Abstract
Five types of cytolethal distending toxin (CDT-I to CDT-V) have been identified in Escherichia coli. In the present study we cloned and sequenced the cdt-IV operon and flanking region from a porcine extraintestinal pathogenic E. coli (ExPEC) strain belonging to serogroup O75. We confirmed that similar to other CDTs, CDT-IV induced phosphorylation of host histone H2AX, a sensitive marker of DNA double-strand breaks, and blocked the HeLa cell cycle at the G2-M transition. The cdt-IV genes were framed by lambdoid prophage genes. We cloned and sequenced the cdt-I operon and flanking regions from a human ExPEC O18:K1:H7 strain and observed that cdt-I genes were also flanked by lambdoid prophage genes. PCR studies indicated that a gene coding for a putative protease was always associated with the cdtC-IV gene but was not associated with cdtC genes in strains producing CDT-I, CDT-III, and CDT-V. Our results suggest that the cdt-I and cdt-IV genes might have been acquired from a common ancestor by phage transduction and evolved in their bacterial hosts. The lysogenic bacteriophages have the potential to carry nonessential “cargo” genes or “morons” and therefore play a crucial role in the generation of genetic diversity within ExPEC.
Cytolethal distending toxins (CDTs) were the first bacterial toxins that block the eukaryotic cell cycle that were described, and they suppress cell proliferation and eventually lead to cell death. CDTs represent an emerging toxin family, which was initially described for Escherichia coli but is actually widely distributed among a variety of pathogenic bacteria, including Shigella dysenteriae, Salmonella enterica serovar Typhi, Campylobacter spp., Helicobacter spp., Aggregatibacter (formerly Actinobacillus) actinomycetemcomitans, and Haemophilus ducreyi (31, 34).
A CDT is a tripartite holotoxin (28) in which CdtB is the active subunit and CdtA and CdtC form a heterodimeric subunit apparatus required for delivery of CdtB into the cell (8, 23, 24). The toxin of S. enterica serovar Typhi is an exception. This organism does not encode CdtA or CdtC; instead, the enzymatic CdtB is delivered directly into host cells by a bacterial internalization pathway (14). Nuclear entry of CdtB, which relies on an atypical nuclear localization signal, is crucial for the cytotoxic activity (25, 27, 29). CdtB has DNase I-like activity (10, 15, 23). Once in the host cell nucleus, CdtB induces DNA double-strand breaks in both proliferating and nonproliferating cells (13, 15, 26). Although pathogenic roles of CDT have been shown for chronic infection in mouse models (12, 39), CDT has not played a significant role in acute infection models tested to date (25, 40, 48, 49). Nevertheless, CDT appears to be a virulence factor; accordingly, studying its spread is important.
So far, five different CDTs have been reported for E. coli, and they were designated in order of publication. CDT-I (44) and CDT-II (38) were identified in enteropathogenic E. coli (EPEC) serotype O86:H34 and O128:NM strains, respectively. CDT-III was cloned from an E. coli serotype O15:H21 strain isolated from a septicemic calf and sequenced (36). CDT-III is encoded by pVir, a conjugative plasmid which also codes for another toxin, cytotoxic necrotizing factor type 2 (35). CDT-V was identified in the sorbitol-fermenting Shiga toxin (Stx)-producing E. coli (STEC) serotype O157:NM strain 493/89 (18) and was detected in several other non-O157 STEC strains (5). We recently identified and characterized CDT-IV in human- and animal-pathogenic E. coli strains of intestinal and extraintestinal origin (46); however, the complete sequence of the cdt-IV gene cluster was not determined. In the present study we confirmed the genotoxic activity of CDT-IV, sequenced the cdt-IV and cdt-I gene clusters and flanking regions, and investigated the dissemination of these clusters in different types of CDT-producing E. coli strains.
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains used in this study are listed in Table 1. The CDT-I-producing prototype strain E6468/62 (serotype O86:H34) was kindly provided by James. B. Kaper. CDT-V-producing reference strain 493/89 and CDT-V producing strains 702/88 and 703/88 (serotype O157:NM) were kindly provided by Helge Karch.
TABLE 1.
