Abstract
A new lipase, LipEH166, isolated from an intertidal flat metagenome, showed no amino acid similarity to any known lipolytic enzyme except in the consensus region. This suggested that LipEH166 and its homologues belong to a new family of lipolytic enzymes. Partial characterization indicated that LipEH166 is a novel cold-adapted alkaline lipase.
Bacterial diversity is tremendous, at no less than 103 to 106 distinct prokaryotic taxa per gram of pristine soil or marine sediment (2, 15, 16), and this likely pertains to the immensely diverse number of metabolic enzyme genes as well. Through screening based on functions or sequence homology, numerous distinct enzymes have been identified from metagenome libraries of various environments such as soil, water, sediment, and extreme environments (3, 7, 12, 14). Intertidal flat sediments possess remarkable and unique bacterial diversity, because dynamic physicochemical conditions, such as periodic flood tides, high degrees of change in salinity, and fluctuations in water temperature, considerably affect the habitat (10). Therefore, intertidal flat sediments are likely a valuable source of novel biocatalysts, including lipases (11).
Lipases (EC 3.1.1.3) are carboxylesterases that catalyze the hydrolysis and synthesis of long-chain triacylglycerol. Lipase is a versatile enzyme that is the one of the most important biocatalysts in the laundry, food, chemical, and pharmaceutical industries (6, 8, 9). In order to find valuable lipolytic enzymes, the metagenome from the intertidal flat sediments of the coastal regions of Saemankum, located in the west of South Korea, was extracted as previously described (17). A metagenomic library was constructed using a CopyControl fosmid library production kit (Epicentre) according to the manufacturer's instructions. This was followed by the screening of the lipolytic activity on a tributyrin agar plate. As a result, a positive clone showing the highest lipolytic activity among approximately 6,000 colonies was selected. Sequence analysis of the short insert DNA obtained by subsequent subcloning experiments revealed the presence of one open reading frame consisting of 1,143 nucleotides, encoding a protein (LipEH166) with a molecular mass of 42 kDa.
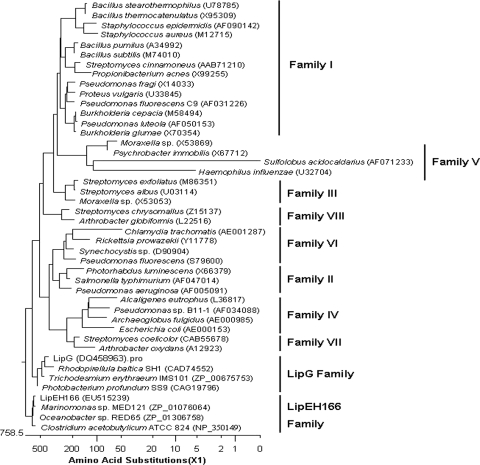
Comparison of the predicted amino acid sequence of LipEH166 with other amino acid sequences in a nonredundant protein sequence database (http://ncbi.nlm.nih.gov) and the Lipase Engineering Database (http://www.led.uni-stuttgart.de/) (5) using the BLAST program showed that LipEH166 is aligned to only three putative open reading frames (E value, < 10−1) and no known lipolytic enzymes (see Table S1 in the supplemental material). One of these uncharacterized secreted proteins is from Clostridium acetobutylicum ATCC 824, isolated from soil (GenBank accession number NC_350149; identity, 34%; similarity, 52%), and the others are from the marine microorganisms Oceanobacter sp. strain RED65 (ZP_01306758; identity, 65%; similarity, 77%) and Marinomonas sp. strain MED121 (ZP_01076064; identity, 58%; similarity, 73%). LipEH166 and the homologous uncharacterized secreted proteins have a conserved active-site motif consisting of the pentapeptide GXSXG, aspartate, and histidine. We selected 36 lipolytic enzymes representing eight different families classified by Arpigny and Jaeger (1) and 4 other lipolytic enzymes, belonging to the LipG family (11), for phylogenetic analysis. The results showed that LipEH166 and its homologues can be classified as a novel lipolytic enzyme family (Fig. 1; see also Fig. S1 in the supplemental material).
FIG. 1.
