Abstract
Neisseria meningitidis is an exclusively human pathogen that has evolved primarily to colonize the nasopharynx rather than to cause systemic disease. Colonization is the most frequent outcome following meningococcal infection and a prerequisite for invasive disease. The mechanism of colonization involves attachment of the organism to epithelial cells via bacterial type IV pili (Tfp), but subsequent events during colonization remain largely unknown. We analyzed 576 N. meningitidis mutants for their capacity to colonize human nasopharyngeal tissue in an organ culture model to identify bacterial genes required for colonization. Eight colonization-defective mutants were isolated. Two mutants were unable to express Tfp and were defective for adhesion to epithelial cells, which is likely to be the basis of their attenuation in nasopharyngeal tissue. Three other mutants are predicted to have lost previously uncharacterized surface molecules, while the remaining mutants have transposon insertions in genes of unknown function. We have identified novel meningococcal colonization factors, and this should provide insights into the survival of this important pathogen in its natural habitat.
The first step in the pathogenesis of most infectious diseases is the colonization of a susceptible individual with a virulent microorganism. For most bacteria, colonization allows replication of the pathogen before it spreads to other tissues or disseminates to other hosts (43). Many pathogens have evolved first and foremost to colonize their hosts rather than to cause disease, with invasive infection being an accidental event that is often irrelevant to the natural life cycle of the microbe (2). Understanding the molecular mechanisms that pathogenic bacteria use to colonize their hosts is crucial since it may aid in the design of interventions to prevent carriage, block disease, and eradicate infection. However, the process of colonization can be difficult to investigate, particularly for human-adapted pathogens that utilize host-specific nutrients and/or receptors, limiting the availability of biologically relevant models for experimental studies (56).
Neisseria meningitidis is an obligate human pathogen that is a leading cause of bacteremia and meningitis in developed and developing countries, affecting mainly children and young adults (53). Although systemic disease can progress rapidly and is fatal in up to 20% of cases (53), the most frequent outcome of infection with N. meningitidis is asymptomatic carriage, with up to 40% of healthy adults harboring the bacterium in the nasopharynx (6). The human nasopharynx is the only known reservoir of infection, and colonization is the first step in the pathogenesis of meningococcal disease and is essential for the successful propagation of the bacterium through human populations.
The interaction between N. meningitidis and the human host during the colonization process is incompletely understood. Previous work has mainly focused on the attachment of the bacterium to human epithelial cells, one of the initial steps in colonization (10, 28). The bacterium expresses type IV pili (Tfp), filamentous processes that extend beyond its surface, which enable it to adhere to epithelial cells in the nasopharynx (32). After attachment, the bacterium traverses the epithelial cell barrier and reaches the subepithelial compartment. Immunohistochemical analysis of tonsillar tissue from healthy carriers has demonstrated that colonizing meningococci can be found on the epithelial cell surface, in association with epithelial cells and in the tissue underlying the mucosal surface (42). The route of traversal has not been defined, although the bacterium has been detected in an intracellular compartment in epithelial cells (33, 45, 47).
Early interactions between the host and pathogen are critical since they may dictate the outcome of infection. Most cases of meningococcal sepsis occur within a few days or weeks of exposure to the bacterium (9, 16), and the ability of strains to penetrate nasopharyngeal tissue directly correlates with their propensity to cause systemic disease (51).
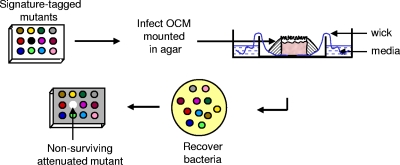
Here we describe the identification of novel N. meningitidis genes required for early interactions with nasopharyngeal tissue using a human organ culture model (OCM). Upper airway OCMs consist of explants of human nasopharyngeal tissue and present bacteria with the physical epithelial barrier (consisting of polarized columnar cells joined by tight junctions) and subepithelial layer present in the upper airway. In addition, active cilia on the apical epithelial surface preserve mucociliary transport in OCMs, which express other mucosal innate immune factors (35). In OCMs, N. meningitidis must also utilize the same repertoire of carbon energy sources and other micronutrients that are present in the human nasopharynx (12). Therefore, tissue explants provide the complex organization of cell types and tissues relevant for studying meningococcal colonization, which cannot be reproduced using current cell culture techniques. We used signature-tagged mutagenesis (STM) to identify genes necessary for survival of N. meningitidis in OCMs. STM was developed for high-throughput analysis of mutants (18) and is a powerful method that allows unbiased genetic screening to be performed with animal models of disease (26, 39). This is the first time that STM has been used to examine the interaction of a pathogen with human tissue.
MATERIALS AND METHODS
Bacterial strains and growth.
The clinical N. meningitidis isolate C311 (B;NT;NT) (54) was grown on brain heart infusion medium with 5% Levanthal's supplement. The generation of N. meningitidis STM mutants has been described previously (49). For infecting OCMs, bacteria were grown for 16 h on solid media, resuspended in phosphate-buffered saline (PBS), and then enumerated by measuring the A260 of the suspension in 1% sodium dodecyl sulfate-0.1 M NaOH; the number of CFU was confirmed by plating. Escherichia coli was grown in Luria-Bertani media. Antibiotics were used at the following concentrations: kanamycin at 50 and 75 μg/ml and erythromycin at 200 and 2 μg/ml for E. coli and N. meningitidis, respectively.
Nasopharyngeal organ culture model.