E. coli strains used in this study and occurrence of two cdt-IV flanking genes (orf5 and rorf1) of ExPEC strain 28C and their association with cdt genes in CDT-producing E. coli strains
| Strain | Origin | Serotype or serogroup | Genotype | cdt type | orf5 | orf6 | orf5/cdtA | rorf1 | cdtC/rorf1a | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| E6468/62d | Human | O86:H34 | eae cdt | I | − | − | − | − | − | 44 |
| ICC95 | Human | O86 | eae cdt | I | − | − | − | − | − | 2 |
| E990 | Human | O86:NM | eae cdt | I | − | − | 6 | |||
| EF142 | Pigeon | O128:NM | eae cdt | I | − | − | − | − | − | This study |
| ED177 | Human | O157:H7 | eae stx1stx2cdt | I | − | − | − | − | − | This study |
| S5 | Bovine | O78 | cdt cnf2 | III | − | − | − | − | − | 36 |
| EF56 | Human | O55:H21 | cdt cnf2 | III | − | − | − | − | − | This study |
| EF147 | Roe deer | O26 | eae cdt cnf2 | III | − | − | − | + | − | This study |
| ED1451 | Dairy cow | O88 | cdt | III | − | − | − | + | − | This study |
| 28c | Porcine | O75 | cnf1 cdt hly | IV | + | + | + | + | + | 46 |
| H173 | Human | O2 | cdt | IV | + | + | + | + | + | 46 |
| H83 | Human | O75 | cnf1 cdt hly | IV | + | + | + | + | + | 46 |
| H62 | Human | O75 | cnf1 cdt hly | IV | + | + | + | + | + | 46 |
| H78 | Human | O75 | cnf1 cdt hly | IV | + | + | + | + | + | 46 |
| AI-9 | Porcine | O164 | cnf1 cdt hly | IV | + | + | + | + | + | 46 |
| H193 | Human | O170 | cdt hly | IV | + | + | + | + | + | 46 |
| H104 | Human | O rough | cnf1 cdt hly | IV | + | + | + | + | + | 46 |
| H29 | Human | O6 | cnf1 cdt hly | IV | − | − | − | + | + | 46 |
| E250 | Poultry | O115 | cdt | IV | − | − | − | + | + | 46 |
| E253 | Poultry | O115 | cdt | IV | − | − | − | + | + | 46 |
| AII-40 | Porcine | O6 | cnf1 cdt hly | IV | − | − | − | + | + | 46 |
| H155 | Human | O rough | cdt | IV | − | − | − | + | + | 46 |
| 493/89b | Human | O157:NM | eae stx2cdt | V | − | − | − | + | − | 5 |
| 703/88b | Human | O157:NM | eae stx2cdt | V | − | − | − | + | − | This study |
| 702/88b | Human | O157:NM | eae stx2cdt | V | − | − | − | + | − | This study |
| C600c | Laboratory | K-12 | − | − | − | − | − | 42 |
The cdtC-III/V-s-rorf1-as1 (Table 2) and P105(5)/rorf1-as2 primer pairs were used for detection of the potential association between cdtC and rorf1 in strains EF147, ED1451, 493/89, 703/88, and 702/88.
E. coli O157:NM strains 493/89, 703/88, and 702/88 strains were kindly provided by H. Karch, University of Münster, Münster, Germany.
E. coli C600 was used as a negative control in all PCRs.
Bold type indicates cdt reference strains.
DNA techniques, PCR, and nucleotide sequencing.
Cosmid libraries were constructed from CDT-IV-producing extraintestinal pathogenic E. coli (ExPEC) strain 28C (serogroup O75) (9, 46) and from CDT-I producing ExPEC strain IHE3034 (serotype O18:K1:H7) (21, 30). Both libraries were created by using the same strategy. Briefly, genomic DNA samples were partially digested with Sau3AI and cloned into the BamHI sites of the SuperCos-1 vector (Stratagene). Following in vitro packaging and introduction into E. coli XL1-Blue, about 2,000 ampicillin-resistant colonies of each library were collected. The libraries were screened by PCR using cdtB-IV- and cdtB-I-specific primers, respectively (46). The CDT production of the PCR-positive cdt-IV and cdt-I clones was investigated by using nonconfluent HeLa cell monolayers in 96-well plates as described previously (35, 36).
Typing of cdt genes was conducted by using previously described PCR primers and assay conditions (5, 46). The dissemination of cdt-IV flanking genes in CDT-producing strains and their association with cdt genes were investigated by performing PCR with primers listed in Table 2. The positions of these PCR primers are shown in Fig. 3.
TABLE 2.
