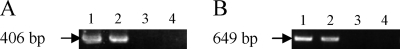Abstract
Legionella organisms are prevalent in manmade water systems and cause legionellosis in humans. A rapid detection method for viable Legionella cells combining ethidium monoazide (EMA) and PCR/real-time PCR was assessed. EMA could specifically intercalate and cleave the genomic DNA of heat- and chlorine-treated dead Legionella cells. The EMA-PCR assay clearly showed an amplified fragment specific for Legionella DNA from viable cells, but it could not do so for DNA from dead cells. The number of EMA-treated dead Legionella cells estimated by real-time PCR exhibited a 104- to 105-fold decrease compared to the number of dead Legionella cells without EMA treatment. Conversely, no significant difference in the numbers of EMA-treated and untreated viable Legionella cells was detected by the real-time PCR assay. The combined assay was also confirmed to be useful for specific detection of culturable Legionella cells from water samples obtained from spas. Therefore, the combined use of EMA and PCR/real-time PCR detects viable Legionella cells rapidly and specifically and may be useful in environmental surveillance for Legionella.
Legionellae are gram-negative, rod-shaped bacteria that are ubiquitous inhabitants of aquatic environments and moist soil, replicating as intracellular parasites of protozoa (6, 22, 23). The bacterium causes legionellosis in humans. Hot springs, public baths, and cooling towers are the most probable sources of legionellosis. In Japan, several legionellosis outbreaks caused by Legionella pneumophila have been reported (13, 16, 17, 19, 29, 33). To prevent this infectious disease, surveillance investigations of manmade water systems, such as cooling towers, showerheads, and water distribution pipelines, should be carried out regularly. Because it takes 4 to 7 days to isolate viable Legionella organisms from environmental and clinical samples, the development of a rapid detection and isolation method is indispensable for identification of sources and routes of incidents at an early stage.
PCR/real-time PCR is the most widely applied technology for direct detection and quantification of pathogens in foods and environmental or clinical samples. PCR/real-time PCR assays targeting the 16S rRNA or 5S rRNA genes specific for Legionella and the macrophage infectivity potentiator (mip) gene specific for L. pneumophila have been developed for detection and identification of the bacterium (14, 27, 32; EnviroAmp Legionella kit package insert [Perkin-Elmer Corporation]). However, a lack of differentiation of DNAs from living and dead Legionella cells has seriously hampered the implementation of DNA diagnostics in routine applications. Since chlorine is routinely added to water distribution systems to kill Legionella and other bacteria, the resulting bacterial death and lysis release copious amounts of genomic DNA into the water. DNAs from dead Legionella strains act as a major obstacle in PCR/real-time PCR detection of viable bacteria. It is conceivable that PCR/real-time PCR can be utilized more extensively for detection if this problem can be cleared up.
Ethidium monoazide (EMA) is a dye that allows microscopic differentiation between viable and dead cells (1, 21). Specifically, the phenanthridinium DNA/RNA-intercalating agent enters only those bacteria that have compromised cell walls and membranes and subsequently covalently links to the DNA within the cells (2, 4, 9, 31). Photolysis of EMA by visible light produces a nitrene that covalently links to genomic DNA, cleaving it into small pieces upon photoactivation (9, 24, 26). Contrastingly, unbound EMA, which remains free in solution, is simultaneously inactivated by reaction with water molecules and no longer capable of covalently binding to DNA (4, 10). Thus, DNA from viable cells, protected from reactive EMA by an intact cell wall/cell membrane, should not be affected by the inactivated EMA after cell lysis during DNA extraction.
EMA can selectively enter the cytoplasm of dead cells and cleave the DNA via photoactivation (24, 26). Therefore, cleaved DNA from damaged and/or dead cells cannot be amplified by PCR/real-time PCR. Thus, the combination of EMA and PCR/real-time PCR may potentially distinguish the DNA of viable cells from the DNA of dead cells. In the present study, we assessed this potential for the specific detection of DNA of viable Legionella.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Legionella strains used in this study are listed in Table 1. All strains were grown at 37°C on buffered charcoal yeast extract (BCYE) agar (Becton Dickinson, Sparks, MD), using standard protocols. Pseudomonas aeruginosa strain PAO1, Escherichia coli K-12 strain XL1-Blue, Serratia marcescens strains E1 and E46, and Brevundimonas nasdae NIIB2318 were incubated at 37°C overnight on LB plates (Becton Dickinson). Sphingomonas paucimobilis (JCM 7516), Caldimonas manganoxidans (JCM 10698), Porphyrobacter sanguineus (JCM 20691), Microbacterium lactium (JCM 1379), Bacillus megaterium (JCM 2506), Tepidimonas arfidensis (JCM 13232), Methyloversatilis universalis (JCM 13912), and Rhizobium radiobacter (JCM 20371) strains were purchased from the Japan Collection of Microorganisms (JCM) (RIKEN BioResource Center, Saitama, Japan) and were incubated as recommended by JCM. After incubation, separate suspensions of each strain were made in sterile normal saline. Bacterial counts were determined by plating cells on appropriate plates after serial 10-fold dilutions.
TABLE 1.
