Abstract
A triphasic process was developed for the production of β dipeptides from cyanophycin (CGP) on a large scale. Phase I comprises an optimized acid extraction method for technical isolation of CGP from biomass. It yielded highly purified CGP consisting of aspartate, arginine, and a little lysine. Phase II comprises the fermentative production of an extracellular CGPase (CphEal) from Pseudomonas alcaligenes strain DIP1 on a 500-liter scale in mineral salts medium, with citrate as the sole carbon source and CGP as an inductor. During optimization, it was shown that 2 g liter−1 citrate, pH 6.5, and 37°C are ideal parameters for CphEal production. Maximum enzyme yields were obtained after induction in the presence of 50 mg liter−1 CGP or CGP dipeptides for 5 or 3 h, respectively. Aspartate at a concentration of 4 g liter−1 induced CphEal production with only about 30% efficiency in comparison to that with CGP. CphEal was purified utilizing its affinity for the substrate and its specific binding to CGP. CphEal turned out to be a serine protease with maximum activity at 50°C and at pH 7 to 8.5. Phase III comprises degradation of CGP to β-aspartate-arginine and β-aspartate-lysine dipeptides with a purity of over 99% (by thin-layer chromatography and high-performance liquid chromatography), employing a crude CphEal preparation. Optimum degradation parameters were 100 g liter−1 CGP, 10 g liter−1 crude CphEal powder, and 4 h of incubation at 50°C. The overall efficiency of phase III was 91%, while 78% (wt/wt) of the used CphEal powder with sustained activity toward CGP was recovered. The optimized process was performed with industrial materials and equipment and is applicable to any desired scale.
Cyanophycin granule polypeptide (CGP), or multi-l-arginyl-poly(l-aspartic acid), was discovered in cyanobacteria about 130 years ago (6). The branched polymer consists of a poly(aspartic acid) backbone with arginine moieties linked to the β-carboxyl group of each aspartic acid by its α-amino group (34, 42) and accumulates during the transition from the exponential to the stationary growth phase (23, 40) and under limiting conditions, including low temperature, low light intensity, and phosphorus or sulfur limitation (44). CGP functions as a temporary nitrogen, energy, and possibly also carbon reserve (10, 21). Because CGP contains five nitrogen atoms in every building block, it is an ideal intracellular nitrogen reserve (43). Most genera of cyanobacteria (5, 22, 23, 43, 48) and some heterotrophic bacteria (12, 16, 51) harbor a cyanophycin synthetase gene (cphA) and synthesize CGP. The polymer is insoluble at neutral pH as well as at physiological ionic strength (4). In cyanobacteria, CGP has a molecular mass of 25 to 100 kDa (41), while in recombinant strains it exhibits a molecular mass of 25 to 30 kDa and a lower polydispersity and contains lysine, which partially replaces arginine (2, 50).
CGP degradation (intra- or extracellularly) leads mainly to the release of dipeptides. Intracellular degradation of CGP is catalyzed by cyanophycinases (CphB); the first cyanophycinase was described for Anabaena cylindrica by Gupta and Carr (14). The enzyme is a monomeric, 29.4-kDa, serine-type CGP-specific exopeptidase with an α-cleavage mechanism (35). In the last few years, aerobic and anaerobic bacteria capable of degrading CGP by extracellular cyanophycinases (CphE) have been identified (28, 29, 31, 36). Similar to CphB, the extracellular CGPases CphEPa and CphEBm, from Pseudomonas anguilliseptica B1 and Bacillus megaterium BAC19, respectively, were identified as serine-type CGP-specific enzymes and produced mainly CGP dipeptides as degradation products. Labeling studies of CphEPa showed that the enzyme hydrolyzes CGP at the carboxyl terminus and successively releases β-Asp-Arg dipeptides from the degraded polymer chain end (30). Moreover, extracellular CGPases were also found in strictly and facultatively anaerobic bacteria, such as Sedimentibacter hongkongensis KI and Pseudomonas alcaligenes DIP1, respectively (31, 36).
Until recently, no practical applications for CGP itself or for the dipeptides derived from it were known. In contrast, economically important applications have been established for poly(aspartic acid) as a substitute for nonbiodegradable polyacrylates (38) or as an additive in the paper, paint, and oil industries (15). Biomedical applications have also been described for poly(aspartic acid) (20, 49). Only recently were biomedical applications for CGP dipeptides and possibly CGP itself proposed; these applications depend on the astonishingly widespread occurrence of CGP-degrading bacteria in the digestive tracts of various vertebrates (A. Sallam and A. Steinbüchel, submitted for publication), thus indicating that CGP is probably degraded in these habitats. On the other hand, di- and tripeptides are more efficiently utilized than intact proteins or free amino acids, have a greater nutritional value, and are better absorbed (24). Moreover, administration of amino acids as dipeptides or in mixtures is clinically approved, and products containing these preparations have been commercialized (7, 9, 19, 39). Thus, CGP and/or its dipeptides are considered potential natural food additives and/or therapeutics (Sallam and Steinbüchel, submitted for publication).
The production and efficient isolation of CGP in gram amounts were established only during recent years. Several recombinant strains of Escherichia coli, Ralstonia eutropha, Pseudomonas putida, and Acinetobacter baylyi strain ADP1 were used; the latter showed a maximum CGP content of about 46% (wt/wt) (32). To provide enough CGP dipeptides for in vivo experiments and to approach industrial-scale production, we developed an economical large-scale process which provides pure CGP dipeptides from CGP-containing biomass. Furthermore, the last two phases of the initial triphasic process were largely optimized for future applications, CphEal was technically purified from a powder obtained from lyophilized crude supernatant, and the biochemical characteristics thereof were revealed.
MATERIALS AND METHODS
Media.
The following simplified medium (SM) was used for fermentative production of CphEal by P. alcaligenes DIP1: 1.0 g NH4Cl, 5.0 g KH2PO4, 1 g MgSO4·7H2O (sterilized and added separately), and 10 ml of trace element stock solution (8) per liter of tap water. The pH of the medium was adjusted to 7.0, and the medium was subsequently sterilized by autoclaving at 121°C for 20 min. For optimization experiments and for easier preparation, a trace element solution containing the following compounds was used (13): KI (0.02 g liter−1), CoCl2·6H2O (0.8 g liter−1), MnSO4·H2O (0.64 g liter−1), FeSO4·7H2O (0.4 g liter−1), ZnSO4·7H2O (0.24 g liter−1), CuSO4 (0.038 g liter−1), H3BO3 (0.060 g liter−1), Na2MoO4·2H2O (0.06 g liter−1), and CaCl2·2H2O (0.46 g liter−1). The composition of SM proved to be suitable for monitoring turbidity changes which occur due to degradation of the inductor (CGP), thereby representing a definite sign for the release of CphEal.
Additionally, Luria-Bertani (LB) medium (37) was used for maintenance of viable cultures. CGP-overlaid agar plates were prepared using a sterile mixture of CGP and 1.2% (wt/vol) Bacto agar, which was poured as thin layers onto SM agar plates (36).
Cultivations on a 500-liter scale.
Cultivations on a 500-liter scale were done in a Biostat D650 stainless steel bioreactor (B. Braun Biotech International, Melsungen, Germany) with a total volume of 650 liters (64-cm inner diameter and 198-cm height) and a d/D value (stirrer diameter/vessel diameter) of 0.375. This bioreactor was equipped with three stirrers, each containing six paddles, and a Funda-Foam mechanical foam destroyer (B. Braun Biotech International, Melsungen, Germany). Sterilizable ports were used to measure dissolved oxygen (pO2) (model 25; Mettler-Toledo, Steinbach, Switzerland), pH (model Pa/25; Mettler-Toledo), foam (model L300/Rd. 28; B. Braun Biotech International, Melsungen, Germany), temperature (pt 100 electrode; M. K. Juchheim, Fulda, Germany), and optical density at 850 nm (OD850) (model CT6; Sentex/Monitek Technology, CA). Operations were controlled and recorded by a digital control unit in combination with the MFCS/win software package (B. Braun Biotech International, Melsungen, Germany).