PCR primers used in this study
| Primer | Target | Nucleotide sequence | Position | Size (bp) | GenBank accession no. | Reference |
|---|---|---|---|---|---|---|
| orf5-s1 | orf5 | 5′-GGGAGACATCACCACGCTAAAG-3′ | 4028-4049 | 450 | AY578329 | This study |
| orf5-as1 | 5′-TCCTACTTCCTCGGCAATCAATC-3′ | 4456-4478 | ||||
| orf5-s2 | orf5/orf6/cdtA-IV | 5′-GCTAACGAAGATGATGCACC-3′ | 4512-4531 | 1,131 | AY578329 | This study |
| cdtA-IV-as1 | 5′-CCATTTGAAGCAACAGCGAC-3′ | 5625-5643 | ||||
| orf5-s2 | orf5/orf6/cdtA-IV | 5′-GCTAACGAAGATGATGCACC-3′ | 4512-4532 | 1,381 | AY578329 | This study |
| cdtA-IV-as2 | 5′-ATCACCCCGTTTTTAAAGGC-3′ | 5874-5893 | ||||
| cdtC-IV-s1 | cdtC-IV/rorf1 | 5′-GAGCTGTGCAAATCAAGTCG-3′ | 7418-7437 | 632 | AY578329 | This study |
| rorf1-as2 | 5′-GAGAAAATCCCGAAAAGACG −3′ | 8031-8050 | ||||
| rorf1-s1 | rorf1 | 5′-TGACAATCCCCTGTGAGAACTGGC-3′ | 8163-8186 | 401 | AY578329 | This study |
| rorf1-as1 | 5′-CCGTGGCGTATTCGTATGGAACAC-3′ | 8564-8541 | ||||
| cdtC-III/V-s | cdtC-III/cdtC-V/rorf1 | 5′-ATCAAAAGTGTTCTGTCTGG-3′ | 7168-7187 | 1,396a | AY578329 | This study |
| 3061-3080 | ||||||
| 2089-2109 | U89305 | |||||
| 2089-2109 | AJ508930 | |||||
| 2089-2109 | AY365045 | |||||
| 1997-2016 | AY365042 | |||||
| rorf1-as1 | 5′-CCGTGGCGTATTCGTATGGAACAC-3′ | 8564-8541 | AY365044 | This study | ||
| c338f | cdtA-V | 5′-AGCATTAAATAAAAGCACGA-3 | 1,329 | AY578329 | 5 | |
| c2135r | 5′-TACTTGCTGTGGTCTGCTAT-3′ | |||||
| P105 | cdtC-V | 5′-GTCAACGAACATTAGATTAT-3′ | 748 | 5 | ||
| c2767r | 5′-ATGGTCATGCTTTGTTATAT-3′ |
Predicted size, calculated on the basis of the position of the homologous sequence (14/20) in 28C (positions 7168 to 7187).
FIG. 3.
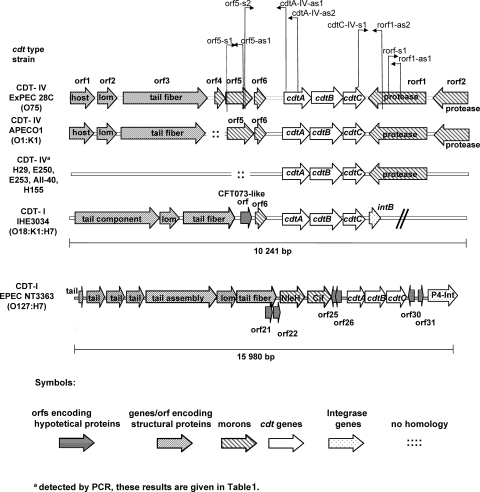
Genomic comparison of cdt loci and flanking regions in E. coli 28C (cdt-IV), APEC O1 (cdt-IV), and IHE3034 (cdt-I) and CDT-IΦ. Searches for homologous sequences were performed by using BLAST software (3), and the results were used to align sequence data. The cdt-I and cdt-IV loci of strains IHE3034 and 28C were sequenced in the present study. The E. coli APEC O1 (accession number NC_008563) and lambdoid phage CDT-1Φ (accession number AB285204) sequences used were deposited previously in the database. Progressive loss of upstream flanking genes was observed in the cdt-IV region in strains H29, E250, E253, AII-40, and H155, as demonstrated by PCR, but no losses of the downstream flanking gene were observed. PCR primers for detection of cdt flanking genes present in E. coli 28C cdt-IV loci and their association with cdt-IV genes were designed. The PCR primer positions in the cdt-IV loci are indicated by arrows. The sequences of these primers and all other primers used are listed in Table 2.
For sequencing, DNA samples from a CDT-IV clone and a CDT-I clone were purified with a QIAquick genome purification kit (Qiagen) and used as templates for sequencing with an ABI PRISM 377 automated sequencer. Sequencing was performed by the chain termination method with a dye terminator kit (Applied Biosystems) using T3, T7, or synthetic primers.
Searches for open reading frames (ORFs) and prediction of translation start positions were performed by using Vector NTI software.