Summary of Legionella strains used in this studya
| Strain | Species | Serogroup | Alternate strain name or source |
|---|---|---|---|
| 80-045 | L. pneumophila | 1 | Clinical isolate |
| Philadelphia-1 | L. pneumophila | 1 | ATCC 33152; clinical isolate |
| NIIB0733 | L. pneumophila | 1 | Bathtub |
| NIIB0805 | L. pneumophila | 1 | Bathtub |
| NIIB0744 | L. pneumophila | 1 | Cooling tower |
| NIIB0802 | L. pneumophila | 1 | Cooling tower |
| NIIB0784 | L. pneumophila | 5 | Bathtub |
| NIIB0797 | L. pneumophila | 5 | Bathtub |
| NIIB0792 | L. pneumophila | 6 | Bathtub |
| NIIB0864 | L. pneumophila | 6 | Bathtub |
| NIIB0794 | L. pneumophila | 7 | Cooling tower |
| NIIB0806 | L. pneumophila | 7 | Cooling tower |
| NIIB0008 | L. micdadei | ATCC 33218; clinical isolate | |
| NIIB0009 | L. bozemanii | 1 | ATCC 33217; clinical isolate |
| NIIB0010 | L. dumoffii | ATCC 33343; clinical isolate | |
| NIIB0012 | L. longbeachae | 2 | ATCC 33484; clinical isolate |
| NIIB0052 | L. feeleii | 2 | ATCC 35849; clinical isolate |
| NIIB0234 | L. gormanii | ATCC 33297; soil isolate |
ATCC, American Type Culture Collection; NIIB, National Institute of Infectious Diseases, Department of Bacteriology.
Heat and chlorine treatments for Legionella strains.
Dead Legionella cells were prepared by treatment with heat or sodium hypochlorite (Sigma-Aldrich, St. Louis, MO). Heat treatment was performed at 95°C for 2 min. Sodium hypochlorite treatment was performed at an available chlorine concentration of 0.5 or 1.0 ppm, followed by incubation for 30 min at room temperature. The residual chlorine concentration was assayed using Rapid DPD liquid (Kanto Chemical, Tokyo, Japan). After either treatment, Legionella cells were pelleted and resuspended in the original volume of normal saline before being subjected to EMA treatment. Death of the heat- and chlorine-treated Legionella cells was confirmed by using a BacLight Live/Dead bacterial viability kit (Molecular Probes, Leiden, The Netherlands).
EMA treatment and visible light irradiation of Legionella strains.
EMA purchased from Sigma-Aldrich was prepared at a concentration of 10 mg/ml. EMA was added to Legionella suspensions at various concentrations and kept at 4°C for 10 min in the dark. Subsequently, each suspension was set on ice and exposed to visible light for 5 min (24).
Preparation of a mock sample of environmental conditions.
To prepare a mock sample for use as an environmental model, isolated Legionella cells were added to tap water along with sodium thiosulfate (0.05%) to inactivate the chlorine already present in the tap water. Two hundred milliliters of water to which viable or chlorine-treated Legionella cells had been added was centrifuged for 15 min, and the pellets were resuspended in 2 ml normal saline. One milliliter of the suspension was treated with a low-pH buffer (0.2 M KCl-HCl buffer, pH 2.2) to reduce the number of environmental bacteria other than Legionella, and 100 μl of each dilution was plated on BCYE agar to determine the number of living Legionella cells. The genomic DNA of the remaining sample, with or without EMA treatment, was purified and used for real-time PCR.
Collection of water samples from public and model spas.
A total of 25 samples, 9 from public spas and 16 from a model spa system (18, 28), were collected. In the model spa, no chlorine disinfection was performed for 10 days after men took baths to allow for Legionella contamination and growth in the bathtub and filter tank. Water samples (samples 10 to 17) from the bathtub were collected from day 3 to day 10 after the bath. On day 10, after one sample (sample 18) was obtained from the filter tank, a high concentration of chlorine was swiftly added into and circulated reversely throughout the filter tank to prepare chlorine-treated Legionella cells (18, 28). Water samples (samples 19 to 25) were separately collected from the filter tank 0, 1, 2, 3, 5, 6, and 7 min after the addition of chlorine. A solution of 500 milliliters of each sample was collected, and chlorine was inactivated by sodium thiosulfate. The samples were centrifuged at 7,500 rpm for 15 min, and the pellets were resuspended in 5 ml normal saline. One milliliter of the suspension was treated with 0.2 M KCl-HCl buffer (pH 2.2) to reduce the number of environmental bacteria other than Legionella and then plated on GVPC agar (Oxoid, Hampshire, United Kingdom) to determine the number of living Legionella cells. Five hundred microliters of each sample, with or without EMA treatment at 1, 5, 10, and 20 μg/ml, was exposed to visible light as described above. After photoactivation, the bacteria were collected by centrifugation and their genomic DNAs were purified for real-time PCR.
PCR.
The genomic DNAs of bacteria were purified using a High Pure PCR template preparation kit (Roche Diagnostics, Mannheim, Germany). Oligonucleotide primers LEG448A and LEG854B, targeting the 16S rRNA gene (32), an EnviroAmp primer targeting the 5S rRNA gene (EnviroAmp Legionella kit package insert; Perkin-Elmer, Waltham, MA), and primers LmipL920 and LmipR1548, targeting the L. pneumophila mip gene (14), were used for PCR amplifications. The amplifications were carried out with Ex Taq polymerase (Takara Bio, Otsu, Japan), using a GeneAmp PCR system 9700 instrument (Applied Biosystems, Foster City, CA). A 20-μl PCR preparation was subjected to 30 cycles of denaturation at 98°C for 10 s, annealing at 63.5°C for 30 s, and extension at 72°C for 60 s. Ten-microliter solutions with the PCR-amplified DNA fragments were separated in 2% agarose gels (Takara Bio).
Real-time PCR.