Cell separation and concentration and desalting of supernatant proteins.
Cells were harvested by centrifugation with a type Z41 or type Z61 CEPA continuous centrifuge (Carl Padberg Zentrifugenbau GmbH, Lahr, Germany). Proteins were concentrated and desalted using a cross-flow Sartocon polyethersulfone cassette with a cutoff point (COP) of 30 kDa and a Sartocon 2 Plus stainless steel holder (Sartorius AG, Göttingen, Germany). To identify a suitable filtration system for CGP dipeptides, several 20-ml aliquots were filtered with different membranes having the following COPs: 30, 10, 5, 1, and 0.5 kDa.
Sources of CGP.
To obtain a large amount of CGP for preliminary tests and for process design, 4,776 g cell dry biomass of CGP-containing cells of R. eutropha H16-PHB-4-Δeda (pBBR1MCS-2::cphA6308/edaH16) were used; these cells were previously produced during one 500-liter fermentation and had a CGP content of 32% (wt/wt) (46). Additionally, several other lyophilized biomass batches from lab-scale fermentations of R. eutropha H16-PHB-4-Δldh/ΩKm-cphA6308 were mixed (a total of 2,490 g) and treated separately. After CGP extraction according to a modified acid extraction method (11; see below), 621 and 82 g of dry CGP were obtained from both charges, respectively.
Fermentative production of CGP on a 500-liter scale.
For optimization experiments for the designed technical process, CGP was produced using the recombinant strain E. coli DH1(pMa/c5-914::cphAPCC6803) (11) in a 500-liter fermentation according to the method of Elbahloul et al. (10). For this process, 7% (vol/vol) protamylasse (Avebe, Veendam, The Netherlands), a residual compound of industrial starch production, was used. Microscopic examination of samples taken each hour revealed progressive intracellular accumulation of CGP (Fig. 1B), with a maximum after 13 h. When the OD600 was 18.3, after 15 h, fermentation was terminated. Analysis revealed a maximum CGP content of the cells of 13% (wt/wt of cell dry mass) after 13 h of fermentation, which dropped to about 10% (wt/wt) during the next 2 hours. This extraction yielded 135 g of CGP powder from the resulting 4,626 g wet mass (1,372 g cell dry mass).
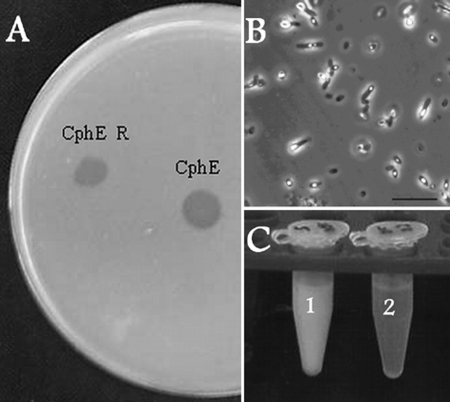
FIG. 1.
(A) Degradation halos caused by the extracellular CGPase from P. alcaligenes strain DIP1 on a CGP-overlaid agar plate. CphE, the crude enzyme powder before the degradation phase (phase III); CphE R, the recovered powder after the degradation phase. (B) Phase-contrast micrograph of cells of E. coli DH1(pMA/c5.914::cphAPCC6803) after 15 h of fermentation in a Biostat D650 reactor in 7% (vol/vol) protamylasse medium. CGP grana appear as light-reflecting accumulations in the cells. Bar, 10 μm. (C) CGP degradation in a 100 g liter−1 water suspension of the polymer by the action of 10 g liter−1 crude CphEal at 50°C. Tube 1, control suspension (no CphEal); tube 2, after CGP degradation.
Technical-scale isolation of CGP from biomass.
To isolate and purify CGP on larger scales, the acid extraction method (11) was optimized as follows. The CGP-containing dry mass was suspended in tap water to give a final concentration of 100 g liter−1. The pH was reduced to 1 with concentrated HCl (32%), and the suspension was stirred overnight and then centrifuged at 17,000 rpm, using a CEPA Z61 continuous centrifuge (Carl Padberg Zentrifugenbau GmbH, Lahr, Germany). The pellet was resuspended in 20 liters of HCl (0.1 N), stirred for 1 h, and centrifuged again, and the supernatant was added to the first charge, while the pellet was discarded. The CGP-containing supernatant was neutralized (pH 7.3) with NaOH (50%), and CGP was precipitated. The milky suspension was left to settle overnight at 4°C before the supernatant was discarded. CGP was dissolved and neutralized three more times to remove all impurities which were insoluble in diluted HCl. The resulting CGP was resolved in 30 liters of 0.1 N HCl and was then passed through a 0.2-μm type 00 Sartobran-P filter unit (Sartorius AG, Göttingen, Germany). The solution was neutralized again (pH 7.3) with NaOH and left overnight to settle, whereas the supernatant was discarded. To remove any water-soluble impurities and to desalt CGP, the pellet was washed three successive times with five bed volumes of distilled water. Finally, the CGP pellet was centrifuged at 20,000 rpm (CEPA Z41 continuous centrifuge), frozen at −30°C, and lyophilized in a BETA 1-16 freeze-dryer (Christ Gefriertrocknungsanlagen, Osterode, Germany). CGP was sterilized by diethyl ether treatment or by solvation in 0.1 N HCl and sterile filtration as described before (36).
Substrate utilization.
Substrate utilization was investigated in 100-ml Klett flasks with baffles; each flask contained 10 ml of SM and 1 g liter−1 of one of the following substrates (sterile filtered [pore size, 0.2 μm]; Millipore GmbH, Eschborn, Germany): lactate, citrate, succinate, acetate, propionate, gluconate, glucose, fructose, sucrose, and glycerol. Experiments were performed in duplicate over an incubation period of 24 h at 30°C and were inoculated from a preculture grown under the same test conditions. The following potential inductors were tested: CGP, β-dipeptides from CGP, synthetic dipeptides (α-arginine-aspartate, α-lysine-aspartate, and α-ornithine-aspartate) (Sigma-Aldrich, St. Louis, MO), α-polyaspartate (Bayer AG, Leverkusen, Germany), poly-ɛ-lysine (Chisso, Tokyo, Japan), the amino acids l-aspartate, l-arginine, l-lysine, l-citrulline, and l-ornithine, and the aspartate analogues N-acetyl-aspartate, ureidosuccinic acid, and N-carbamoyl-aspartate.
Analytical techniques.