Phylogenetic tree of LipEH166 and closely related proteins. Phylogenetic analysis was performed using Megalign in the Lasergene program (version 6.0; DNAStar Inc.). Amino acid sequences were aligned by Clustal V. Except for LipEH166 and related proteins, the protein sequences for families of bacterial lipolytic enzymes previously classified by Arpigny and Jaeger (1) and for the LipG family (11) were retrieved from GenBank (http://www.ncbi.nlm.nih.gov).
In order to characterize the biochemical properties of the enzyme, we subcloned the lipEH166 gene, which was amplified by PCR with two primers (5′-GGAATTCCATATGATGCTAAGACAATTTCGAATTC-3′ and 5′-CCGCTCGAGATAA TTTTTGCGTTCAAACTGG-3′; underlined letters indicate the NdeI and XhoI recognition sites, respectively), into pET-22(b) (Novagen). LipEH166 was overexpressed, with a six-histidine tag fused at its C terminus, in Escherichia coli C43(DE3) and was purified to homogeneity in 50 mM Tris-HCl buffer (pH 8.0) with 300 mM NaCl by gel filtration chromatography using a Superdex 200 gel filtration column (GE Healthcare) after metal affinity chromatography using a nickel-nitrilotriacetic acid column (Qiagen). A standard assay was carried out as previously described (11) for 3 min using 10 mM p-nitrophenyl caprate as a substrate.
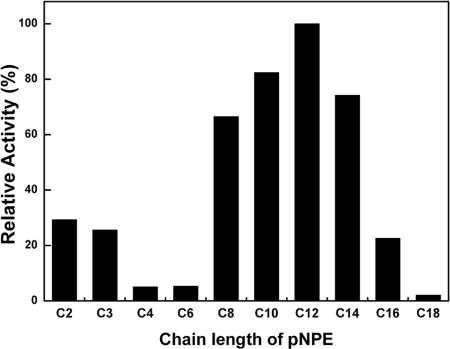
The relative lipase activities in the presence of various p-nitrophenyl esters (pNPEs) were examined. LipEH166 showed the highest activity with p-nitrophenyl laurate (C12), while pNPEs with acyl groups shorter than C8 or longer than C16 were poor substrates (Fig. 2). The influence of divalent metal ions on the activity of LipEH166 was tested by adding 1, 5, or 10 mM CaCl2, CuSO4, MgSO4, FeSO4, ZnSO4, NiSO4, MnSO4, or CoCl2. The activity was enhanced by 30% in 5 mM CaCl2 and was slightly increased by MgSO4 and MnSO4. The presence of CuSO4 or ZnSO4 strongly inhibited lipolytic activity. Moreover, the chelating agent effectively inhibited the enzyme activity. These results indicated that divalent ions, especially Ca2+, appear to play a role in the catalytic reaction. In addition, LipEH166 activity was inhibited by 0.1% (vol/vol) and 1% (vol/vol) nonionic and ionic detergents (data not shown).
FIG. 2.
Substrate specificity of LipEH166 for pNPEs. The lipase activity of the purified LipEH166 enzyme toward various pNPEs was assayed at 25°C and pH 8.0. The chain length of pNPEs is expressed as the carbon number of esters. Data are averages from duplicate experiments.