Human nasopharyngeal mucosal explants were derived from inferior turbinates resected from patients with nonallergic nasal obstruction who had given informed consent. Tissue was immersed in minimal essential medium (MEM; Gibco, United Kingdom) containing penicillin at 50 U/ml, streptomycin at 50 μg/ml, and gentamicin at 50 μg/ml for 4 h and then dissected to produce 3- to 4-mm squares of full-thickness mucosa (51). Next, the tissue was transferred to MEM without antibiotics for 1 h, supported in 4-cm petri dishes with the epithelial surface projecting above a bed of non-nutrient agar (Fig. 1). The petri dishes containing the agar-embedded explant were placed inside 10-cm petri dishes containing a reservoir of MEM, which perfused the OCM via a wick, and incubated in a humidified atmosphere of 5% CO2 in air at 37°C. Ethical approval was obtained from the Central Oxford Research Ethics Committee (COREC no. C00.0085) and from the South Sheffield Research Ethics Committee (SS/01/055).
FIG. 1.
Pools of signature-tagged N. meningitidis mutants were used to infect OCMs and recovered 18 h later after treatment with sodium taurocholate to kill extracellular bacteria. Colonization-defective mutants fail to survive in the tissue and are not recovered from the OCMs.
Measurement of meningococcal survival within nasopharyngeal tissue.
Pools of 95 mutants were screened in triplicate using explants from a single donor. To assess the survival of N. meningitidis in the tissue, bacteria in 100 μl of PBS were placed upon the surface of each explant. After a 6- or 18-h incubation, explants were washed, weighed, and then homogenized in a modified French press (Constant Systems, Warwick, United Kingdom) at 10 lb/in2, and viable bacteria were recovered by plating. To estimate the invasion of OCMs, explants were immersed in 0.25% sodium taurocholate (Sigma, United Kingdom) for 30 s prior to homogenization to kill extramucosal bacteria as described previously (51). To assess the sensitivity of mutants to bile salts, 107 CFU were suspended in 100 μl of PBS and then exposed to sodium taurocholate (from 0.015 to 0.5% final concentration) for 1 h at 37°C. The number of surviving bacteria was determined by plating.
Explants from at least six donors were used to analyze the colonization capacity of individual mutants compared to the wild-type strain. Bacteria were grown overnight, and 5 × 107 CFU used to challenge separate OCMs. Bacteria were recovered by the same method as for screening the library of mutants.
Molecular methods.
Chromosomal and plasmid DNA was extracted by standard methods, and signature tags were detected by hybridization as previously described (49). Transposon insertion sites were recovered by marker rescue. In brief, genomic DNA was digested with DraI, EcoRV, and HpaII and then self-ligated in a 250-μl volume with T4 DNA ligase for 16 h at 16°C. The ligation reactions were used to transform E. coli DH5α to kanamycin resistance. The nucleotide sequence of plasmids was determined with the oligonucleotide NG62 (5′-TTGGTTAATTGGTTGTAACACTGG-3′). Sequence searches were performed by using BLAST algorithms against the N. meningitidis genome databases (http://www.sanger.ac.uk/Projects/N-meningitidis and http://www.tigr.org). Genomic DNA was recovered from all colonization-defective mutants and used to reconstruct the mutants by transforming the parental strain. This was undertaken to exclude potential effects caused by spontaneous on-off expression of surface molecules by N. meningitidis.
For complementation, NMB1829 was amplified from genomic DNA from strain C311 with High Fidelity Expand Taq (Roche Applied Science) using the primers 5′-GAGGATCCAAACTGTAACGCAGGTTTGCC-3′ and 5′-CAGCTAGCAGCAGGTTGGTTGCAGTAAA-3′, containing BamHI and NheI restriction sites (underlined), respectively. The 2.29-kb product was ligated into pCRTopo2.1 (Invitrogen), excised, and then introduced into a multiple cloning site in pYHS25 (57). This vector contains the erythromycin resistance gene flanked by fragments of NMB0102 and NMB0103. The vector was used to transform C311, introducing a single chromosomal copy of the complementing gene in the intergenic region between NMB0102 and NMB0103, open reading frames orientated in a tail-to-tail fashion. Transformants were analyzed by Southern hybridization and PCR with primers annealing to regions upstream of NMB0102 and the upstream region of NMB1829 (5′-GGTCGGGTAAAATGAAAGTT-3′ and 5′-CGGCTTTATATTGTTTGTGC-3′, respectively) and with primers based on NMB0103 and the downstream region of NMB1829 (5′-ATATCTTTGAAATCGGCTCA-3′ and 5′-ATTCCGGCATAGTAAAACAA-3′, respectively). Genomic DNA from the mutant was used to transform the wild-type strain and mutant, and multiple transformants were collected to exclude effects of phase variation.
Sensitivity to antimicrobial peptides.
The antimicrobial activity of synthetic human LL-37 (supplied by Jan Pohl, Emory University) was examined in assays based on previously described methods (41). Briefly, bacteria were grown overnight on solid media and then grown in liquid GC media (Difco) supplemented with 0.43% NaHCO3, 0.4% glucose, and 0.68 mM Fe(NO3)3 for 3 h at 37°C with shaking at 100 rpm. The optical density at 600 nm was measured and adjusted to 0.2, and the suspension was diluted 1 in 100 in 0.2× GC broth. A 90-μl aliquot of the suspension was added to wells of a 96-well plate containing 10 μl of peptide diluted in 0.01% acetic acid; acetic acid without peptide was added to control wells. Bacteria were incubated for 45 min at 37°C in 5% CO2, and then aliquots were plated to solid media. All assays were performed in triplicate.
Western blot analysis and adhesion assays.
For Western blot analysis, bacteria were grown overnight on solid media and then resuspended in loading buffer at a concentration of 1010 CFU/ml, and the proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis before transfer to membranes. Membranes were washed in 0.5% milk in PBS and then incubated with the following antibodies at a 1:10,000 dilution: SM1, anti-pilin; B33, anti-Opa (kindly provided by M. Virji); and anti-PilC (kindly provided by A.-B. Jonsson). After incubation with a secondary antibody (horseradish peroxidase-conjugated anti-mouse immunoglobulin at 1 in 2,000), cross-reaction was detected by using the ECL Western blotting detection reagent (GE Healthcare).