Bacterial growth and CGP degradation were monitored by measuring changes in turbidity in Klett flasks, using a Klett photometer (Manostat Corporation, NY) or an Eppendorf 1101 M spectrophotometer (578-nm wavelength; Eppendorf, Hamburg, Germany). Otherwise, the OD600 of 1-ml fermentation samples was estimated using a Libra S5 photometer (Biochrom Ltd., Cambridge, United Kingdom). Thin-layer chromatography was done on silica gel 60 plates (Merck, Darmstadt, Germany), and the starting eluent for high-performance liquid chromatography (HPLC) (81% [vol/vol] 50 mM sodium acetate, 19% [vol/vol] methanol) was also used here as a run buffer; for staining, 20% (wt/vol) ninhydrin solution in acetone was used. DNA concentrations were determined photometrically at 260 nm, and the concentrations of carbohydrates were measured by the anthrone reagent method (45).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on 11.5% (wt/vol) gels according to the method of Laemmli (18). In-gel renaturation of proteins was applied according to the method of Lacks and Springhorn (17). Proteins and CGP were visualized by Coomassie blue staining (47) or silver staining (only for proteins) (27). Concentrations of total protein and CGP were determined using Bradford reagent (10). Proteins in cultivation samples were concentrated, desalted, and separated from CGP dipeptides by ultrafiltration, using 10-kDa-membrane Vivaspin tubes (Vivascience AG, Hannover, Germany) or an Amicon chamber (Amicon, Beverly, MA) with 10-kDa membranes (Millipore Corporation, Bedford, MA) for volumes of up to 500 ml. Free amino acids and dipeptides were detected by HPLC (Kontron Instruments, Neufahrn, Germany) after precolumn derivatization with orthophthaldialdehyde (OPA) as described before (1, 36). CGP samples were subjected in advance to acid hydrolysis (6 N HCl, 95°C, overnight).
Determination of enzyme activity and concentration.
The activity of cyanophycinase (CphEal) was inspected by the formation of degradation halos on CGP-overlaid agar plates. For this assay, 5-ml culture samples were centrifuged at 2,800 × g for 30 min (Megafuge 1.0 R; Heraeus Sepatech GmbH, Osterode, Germany), and 4 ml of the supernatant was concentrated 100-fold. For quantitative determination of the enzyme in culture samples, a photometric method was developed as follows. Two microliters of the concentrated culture supernatant was added to 1 ml of a CGP suspension (100 mg liter−1) and incubated for 30 min at 30°C in a tube rotator (3 rpm). Finally, the OD600 of samples indicated the decrease in CGP, which in turn determined the concentration of CphEal through a respective calibration curve.
Optimal concentrations and conditions for CGP degradation.
To determine the optimum ratio between CGP and crude CphEal concentrations in relation to the incubation time, a series of dilutions (10 to 100 g liter−1) of pure CGP suspensions (in water, pH 7.3) were prepared in plastic tubes with a total volume of 1 ml each. Different amounts of crude CphEal powder (4.6% [wt/wt] CphEal content) were added to each tube, and the tubes were incubated at 30°C in a tube rotator (3 rpm) to reveal the required incubation periods for complete CGP degradation. Experiments to determine the optimum pH for CGP degradation were conducted using plastic tubes containing 1 ml CGP suspension (100 g liter−1) with pH values between 5.0 and 9.0 in addition to 10 g liter−1 crude CphEal, and the tubes were incubated at 30°C for 30 min. Reaction mixtures for determination of the optimum degradation temperature contained similar concentrations of CGP (pH 7.0) and CphEal and were incubated for 30 min at 15, 20, 25, 30, 35, 37, 40, 50, 60, or 70°C. After both experiments, CGP degradation activities in all tubes were calculated as percentages and compared.
Purification of CphEal with organic solvents and ammonium sulfate precipitation.
Each 1 ml of cold ethanol, acetone, or methanol at a concentration of 10 to 100% (vol/vol) was added to a 50-μl aliquot of concentrated crude CphEal solution (14 g liter−1) in a plastic tube and incubated for 60 min at −20°C. After 5 min of centrifugation at 16,000 × g, the pellets were dried and resuspended in 50 μl sodium phosphate buffer (50 mM, pH 7.0). Ammonium sulfate fractionation was done by stepwise increasing the solubility saturations from 10 to 100% in 10 ml of crude CphEal solution (3.5 g liter−1). Tubes were incubated at room temperature for 30 min and centrifuged for 10 min at 16,000 × g. Pellets were suspended in sodium phosphate buffer (pH 7.0) and then desalted by ultrafiltration. All pellets were assayed for CGP degradation on CGP-overlaid agar plates as well as for protein content by SDS-PAGE.
Substrate affinity purification of CphEal.
A purification method for CphEal was developed which depended on the strong affinity of the enzyme in the crude extract for its insoluble substrate (CGP). To determine the time required for complete binding of the enzyme to CGP at pH 7.0, 0.5 ml of concentrated crude CphEal solution (14 g liter−1) was added to 0.5 ml of a CGP suspension (100 g liter−1). Samples were withdrawn every 10 min, and 2-μl aliquots of the supernatant were tested for activity. The degree of diminishment of degradation halos on CGP-overlaid agar plates indicated the extent of binding of CphEal to CGP. The actual purification process was performed in a similar 1-ml reaction mixture and proceeded as follows: after complete binding of CphEal to CGP, the mixture was centrifuged for 0.5 min (16,000 × g), the supernatant was discarded, and the pellet was washed five times with 10 ml of sodium phosphate buffer. Afterwards, the pellet was suspended in 1 ml phosphate buffer and incubated overnight at 30°C with rotation (3 rpm) until complete degradation of CGP had occurred. The mixture was centrifuged (5 min, 16,000 × g), CGP dipeptides were then removed by ultrafiltration, and the concentrated protein fraction of the supernatant was analyzed for purity by SDS-PAGE.
Characterization of CphEal.
To determine the temperature stability of purified CphEal, 10-μl aliquots were incubated for 20 min at different temperatures (10 to 80°C); 3 μl thereof was then tested for degradation activity on CGP-overlaid agar plates at 30°C. For the optimum degradation temperature, 3-μl aliquots of the purified CphEal solution and 1-ml CGP suspensions (100 g liter−1 in sodium phosphate buffer, pH 7.0) were mixed and incubated for 20 min at different temperatures (10 to 60°C), whereas the optimum pH was determined in similar mixtures with different pH values (5 to 9), which were incubated for 30 min at 30°C. Finally, CGP degradation was determined photometrically for both experiments, as described above.
The substrate specificity of purified CphEal was tested as described before (29), employing the following polypeptide substrates: CGP, bovine casein (Hammersten-grade; Merck, Darmstadt, Germany), bovine serum albumin (BSA) (Roth, Karlsruhe, Germany), and poly(α,β-d/l-aspartic acid) (Bayer AG, Leverkusen, Germany). To reveal the effects of enzyme inhibitors, 50-μl aliquots of the purified enzyme solution were added to 450 μl of sodium phosphate buffer and incubated for 2 h at 30°C in the presence of one of the following group-specific inhibitors: leupeptin (thiol proteases), EDTA (metalloproteases), Pefabloc (serine proteases), phenylmethylsulfonyl fluoride (PMSF) (serine proteases), or N-bromosuccinimide (tryptophan residues). Five-microliter samples of each reaction mixture were assayed for activity on CGP-overlaid agar plates. Afterwards, 5-μl aliquots of a CGP suspension (50 g liter−1) were added to the reaction tubes, incubated for a further 15 min, centrifuged, and screened for degradation products via HPLC.
RESULTS
A triphasic technical process was designed and optimized for the production of β-dipeptides from highly concentrated CGP suspensions. Phase I provided CGP and comprised large-scale isolation and purification of CGP from biomass. Phase II provided cyanophycinase and comprised fermentative production of crude CphEal powder from P. alcaligenes strain DIP1. In phase III, the products of phases I and II were combined, and degradation of CGP in highly concentrated CGP suspensions to its dipeptides was achieved.
Phase I (large-scale isolation and purification of CGP).
The optimized extraction method was applied to CGP-containing dry biomass of R. eutropha (7,266 g) and E. coli (1,372 g). Total amounts of 704 g and 135 g of purified CGP, respectively, were obtained. HPLC analysis of the individual amino acid constituents of the isolated CGPs revealed that the polymer was highly pure and consisted of aspartate, arginine, and lysine. The latter comprised approximately 6.8 mol% of the polymer constituents. SDS-PAGE analysis of the isolated CGP showed a molecular mass of about 25 to 30 kDa. These characteristics are in strong agreement with those of CGP previously produced by both strains (10, 11, 46).