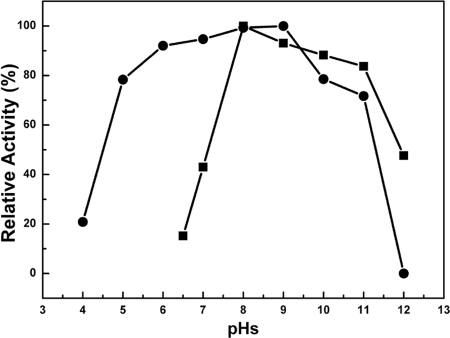
The enzyme showed a relatively low apparent optimum hydrolytic activity around 30°C and retained 47% of the activity at 5°C (Fig. 3A). To determine the conformational stability of LipEH166, the change in ellipticity at 222 nm of 1.6 mg/ml of the protein in 2.5 mM Tris-HCl buffer (pH 8.0) with 15 mM NaCl was observed using a Jasco-815 spectropolarimeter (Jasco Inc., Japan) with a thermostated 1-cm cuvette as the temperature was increased by 1°C/min and held for 2 min at intervals of 1°C. The signal was averaged over five measurements at each temperature. The thermal unfolding curve showed that LipEH166 started to be unfolded around 25°C and was fully denatured at 55°C (Fig. 3B). In addition, the activity of LipEH166 was measured at various pHs. The enzyme was active in the broad alkaline range, had the highest activity at pH 8.0, and maintained more than 80% activity in the pH range of 5 to 11 after incubation for 1 h (Fig. 4). From these results, we concluded that LipEH166 was a novel cold-adapted alkaline lipase, probably derived from a psychrophilic organism. The fact that the enzyme was active and stable under broad alkaline conditions is difficult to explain by the environment from which it was acquired. It has been reported that esterases from metagenomes of soil (4), drinking water biofilm (4), and a deep-sea sediment (13) showed remarkable activities and stabilities under alkaline conditions that were not directly linked to their environments. It seems that distinctive enzymes that are active and able to tolerate extreme conditions can be more easily obtained through screening of a metagenomic library from a dynamic environment such as an intertidal flat region than has been generally predicted. The present findings of this new cold-adapted alkaline lipase reemphasize the importance of the diversity of genomes as a source of the isolation of novel characteristic genes.
FIG. 3.
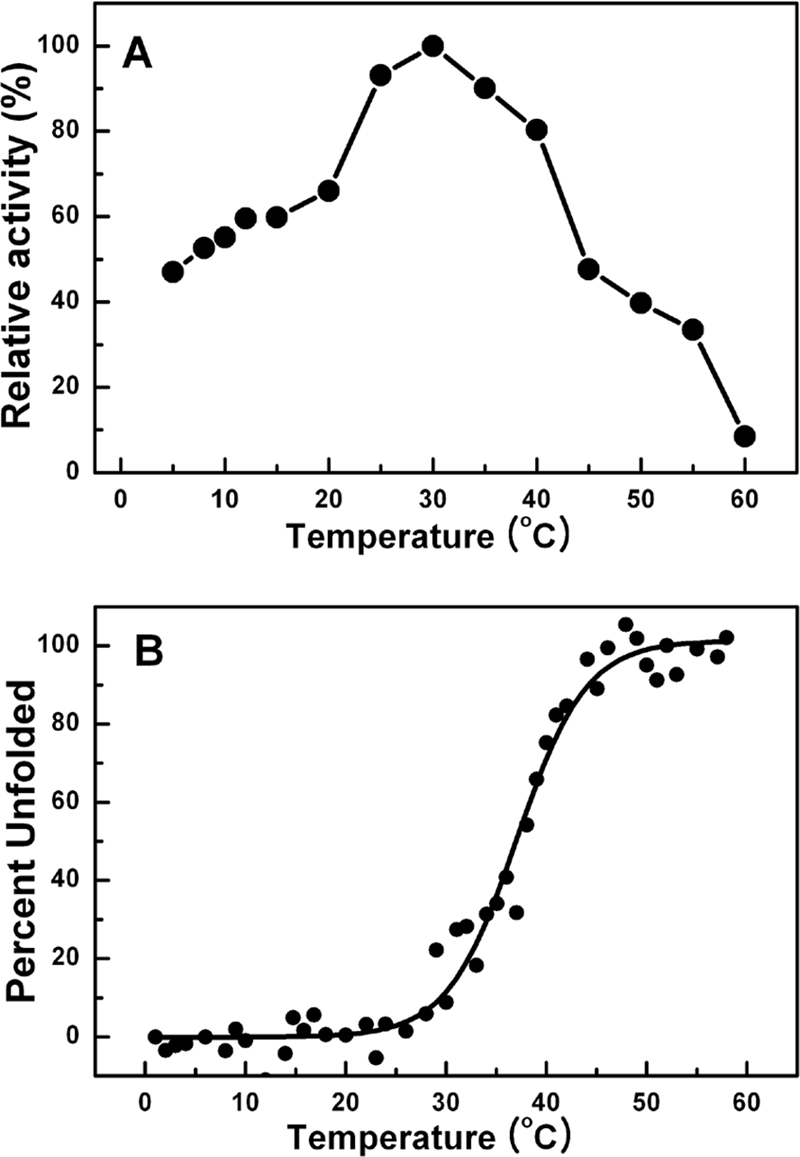
Effect of temperature on LipEH166. (A) Enzyme activity was analyzed at each temperature under standard assay conditions. (B) The proportion of unfolded LipEH166 is displayed as a function of temperature. Each percentage was calculated from the change in ellipticity at 222 nm, obtained by circular dichroism using a spectropolarimeter. Data are averages from triplicate experiments.