To examine the adhesive capacity of strains, Chang human epithelial cells were incubated with bacteria at a ratio of 100 bacteria per cell for 3 h at 37°C in the presence of 5% CO2 and then washed in Hanks buffered saline solution before being lysed in 1% saponin for 10 min at 37°C. Cell-associated bacteria were measured by plating dilutions of the lysed cells to media. Assays were performed in triplicate on three separate occasions, and the results were expressed as a percentage adhesion of the mutants compared to the wild-type strain.
Electron microscopy.
OCMs infected for 18 h with pools of STM N. meningitidis mutants were prepared for transmission electron microscopy or scanning electron microscopy analysis using standard protocols as described previously (36).
Statistical analysis.
Comparisons between the survival of the mutants and the wild-type strain were screened by Kruskal-Wallis analysis, and where the null hypothesis was rejected, comparisons between the mutants and wild-type strain were tested by Mann-Whitney analysis (SPSS, version 11).
RESULTS
Establishing conditions for screening N. meningitidis mutants in OCMs.
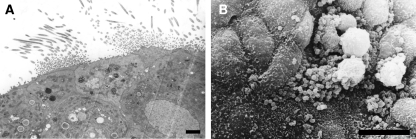
Different inoculum sizes of N. meningitidis and incubation times were initially assessed to ensure that reproducible results were obtained when separate explants were inoculated with the same pool of mutants. Explants were infected with between 106 and 108 bacteria, left for 6 or 18 h, after which they were exposed to sodium taurocholate to kill extracellular bacteria on the mucosal surface; sodium taurocholate is only taken up by cells in the terminal ileum during the enterohepatic circulation of bile salts (1) and so does not kill bacteria in respiratory epithelial cells (51). Furthermore, bacteria are not recovered from taurocholate-treated OCMs exposed to cytochalasin which blocks actin polymerization and thereby uptake of bacteria (51). Bacteria were then harvested from the tissue after homogenization. In initial experiments, the recovery of bacteria from explants inoculated with <107 CFU was inconsistent. When OCMs were challenged with between 107 and 108 CFU, only a few bacteria were recovered from tissue homogenates treated with sodium taurocholate after 6 h, but by 18 h between 103 and 104 bacteria were recovered. With this level of inoculum, consistent hybridization patterns were obtained from bacteria recovered from separate OCMs receiving the same pool of 96 mutants (Fig. 1). Therefore, for screening the library of mutants, the OCMs were inoculated with 5 × 107 CFU of N. meningitidis, and bacteria were recovered 18 h later. Examination of the infected tissue under light microscopy demonstrated the presence of active cilia (results not shown). Both transmission and scanning electron microscopy analysis showed that the epithelium was intact and ciliated (Fig. 2A) and adherent bacteria were detected on the surface of explants (Fig. 2B) with no degradation of the architecture of the OCM.
FIG. 2.
Electron microscopic analysis of integrity of OCMs infected with N. meningitidis for 18 h. (A) Transmission electron microscopy image of infected tissue demonstrates presence of cilia and an intact epithelium. Magnification, ×2,500. Scale bar, 2 μm. (B) Scanning electron microscopy image of infected explants. Bacteria are visible adhering to nonciliated cells and mucous globules. Scale bar, 10 μm.
Tfp are required for colonization of OCMs.
We examined a total of 576 N. meningitidis mutants for their ability to colonize OCMs derived from human nasopharyngeal tissue. Colonization-defective mutants were defined as those that were absent from explants treated with sodium taurocholate and yet present in the inoculum and untreated homogenates. From the library, eight colonization-defective mutants were identified. To exclude the possibility that mutants were not recovered from the initial screen because they were sensitive to sodium taurocholate, each mutant was tested for its sensitivity to this bile salt. The sensitivity of all mutants was similar to that of the wild-type strain, with a minimum growth inhibitory concentration of 0.125%.
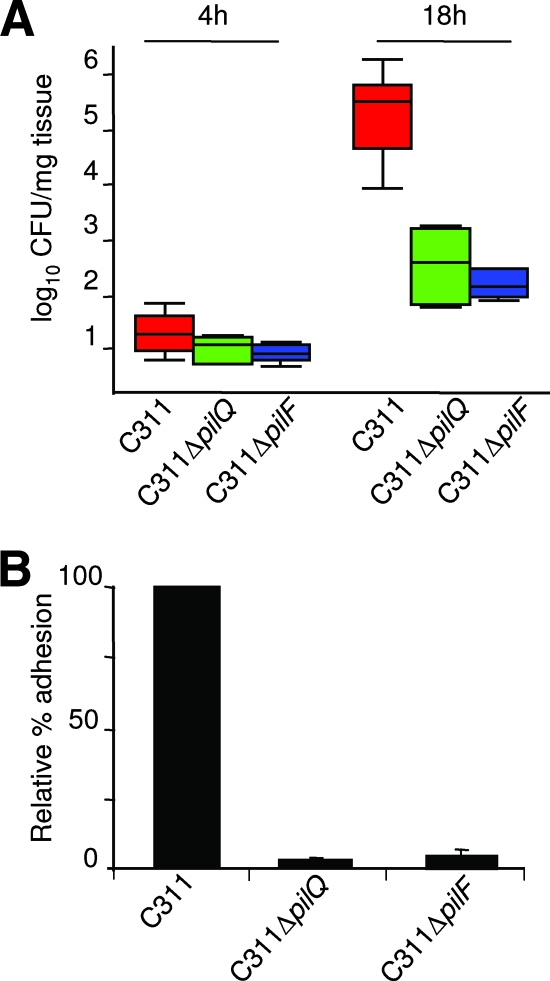
The transposon insertion sites in the eight colonization-defective mutants were recovered by marker rescue, and sequences flanking the insertion were used to search the serogroup B N. meningitidis genome database (www.tigr.org) by BLASTP. The results are shown in Table 1. Two mutants, C311ΔpilQ and C311ΔpilF, are unable to express Tfp, the principal adhesin of N. meningitidis (5). Multimers of PilQ form a pore in the outer membrane that allow the export of Tfp to the bacterial surface (8), while PilF is necessary for pilus assembly (14). The identification of two mutants defective for the biogenesis of Tfp provides proof-in-principle for our approach since it has been shown previously that variants lacking Tfp are defective for survival in nasopharyngeal tissue (34). To confirm the requirement for these genes during colonization of OCMs, the C311ΔpilQ and C311ΔpilF mutants were analyzed individually in explants. Both mutants were recovered in significantly lower numbers than the wild-type strain from tissue before treatment with sodium taurocholate at 18 h after inoculation (P < 0.05, Fig. 3A). After treatment with this bile salt, neither mutant could be detected. We also assessed the adherence of the mutants to Chang epithelial cells. As expected, C311ΔpilQ and C311ΔpilF exhibited marked defects for adhesion, binding at <10% the level of the wild-type strain (P < 0.01, Fig. 3B). The failure of the mutants to attach to epithelial cells in the upper airway is likely to account for their colonization defect in OCMs.