Phase II (large-scale production of crude CphE powder).
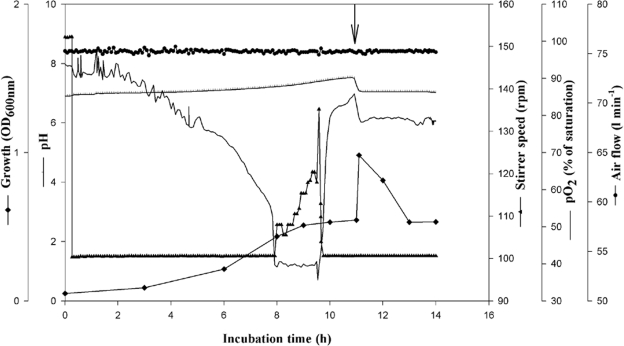
To obtain enough crude CGPase for CGP degradation (phase III), P. alcaligenes DIP1 (36) was cultivated on a 500-liter scale. A preculture was cultivated in 2-liter baffled flasks containing 1 liter of SM with 1 g liter−1 sodium citrate; the flasks were incubated with shaking for 12 h at 30°C. The bioreactor was filled with 420 liters of the same medium, sterilized, and inoculated with 4% (vol/vol) preculture. The initial pH was adjusted to 6.9, and an increase to 7.5 during growth was tolerated. The pO2 was adjusted to exceed 40% saturation in the medium and was automatically controlled by stirring; the aeration rate was kept constant at 0.7 volume per volume per minute. During fermentation, cells of strain DIP1 started to grow after 1 hour and reached a maximum OD600 value of 0.4 after 9 h. Excretion of CphEal was induced when 0.25 g liter−1 sterile CGP suspension was added after 11 h of incubation. CGP increased the OD600 immediately from 0.55 to 0.98 and was completely degraded within 2 h. About 1 hour later, cells started to grow slightly on the released CGP dipeptides. The fermentation was terminated after a total cultivation period of 14 h (Fig. 2).
FIG. 2.
Batch fermentation for CphEal production from P. alcaligenes DIP1 in a Biostat D650 stirred tank reactor containing 420 liters SM with 1 g liter−1 sodium citrate. The reactor was inoculated with 4% (vol/vol) of a preculture that was cultivated in 2-liter baffled flasks containing 1 liter of the same medium and was incubated for 12 h at 30°C. The fermentation parameters and cultivation conditions in the Biostat D650 reactor were as follows: pH of 6.9 to 7.5, temperature of 30°C, and aeration rate of 0.2 volume per volume per minute. pO2 was set to a minimum of 40% and was adjusted automatically by increasing the stirring rate, which otherwise was kept at 100 rpm. The arrow indicates the time of induction by CGP, which caused a sudden increase in medium turbidity followed by a return to the initial value after the release of CphEal and CGP degradation. ⧫, OD600; ⊥, pH; ▴, stirrer speed (rpm); −, pO2 (% of saturation); •, airflow (liters min−1).
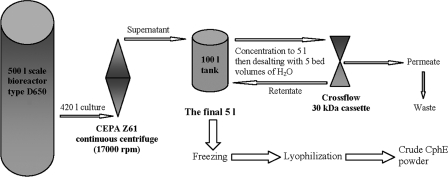
For harvesting, concentration, and desalting of proteins in the cell-free supernatant, including the excreted cyanophycinase, a continuous process was installed (Fig. 3). Cells were continuously separated in a CEPA Z41 continuous centrifuge. The supernatant was collected in a 100-liter tank and was simultaneously concentrated by a cross-flow unit with a 30-kDa filter cassette. The temperature of the solution was kept at 20°C or below by adding ice to the tank. The final 5 liters of the concentrated supernatant was desalted with 25 liters of H2O, frozen at −30°C, and lyophilized to obtain 17.5 g of a crude protein powder.
FIG. 3.
Continuous system for harvesting, concentration, and desalting of proteins in the supernatants of fermentation broth of P. alcaligenes strain DIP1 (phase II). For harvesting, a CEPA Z41 continuous centrifuge was used to separate the cells from the medium. The supernatant was collected in a central 100-liter tank. For concentration, a cross-flow unit with a 30-kDa cassette was connected to the central tank; the concentrated retentate was pumped back into the tank, while the permeant material was directly discarded. The flow rate of the cross flow was adjusted to maintain only 50 liters in the tank. The final concentrated enzyme solution of 5 liters was desalted with 5 bed volumes of H2O, frozen at −30°C, and lyophilized.
Phase III (degradation of purified CGP to its dipeptides).
Large-scale degradation of CGP to its dipeptides was started from 250 g purified CGP in tap water (final concentration, 50 g liter−1) and 10 g of the crude CphEal powder (final concentration, 2 g liter−1). The mixture was stirred slowly at 30°C to ensure complete degradation within 12 h, as estimated from the results of small-scale experiments (Fig. 4). Afterwards, the dipeptide solution was sterilized by filtration, separated from the crude protein components by use of the same cross-flow system as in phase II, frozen at −30°C, and finally lyophilized. Loss in the resulting dry weight with different tested filtration systems increased with a decreasing COP of the filter. This indicated that the applied cross-flow system clearly provided the most suitable COP to purify CGP dipeptides and also to recover the crude CphE portion of the degradation mixture. The recovery rate for the crude enzyme was 78% (wt/wt), and the regained protein powder showed retained cyanophycinase activity on CGP-overlaid agar plates (Fig. 1A).
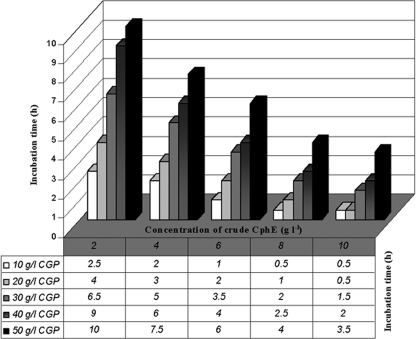
FIG. 4.
Incubation periods required for complete degradation of CGP at different concentrations (10 to 50 g liter−1) by crude CphEal powder applied at different concentrations (1 to 10 g liter−1). The reaction tubes were incubated at 30°C in a tube rotator rotating at 3 rpm. Suspensions with a CGP concentration of 50 g liter−1 could be degraded within 10 h in the presence of 2 g liter−1 crude CphEal powder. Each value represents a mean time period measured for two parallel experimental repetitions, with an estimated standard deviation maximum of 10%.
Quality control.
For final quality control of the resulting dipeptide powder, samples were examined, directly and also after acid-catalyzed hydrolysis, by thin-layer chromatography and HPLC. Both methods revealed the amino acids aspartate, arginine, and lysine as the only constituents in the analyzed samples. Because DNA and total carbohydrate contents did not exceed 0.0002 × 10−5% ± 4 × 10−5% and 0.05 × 10−3% ± 8 × 10−3%, respectively, the purity of the dipeptides in the obtained powder was estimated to be over 99%. The process yielded 227.5 g purified dipeptides from 250 g CGP, representing an overall efficacy of the process of 91%.
Process optimization.
The conditions applied during phase II and phase III were further investigated in order to enhance the parameters and effectiveness of the process.
Optimum conditions for growth and CphEal production by P. alcaligenes DIP1.