Searches for homologous DNA sequences were conducted using BLAST software (3) and the GenBank database (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Nucleotide sequences of cdtABC genes with all the available E. coli cdt alleles and the cdt genes of one Campylobacter reference strain were aligned by using the ClustalW multiple-sequence alignment program (45). The phylogenetic analysis was carried out with the MEGA (Molecular Evolutionary Genetics Analysis) 3.1 program (22) using the neighbor-joining method (41). Distances were calculated using the Kimura two-parameter nucleotide substitution model with pairwise deletion of gaps. The branches were tested by bootstrap analysis (11) of 300 replicates. The tree was rooted with the cdtABC sequences of Campylobacter jejuni M1221 (accession number NC_003912).
CDT-IV genotoxic effects.
Experiments and preparation of bacterial lysates were performed as described previously (30, 36). Briefly, E. coli strains were grown at 37°C in tryptic soy broth with vigorous (200 rpm) shaking for 2 days. Bacterial cells were sonicated, and the sonic lysates were sterile filtered separately by using 0.22-μm-pore-size filters. HeLa cells were treated with sterile sonic lysates of CDT-IV-producing E. coli strain 28C. After 72 h of interaction at 37°C in a 5% CO2 atmosphere, the infecting material was removed by washing the HeLa cell monolayers several times. The morphological changes characteristic of CDT were investigated after staining with anti-phospho-histone H2AX (γH2AX) antibodies (Cell Signaling Technology), followed by rhodamine-conjugated secondary antibodies, or with Giemsa stain, as described previously (30). The cell cycle distribution of 20,000 cells was determined as described previously (36) by flow cytometry. Lysates of CDT-I-producing strain E6468/62 were used as positive controls, and lysates of E. coli C600 were used as negative controls.
Nucleotide sequence accession number.
The sequence of the E. coli 28C cdt-IV gene cluster and neighboring region (see below) has been deposited in the GenBank database under accession number AY578329.
RESULTS AND DISCUSSION
Cloning and genotoxic activity of CDT-IV from ExPEC strain 28C.
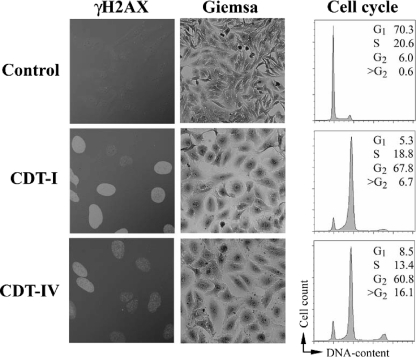
To clone cdt-IV genes, a cosmid library was constructed using porcine E. coli strain 28C (9), in which the CDT-IV toxin was identified for the first time (46). The library was screened by PCR using cdtB-IV-specific primers (46), and CDT-IV production of the PCR-positive clones was tested by using HeLa cell cultures. Sonic lysates of PCR-positive clones altered HeLa cells like sonic lysates of parental strain 28C and CDT-I-producing prototypic strain E6468/62. Three days following treatment, HeLa cells showed morphological changes that included cell body distension, giant nuclei, and G2 cell cycle arrest. Similar to CDT-I, CDT-IV induced phosphorylation of the histone H2AX, a sensitive marker for double-strand DNA breaks. In contrast, a lysate of a laboratory E. coli strain did not cause morphological changes, cell cycle arrest, and H2AX phosphorylation (Fig. 1 and data not shown). This observation confirmed that like other CDTs, CDT-IV inflicted DNA damage in eukaryotic cells.
FIG. 1.
CDT-IV genotoxic effects. HeLa cells were treated with sterile filtered sonic lysates of bacteria producing CDT-I (E6468/62) or CDT-IV (28C). Seventy two hours following treatment the cells were stained as described elsewhere (28) with anti-phospho-histone H2AX (γH2AX) antibodies, followed by rhodamine red-conjugated secondary antibodies (left panels), or with Giemsa stain (middle panels). The cell cycle distribution of 20,000 cells was determined as described previously (34) by flow cytometry (right panels). The percentages of cells in each cell cycle phase are indicated. CDT-I- and CDT-IV-treated cells exhibited nuclear γH2AX (indicating host DNA double-strand breaks), enlarged nuclei and cell bodies, and an absence of mitotic figures. The majority of CDT-I- and CDT-IV-treated cells had a 4n DNA content, indicating that these cells were blocked at the G2-M transition in response to DNA damage.
Comparison of nucleotide sequences of cdt-IV gene clusters in ExPEC strains 28C and APEC O1.