Real-time PCR targeting the 16S rRNA gene of Legionella was performed using an ABI Prism 7000 machine (Applied Biosystems). The 25-μl reaction volume contained 2 μl of DNA purified from each sample. Real-time quantification for SYBR green detection was performed with SYBR green PCR master mix (Applied Biosystems). The primers used were LEG427F (5′-GTAAAGCACTTTCAGTGGGGAG-3′) and LEG880R (5′-GGTCAACTTATCGCGTTTGCT-3′). The amplification reaction was performed with an initial 10-min denaturation step at 95°C followed by 40 cycles of repeated denaturation at 95°C for 15 s and annealing and polymerization at 63.5°C for 60 s. Premix Ex Taq (Takara Bio) was used for fluorescent probe-based real-time PCR. The quantification was performed with primers LEG427F and LEG880R and the molecular beacon probe P1 (5′-6-carboxyfluorescein-ACTGGACGTTACCCACAGAAGAAG-6-carboxytetramethylrhodamine-3′) (Takara Bio), designed for detection of the Legionella 16S rRNA gene. The amplification reaction was performed with 40 cycles of repeated denaturation at 95°C for 10 s and annealing and polymerization at 63.5°C for 60 s after a denaturation step at 95°C for 30 s.
Purified genomic DNA from 2 × 108 CFU of L. pneumophila 80-045 was used as an external standard. For each real-time PCR, the purified DNA was thawed and serially diluted to prepare four to six dilution points ranging from 1 × 107 to 1 × 102 Legionella cells as an external standard. A negative extraction control (PCR-grade water), a positive control, and the test samples were run in duplicate.
Statistical analyses.
All experiments were carried out more than twice. The significance of the results was analyzed using Student's t test. Differences were considered significant at P values of <0.05.
RESULTS
Heat and chlorine treatment of Legionella strains.
Eighteen Legionella strains, comprising 12 L. pneumophila and 6 non-L. pneumophila strains, were used (Table 1). The 12 L. pneumophila strains, which belonged to serogroups 1, 5, 6, and 7 (data not shown), were isolated from patients, water from cooling towers, or bathtubs. The six non-L. pneumophila strains comprised different Legionella species (Table 1) that are known as human pathogens.
All Legionella strains were suspended in sterile normal saline at approximately 1 × 107 CFU/ml and treated with heat or chlorine. After heat treatment at 95°C for 2 min, no colonies were detected in any suspensions of the Legionella strains plated on BCYE agar (data not shown). Chlorine treatment was initially performed at a concentration of 0.5 ppm of free residual chlorine. After 30 min of incubation at room temperature, the residual chlorine concentration became 0.3 ppm. No colonies were cultured, with the exception of strain 80-045, where approximately 5 × 104 cells remained culturable. Use of 1.0 ppm chlorine, which produced a residual concentration of 0.6 ppm, resulted in no detectable growth of strain 80-045. Therefore, 1.0 ppm chlorine was used in further experiments. By using a BacLight Live/Dead bacterial viability kit, >99% of the heat- and chlorine-treated Legionella cells were determined to be in a nonviable state (dead), while >99% of the Legionella cells without treatment were in a viable state.
Combined use of EMA and PCR to detect viable Legionella cells.
We examined whether the combined use of EMA and PCR could specifically detect viable Legionella cells. Viable, heat-treated, and chlorine-treated Legionella cells were treated with 10 μg/ml, 20 μg/ml, and 50 μg/ml of EMA. Viable Legionella cells that were not treated with EMA were used as a control. Genomic DNA was purified and used as a template for PCR.
The results of the EMA-PCR assay using 20 μg/ml EMA are depicted in Fig. 1. PCR products targeting the 16S rRNA (Fig. 1A) and mip (Fig. 1B) genes in genomic DNA from viable cells of L. pneumophila strain 80-045, with or without treatment of EMA, displayed similar agarose gel electrophoretic patterns (Fig. 1, lanes 1 and 2). However, no PCR products for DNAs from the heat- and chlorine-killed cells with EMA treatment were observed (Fig. 1, lanes 3 and 4). Amplified fragments of Legionella DNA from the heat- and chlorine-killed cells without EMA treatment were detected and were similar to those from viable cells (data not shown). Similar results were also obtained by the use of the other 11 L. pneumophila strains for the detection of 16S rRNA and mip genes and the 6 non-L. pneumophila strains for detection of the 16S rRNA gene (data not shown). When 10 μg/ml of EMA was used, the intensity of the amplified fragments from the heat- and chlorine-killed Legionella cells was weaker than that for the viable cells, although amplified bands were still visible on the gel (data not shown). Conversely, the amplified fragments from the heat- and chlorine-killed Legionella cells were undetectable when 50 μg/ml EMA was used, while the intensity of fragments from EMA-treated viable Legionella cells was significantly weaker than that for untreated cells (data not shown). Therefore, 20 μg/ml EMA was used in further experiments, except for treatment of water samples from spas.
FIG. 1.
PCRs targeting the 16S rRNA (A) and mip (B) genes of Legionella strain 80-045. The sizes of the amplified fragments from the 16S rRNA and mip genes are 406 and 649 bp, respectively. Lanes 1, viable Legionella cells without EMA treatment; lanes 2, viable Legionella cells with EMA treatment; lanes 3, heat-killed Legionella cells with EMA treatment; lanes 4, chlorine-killed Legionella cells with EMA treatment. EMA was used at a concentration of 20 μg/ml.
PCR targeting the 5S rRNA gene was also performed. Although the intensity of PCR fragments for the DNAs from the heat- and chlorine-treated cells became weaker than that for the viable cells with EMA treatment, the bands were clearly observed on the gel (data not shown).
Combined use of EMA and real-time PCR to detect viable Legionella cells.
To quantify the DNA purified from the bacteria treated with EMA, real-time PCR targeting the 16S rRNA gene was performed with SYBR green PCR master mix. Approximately 1 × 106 to 1 × 107 CFU/ml of strains 80-045, Philadelphia-1, NIIB0008, and NIIB0009 was used. The detection limit for L. pneumophila was 1 CFU/reaction (data not shown).