FIG. 4.
Effect of pH on LipEH166. Enzyme activity was measured at each pH under standard assay conditions (▪). In addition, the enzyme was preincubated at the indicated pH, and the remaining activity was determined (•). Data are averages from triplicate experiments.
Nucleotide sequence accession number.
The nucleotide sequences obtained in this study have been deposited in the GenBank database under accession number EU515239.
Supplementary Material
Acknowledgments
We thank B. Kim of the research supporting team in the Korea Advanced Institute for Science and Technology for technical support for circular dichroism analysis.
This work was supported by the 21C Frontier Program of Microbial Genomics and Applications (grant MG05-0401-2-0) of the Ministry of Education, Science and Technology (MEST) of the Republic of Korea.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Arpigny, J. L., and K.-E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343:177-183. [PMC free article] [PubMed] [Google Scholar]
- 2.Curtis, T. P., and W. T. Sloan. 2005. Exploring microbial diversity—a vast below. Science 309:1331-1333. [DOI] [PubMed] [Google Scholar]
- 3.Daniel, R. 2004. The soil metagenome—a rich resource for the discovery of novel natural products. Curr. Opin. Biotechnol. 15:199-204. [DOI] [PubMed] [Google Scholar]
- 4.Elend, C., C. Schmeisser, C. Leggewie, P. Babiak, J. D. Carballeira, H. L. Steele, J.-L. Reymond, K.-E. Jaeger, and W. R. Streit. 2006. Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl. Environ. Microbiol. 72:3637-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer, M., and J. Pleiss. 2003. The Lipase Engineering Database: a navigation and analysis tool for protein families. Nucleic Acids Res. 31:319-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta, R., N. Gupta, and P. Rathi. 2004. Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 64:763-781. [DOI] [PubMed] [Google Scholar]
- 7.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaeger, K.-E., and M. T. Reetz. 1998. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 16:396-403. [DOI] [PubMed] [Google Scholar]
- 9.Jaeger, K.-E., and T. Eggert. 2002. Lipases for biotechnology. Curr. Opin. Biotechnol. 13:390-397. [DOI] [PubMed] [Google Scholar]
- 10.Kim, B., H. Oh, H. Kang, S. Park, and J. Chun. 2004. Remarkable bacterial diversity in the tidal flat sediment as revealed by 16S rDNA analysis. J. Microbiol. Biotechnol. 14:205-211. [PubMed] [Google Scholar]
- 11.Lee, M. H., C. H. Lee, T. K. Oh, J. K. Song, and J. H. Yoon. 2006. Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl. Environ. Microbiol. 72:7406-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorenz, P., and J. Eck. 2005. Metagenomics and industrial application. Nat. Rev. Microbiol. 3:510-516. [DOI] [PubMed] [Google Scholar]
- 13.Park, H. J., J. H. Jeon, S. K. Kang, J. H. Lee, S. A. Lee, and H. K. Kim. 2007. Functional expression and refolding of new alkaline esterase, EM2L8 from deep-sea sediment metagenome. Protein Expr. Purif. 52:340-347. [DOI] [PubMed] [Google Scholar]
- 14.Schmeisser, C., H. Steele, and R. Streit. 2007. Metagenomics, biotechnology with non-culturable microbes. Appl. Microbiol. Biotechnol. 75:955-962. [DOI] [PubMed] [Google Scholar]
- 15.Torsvik, V., J. Gorsøry, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torsvik, V., L. Øvreås, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 17.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.