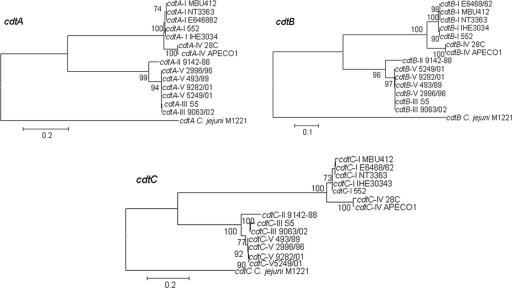
The E. coli 28C cdt-IV gene cluster and neighboring region (see below) were sequenced (GenBank accession number AY578329). A similar cdt-IV operon (GenBank accession number NC008563) is also present in the recently sequenced genome of avian pathogenic E. coli serotype O1:K1:H7 strain APEC O1 (19). The nucleotide sequences of the cdtA-IV, cdtB-IV, and cdtC-IV genes are 99, 99, and 98% identical in strains 28C and APEC O1. Nucleotide sequences of the cdtABC genes for all deposited E. coli alleles were then aligned by using the ClustalW multiple-sequence alignment program (45). In order to facilitate the sequence comparison, we used homologous sequences of cdtABC from C. jejuni as an outgroup. The levels of homology between cdtB genes were higher than the levels of homology between the cdtA and cdtC genes, consistent with the hypothesis that CdtB is the active enzymatic subunit of the holotoxin (data not shown). A neighbor-joining tree based on the multiple-sequence alignments was constructed, and bootstrap values were generated (Fig. 2). Two different phylogenetic lineages were observed for cdtABC alleles from E. coli. The cdt-I and cdt-IV genes appeared to belong to the same phylogenetic lineage, whereas the cdt-II, cdt-III, and cdt-V genes clustered together in an another lineage. These data suggest that the cdt genes were acquired by horizontal transfer events at some point and evolved separately since then.
FIG. 2.
Phylogenetic trees for E. coli cdtABC I-V and C. jejuni cdt genes. The neighbor-joining trees were constructed with MEGA3.1 using the Kimura two-parameter nucleotide substitution model with pairwise deletion of gaps. The levels of bootstrap support for the branches were calculated using 300 replicates, and only values greater than 70% are indicated at the nodes. The alignments of the cdtA, cdtAB, and cdtAC genes were 707, 812, and 582 nucleotides long, respectively. The bars indicate 10 and 20 differences in 100 nucleotides. The following reference sequences were used: for CDT-I, sequences of E. coli E6468/62 (serotype O86:H34; accession number U03293) and 552 (serotype O2:H12; accession number AB258385), CDT-I phage in EPEC strain NT3363 (accession number AB285204), and MBU 412 (Ont; accession number AF373206); for CDT-II, E. coli 9142-88 (serotype O128:NM, accession number U04208); for CDT-III, E. coli S5 (serotype O115:K?:H21; accession number U89305) and E. coli 9063/02 (O153:H18, accession number AY365044); for CDT-IV, E. coli 28C (serotype O75:K95; accession number AY578329) and E. coli APEC O1 (serotype O1:K1; accession number CP000468); and for CDT-V, E. coli 2996/96 (serotype O73:H18; accession number AY365045), E. coli 9282/01 (serotype O91:H21; accession number AY365042), E. coli 5249/01 (serotype O113:H21; accession number AY365043), E. coli 493/89 (serotype O157:NM; accession number AJ508930), and C. jejuni M1221 (accession number NC_003912).
cdt-IV gene clusters in ExPEC strains 28C and APEC O1 are framed by lambdoid prophage genes.
A DNA homology search indicated that the cdt-IV operon of strain 28C is flanked on both sides by prophage-related ORFs encoding putative prophage-related proteins. These putative proteins include a lambdoid prophage host specificity protein (orf1), a Lom-like protein (orf2), a putative tail fiber protein (orf3), a putative protease encoded in enterohemorrhagic E. coli (EHEC) EDL933 prophage CP-933 (rorf1), and a putative OmpT-like outer membrane protease (rorf2). Seven of the eight ORFs that were identified in the cdt-IV locus of E. coli strain 28C were also present in the APEC O1 strain, and all of them were grouped in the same way in these two strains (Fig. 3). As the APEC O1 genome was only partially annotated at the beginning of this study, the present nucleotide sequence analysis of cdt-IV flanking ORFs of strain 28C represents an alternative annotation for the cdt-IV locus in the APEC O1 strain. Sequence comparison of the cdt alleles in ExPEC strain 28C (CDT-IV), APEC O1 (CDT-IV), IHE3034 (CDT-I), and a recently described (4) inducible CDT-I-encoding lambdoid prophage (CDT-IΦ) genome in EPEC revealed that in all cases the cdt genes were located downstream of the prophage head and tail genomic regions. Between the structural-protein-encoding genes and the cdt genes there is a variable region that spans the tail fiber and orf6 genes (Fig. 3). The main differences between the sequences of strains 28C, APEC O1, and IHE3034 were restricted to orf4 and orf5. orf4 is replaced by a 2-kb region in APEC O1 and by the c3149 ORF in IHE3034, while orf5 characterizes the CDT-IV 28C and APEC O1 ExPEC strains. The fact that the c3149 ORF and orf4 are similar but not identical could suggest that they have similar functions or the same function. The cdt downstream region is identical in CDT-IV strains, and the cdtC genes in the 28C and APEC O1 strains are flanked by two morons. These DNA sequence comparison data indicate that the CDT-IV-encoding locus may have undergone recombination events, resulting in exchange of DNA regions in different parts of the phage genome.