Optimum growth for strain DIP1 occurred in the presence of 6 g liter−1 citrate and at pH 6.5 and 37°C. However, no CphEal production was observed at citrate concentrations below 0.5 or above 4 g liter−1. Because the maximum CphEal production was monitored at 2 g liter−1 citrate, the following conditions were considered optimal: SM with 2 g liter−1 citrate, pH 6.5, 37°C, induction after 13 h, and harvesting after 3 h of further incubation.
Optimum induction of CphEal with CGP and alternative substances.
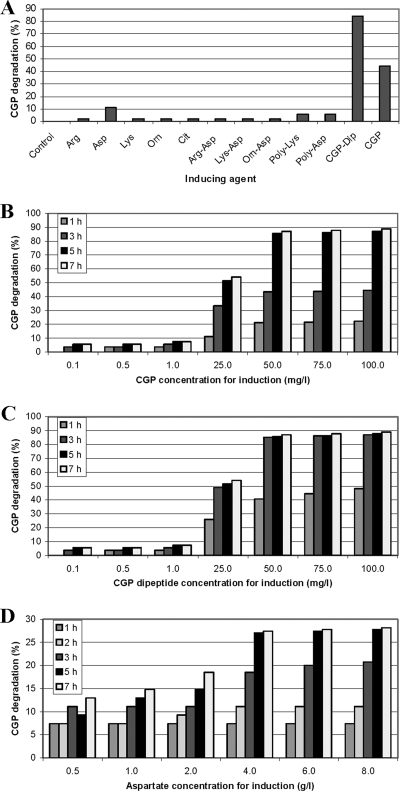
All potential inductors (see Materials and Methods) were tested first at a concentration of 0.25 g liter−1. Only CGP, its dipeptides, and aspartate induced the formation of significant amounts of CphEal (Fig. 5A). At CGP concentrations between 0.001 and 3.0 g liter−1, the inductive effect increased with concentrations of up to 0.05 g liter−1, which led to maximum CphEal production after 5 h of induction (Fig. 5B). CGP dipeptides used in the same concentration range as CGP showed similar results; however, maximum CphEal production was obtained after only 3 h (Fig. 5C). Maximum induction by aspartate was obtained at 4 g liter−1 after 5 h of induction (Fig. 5D), but with a lower productivity than that with the optimum CGP concentration.
FIG. 5.
CGP degradation by concentrated supernatant samples from cultures of P. alcaligenes DIP1 after induction. CGP degradation was determined photometrically and was used as an indicator of the CphEal concentration. (A) Samples from cultures induced with different potential inductors, indicated on the x axis, at a concentration of 0.25 g liter−1. (B) Samples from cultures induced with different concentrations of CGP. (C) Samples from cultures induced with different concentrations of CGP dipeptides. (D) Samples from cultures induced with different concentrations of aspartate. CGP, cyanophycin; Arg-Asp, α-arginine-aspartate; Lys-Asp, α-lysine-aspartate; Orn-Asp, α-ornithine-aspartate; CGP-Dip, CGP dipeptides; poly-Asp, poly(aspartic acid); poly-Lys, poly(lysine); Asp, l-aspartate; Lys, l-lysine; Cit, l-citrulline; Orn, l-ornithine; Arg, l-arginine; control, noninduced culture.
Optimum conditions for CGP hydrolysis to dipeptides by crude CphEal.
To decrease the reaction volumes and the duration of the third process phase (CGP degradation), the time required for CGP hydrolysis starting with significantly higher concentrations (50 to 100 g liter−1) of the polymer was investigated at 30°C. These studies showed an increase in degradation time with decreasing concentrations of crude CphEal and with increasing concentrations of CGP. In the presence of 10 g liter−1 crude CphEal, complete degradation of CGP applied at concentrations of 50 g liter−1 and 100 g liter−1 was observed after 4 and 16 h of incubation, respectively. Maximum CGP degradation occurred at pH 6.5, but at pH values between 5.5 and 7.5, the degradation rates were not much lower. CGP degradation increased with increasing temperature, with a maximum at 50°C. Applying these parameters (50°C, pH 6.5) (Fig. 1C) reduced the degradation time for 50 and 100 g liter−1 CGP to only 1 and 4 h, respectively.
Purification of CphEal from crude supernatant powder, utilizing specific substrate binding.
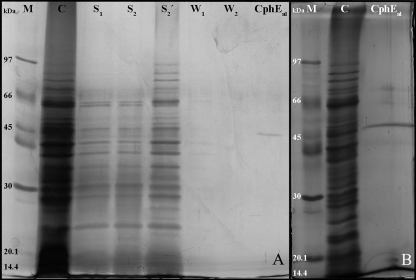
Standard methods for technical protein purification using ammonium sulfate or solvents gave only low recovery rates and low purification factors and were therefore not satisfactory. In contrast, using the insoluble CGP as a matrix to specifically bind CphEal gave very promising results. Initial experiments demonstrated that an incubation time of 5 min was enough to bind CphEal quantitatively to the CGP matrix. SDS-PAGE of protein samples from all purification steps showed a gradual purification of the enzyme (Fig. 6A) and that the first two washing steps are necessary to remove other proteins. The purified enzyme exhibited an apparent molecular mass of 45 kDa by SDS-PAGE and regained activity by in-gel renaturation. The concentration of the purified CphEal was 43.2 μg ml−1, while the total protein content in the initial crude solution was 944 μg ml−1. SDS-PAGE analysis of highly concentrated samples and intensive silver staining revealed the presence of only a few other protein bands, occurring at much lower concentrations (Fig. 6B).
FIG. 6.
SDS-PAGE of samples obtained during purification of CphEal from P. alcaligenes strain DIP1 via specific binding of CGP. (A) SDS-polyacrylamide gel stained by the silver nitrate method. M, molecular mass standard proteins; C, control of crude CphEal; S1, supernatant sample obtained immediately after mixing CGP and crude CphEal; S2, supernatant sample obtained after 6 min of binding time; S2′, same as S1 but after 10-fold concentration; W1 and W2, supernatant samples obtained after the two washing steps. (B) SDS-polyacrylamide gel with triple the volume of purified CphEal as that used for panel A and extended staining with silver nitrate. Only a few other low-concentration protein bands can be observed in addition to that of CphEal at 45 kDa.
Determination of CphEal content in fermentation supernatants.
If an application of CphEal is desired, then the concentration of this CGPase in the produced supernatants must be known. For this purpose, an easy and rapid assay was developed in which 3-μl solutions containing different concentrations (2.7 to 43.2 μg ml−1) of the purified enzyme were added to 1 ml of CGP suspension with a fixed concentration (100 mg liter−1, corresponding to an OD600 of 0.215). Suspensions were incubated for 60 min at 30°C, and the extent of CGP degradation was determined by measuring the OD600. The following formula could be applied if test parameters were kept constant and if measurements were made in an OD600 range of 0.05 to 0.215: E = (0.2404 − D)/0.0017, where E is the CphEal content in crude extract (μg ml−1) and D is the OD600 after 60 min.
Required CGP amounts for technical purification of CphEal.
To integrate the enzyme purification procedure into the main production process, the amount of CGP which is necessary to bind all CphEal (in supernatants with known CphEal content) has to be determined. In a total volume of 0.5 ml, different concentrations of purified CphEal solutions (1.4 to 19 μg ml−1) in 50 μl were added to different CGP concentrations (0.1 to 6 g liter−1). The reaction mixtures were then incubated for 10 min at 30°C and slowly rotated at 3 rpm. After 2 min of centrifugation at 16,000 × g, the supernatants were screened for activity on CGP-overlaid agar plates to detect the minimum CGP concentration which did not induce degradation halos (i.e., complete binding). The determined data were integrated in the following formula: C = (E − 2.4392)/0.9432, where C is the required CGP concentration (g liter−1) and E is the CphEal content in crude extract (μg ml−1).