TABLE 1.
Colonization factors identified by STM
| Gene affected (TIGR gene no.) | Predicted function |
|---|---|
| 0329 | Pilus biogenesis, PilF |
| 1812 | Pilus biogenesis, PilQ |
| 0103 | Putative bacteriocin immunity protein |
| 0313 | OmpU homologue |
| 1829 | Putative TonB-dependent receptor |
| 0541 | Function unknown |
| 1012 | Function unknown |
| 1726 | Function unknown |
FIG. 3.
(A) Colonization capacity of the pilQ (green boxes) and pilF (blue boxes) mutants compared to the wild-type strain (red boxes) at 4 and 18 h after inoculation of OCMs. The graph shows the median and 25th/75th percentile of CFU/mg of bacteria recovered from explants; P = 0.015 and P = 0.041 for the wild-type strain versus C311ΔpilF and C311ΔpilQ at 18 h, respectively. (B) Relative adhesion to human epithelial cells of the mutants compared to the wild-type strain. Error bars show the standard error of the mean.
Identification of novel colonization factors.
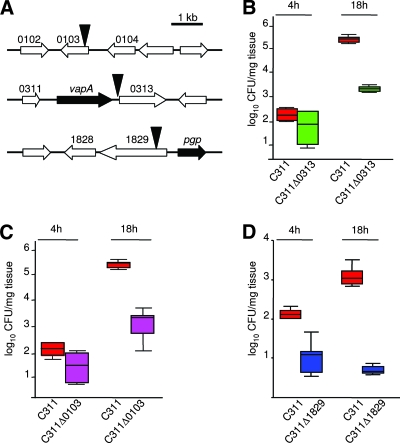
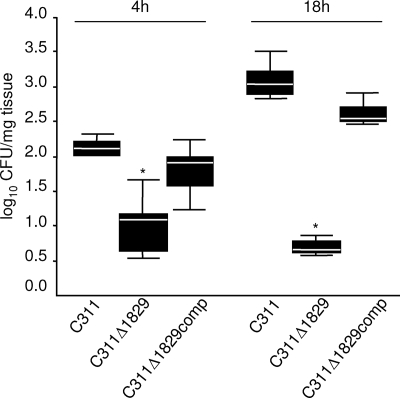
Three of the six remaining colonization-defective mutants had transposon insertions in genes predicted to encode surface-expressed proteins, NMB0103, NMB0313, and NMB1829 (Fig. 4A, numbering according to www.tigr.org), by homology with known surface proteins and/or bioinformatic analysis (PSORTb v2.0 [www.psort.org] and Tmpred [www.ch.embnet.org/software/TMPRED-form.html]). These genes are of interest since many key host-pathogen interactions are mediated by bacterial surface molecules. The impact of the loss of these three genes on colonization was assessed by inoculating OCMs with each mutant individually (Fig. 4B, C, and D). All three mutants were recovered from the OCMs at significantly lower levels than the wild-type strain at 18 h after challenge (P = 0.009, 0.002, and 0.004 for C311Δ0103, C311Δ0313, and C311Δ1829, respectively), confirming their role during colonization of the tissues of the upper airway. Examination of the orientation of open reading frames around the transposon insertions in these mutants indicate that polar effects could only account for the attenuation of the NMB1829 mutant (Fig. 4A). Therefore, this mutant was complemented in trans with a single copy of the wild-type gene in an ectopic chromosomal location to generate C311Δ1829comp; this reverted the colonization defect of the mutant to wild-type levels (Fig. 5).
FIG. 4.
(A) Gene organization around the transposon insertions (indicated by solid triangles) in colonization-defective mutants. Gene designation: open block arrows, gene of unknown function; vapA, virulence associated protein (authentic frameshift); pgp, phosphoglycolate phosphatase. (B to D) Colonization capacity of the mutants and the wild-type strain (C311) at 4 and 18 h after inoculation of OCMs. The data shows median and 25th/75th percentile of the number of bacteria recovered from explants/mg of tissue. The mutants were all defective for colonization; P = 0.002, 0.009 and 0.004 for C311 versus C311Δ0313 (B), C311Δ0103 (C), and C311Δ1829 (D) at 18 h, respectively.
FIG. 5.
Complementation of C311Δ1829 restores the colonization capacity to wild-type levels. Individual OCMs were infected with the wild-type, C311Δ1829 and the complemented strain (C311Δ1829comp), and bacteria recovered after 4 and 18 h. The data show the median and 25th/75th percentile of the CFU recovered from untreated explants/mg of tissue. The C311Δ1829 mutant is defective for colonization at 4 and 18 h (*, P < 0.001). The complemented strain was recovered at wild-type levels after 4 h and at levels only slightly lower than the wild-type strain at 18 h.