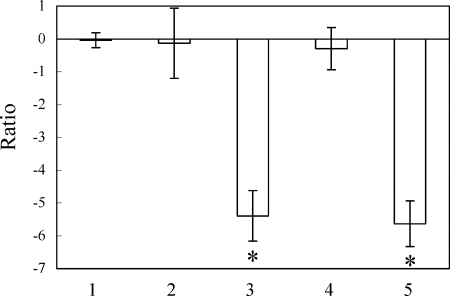
Results of real-time PCR are shown in Fig. 2. When the amount of DNA detected by real-time PCR was calculated as the cell count, the number of viable bacteria treated with EMA, heat-killed bacteria with or without EMA treatment, or chlorine-killed bacteria with or without EMA treatment was compared to the number of viable bacteria without EMA treatment and expressed as a ratio. The ratio of viable to EMA-treated 80-045 cells was −0.04 ± 0.23 log10 (Fig. 2, bar 1). No significant difference was evident between the amounts of DNA of untreated and EMA-treated viable Legionella cells, because EMA could not intercalate and cleave the genomic DNA of viable Legionella cells. The numbers of heat- and chlorine-killed Legionella cells without EMA treatment, estimated by real-time PCR, did not obviously decrease compared to that of the viable cells. The ratios of heat- and chlorine-killed 80-045 cells were −0.13 ± 1.06 log10 (Fig. 2, bar 2) and −0.30 ± 0.64 log10 (Fig. 2, bar 4), respectively. These results indicate that the heat and chlorine treatments performed here left the DNAs intact. Conversely, after EMA treatment, the amounts of amplifiable DNA in heat- and chlorine-killed Legionella cells significantly decreased compared to that in the viable cells. The ratios of heat- and chlorine-killed 80-045 cells were −5.39 ± 0.77 log10 (Fig. 2, bar 3) and −5.64 ± 0.70 log10 (Fig. 2, bar 5), respectively. Comparable results were also yielded when the Philadelphia-1, NIIB0008, and NIIB0009 strains were used (data not shown). Altogether, these results are consistent with the ability of EMA to cleave genomic DNA and to decrease the amount of intact DNA of the heat- and chlorine-treated Legionella cells to the level of approximately 4 to 5 log10 fewer cells.
FIG. 2.
EMA and real-time PCR combined analyses of viable or dead cells of L. pneumophila strain 80-045. The number of bacteria was estimated from the amount of DNA detected by real-time PCR with SYBR green as the reporter dye. The number of viable cells without EMA treatment was set as 1. The numbers of EMA-treated viable cells (bar 1), untreated heat-killed cells (bar 2), EMA-treated heat-killed cells (bar 3), untreated chlorine-killed cells (bar 4), and EMA-treated chlorine-killed cells (bar 5) are described as ratios against the number of untreated viable cells [i.e., ratio = log10 (number of test cells/number of untreated viable cells)]. The error bars represent standard deviations from more than three independent experiments. Asterisks indicate significant decreases in the numbers of EMA-treated samples compared to those of untreated samples.
Discrimination of viable and chlorine-treated Legionella cells from mock environmental samples by the combined use of EMA and real-time PCR.
In order to investigate the possible utility of EMA and real-time PCR for the detection of viable Legionella cells in environmental samples, we prepared a mock sample by the suspension of Legionella strains in tap water. Approximately 1 × 106 to 1 × 107 CFU of viable or chlorine-treated Legionella were added to tap water. The number of bacteria was determined by plating on BCYE agar and estimated by the combined EMA and real-time PCR assay. To avoid the possible contamination of the assay by nonlegionella bacteria present in tap water, fluorescently probed real-time PCR targeting the 16S rRNA gene was performed with the LEG427F and LEG880R primers and probe P1. The detection limit was 10 CFU/reaction (data not shown).
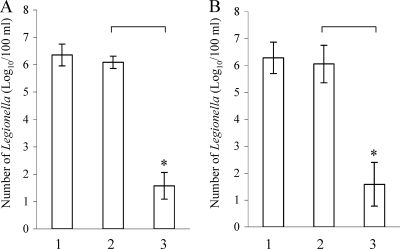
The numbers of Legionella cells detected in 100 ml water are shown in Fig. 3. Approximately 6.35 ± 0.40 log10 CFU of viable 80-045 cells (Fig. 3A, bar 1) and 6.29 ± 0.58 log10 CFU of viable NIIB0009 cells (Fig. 3B, bar 1) were detected by plating. After treatment with 20 μg/ml of EMA, 6.09 ± 0.22 log10 80-045 cells (Fig. 3A, bar 2) and 6.06 ± 0.70 log10 NIIB0009 cells (Fig. 3B, bar 2) were detected by real-time PCR. When the chlorine-treated above Legionella solution (after chlorine treatment, the number of cultivable cells became <10 CFU/100 ml by plating) was treated with EMA, only 1.57 ± 0.49 log10 80-045 cells (Fig. 3A, bar 3) and 1.59 ± 0.81 log10 NIIB0009 cells (Fig. 3B, bar 3) were detected by real-time PCR. The number of EMA-treated dead Legionella cells, estimated by real-time PCR in this experiment, was approximately 4.5 log10 less than that of the viable cells.
FIG. 3.
Number of L. pneumophila strain 80-045 cells (A) and L. bozemanii strain NIIB0009 cells (B) isolated from 100-ml mock samples, determined by incubation or estimated by real-time PCR. 6-Carboxyfluorescein was used as the reporter dye for real-time PCR. Bars 1, real numbers of viable Legionella cells determined by plating on BCYE plates; bars 2, numbers of EMA-treated viable Legionella cells determined by fluorescent probe-based real-time PCR; bars 3, numbers of EMA-treated chlorine-killed Legionella cells determined by real-time PCR. No viable cells of chlorine-killed 80-045 and NIIB0009 were detected by plating on BCYE agar. The experiments were repeated separately more than five times. Asterisks indicate significant decreases in the number of EMA-treated chlorine-killed Legionella cells determined by real-time PCR.
Specificity of real-time PCR.