Biochemical characterization of purified CphEal.
The purified enzyme showed maximum CGP degradation activity at 50°C and complete inactivation at 68°C. The optimum pH range for CGP degradation was 7 to 8.5, with an optimum at 8.5. Among the tested substrates, the purified CphEal showed the highest degradation activity with CGP. However, BSA was also partially degraded (65% in comparison to CGP). Bovine casein (17%) and poly(aspartic acid) (1.5%) were minimally affected by the enzymatic activity of the purified CphEal.
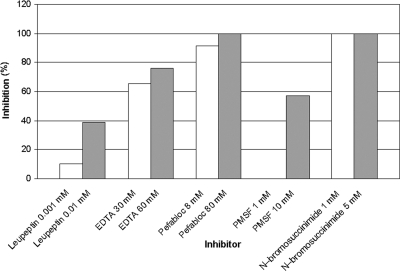
Inhibition experiments employing different group-specific inhibitors (Table 1) revealed that extracellular CphEal is strongly inhibited by the serine protease inhibitors Pefabloc and PMSF. Also, N-bromosuccinimide (tryptophan oxidant) led to a total inhibition of the enzyme. In contrast, CphEal was not negatively affected by the thiol protease inhibitor leupeptin or by the metalloprotease inhibitor EDTA. HPLC analysis of CGP degradation products (Fig. 7) confirmed these results, except for samples that were treated with EDTA, which showed inhibition of CphEal (75%).
TABLE 1.
Effects of several group-specific inhibitors on purified CphEal from P. alcaligenes strain DIP1
| Inhibitor specificity | Inhibitor | Low concn-high concn (mM) | % Inhibition by HPLC analysisb
|
|
|---|---|---|---|---|
| Low inhibitor concn | High inhibitor concn | |||
| Thiol proteases | Leupeptin | 0.001-0.01 | 10 | 39 |
| Metalloproteases | EDTA | 30-60 | 65c | 75c |
| Serine proteases | Pefablocd | 8-80 | 91 | 100 |
| Serine proteases | PMSF | 1-10 | 0 | 57 |
| Tryptophan residues | N-bromosuccinimide | 1-5 | 100 | 100 |
| Controla | 0 | 0 | ||
Control without inhibitor.
The values for inhibition are relative to the control, that is, the activity of CGPase in the absence of an inhibitor.
Values are not due to inhibition but due to the formation of precipitates during OPA derivatization prior to HPLC analysis.
Pefabloc, 4-(2-aminoethyl) benzenesulfonylfluoride·HCl.
FIG. 7.
Effects of different protease inhibitors on CphEal are indicated by the decrease in CGP degradation products in comparison to the control (without inhibitor), as determined by HPLC.
DISCUSSION
Economic factors are generally of great importance for technical processes. Strains of P. alcaligenes, including strain DIP1, are known to grow on simple media and on minimal amounts of a wide range of substrates (36). Citrate is a cheap substrate, that is available in technical quantities; it was previously used for the fermentative production of extracellular lipases from P. alcaligenes. This, in addition to the high enzyme productivity of P. alcaligenes strains, was the reason for their application in fermentative production processes (3, 13, 26). Besides these characteristics, the high stability and activity of the CGPase from strain DIP1 (36) rendered this strain ideal for the designed technical process.
The CGP acid extraction method of Frey et al. (11) was optimized for kilogram amounts of biomass. Optimization comprised the logistics applied and an integrated filtration step which improved CGP purity. In contrast, increasing the number of other purification steps (with diluted acid and water) may unnecessarily increase the loss of CGP. Alternatively, the application of an effective instrument, such as in cross-flow filtration, would increase the overall productivity of this extraction method. However, the relatively high prices and limited life of such ultrafiltration cassettes leave their application to be a matter of cost.
The identical purity grade of the resulting CGP dipeptides before and after filtration is clearly due to the initial lack of impurities in the resulting dipeptide solution. This represents an advantage for applying a defined enzymatic process in comparison to direct cultivation strategies with bacterial cells. On the other hand, the quantitative loss of dipeptides during filtration was expected and, unfortunately, inevitable. Several general factors, including filter material, COPs, saturation of the membrane surface, and/or the characteristics of the filtered substance itself, are known to cause such losses. This also explains the loss of 9% of CGP and 22% of crude CphEal during the degradation phase (phase III).
Maximum production of CphEal by strain DIP1 in the presence of 2 g liter−1 citrate and a lack of enzyme formation in the presence of 6 g liter−1 citrate (which was optimal for growth) indicated that the enzyme is induced only during substrate limitation. Furthermore, the fact that the dipeptides can render the same inductive effect (Fig. 5C), but in a much shorter time (3 h) than that with CGP (5 h) (Fig. 5B), indicated that CGP dipeptides are the actual inductors for CphEal. On the other hand, the ability of aspartate to induce CphEal, but with a lower efficiency (Fig. 5D), makes the choice of the inductor case and cost dependent.
The developed procedure for purification of CphEal by specific binding to CGP proved to be highly effective and has the advantage of separating CphEal from other proteins in crude solutions by use of one substance (CGP). The purification method ends with the degradation of the CGP matrix to its dipeptides. At the same time, these are the valuable end products of the process and can be directed further to the main production stream (no loss). The purification method is easy to scale up and to integrate into the process if desired.
The efficiency of the second phase of the process and the possible purification of the enzyme could be enhanced by the application of two formulas that were developed in this study. The first formula depends on the described photometric test and enables rapid determination of CphEal content in crude supernatants. The determined enzyme concentration can then be integrated into the second formula to estimate the required amount of CGP to bind the complete content of CphEal in the supernatant for purification. The scheme provides a reliable instrument for future batches of crude CphEal, which might differ in their protein composition.
The third phase of the process was found to be much more effective at 50°C than at 30°C. This optimization rendered much higher concentrations of CGP (up to 100 g liter−1) to be easily degraded in only about 25% of the time required at 30°C. Also, the volume of degradation mixtures and the risk of their contamination are minimized at the elevated temperature. Apart from temperature, degradation time showed a colinear increase with decreasing concentrations of crude CphEal and with increasing CGP concentrations. Thus, the respective formula will be helpful to apply optimum degradation parameters. The following formula was calculated for applications at 50°C and is suitable for CGP concentrations of up to 100 g liter−1: P = 17.55 − 29.891E, where P is the degradation time at 50°C and E is the desired concentration of pure CphEal (g liter−1).
Substrate specificity experiments employing purified CphEal revealed that the enzyme degraded not only CGP but also BSA, but to a lesser extent. This indicated that the CGPase from strain DIP1 might be less specific than the previously characterized CphEPa and CphEBm enzymes, from P. anguilliseptica B1 and B. megaterium BAC19, respectively. Another cause for this unspecificity may be the presence of a few other proteins in the purified enzyme solution. Although these proteins were present at very low concentrations, even a small amount of an unspecific protease might have partially degraded BSA.
The inhibition of CphEal by serine protease inhibitors indicates that this CGPase most probably also belongs to the serine-type proteases, as do the previously characterized CphB, CphEPa, and CphEBm enzymes. Moreover, total inhibition of CphEal by N-bromosuccinimide shows that a tryptophan residue might be involved in its catalytic mechanism, in accord with the case for CphEPa and CphEBm. CphEal samples which were treated with leupeptin or EDTA showed no activity inhibition on CGP-overlaid agar plates; the positive results of HPLC analysis (Fig. 7) were due to precipitate formation during OPA derivatization (28, 29).