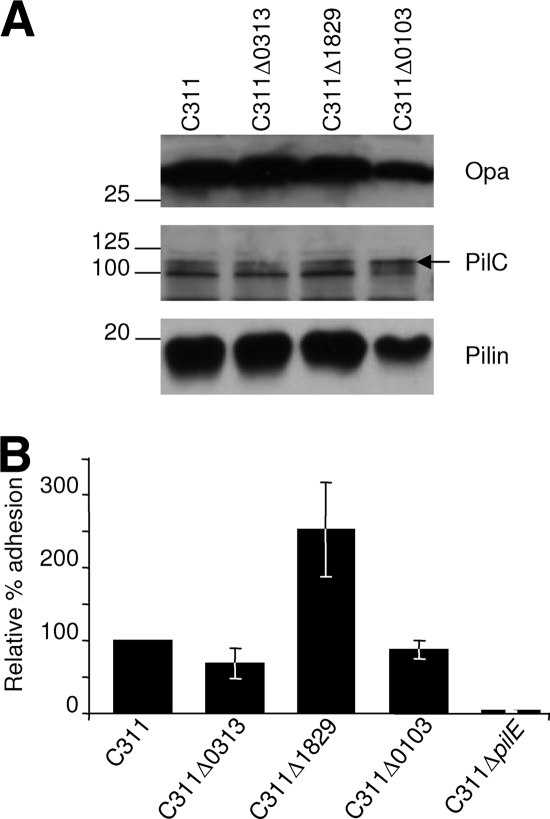
To further characterize the basis of the colonization defect of these three mutants, we examined their expression of characterized meningococcal adhesins and compared their capacity to adhere to human epithelial cells with the wild-type strain. As well as Tfp, members of the Opa family of proteins contribute to the initial interactions of N. meningitidis with human epithelial cells (32). However, none of these mutants had a significant defect for the expression of adhesins or for binding to epithelial cells (Fig. 6A and B, respectively). It was also possible that these mutants were attenuated due to increased sensitivity to antimicrobial peptides, since alterations in surface structures can render bacteria more susceptible to peptide-mediated killing (27). We determined whether the mutants were sensitive to LL-37, a human neutrophil-derived antimicrobial peptide that is found in the upper airways. However, all three mutants exhibited the same MIC to LL-37 as the wild-type strain (Table 2).
FIG. 6.
(A) Western blot analysis for the expression of adhesins in whole-cell lysates. The adhesin is shown next to each panel and molecular mass given in kilodaltons. The strains are indicated above each lane. (B) Relative adhesion of colonization defective mutants to Chang epithelial cells. The data are presented as the mean relative adhesion ± the standard deviation. None of the mutants shows a significant reduction in adhesion capacity compared to the wild-type strain C311, indicating that their inability to survive in OCMs was unlikely to be due to reduced adhesion to epithelial cells. The C311Δ1829 mutant showed enhanced adhesion, while C311ΔpilE, which is unable to express Tfp, was recovered at <4% compared to the wild-type strain.
TABLE 2.
Colonization defective mutants are not more sensitive to the antimicrobial peptide, LL-37, than the wild type
| Assay | Minimum inhibitory growth concn (μg/ml)a for strain:
|
|||
|---|---|---|---|---|
| C311 | C311Δ0103 | C311Δ0313 | C311Δ1829 | |
| 1 | 7.8 | 15.6 | 15.6 | 15.6 |
| 2 | 15.6 | 15.6 | 15.6 | 15.6 |
| 3 | 15.6 | 15.6 | 15.6 | 15.6 |
The minimum growth inhibitory concentration of LL-37 was determined by incubating bacteria with a range of concentrations of peptide from 128.4 to 0.98 μg/ml.
The remaining three colonization-defective mutants had transposon insertions in genes of entirely unknown function (NMB0541, NMB1012, and NMB1726). Analysis of these mutants demonstrated that none had a significant defect for adhesion to human epithelial cells or in the expression of known adhesins (not shown).
DISCUSSION
N. meningitidis is specifically adapted to colonize the human nasopharynx, with evolution selecting for traits that promote its survival in this environment. Defining those traits should be informative about how N. meningitidis successfully inhabits this ecological niche and help to design interventions to prevent disease. However, a full appreciation of the mechanisms underlying this key stage in the bacterium's life cycle can only be gained using models, such as OCMs, that accurately mimic conditions in its natural habitat in the human host.
Explants of human tissue can be maintained in vitro as OCMs which provide physiologically relevant systems for studying host-pathogen interactions. Early studies using organ culture examined the interactions between Neisseria gonorrhoeae with fallopian tube explants (55), Bordetella pertussis with tracheal tissue (7), and Chlamydia with conjunctival tissue (30). More recently, biopsies of tissue have been used to study the importance of the mucus layer during the initial stages of adhesion of Mycobacterium to the human upper respiratory tract (29), and the interaction of C. jejuni with explanted intestinal mucosa (15). OCMs using adenoid or turbinate tissue have previously been used to investigate the behavior of N. meningitidis (34, 35, 47) but not for the identification of colonization factors. The advantage of using nasopharyngeal tissue is that it avoids the use of animal models which may lack human specific factors such as CD46 that are relevant for colonization (20).
Although this was not an exhaustive screen, we analyzed 576 N. meningitidis mutants in nasopharyngeal explants to identify genes required for survival in this tissue as a model of colonization of the human airway. Since it was not feasible to study this number of mutants individually in OCMs, we used STM to facilitate large-scale analysis (18). STM has been mainly used to identify genes required for virulence in animal models of infection (26). As far as we are aware, this is the first time that STM has been applied to examine the behavior of a bacterium in human tissue.