Surveillance performed in Japan has detected over 20 species of nonlegionella bacteria in bathtub water by using a PCR-denaturing gradient gel electrophoresis method (5). In addition to these bacterial species, E. coli, S. marcescens, and Brevundimonas species have been reported to usually be present in water and/or soil of the external environment (3, 8, 11, 15, 25). Thirteen appropriate nonlegionella strains (Table 2) were used to assess PCR specificity. Genomic DNAs purified from these strains were used as templates, and real-time PCR was performed with primers LEG427F and LEG880R and probe P1. After 40 PCR cycles, no amplification signals could be detected (Table 2). The results indicate that real-time PCR has a high specificity for Legionella species.
TABLE 2.
Nonlegionella bacteria used to test the specificity of real-time PCRa
| Species | Strain or sourceb | No. of bacteria (CFU/ml)c |
|---|---|---|
| Pseudomonas aeruginosa | Clinical isolate | 1.6 × 106 |
| Escherichia coli | K-12 | 1.3 × 106 |
| Serratia marcescens E1 | Environmental isolate | 8.6 × 105 |
| Serratia marcescens E46 | Environmental isolate | 3.8 × 105 |
| Brevundimonas nasdae | Environmental isolate | 1.6 × 107 |
| Sphingomonas paucimobilis | JCM7516; environmental isolate | 1.0 × 106 |
| Caldimonas manganoxidans | JCM10698; environmental isolate | 3.1 × 106 |
| Porphyrobacter sanguineus | JCM20691; environmental isolate | 1.5 × 106 |
| Microbacterium lactium | JCM1379; environmental isolate | 1.0 × 106 |
| Bacillus megaterium | JCM2506; environmental isolate | 7.0 × 105 |
| Tepidimonas arfidensis | JCM13232; clinical isolate | 8.0 × 105 |
| Methyloversatilis universalis | JCM13912; environmental isolate | 1.0 × 106 |
| Rhizobium radiobacter | JCM20371; environmental isolate | 1.2 × 106 |
Real-time PCR was performed with primers LEG427F and LEG880R and probe P1. No products were detected for any of the organisms tested.
JCM, Japan Collection of Microorganisms.
The number of bacteria of each strain was determined by plating cells on suitable plates, and the number indicated was used for real-time PCR.
Detection of Legionella in water samples collected from spas by the combined use of EMA and real-time PCR.
In order to investigate the utility of the proposed method for the specific detection of culturable Legionella from the environment, 25 water samples were analyzed in this study. Samples 1 to 9 were collected from public spas, whereas samples 10 to 25 were from a model spa (18, 28), as described in Materials and Methods.
The results are shown in Table 3. Samples 1 and 2 were collected from jetted and outdoor bathtubs, respectively, at the same public spa facility. The number of Legionella cells in sample 1, as estimated by real-time PCR assay, was higher than that determined by plating on GVPC agar. After treatment with EMA at 20 μg/ml, the number of Legionella cells estimated by real-time PCR was similar to that determined by plating. These results indicate that sample 1 contained DNA and/or uncultivable cells of Legionella which are sensitive to treatment with EMA. On the other hand, the number of Legionella cells in sample 2, as estimated by real-time PCR, was similar to that determined by plating. This result suggested that almost all of the Legionella cells in the sample were culturable. After treatment with 20 μg/ml of EMA, the number of Legionella cells estimated by real-time PCR was smaller (1.1 log10 CFU/100 ml) than that determined by plating. It seems that EMA cleaves a part of the genomic DNA of viable cells under environmental conditions. All of these results suggested that the appropriate concentrations of EMA were different among water samples. Therefore, EMA was used at 1, 5, 10, and 20 μg/ml for the other 23 samples.
TABLE 3.
Comparison of results of plating and real-time PCR for water samples from spas
| Sample no.a | Free chlorine concn (ppm) | No. of Legionella cells by plating (log10 CFU/100 ml)b | No. of Legionella cells estimated by real-time PCR (log10 CFU/100 ml)c
|
||||
|---|---|---|---|---|---|---|---|
| No EMA treatment | Treatment with EMA
|
||||||
| 1 μg/ml | 5 μg/ml | 10 μg/ml | 20 μg/ml | ||||
| 1 | 1.0 | 1.3 | 3.2 | ND | ND | ND | 1.4 |
| 2 | 0.1 | 2.4 | 2.2 | ND | ND | ND | 1.3 |
| 3 | 0.5 | 1.3 | 2.8 | 1.9 | 1.3 | — | — |
| 4 | 0 | 3.0 | 3.2 | 2.9 | 2.4 | 2.2 | 1.9 |
| 5 | 0.1 | 1.3 | — | — | — | — | — |
| 6 | 3 | <1 | — | — | — | — | — |
| 7 | 2 | <1 | — | — | — | — | — |
| 8 | 0 | 1 | — | — | — | — | — |
| 9 | 2.4 | <1 | — | — | — | — | — |
| 10 | 0 | <1 | — | — | — | — | — |
| 11 | 0 | <1 | — | — | — | — | — |
| 12 | 0 | <1 | — | — | — | — | — |
| 13 | 0 | <1 | — | — | — | — | — |
| 14 | 0 | 2.9 | 3.0 | 3.1 | 3.0 | 2.7 | 1.9 |
| 15 | 0 | 3.8 | 4.0 | 3.9 | 3.8 | 3.1 | 3.0 |
| 16 | 0 | 4.8 | 5.2 | 4.7 | 3.8 | 3.1 | — |
| 17 | 0 | 4.8 | 5.4 | 4.3 | 3.5 | 2.9 | 2.3 |
| 18 | 0 | 4.6 | 5.2 | 4.4 | 3.5 | 2.5 | 2.2 |
| 19 | 0.01 | 5.4 | 5.9 | 5.5 | 5.5 | 4.0 | 3.0 |
| 20 | 2.5 | 3.6 | 5.5 | 4.8 | 3.8 | 3.1 | 2.8 |
| 21 | 3.5 | 1.8 | 5.2 | 3.3 | 2.6 | 2.1 | 1.6 |
| 22 | 6.4 | 1.5 | 4.0 | 3.6 | 3.4 | 1.6 | — |
| 23 | 8.1 | <1 | 3.3 | 1.4 | 1.3 | 0.7 | 0 |
| 24 | 8.2 | <1 | 3.0 | 1.2 | 1.3 | 0.7 | — |
| 25 | 8.3 | <1 | 3.8 | 3.6 | 1.1 | 1 | — |
Samples 1, 2, and 7 to 9 were obtained from bathtubs, samples 3 to 5 were from filter tanks, and sample 6 was from a pipeline of public spas. Samples 10 to 17 were obtained from the bathtub and samples 18 to 25 were from the filter tank of a model spa.