Although several characteristics of the purified CphEal were relatively similar to those of CphEPa and CphEBm (Table 2), some relevant differences were observed. Among these, the optimum temperature for CphEal (50°C), which is the highest temperature optimum for all known CGPases, provides a great advantage for large-scale application. The shift in optimum pH between the purified enzyme (7 to 8.5, with an optimum at 8.5) and the enzyme in crude preparations (5.5 to 7.5, with an optimum of 6.5) is most probably due to the presence of many other proteins in the crude preparations, representing a complex milieu. Interactions within such milieus may in turn affect the structure and/or properties of the CGPase.
TABLE 2.
Comparison of biochemical characteristics of CphEal from P. alcaligenes strain DIP1 and previously characterized extracellular cyanophycinases
| Bacterium (enzyme) | Enzyme properties
|
||||||
|---|---|---|---|---|---|---|---|
| Localization | Molecular size (kDa) | Optimum temp (°C) | Optimum pH range | CGP degradation product(s) | Mechanism | Inhibitorse | |
| Synechocystis sp. strain PCC 6803 (CphB)a | Intracellular | 28 | 35 | 7-8 | β-Asp-Arg | Exopeptidase | Pefabloc, PMSF, 3,4-dichloroisocoumarin |
| P. anguilliseptica strain BI (CphEPa)b | Extracellular | 43 | NDf | 8-8.5 | β-Asp-Arg | Exopeptidase | Pefabloc, PMSF, N-bromosuccinimide |
| B. megaterium strain BAC19 (CphEBm)c | Extracellular | 37 | 30-42 | 8-8.5 | β-Asp-Arg + (β-Asp-Arg)2 | ND | Pefabloc, PMSF, N-bromosuccinimide |
| P. alcaligenes strain DIP1 (CphEal)d | Extracellular | 45 | 50 | 7-8.5 | β-Asp-Arg | ND | Pefabloc, PMSF, N-bromosuccinimide |
Besides the ecological importance of enzymatic processes in general, and in contrast to direct cultivation strategies with bacterial cells, defined enzymatic processes such as the one described in this article have several advantages, including small losses of end product (consumption during growth is excluded), smaller reaction volumes, fewer contaminants, and time savings. On the other hand, the commercial value of CGP dipeptides is directly related to the production costs of CGP itself. Over the last few years, increasing CGP contents have been achieved using new and more suitable economical substrates (25); these developments seem to move fast toward commercial CGP production. Such a development was also needed for other known bacterial poly(amino acids), such as poly(glutamic acid) and poly(ɛ-lysine), to become commercialized (30, 33). Until that point, the biomedical value of CGP dipeptides, currently under investigation (Sallam and Steinbüchel, submitted for publication), may in fact provide a balanced relationship to the current production costs of CGP.
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbüchel. 2000. Molecular characterization of the cyanophycin synthetase from Synechocystis sp. strain PCC6308. Arch. Microbiol. 174:297-306. [DOI] [PubMed] [Google Scholar]
- 2.Aboulmagd, E., I. Voss, F. B. Oppermann-Sanio, and A. Steinbüchel. 2001. Heterologous expression of cyanophycin synthetase and cyanophycin synthesis in the industrial relevant bacteria Corynebacterium glutamicum and Ralstonia eutropha and in Pseudomonas putida. Biomacromolecules 2:1338-1342. [DOI] [PubMed] [Google Scholar]
- 3.Aehle, W., G. Gerritse, and H. B. Lenting. 1995. Lipases with improved surfactant resistance. International patent WO 95/30744.
- 4.Allen, M. M., M. F. Hutchinson, and P. J. Weathers. 1980. Cyanophycin granule polypeptide formation and degradation in the cyanobacterium Aphanocapsa 6308. J. Bacteriol. 141:687-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen, M. M. 1988. Inclusions: cyanophycin. Methods Enzymol. 167:207-213. [Google Scholar]
- 6.Borzi, A. 1887. Le comunicazioni intracellulari delle Nostochinee. Malpighia 1:28-74. [Google Scholar]
- 7.De-Aloysio, D., R. Mantuano, M. Mauloni, and G. Nicoletti. 1982. The clinical use of arginine aspartate in male infertility. Acta Eur. Fertil. 13:133-167. [PubMed] [Google Scholar]
- 8.Dufresne, A., and E. Samain. 1998. Preparation and characterization of a poly(β-hydroxyalkanoate) latex produced by Pseudomonas oleovorans. Macromolecules 31:6426-6433. [Google Scholar]
- 9.Duruy, A. 1966. Expertise clinique de l'aspartate d’arginine. Med. Int. 1:203. [Google Scholar]
- 10.Elbahloul, Y., K. Frey, J. Sanders, and A. Steinbüchel. 2005. Protamylasse, a residual compound of industrial starch production, provides a suitable medium for large-scale cyanophycin production. Appl. Environ. Microbiol. 71:7759-7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey, K. M., F. B. Oppermann-Sanio, H. Schmidt, and A. Steinbüchel. 2002. Technical-scale production of cyanophycin with recombinant strains of Escherichia coli. Appl. Environ. Microbiol. 68:3377-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Füser, G., and A. Steinbüchel. 2007. Analysis of genome sequences for genes of cyanophycin metabolism: identifying putative cyanophycin metabolizing prokaryotes. Macromol. Biosci. 7:278-296. [DOI] [PubMed] [Google Scholar]
- 13.Gerritse, G., R. W. Hommes, and W. J. Quax. 1998. Development of a lipase fermentation process that uses a recombinant Pseudomonas alcaligenes strain. Appl. Environ. Microbiol. 64:2644-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, M., and N. G. Carr. 1981. Enzyme activities related to cyanophycin metabolism in heterocysts and vegetative cells of Anabaena spp. J. Gen. Microbiol. 125:17-23. [Google Scholar]
- 15.Joentgen, W., N. Müller, A. Mitschker, and H. Schmidt. 2003. Polyaspartic acids, p. 175-199. In S. R. Fahnestock and A. Steinbüchel (ed.), Biopolymers, vol. 7. Wiley, Weinheim, Germany. [Google Scholar]
- 16.Krehenbrink, M., F. B. Oppermann-Sanio, and A. Steinbüchel. 2002. Evaluation of non-cyanobacterial genome sequences for occurrence of genes encoding proteins homologous to cyanophycin synthetase and cloning of an active cyanophycin synthetase from Acinetobacter sp. strain DSM 587. Arch. Microbiol. 177:371-380. [DOI] [PubMed] [Google Scholar]
- 17.Lacks, S. A., and S. S. Springhorn. 1980. Renaturation of enzymes after polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulphate. J. Biol. Chem. 225:7467-7473. [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lamm, S., F. Schönlau, and P. Rohdewald. 2003. Prelox for improvement of erectile function: a review. Eur. Bull. Drug Res. 11:29-37. [Google Scholar]
- 20.Leopold, C. S., and D. R. Friend. 1995. In vivo pharmacokinetics study for the assessment of poly(l-aspartic acid) as a drug carrier for colon-specific drug delivery. J. Pharmacokinet. Biopharm. 4:397-406. [DOI] [PubMed] [Google Scholar]
- 21.Li, H., D. M. Sherman, S. Bao, and L. A. Sherman. 2001. Pattern of cyanophycin accumulation in nitrogen-fixing and non-nitrogen-fixing cyanobacteria. Arch. Microbiol. 176:9-18. [DOI] [PubMed] [Google Scholar]