The main disadvantages of OCMs are (i) that there is considerable host-to-host variation in the level of colonization after challenge (51) and (ii) the limited availability of tissue. The former problem was circumvented during screening the mutant library by inoculating OCMs with pools of mutants. However, to confirm the colonization defect of selected mutants, OCMs were challenged separately with mutant and wild-type strains on six separate occasions with tissue from different individuals. Therefore, rescreening of mutants was limited to those required for the biogenesis of Tfp and those with insertions in genes predicted to encode surface factors, and complementation analysis was restricted to where there was the possibility of polar effects (i.e., NMB1829; Fig. 4A).
It has been shown previously that selected meningococcal variants lacking Tfp are defective for survival in adenoidal tissue (34). Our work with two isogenic mutants confirms the importance of Tfp during colonization of the human upper airway. Tfp are likely to contribute to survival during interactions with epithelial cells, either acting as an adhesin or by promoting aggregation (17). Interestingly, both the pilF and pilQ mutants were recovered from OCMs at the 4-h time point at levels equivalent to that of the wild-type strain, suggesting that bacterial adhesion to and entry into epithelial cells occurs after this time. This is consistent with the finding that inhibition of actin polymerization with cytochalasin has no discernible effect on colonization of OCMs at early time points (51) and is distinct from interactions of the meningococcus observed with isolated epithelial cells, when pilus-negative mutants are at a significant disadvantage within 2 to 3 h of challenge.
We also isolated mutants affected in genes not previously characterized as important for meningococcal colonization. This highlights the advantages of using OCMs as these genes would have not have been identified through studies with epithelial cells alone since none of these mutants were defective for cell adhesion. The expected product of NMB0103 has 37% amino acid identity with HmcC, a bacteriocin processing protein from Haemophilus influenzae (31), another pathogenic member of the nasopharyngeal flora. Bacteriocins are antimicrobial compounds that are lethal for members of the same or related species (21, 31). Although the explanted OCM tissue is treated with antibiotics prior to infection with N. meningitidis, it is possible that residual bacteriocins could be present in the tissue and inhibit the survival of the NMB0103 mutant. In addition, bacteriocin immunity proteins can mediate resistance against a variety of antimicrobial agents, including antimicrobial peptides (25). However, the strain lacking NMB0103 did not exhibit increased susceptibility to LL-37, an antimicrobial peptide detected in the human upper airway which has proven activity against N. meningitidis (52).
NMB0313 encodes a protein of unknown function which has 94% amino acid identity with a protein from a serogroup C N. meningitidis strain that has been designated as an outer membrane protein OmpU in the EMBL-EBI database (accession no. AF118122.1). In Vibrio cholerae OmpU may act as an adhesion (44), but it also mediates resistance against antimicrobial peptides (24). Amino acid alignments demonstrate that NMB0313 has <20% amino acid sequence identity with OmpU from V. cholerae (data not shown), and our experimental data indicate that the OmpU homologue in N. meningitidis does not contribute to adhesion to human epithelial cells or defense against LL-37.
Iron is an essential element required for key metabolic processes, and pathogenic bacteria possess several systems for iron acquisition from host tissues (13). NMB1829 is predicted to encode a TonB-dependent receptor, a member of a family of outer membrane proteins involved in acquisition of iron and other molecules (3, 37, 40). These proteins rely on TonB, an inner membrane protein that provides energy for iron transport across the outer membrane (48). N. meningitidis possesses several TonB-dependent receptors which bind transferrin, lactoferrin, and hemoglobin/haptoglobin or siderophores (37). Siderophores are small molecules secreted by microbes to chelate iron in the environment and deliver it to the bacterium via cognate surface receptors. Of note, N. meningitidis does not itself produce siderophores. However, to maximize fitness and compete within its ecological niche, the meningococcus may utilize NMB1829 to acquire siderophores secreted by other microorganisms and/or acquire host iron sources such as ferritin, which is necessary for the intracellular replication of N. meningitidis (23). Thus far, we have no evidence that NMB1829 is required for intracellular replication (not shown), and the gene is required early during colonization (within 4 h of challenge), suggesting that it is required by the bacterium before interaction with epithelial cells.
A consistent feature of the expanding database of sequenced bacterial genomes is the large number of entirely uncharacterized genes which account for around half the open reading frames in many pathogens (38); N. meningitidis contains around 900 genes with no function ascribed (50). A major challenge is to decipher the activity of these recently discovered genes since this could prove valuable for the development of novel antimicrobials and vaccines (4, 38). This work identified several genes of unknown function whose role in colonization could not have been foreseen by examination of their predicted amino acid sequences. Microarray analysis demonstrates that these three genes are conserved in the major hypervirulent lineages and serogroups of N. meningitidis that cause human disease (46), with NMB0541 and NMB1726 also present in isolates of the commensal species Neisseria lactamica, Neisseria sicca, and Neisseria polysaccharea.
Most vaccines against bacterial infection target antigens (such as capsules and toxins) that are expressed during systemic infection. More recently, strategies are also being developed to block the adhesion of pathogens to host cells (19, 22). Therefore, further understanding of the bacterial factors required for meningococcal survival in the nasopharynx and the nature of the local immune responses (11) could not only be helpful in defining the molecular basis of pathogenesis but also be used for designing vaccines to prevent disease caused by this important human pathogen.
Acknowledgments
We are grateful to Eva Mowe for technical assistance and to Sara Marshall for critical reading of the manuscript.
This work was supported by the Meningitis Research Foundation. R.M.E. is a Leverhulme Trust Early Career Fellowship holder.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 20 October 2008.