The number of bacteria was determined by plating cells on GVPC plates.
Real-time PCR was performed with primers LEG427F and LEG880R and probe P1. ND, not done; —, not detected.
Among the 25 water samples, no amplification signals were detected in 9 samples (samples 5 to 9 and 10 to 13), with or without treatment of EMA, by using real-time PCR (Table 3). By plating on GVPC agar, no colonies were isolated from seven of the nine samples. Only a few colonies (10 CFU/100 ml and 20 CFU/100 ml) were detected in the other two samples (samples 5 and 8), which almost corresponded to the results obtained by real-time PCR (Table 3). More than 2 log10 CFU/100 ml of Legionella cells, estimated by real-time PCR, existed in the remained 16 water samples (Table 3). The numbers of Legionella cells in nine samples (samples 3, 4, and 14 to 20), estimated by the combined EMA and real-time PCR assay, were similar to those determined by plating when EMA treatment was performed at 1 or 5 μg/ml (Table 3). When these samples were treated with EMA at 10 and 20 μg/ml, the numbers of Legionella cells estimated by real-time PCR were smaller than those determined by plating. Meanwhile, 10 or 20 μg/ml of EMA treatment was needed for the remaining six samples (samples 1 and 21 to 25) in order to obtain similar results between the numbers of Legionella cells estimated by real-time PCR and those determined by plating (Table 3). The numbers of Legionella cells, estimated by real-time PCR, in the water samples (samples 19 to 25) without EMA treatment were gradually decreased after the addition of chlorine (Table 3). Although the precise cause has not yet been elucidated, it may be attributed to breaking of the genomic DNA from the uncultivable Legionella cells by chlorine.
Taking all of these results together, EMA treatment could selectively amplify the genomic DNA of the culturable Legionella cells in the water samples by real-time PCR assay. However, EMA concentrations that were effective were different among environmental samples and seemed to be related to the sensitivity of Legionella cells to EMA or the chlorine concentrations of the samples. A low concentration of EMA (1 or 5 μg/ml) was enough for eight samples (samples 3, 4, and 14 to 19) in which chlorine was not detected or detected at a low concentration (≤0.5 ppm). On the other hand, a high concentration of EMA (10 or 20 μg/ml) was needed for six samples (samples 1 and 21 to 25) in which chlorine was detected at high concentrations (≥1.0 ppm). A large number of uncultivable cells killed by chlorine in the six samples may be one of the reasons that EMA must be used at high concentrations for the number of Legionella cells determined by plating to match that estimated by real-time PCR. All of the results suggested that a high concentration of EMA is needed to cleave genomic DNA of uncultivable Legionella cells treated with chlorine at high concentrations. In the case of sample 20, the water sample was collected immediately after chlorine treatment, so 5 μg/ml of EMA was probably sufficient.
DISCUSSION
In the present study, we demonstrate the combined use of EMA and PCR/real-time PCR for rapid detection of viable Legionella cells. The results reveal that EMA can specially enter and cleave the genomic DNA of heat- and chlorine-treated Legionella cells. After treatment with EMA, PCR could not detect the DNA present in the dead cells and the amount of DNA significantly decreased compared to that for the viable cells in the real-time PCR assay. The assay was also useful for detection of culturable Legionella in water samples. These results show that the combined use of EMA and PCR/real-time PCR is sufficient to detect viable Legionella cells.
Approximately 1 × 106 to 1 × 107 CFU/ml of Legionella strains was used in the EMA real-time PCR assay. The detected decrease of DNA in the dead cells by EMA treatment was approximately 4 to 5 log10 at 20 μg/ml EMA (Fig. 3); therefore, part of the genomic DNA, corresponding to approximately 102 CFU/ml, still remained. However, the number of uncultivable Legionella cells estimated by real-time PCR with 68 environmental samples was <105 CFU/100 ml, and that with 66 samples (97%) was <104 CFU/100 ml in our preliminary experiment (unpublished data). In Japan, the guideline by the Ministry of Health, Labor, and Welfare for prevention of Legionnaires' disease specifies that the detection limit of culturable Legionella from bath water must be <10 CFU/100 ml. Therefore, the decrease of 4 to 5 log10 CFU with EMA treatment is theoretically sufficient to place environmental samples with low levels of putative viable but uncultivable Legionella in a low-risk category according to Japanese guidelines. However, plating for detection of viable Legionella cells should be performed at the same time because the possibility of false-positive results cannot entirely be eliminated.