cdt-I gene cluster in ExPEC strain IHE3034 is flanked by lambdoid prophage genes.
On the basis of the known homology between cdt-I and cdt-IV, we decided to compare their flanking regions as well. However, the flanking sequences of the cdt-I operon in prototypic EPEC strain E6468/62 were not available (GenBank accession number U03293). Thus, we cloned and sequenced the cdt-I genes and their flanking regions in the CDT-I-producing O18:K1:H7 ExPEC strain IHE3034 (21, 30). A genomic DNA library was constructed and screened by PCR for the cdtB-I gene. From the PCR-positive cdtB-I clones, one clone producing CDT-I was selected for sequencing. An 18.5-kb DNA fragment was sequenced. Nucleotide sequence comparison revealed that the cdtABC genes of strain IHE3034 were 99% identical to the genes of EPEC prototypic strain E6468/62. The cdt-I genes in strain IHE3034 were also flanked by prophage-related ORFs. The cdt-I-flanking ORFs were homologous to several flanking ORFs identified in the cdt-IV loci of strains 28C and APEC O1. Upstream of the cdtA-I gene three 28C-related genes, orf2, orf3, and orf6, encoding the putative Lom-like and tail fiber proteins and a CFT073-like hypothetical protein (c3147), were localized (Fig. 3). Downstream of the cdtC-I gene a P4-like prophage integrase gene (intB) was identified. This integrase gene was previously found in EHEC serotype O157:H7 reference strains EDL933 and Sakai and was found in the APEC O1 genome as well (16, 19, 37). However, intB was not associated with the cdt-IV operon in the APEC O1 strain.
A database search conducted with all other cdt-I flanking ORFs of strain IHE3034 revealed that the sequenced fragment contained only prophage genes. Furthermore, the nucleotide sequence comparison allowed further annotation of the cdt-IV locus in APEC O1. The prophage genes upstream of the cdt-I and cdt-IV operons encode several putative tail, minor tail, and tail assembly proteins. These genes are homologous and grouped similarly in IHE3034 and APEC O1. Interestingly, the last six genes at the left end of the IHE3034 fragment correspond to six genes which so far have been observed only in APEC O1 (data not shown).
Comparison of the cdt-I and cdt-IV genes and their flanking regions revealed that not only are these cdt operons similar but the cdt upstream flanking regions contain similar prophage genes. These cdt genes and their flanking genes might have been acquired from a common ancestor and after chromosomal integration in different bacteria could have evolved in slightly different ways, resulting in minor sequence variations and some deletions. Similarly to our findings, Janka et al. (18) previously demonstrated that in EHEC O157:NM strain 493/89 the cdt-V operon is framed by P2 prophage sequences. However, the cdt-IV and cdt-I flanking genes are not related to P2 phage genes.
Recently, an inducible lambdoid phage (CDT-IΦ) which codes for CDT-I in EPEC strains was sequenced (4). The genome of this CDT-I-converting phage comprises 47,021 nucleotides in 60 predicted ORFs, which are organized into six genomic regions coding for the head and tail, passenger virulence traits (morons), and integrase, as well as unknown regulatory and lysis functions. The CDT-1Φ genome has a highly mosaic structure and shows homology with sequences derived from several lambdoid prophages, including the UTI89 prophage of uropathogenic strains and an EPEC B171 prophage-like element of an EPEC and the serotype-converting SfV phage of Shigella flexneri (4). However, none of these phages have a cdt gene cluster in their passenger regions. Nonetheless, CDT-1Φ and the other prophages are very similar in their overall genomic organization. The nucleotide sequence data for E. coli 28C and IHE3034 did not allow identification of the integration sites of the CDT-I and CDT-IV phages, but the genome sequence of the APEC O1 strain (19) revealed that the cdt-IV operon is framed by two prophages and pathogenicity island-associated DNA sequences, including integrase and tRNA genes. In the present study we also observed genetic mosaic organization among the sequenced cdt-I and cdt-IV alleles carried by ExPEC strains IHE3034 and 28C, respectively. This phenomenon could also be a result of extensive genetic exchanges among different phages, which might even occur in the mammalian intestine.