REFERENCES
- 1.Agellon, L. B., and E. C. Torchia. 2000. Intracellular transport of bile acids. Biochim. Biophys. Acta 1486198-209. [DOI] [PubMed] [Google Scholar]
- 2.Bull, J. J. 1994. Perspective: virulence. Evolution 481423-1437. [DOI] [PubMed] [Google Scholar]
- 3.Cadieux, N., P. G. Phan, D. S. Cafiso, and R. J. Kadner. 2003. Differential substrate-induced signaling through the TonB-dependent transporter BtuB. Proc. Natl. Acad. Sci. USA 10010688-10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capecchi, B., D. Serruto, J. Adu-Bobie, R. Rappuoli, and M. Pizza. 2004. The genome revolution in vaccine research. Curr. Issues Mol. Biol. 617-27. [PubMed] [Google Scholar]
- 5.Carbonnelle, E., S. Helaine, X. Nassif, and V. Pelicic. 2006. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization, and export of type IV pili. Mol. Microbiol. 611510-1522. [DOI] [PubMed] [Google Scholar]
- 6.Cartwright, K. A., J. M. Stuart, D. M. Jones, and N. D. Noah. 1987. The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol. Infect. 99591-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier, A. M., L. P. Peterson, and J. B. Baseman. 1977. Pathogenesis of infection with Bordetella pertussis in hamster tracheal organ culture. J. Infect. Dis. 136(Suppl.)S196-S203. [DOI] [PubMed] [Google Scholar]
- 8.Collins, R. F., S. A. Frye, A. Kitmitto, R. C. Ford, T. Tønjum, and J. P. Derrick. 2004. Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 Å resolution. J. Biol. Chem. 27939750-39756. [DOI] [PubMed] [Google Scholar]
- 9.Cooke, R. P., T. Riordan, D. M. Jones, and M. J. Painter. 1989. Secondary cases of meningococcal infection among close family and household contacts in England and Wales, 1984-7. BMJ 298555-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbett, A., R. Exley, S. Bourdoulous, and C. M. Tang. 2004. Interactions between Neisseria meningitidis and human cells that promote colonization and disease. Expert Rev. Mol. Med. 61-14. [DOI] [PubMed] [Google Scholar]
- 11.Davenport, V., T. Guthrie, J. Findlow, R. Borrow, N. A. Williams, and R. S. Heyderman. 2003. Evidence for naturally acquired T cell-mediated mucosal immunity to Neisseria meningitidis. J. Immunol. 1714263-4270. [DOI] [PubMed] [Google Scholar]
- 12.Exley, R. M., L. Goodwin, E. Mowe, J. Shaw, H. Smith, R. C. Read, and C. M. Tang. 2005. Neisseria meningitidis lactate permease is required for nasopharyngeal colonization. Infect. Immun. 735762-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelstein, R. A., C. V. Sciortino, and M. A. McIntosh. 1983. Role of iron in microbe-host interactions. Rev. Infect. Dis. 5(Suppl. 4)S759-S777. [DOI] [PubMed] [Google Scholar]
- 14.Freitag, N. E., H. S. Seifert, and M. Koomey. 1995. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 16575-586. [DOI] [PubMed] [Google Scholar]
- 15.Grant, A. J., J. Woodward, and D. J. Maskell. 2006. Development of an ex vivo organ culture model using human gastro-intestinal tissue and Campylobacter jejuni. FEMS Microbiol. Lett. 263240-243. [DOI] [PubMed] [Google Scholar]
- 16.Greenwood, B. M., M. Hassan-King, and H. C. Whittle. 1978. Prevention of secondary cases of meningococcal disease in household contacts by vaccination. BMJ 11317-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helaine, S., E. Carbonnelle, L. Prouvensier, J. L. Beretti, X. Nassif, and V. Pelicic. 2005. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol. Microbiol. 5565-77. [DOI] [PubMed] [Google Scholar]
- 18.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269400-403. [DOI] [PubMed] [Google Scholar]
- 19.Hewlett, E. L. 1997. Pertussis: current concepts of pathogenesis and prevention. Pediatr. Infect. Dis. J. 16S78-S84. [DOI] [PubMed] [Google Scholar]
- 20.Johansson, L., A. Rytkonen, P. Bergman, B. Albiger, H. Kallstrom, T. Hokfelt, B. Agerberth, R. Cattaneo, and A. B. Jonsson. 2003. CD46 in meningococcal disease. Science 301373-375. [DOI] [PubMed] [Google Scholar]
- 21.Kingsbury, D. T. 1966. Bacteriocin production by strains of Neisseria meningitidis. J. Bacteriol. 911696-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langermann, S., S. Palaszynski, M. Barnhart, G. Auguste, J. S. Pinkner, J. Burlein, P. Barren, S. Koenig, S. Leath, C. H. Jones, and S. J. Hultgren. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276607-611. [DOI] [PubMed] [Google Scholar]
- 23.Larson, J. A., H. L. Howie, and M. So. 2004. Neisseria meningitidis accelerates ferritin degradation in host epithelial cells to yield an essential iron source. Mol. Microbiol. 53807-820. [DOI] [PubMed] [Google Scholar]
- 24.Mathur, J., and M. K. Waldor. 2004. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect. Immun. 723577-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto-Nakano, M., and H. K. Kuramitsu. 2006. Role of bacteriocin immunity proteins in the antimicrobial sensitivity of Streptococcus mutans. J. Bacteriol. 1888095-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazurkiewicz, P., C. M. Tang, C. Boone, and D. W. Holden. 2006. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat. Rev. Genet. 7929-939. [DOI] [PubMed] [Google Scholar]
- 27.Menendez, A., and B. Brett Finlay. 2007. Defensins in the immunology of bacterial infections. Curr. Opin. Immunol. 19385-391. [DOI] [PubMed] [Google Scholar]
- 28.Merz, A. J., and M. So. 2000. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16423-457. [DOI] [PubMed] [Google Scholar]
- 29.Middleton, A. M., M. V. Chadwick, A. G. Nicholson, R. Wilson, D. J. Thornton, S. Kirkham, and J. K. Sheehan. 2004. Interaction between mycobacteria and mucus on a human respiratory tissue organ culture model with an air interface. Exp. Lung Res. 3017-29. [DOI] [PubMed] [Google Scholar]