Part of the genomic DNA, corresponding to approximately 102 CFU/ml, of the heat- and chlorine-treated Legionella cells still remained when the combined EMA (20 μg/ml) and real-time PCR assay was performed (see above) (Fig. 2 and 3), which may be due to the limit of EMA activity at that concentration. When the EMA concentration was increased, the intensity of PCR-amplified fragments in the viable Legionella cells became lower, which would be due to the damage of viable cells caused by EMA. The concentration of EMA used seems to be critical for the maximum discrimination of viable cells from dead cells. The 5S rRNA, 16S rRNA, and mip genes could be amplified by PCR as fragments of 108 bp, 406 bp, and 649 bp, respectively, in our experiment. If EMA randomly binds and cleaves the DNA sequence, a smaller DNA region would not be affected after EMA treatment and could be amplified by PCR. Amplification of the 16S rRNA gene was most effective for the discrimination of viable Legionella cells from dead cells, although we do not know the exact reason that amplification of the 16S rRNA gene was most available in our experiment. EMA might recognize the DNA region of the 16S rRNA gene most effectively, but the problems with amplification should be resolved in the future.
Twenty-five water samples were tested in this study, and the combined EMA-real-time PCR assay was confirmed to be useful for specific detection of viable Legionella cells in these environmental samples. In order to avoid false-positive or -negative results by combined use of EMA and real-time PCR, the concentration of EMA used for water samples may be most critical. The concentration of EMA needed was shown to be related to the residual chlorine concentration in the water samples in this study. Because the investigation in this study was done on a small number of water samples, further confirmation will be required by the use of a large number of water samples from public spas. It is probable that EMA at the high concentration used in some water samples (samples 3, 4, and 14 to 19) (Table 3) could also enter viable Legionella cells and cleave their genomic DNA. It was recently shown that propidium monoazide (PMA) is superior to EMA for avoiding entrance into and/or cleavage of genomic DNA of viable bacterial cells (20). We are planning to compare the effects of EMA and PMA on detection of viable Legionella cells in water samples in our next experiment.
After the first outbreak of legionellosis caused by L. pneumophila in Philadelphia (7), much research was conducted on the behavior and life cycle of Legionella. It is now clear that monitoring and removal of Legionella from waters that come into contact with humans, particularly water from distribution systems, are an effective way to prevent infections caused by Legionella. In recent years, disinfection and cleaning of manmade water systems have been strictly observed in Japan. Surveillance investigations on the water systems in the Kanto area of Japan showed that the number of Legionella-positive samples and the number of Legionella isolates from such samples have been decreasing annually since 2003 (12, 30). We hope that the rapid detection method described here is useful for the control and monitoring of water systems, especially for continuous environmental surveillance at certain points.
Acknowledgments
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (no. 18790316) and from the Ministry of Health, Labour and Welfare (H19-kenki-014 to F.K.).
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Caron, G. N., P. Stephens, and R. A. Badley. 1998. Assessment of bacterial viability status by flow cytometry and single cell sorting. J. Appl. Microbiol. 84:988-998. [DOI] [PubMed] [Google Scholar]
- 2.Coffman, G. L., J. W. Gaubatz, K. L. Yielding, and L. W. Yielding. 1982. Demonstration of specific high affinity binding sites in plasmid DNA by photoaffinity labeling with ethidium analog. J. Biol. Chem. 257:13205-13297. [PubMed] [Google Scholar]
- 3.Davis, E. P., R. Boopathy, and J. Manning. 1997. Use of trinitrobenzene as a nitrogen source by Pseudomonas vesicularis isolated from soil. Curr. Microbiol. 34:192-197. [DOI] [PubMed] [Google Scholar]
- 4.DeTraglia, M. C., J. S. Brand, and A. M. Tometsko. 1978. Characterization of azidobenzamidines as photoaffinity labeling for trypsin. J. Biol. Chem. 253:1846-1852. [PubMed] [Google Scholar]
- 5.Endo, T., T. Kuroki, T. Karasudani, H. Sekine, and S. Izumiyama. 2007. Analysis of water bacterial flora in bathtubs of hot springs by PCR-DGGE method, p. 67-86. In H. Inoue (ed.), Research report on the development of appropriate techniques for public health of single-path-flow hot springs. Ministry of Health, Labour and Welfare, Tokyo, Japan. (In Japanese.)
- 6.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepard, P. S. Brachman, and The Field Investigation Team. 1977. Legionnaires' disease. Description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189-1197. [DOI] [PubMed] [Google Scholar]
- 8.Hejazi, A., and F. R. Falkiner. 1997. Serratia marcescens. J. Med. Microbiol. 46:903-912. [DOI] [PubMed] [Google Scholar]
- 9.Hixon, S. C., W. E. White, and K. L. Yielding. 1975. Selective covalent binding of an ethidium analog to mitochondrial DNA with production of petite mutants in yeast by photoaffinity labeling. J. Mol. Biol. 92:319-329. [DOI] [PubMed] [Google Scholar]
- 10.Kell, D. B., A. S. Kaprelyants, D. H. Weichart, C. R. Harwood, and M. R. Barer. 1998. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie van Leeuwenhoek 73:169-187. [DOI] [PubMed] [Google Scholar]
- 11.Koide, M., T. Miyata, M. Nukina, T. Teramoto, H. Nakanishi, T. Kamiki, B. Umeda, and H. Nakai. 1989. A strain of Pseudomonas vesicularis isolated from shower hose which supports the multiplication of Legionella. Kansenshogaku Zasshi 63:1160-1164. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 12.Kura, F., J. Amemura-Maekawa, A. Suzuki-Hashimoto, B. Chang, S. Izumiyama, M. Ichinose, T. Endo, and H. Watanabe. 2007. Surveillance of Legionella isolates from bathtub water in Japan: an increase of the rate of Legionella pneumophila serogroup 1. In Abstr. 22nd Annu. Meet. Eur. Working Group Legionella Infect., Stockholm and Uppsala, Sweden.