Dissemination of two cdt-IV flanking genes in CDT-producing strains.
We decided to investigate the presence of two 28C-specific flanking prophage genes and their association with cdt genes in a set of CDT-producing E. coli strains. We used a PCR-based strategy (Fig. 3) with primers listed in Table 2. Five of the 25 CDT-producing E. coli strains investigated carried cdt-I genes, 4 strains carried cdt-III genes, 3 strains had cdt-V genes, and 13 strains harbored cdt-IV genes (Table 1). orf5 codes for a hypothetical protein, which was detected only in eight cdt-IV strains, while rorf1 encoding a putative protease similar to ECs1662 in EHEC strain Sakai was detected in all the cdt-IV strains. The rorf1 sequences were also present in two additional cdt-III strains and three additional cdt-V strains. orf5 was uniformly associated with cdtA, and the size of the orf5/cdtA-specific PCR amplicon indicated that all the cdt-IV strains carried orf6 as well. In all the cdt-IV strains rorf1 was located directly downstream of the cdtC gene. At the same time cdtC genes were not immediately upstream of rorf1 in either the cdt-III strains or the cdt-V strains. These results indicate that the protease gene could characterize not only the cdt-IV strains, although this gene is always and exclusively adjacent to the cdtC gene in cdt-IV strains. The fact that orf5 can be absent in some cdt-IV strains demonstrates that the cdt-IV prophages could be divergent. PCR results indicated that there was a progressive loss of upstream flanking genes, while no losses for the downstream prophage genes were observed in the cdt-IV region. Nucleotide sequence analysis verified these observations and revealed further sequence variations in CDT-encoding phages. The DNA fragment of E. coli strain IHE3034 showed great homology to CDT-IV-encoding phages, but in the corresponding fragment orf4 and orf5 were both deleted. In the APEC O1 strain an unrelated sequence was observed instead of orf4 between orf5 and orf3 (Fig. 3).
The orf4, orf5, orf6, cdt, CP-933-like protease (rorf1), and outer membrane protease (rorf2) genes can be defined as morons that are not required for the phage life cycle but may code for fitness factors contributing to the survival of the bacterial host (7). The fact that these “28C-specific” morons are also present in several intestinal E. coli and ExPEC strains, including uropathogenic isolates CFT073, 536, and UTI89, strain APEC O1, and EHEC strains EDL933 and Sakai, indicates that these morons, similar to virulence genes, might have spread by horizontal gene transfer (Table 3).
TABLE 3.
Morons of lambdoid prophage encoding CDT-IV in ExPEC strain 28C
| ORF or gene | Feature or product of sequence | Start position | Stop position | Length (bp) | No. of identical nucleotides/no. of nucleotides (% identity) | Position in clone or genome | Strain or location | GenBank accession no. | Reference |
|---|---|---|---|---|---|---|---|---|---|
| orf4 | Hypothetical protein | 3723 | 3992 | 270 | 270/270 | 3792-3992 | ExPEC 28C | AY578329d | This study |
| orf5 | Conserved hypothetical protein | 4005 | 4688 | 684 | 619/648 (95) | 1512261-1512905 | E. coli APEC O1 | CP000468 | 19 |
| 608/643 (94) | 5046238-5046879 | ||||||||
| orf6 | Hypothetical protein | 4699 | 4989 | 291 | 287/291 (98) | 1512916-1513206 | E. coli APEC O1 | CP000468 | 19 |
| 273/287 (95) | 5046890-5047176 | ||||||||
| 271/288 (94) | 1220158-1220444 | ||||||||
| 271/288 (94) | 2052137-2051851 | ||||||||
| 68/78 (87) | 314824-314901 | ||||||||
| 21/22 (95) | 314784-314803 | ||||||||
| 274/287 (95) | 3022497-3022211 | E. coli CFT073 | AE014075 | ||||||
| 259/269 (96) | 1371361-1371629 | ||||||||
| 270/287 (94) | 1446322-1446608 | ||||||||
| 68/78 (87) | 377759-377836 | ||||||||
| 271/287 (94) | 5029960-5030246 | E. coli ÚTI 89 | CP000243 | ||||||
| 270/287 (94) | 1282862-1283148 | ||||||||
| 68/78 (87) | 313235-313312 | ||||||||
| 270/287 (94) | 1228776-1229062 | E. coli 536 | CP000247 | ||||||