- 30.Moore, J. E., M. S. Griffiths, and J. H. Pearce. 1974. Chlamydial infection of conjunctival tissues in culture. Br. J. Exp. Pathol. 55396-405. [PMC free article] [PubMed] [Google Scholar]
- 31.Murley, Y. M., T. D. Edlind, J. M. Pozsgay, and J. J. LiPuma. 1997. Cloning and characterization of the haemocin immunity gene of Haemophilus influenzae. J. Bacteriol. 1791684-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nassif, X., C. Pujol, P. Morand, and E. Eugene. 1999. Interactions of pathogenic Neisseria with host cells: is it possible to assemble the puzzle? Mol. Microbiol. 321124-1132. [DOI] [PubMed] [Google Scholar]
- 33.Pujol, C., E. Eugene, L. de Saint Martin, and X. Nassif. 1997. Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect. Immun. 654836-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rayner, C. F., A. Dewar, E. R. Moxon, M. Virji, and R. Wilson. 1995. The effect of variations in the expression of pili on the interaction of Neisseria meningitidis with human nasopharyngeal epithelium. J. Infect. Dis. 171113-121. [DOI] [PubMed] [Google Scholar]
- 35.Read, R. C., A. Fox, K. Miller, T. Gray, N. Jones, R. Borrows, D. M. Jones, and R. G. Finch. 1995. Experimental infection of human nasal mucosal explants with Neisseria meningitidis. J. Med. Microbiol. 42353-361. [DOI] [PubMed] [Google Scholar]
- 36.Read, R. C., and L. Goodwin. 2001. Experimental nasopharyngeal colonization by Neisseria meningitidis using explant organ culture, p. 621-634. In A. J. Pollard and M. C. Maiden (ed.), Meningococcal disease: methods and protocols. Humana Press, New York, NY. [DOI] [PubMed]
- 37.Rohde, K. H., and D. W. Dyer. 2003. Mechanisms of iron acquisition by the human pathogens Neisseria meningitidis and Neisseria gonorrhoeae. Front. Biosci. 8d1186-d1218. [DOI] [PubMed] [Google Scholar]
- 38.Rosamond, J., and A. Allsop. 2000. Harnessing the power of the genome in the search for new antibiotics. Science 2871973-1976. [DOI] [PubMed] [Google Scholar]
- 39.Saenz, H. L., and C. Dehio. 2005. Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Curr. Opin. Microbiol. 8612-619. [DOI] [PubMed] [Google Scholar]
- 40.Schauer, K., D. A. Rodionov, and H. de Reuse. 2008. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33330-338. [DOI] [PubMed] [Google Scholar]
- 41.Shafer, W. M., X. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 951829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sim, R. J., M. M. Harrison, E. R. Moxon, and C. M. Tang. 2000. Underestimation of meningococci in tonsillar tissue by nasopharyngeal swabbing. Lancet 3561653-1654. [DOI] [PubMed] [Google Scholar]
- 43.Smith, H. 1998. What happens to bacterial pathogens in vivo? Trends Microbiol. 6239-243. [DOI] [PubMed] [Google Scholar]
- 44.Sperandio, V., J. A. Giron, W. D. Silveira, and J. B. Kaper. 1995. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect. Immun. 634433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spinosa, M. R., C. Progida, A. Tala, L. Cogli, P. Alifano, and C. Bucci. 2007. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect. Immun. 753594-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stabler, R. A., G. L. Marsden, A. A. Witney, Y. Li, S. D. Bentley, C. M. Tang, and J. Hinds. 2005. Identification of pathogen-specific genes through microarray analysis of pathogenic and commensal Neisseria species. Microbiology 1512907-2922. [DOI] [PubMed] [Google Scholar]
- 47.Stephens, D. S., L. H. Hoffman, and Z. A. McGee. 1983. Interaction of Neisseria meningitidis with human nasopharyngeal mucosa: attachment and entry into columnar epithelial cells. J. Infect. Dis. 148369-376. [DOI] [PubMed] [Google Scholar]
- 48.Stojiljkovic, I., and N. Srinivasan. 1997. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in neisseriae. J. Bacteriol. 179805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, Y. H., S. Bakshi, R. Chalmers, and C. M. Tang. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 61269-1273. [DOI] [PubMed] [Google Scholar]
- 50.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 2871809-1815. [DOI] [PubMed] [Google Scholar]
- 51.Townsend, R., L. Goodwin, T. M. Stevanin, P. B. Silcocks, A. Parker, M. C. Maiden, and R. C. Read. 2002. Invasion by Neisseria meningitidis varies widely between clones and among nasopharyngeal mucosae derived from adult human hosts. Microbiology 1481467-1474. [DOI] [PubMed] [Google Scholar]
- 52.Tzeng, Y. L., K. D. Ambrose, S. Zughaier, X. Zhou, Y. K. Miller, W. M. Shafer, and D. S. Stephens. 2005. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol. 1875387-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Deuren, M., P. Brandtzaeg, and J. W. van der Meer. 2000. Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin. Microbiol. Rev. 13144-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Virji, M., H. Kayhty, D. J. Ferguson, C. Alexandrescu, J. E. Heckels, and E. R. Moxon. 1991. The role of pili in the interactions of pathogenic Neisseria with cultured human endothelial cells. Mol. Microbiol. 51831-1841. [DOI] [PubMed] [Google Scholar]
- 55.Ward, M. E., P. J. Watt, and J. N. Robertson. 1974. The human fallopian tube: a laboratory model for gonococcal infection. J. Infect. Dis. 129650-659. [DOI] [PubMed] [Google Scholar]
- 56.West, N. P., P. J. Sansonetti, G. Frankel, and C. M. Tang. 2003. Finding your niche: what has been learnt from STM studies on GI colonization. Trends Microbiol. 11338-344. [DOI] [PubMed] [Google Scholar]
- 57.Winzer, K., Y. H. Sun, A. Green, M. Delory, D. Blackley, K. R. Hardie, T. J. Baldwin, and C. M. Tang. 2002. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect. Immun. 702245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]