- 13.Kura, F., J. Amemura-Maekawa, K. Yagita, T. Endo, M. Ikeno, H. Tsuji, M. Taguchi, K. Kobayashi, E. Ishi, and H. Watanabe. 2006. Outbreak of Legionnaires' disease on a cruise ship linked to spa-bath filter stones contaminated with Legionella pneumophila serogroup 5. Epidemiol. Infect. 134:385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahbubani, M. H., A. K. Bej, R. Miller, L. Haff, J. DiCesare, and R. M. Atlas. 1990. Detection of Legionella with polymerase chain reaction and gene probe methods. Mol. Cell. Probes 4:175-187. [DOI] [PubMed] [Google Scholar]
- 15.Morais, P. V., and M. S. da Costa. 1990. Alterations in the major heterotrophic bacterial populations isolated from a still bottled mineral water. J. Appl. Bacteriol. 69:750-757. [DOI] [PubMed] [Google Scholar]
- 16.Nakadate, T., K. Yamauchi, and H. Inoue. 1999. An outbreak of Legionnaire's disease associated with a Japanese spa. Nihon Kokyuki Gakkai Zasshi 37:601-607. (In Japanese.) [PubMed] [Google Scholar]
- 17.Nakamura, H., H. Yagyu, K. Kishi, F. Tsuchida, S. Oh-ishi, K. Yamaguchi, and T. Matsuoka. 2003. A large outbreak of Legionnaires' disease due to an inadequate circulating and filtration system for bath water—epidemiologic manifestations. Intern. Med. 42:806-811. [DOI] [PubMed] [Google Scholar]
- 18.Ohata, K., K. Sugiyama, M. Suzuki, R. Shimogawara, S. Izumiyama, K. Yagita, and T. Endo. 2006. Growth of Legionella in nonsterilized, naturally contaminated bathing water in a system that circulates the water, p. 431-435. In N. P. Cianciotto, Y. Abu Kwaik, P. H. Edelstein, B. S. Fields, D. F. Geary, T. G. Harrison, C. B. Joseph, R. M. Ratcliff, J. E. Stout, and M. S. Swanson (ed.), Legionella: state of the art 30 years after its recognition. American Society for Microbiology, Washington, DC.
- 19.Okada, M., K. Kawano, F. Kura, J. Amemura-Maekawa, H. Watanabe, K. Yagita, T. Endo, and S. Suzuki. 2005. The largest outbreak of legionellosis in Japan associated with spa baths: epidemic curve and environmental investigation. Kansenshogaku Zasshi 79:365-374. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 20.Pan, Y., and F. Breidt, Jr. 2007. Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl. Environ. Microbiol. 73:8028-8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riedy, M. C., K. A. Muirhead, C. P. Jensen, and C. C. Stewart. 1991. Use of a photolabeling technique to identify nonviable cells in fixed homologous or heterologous cell populations. Cytometry 12:133-139. [DOI] [PubMed] [Google Scholar]
- 22.Rogers, J., and C. W. Keevil. 1992. Immunogold and fluorescein immunolabeling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 58:2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudi, K., B. Moen, S. M. Drømtorp, and A. L. Holck. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savageau, M. A. 1983. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat. 122:732-744. [Google Scholar]
- 26.Soejima, T., K. Iida, T. Qin, H. Taniai, M. Seki, A. Takade, and S. Yoshida. 2007. Photoactivated ethidium monoazide directly cleaves bacterial DNA and is applied to PCR for discrimination of live and dead bacteria. Microbiol. Immunol. 51:763-775. [DOI] [PubMed] [Google Scholar]
- 27.Stølhaug, A., and K. Bergh. 2006. Identification and differentiation of Legionella pneumophila and Legionella spp. with real-time PCR targeting the 16S rRNA gene and species identification by mip sequencing. Appl. Environ. Microbiol. 72:6394-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugiyama, K., K. Ohata, M. Suzuki, R. Shimogawara, S. Izumiyama, K. Yagita, and T. Endo. 2006. Inhibition of Legionella growth in circulating bathing water by filter refreshment method using high concentration chlorine, p. 497-500. In N. P. Cianciotto, Y. Abu Kwaik, P. H. Edelstein, B. S. Fields, D. F. Geary, T. G. Harrison, C. B. Joseph, R. M. Ratcliff, J. E. Stout, and M. S. Swanson (ed.), Legionella: state of the art 30 years after its recognition. American Society for Microbiology, Washington, DC.
- 29.Sugiyama, K., T. Nishio, Y. Goda, K. Masuda, F. Zhang, M. Akiyama, and H. Miyamoto. 2000. An outbreak of legionellosis linked to bath water circulating through a filter at a spa resort, March-April 2000—Shizuoka. Infect. Agents Surveill. Rep. 21:188. (In Japanese.) [Google Scholar]
- 30.Suzuki-Hashimoto, A., J. Amemura-Maekawa, F. Kura, B. Chang, S. Izumiyama, M. Ichinose, H. Watanabe, and T. Endo. 2007. The surveillance of Legionella from cooling towers between 2001 and 2006 in Japan: an increase of the rate of Legionella pneumophila serogroup 1. In Abstr. 22nd Annu. Meet. Eur. Working Group Legionella Infect., Stockholm and Uppsala, Sweden.
- 31.Waring, M. J. 1965. Complex formation between ethidium bromide and nucleic acids. J. Mol. Biol. 13:269-282. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto, H., Y. Hashimoto, and T. Ezaki. 1993. Comparison of detection methods for Legionella species in environmental water by colony isolation, fluorescent antibody staining, and polymerase chain reaction. Microbiol. Immunol. 37:617-622. [DOI] [PubMed] [Google Scholar]
- 33.Yoshikuni, K., K. Nakayama, S. Honda, N. Shinkawa, T. Arima, Y. Yumata, and Y. Ito. 2003. An outbreak of legionellosis presumably due to the circulating water system at a spa resort, August 2002—Kagoshima Prefecture. Infect. Agents Surveill. Rep. 24:31-32. (In Japanese.) [Google Scholar]