- 22.Liotenberg, S., D. Campbell, R. Rippka, J. Houmard, and N. T. de Marsac. 1996. Effect of the nitrogen source on phycobiliprotein synthesis and cell reserves in a chromatically adapting filamentous cyanobacterium. Microbiology 142:611-622. [DOI] [PubMed] [Google Scholar]
- 23.Mackerras, A. H., N. M. de Chazal, and G. D. Smith. 1990. Transient accumulation of cyanophycin in Anabaena cylindrical and Synechocystis 6308. J. Gen. Microbiol. 136:2057-2065. [Google Scholar]
- 24.Meredith, J. W., J. A. Ditesheim, and G. Zaloga. 1990. Visceral protein levels in trauma patients are greater with peptide diet than with intact protein diet. J. Trauma 30:825-828. [DOI] [PubMed] [Google Scholar]
- 25.Mooibroek, H., N. Osterhuis, M. Giuseppin, M. Toonen, H. Franssen, E. Scott, J. Sanders, and A. Steinbüchel. 2007. Assessment of technological options and economical feasibility for cyanophycin biopolymer and high-value amino acid production. Appl. Microbiol. Biotechnol. 77:257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, E. R. B., B. J. Tindall, V. A. P. Martins Dos Santos, D. R. H. Pieper, J. Ramos, and N. J. Palleroni. May 1999, posting date. Pseudomonas: nonmedical. In M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.0. Springer, New York, NY. doi: 10.1007/0-387-30746-X_21. [DOI]
- 27.Nesterenko, M. V., M. Tilley, and S. J. Upton. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 3:239-242. [DOI] [PubMed] [Google Scholar]
- 28.Obst, M., F. B. Oppermann-Sanio, and A. Steinbüchel. 2002. Isolation of cyanophycin degrading bacteria, cloning and characterization of an extracellular cyanophycinase gene (cphE) from Pseudomonas anguilliseptica strain BI—the cphE gene from P. anguilliseptica BI encodes a cyanophycin-hydrolyzing enzyme. J. Biol. Chem. 277:25096-25105. [DOI] [PubMed] [Google Scholar]
- 29.Obst, M., A. Sallam, H. Luftmann, and A. Steinbüchel. 2004. Isolation and characterization of gram-positive cyanophycin-degrading bacteria—kinetic studies on cyanophycin depolymerase activity in aerobic bacteria. Biomacromolecules 5:153-161. [DOI] [PubMed] [Google Scholar]
- 30.Obst, M., and A. Steinbüchel. 2004. Microbial degradation of poly(amino acid)s. Biomacromolecules 5:1166-1176. [DOI] [PubMed] [Google Scholar]
- 31.Obst, M., A. Krug, H. Luftmann, and A. Steinbüchel. 2005. Degradation of cyanophycin by Sedimentibacter hongkongensis strain KI and Citrobacter amalonaticus strain G isolated from an anaerobic bacterium consortium. Appl. Environ. Microbiol. 71:3642-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obst, M., and A. Steinbüchel. 2006. Cyanophycin—an ideal bacterial nitrogen storage material with unique chemical properties, p. 167-194. In J. M. Shively (ed.), Inclusions in prokaryotes, vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 33.Oppermann-Sanio, F. B., and A. Steinbüchel. 2002. Occurrence, functions and biosynthesis of polyamides in microorganisms and biotechnological production. Naturwissenschaften 89:11-22. [DOI] [PubMed] [Google Scholar]
- 34.Oppermann-Sanio, F. B., and A. Steinbüchel. 2003. Cyanophycin, p. 83-106. In S. R. Fahnestock and A. Steinbüchel (ed.), Biopolymers, vol. 7. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 35.Richter, R., M. Hejazi, R. Kraft, K. Ziegler, and W. Lockau. 1999. Cyanophycinase, a peptidase degrading the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartic acid (cyanophycin). Molecular cloning of the gene of Synechocystis sp. PCC6803, expression in Escherichia coli, and biochemical characterization of the purified enzyme. Eur. J. Biochem. 263:163-169. [DOI] [PubMed] [Google Scholar]
- 36.Sallam, A., and A. Steinbüchel. 2008. Anaerobic and aerobic degradation of cyanophycin by the denitrifying bacterium Pseudomonas alcaligenes strain DIP1 and the role of three other co-isolates in a mixed bacterial consortium. Appl. Environ. Microbiol. 74:3434-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schwamborn, M. 1998. Chemical synthesis of polyaspartates: a biodegradable alternative to currently used polycarboxylate homo- and copolymers. Polym. Degrad. Stabil. 59:39-45. [Google Scholar]
- 39.Sellier, J. 1979. Intéret de l’aspartate d’arginine sargenor chez des athletes de compétition en périod d'entrainement intensif. Rev. Med. Toulouse 5:879. [Google Scholar]
- 40.Sherman, D. M., D. Tucher, and L. A. Sherman. 2000. Heterocyst development and localization of cyanophycin in N2-fixing cultures of the cyanobacterium Anabaena sp. PCC7120. J. Phycol. 36:932-941. [Google Scholar]
- 41.Simon, R. D. 1976. The biosynthesis of multi-l-arginyl-poly(l-aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim. Biophys. Acta 422:407-418. [DOI] [PubMed] [Google Scholar]
- 42.Simon, R. D., and P. Weathers. 1976. Determination of the structure of the novel polypeptide containing aspartic acid and arginine which is found in cyanobacteria. Biochim. Biophys. Acta 420:165-176. [DOI] [PubMed] [Google Scholar]
- 43.Simon, R. D. 1987. Inclusion bodies in the cyanobacteria: cyanophycin, polyphosphate, polyhedral bodies, p. 199-225. In P. Fay and C. van Baalen (ed.), The cyanobacteria. Elsevier, Amsterdam, The Netherlands.
- 44.Stephan, D. P., H. G. Ruppel, and E. K. Pistorius. 2000. Interrelation between cyanophycin synthesis, l-arginine catabolism and photosynthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. Z. Naturforsch. 55:927-942. [DOI] [PubMed] [Google Scholar]
- 45.Tamura, S. I., H. Tanaka, R. Takayama, H. Sato, Y. Sato, and N. Uchida. 1985. Break of unresponsiveness to delayed-type hypersensitivity to sheep red blood cells by pertussis toxin. Cell. Immunol. 92:376-386. [DOI] [PubMed] [Google Scholar]
- 46.Voss, I., and A. Steinbüchel. 2006. Application of a KDPG-aldolase gene-dependent addiction system for enhanced production of cyanophycin in Ralstonia eutropha strain H16. Metab. Eng. 8:66-78. [DOI] [PubMed] [Google Scholar]
- 47.Weber, K., and M. Osborn. 1969. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
- 48.Wingard, L. L., S. R. Miller, J. M. Sellker, E. Stenn, M. M. Allen, and A. M. Wood. 2002. Cyanophycin production in a phycoerythrin-containing marine Synechococcus strain of unusual phylogenetic affinity. Appl. Environ. Microbiol. 68:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yokoyama, M., M. Miyauchi, N. Yamada, T. Okano, Y. Sakurai, K. Kataoka, and S. Inoue. 1990. Characterization and anticancer activity of the micelle forming polymeric anticancer drug adriamycin-conjugated poly(ethylene glycol)-poly(aspartic acid) block copolymer. Cancer Res. 6:1693-1700. [PubMed] [Google Scholar]
- 50.Ziegler, K., A. Diener, C. Herpin, R. Richter, R. Deutzmann, and W. Lockau. 1998. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartate (cyanophycin). Eur. J. Biochem. 254:154-159. [DOI] [PubMed] [Google Scholar]
- 51.Ziegler, K., R. Deutzmann, and W. Lockau. 2002. Cyanophycin synthetase-like enzymes of non-cyanobacterial eubacteria: characterization of the polymer produced by a recombinant synthetase of Desulfitobacterium hafniense. Z. Naturforsch. 57c:522-529. [DOI] [PubMed] [Google Scholar]