google Scholar]
- 18.Kojima, K., Y. Takano, A. Yoshimi, C. Tanaka, T. Kikuchi, and T. Okuno. 2004. Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 53:1785-1796. [DOI] [PubMed] [Google Scholar]
- 19.Miller, T. K., S. Renault, and C. P. Selitrennikoff. 2002. Molecular dissection of alleles of the osmotic-1 locus of Neurospora crassa. Fungal Genet. Biol. 35:147-155. [DOI] [PubMed] [Google Scholar]
- 20.Motoyama, T., T. Ohira, K. Kadokura, A. Ichiishi, M. Fujimura, I. Yamaguchi, and T. Kudo. 2005. An Os-1 family histidine kinase from a filamentous fungus confers fungicide-sensitivity to yeast. Curr. Genet. 47:298-306. [DOI] [PubMed] [Google Scholar]
- 21.Noguchi, R., S. Banno, R. Ichikawa, F. Fukumori, A. Ichiishi, M. Kimura, I. Yamaguchi, and M. Fujimura. 2007. Identification of OS-2 MAP kinase-dependent genes induced in response to osmotic stress, antifungal agent fludioxonil, and heat shock in Neurospora crassa. Fungal Genet. Biol. 44:208-218. [DOI] [PubMed] [Google Scholar]
- 22.Oshima, M., S. Banno, K. Okada, T. Takeuchi, M. Kimura, A. Ichiishi, I. Yamaguchi, and M. Fujimura. 2006. Survey of mutations of a histidine kinase gene BcOS1 in dicarboximide resistant field isolates of Botrytis cinerea. J. Gen. Plant Pathol. 72:65-73. [Google Scholar]
- 23.Oshima, M., M. Fujimura, S. Banno, C. Hashimoto, T. Motoyama, A. Ichiishi, and I. Yamaguchi. 2002. A point mutation in the two-component histidine kinase BcOS-1 gene confers dicarboximide resistance in field isolates of Botrytis cinerea. Phytopathology 92:75-80. [DOI] [PubMed] [Google Scholar]
- 24.Ota, I. M., and A. Varshavsky. 1993. A yeast protein similar to a bacterial two-component regulator. Science 262:566-569. [DOI] [PubMed] [Google Scholar]
- 25.Ringle, S. M., R. C. Greenough, S. Roemer, D. Connor, A. L. Gutt, B. Blair, G. Kanter, and M. von Srandtmann. 1977. Ambruticin (W7783) a new antifungal antibiotic. J. Antibiot. (Tokyo) 30:371-375. [DOI] [PubMed] [Google Scholar]
- 26.Shubitz, L. F., J. N. Galgiani, Z.-Q. Tian, Z. Zhong, P. Timmermans, and L. Katz. 2006. Efficacy of ambruticin analogs in a murine model of coccidioidomycosis. Antimicrob. Agents Chemother. 50:3467-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sweigard, J., F. Chumly, A. Caroll, L. Farrall, and B. Valent. 1997. A series of vectors for fungal transformation. Fungal Genet. Newsl. 44:52-53. [Google Scholar]
- 28.Vetcher, L., H. G. Menzella, T. Kudo, T. Motoyama, and L. Katz. 2007. The antifungal polyketide ambruticin targets the HOG pathway. Antimicrob. Agents Chemother. 51:3734-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel, H. J. 1964. Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat. 98:435-446. [Google Scholar]
- 30.Wolanin, P. M., P. A. Thomason, and J. B. Stock. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3:REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimi, A., K. Kojima, Y. Takano, and C. Tanaka. 2005. Group III histidine kinase is a positive regulator of Hog1-type mitogen-activated protein kinase in filamentous fungi. Eukaryot. Cell 4:1820-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]