| 68/78 (87) | 369932-370009 | ||||||||
| 21/22 (95) | 369890-369911 | ||||||||
| Partial sit iron uptake system operon | 269/287 (93) | 4537-4251 | E. coli chromosome | AM072350 | |||||
| Pathogenicity island II right junction | 68/78 (87) | 1131-1208 | E. coli str4787 | AY560916 | |||||
| Unknown genes | 62/61 (85) | 20684-20744 | E. coli str222 | AY151282 | |||||
| S. flexneri 2a insertion sequence | 21/22 (95) | 20644-20663 | |||||||
| cdtA | CDT-IV A subunit | 5502 | 6215 | 714 | 712/714 (99) | 1513719-1514378 | E. coli APEC O1 | CP000468 | 19 |
| 669/714 (93) | 201-914 | E. coli E6468/62 | U03293 | 44 | |||||
| cdtB | CDT-IV B subunit | 6212 | 7033 | 822 | 821/822 (99) | 1514429-1515250 | E. coli APEC O1 | CP000468 | 19 |
| 722/822 (87) | 911-1732 | E. coli E6468/62 | U03293 | 44 | |||||
| cdtC | CDT-IV C subunit | 7030 | 7602 | 573 | 563/573 (98) | 1515247-1515819 | E. coli APEC O1 | CP000468 | 19 |
| 437/496 (88) | 1801-2296 | E. coli E6468/62 | U03293 | 44 | |||||
| rorf1 | Putative protease | 7672 | 9156 | 1485 | 1472/1485 (99) | 1515889-1517373 | E. coli APEC O1 | CP000468 | 19 |
| Encoded in prophage CP-933X | 1417/1485 (95) | 1751300-1752784 | E. coli O157:H7 EDL933 | AE005174 | 37 | ||||
| 1417/1485 (95) | 1659506-1660990 | E. coli O157:H7 Sakai | BA000007 | 16 | |||||
| rorf2 | Outer membrane 3b, protease VII | 9341 | 10024 | 684 | 679/684 (99) | 1517558-1518241 | E. coli APEC O1 | CP000468 | 19 |
| 624/693 (90) | 572657-573343 | ||||||||
| 625/692 (90) | 622998-623684 | E. coli 536 | CP000247 | ||||||
| 624/693 (90) | 571059-571745 | E. coli UTI89 | CP000243 | ||||||
| 623/693 (89) | 1661177-1661863 | E. coli O157:H7 Sakai | BA000007 | 16 | |||||
| Omp precursor | 623/693 (89) | 1752971-1753657 | E. coli O157:H7 EDL933 | AE005174 | 37 | ||||
| 620/692 (89) | 629100-629786 | E. coli CFT073 | AE014075 | ||||||
| DLP12 prophage, outer membrane protease VII | 620/693 (89) | 583903-584589 | E. coli MG1655 | U00096 |
Phages in intestinal E. coli and ExPEC.
CDT-I and CDT-II have been detected in E. coli strains that mainly cause human gastrointestinal infections. CDT-III and CDT-IV were identified in human- and animal-pathogenic E. coli strains of intestinal and extraintestinal origin, and these strains frequently produced cytotoxic necrotizing factors (cytotoxic necrotizing factor type 1 and cytotoxic necrotizing factor type 2) and hemolysin as well. CDT-V was detected in E. coli O157 isolates and non-O157 STEC strains from humans with diarrhea or hemolytic-uremic syndrome (18). Although CDTs are produced by other diverse pathogenic bacterial species, the mechanism associated with possible horizontal transfer of the various cdt genes resulting in the wide distribution of cdt genes among pathogenic bacteria is not adequately known.
Bacteriophages are the major vehicles for transfer of genes, including fitness and virulence genes, between bacteria (7). Analysis of the genome sequences of EHEC O157:H7 strains Sakai and EDL933 revealed the presence of as many as 18 prophages (16, 37). The prophage DNA accounts for one-half of the 1.3 Mb of DNA found in O157 but is not present in the E. coli K-12 reference strain (32). Of the 18 prophages on the O157 Sakai chromosome, 13 are lambda-like phages, and in addition to Shiga toxins (Stx1 and Stx2), they encode other virulence-related proteins, such as Lom homologues, Bor proteins, and zinc/copper-type superoxide dismutases (16). The stx genes are located in the genomes of heterogeneous, lytic (stx2) or cryptic (stx1) lambdoid phages (16, 37). The dissemination of Stx-encoding phages is the most likely mechanism for the emergence of new STEC serotypes that has been demonstrated in vitro (17, 43) and in vivo (1, 47).
Recently, Asakura et al. (4) provided evidence of the existence of an inducible converting cdt-I lambdoid phage (CDT-1Φ) in a serotype O127:H7 EPEC strain. It was demonstrated that the cdt-I gene cluster is transferred by CDT-1Φ to a recipient strain, which then produces biologically active CDT-I toxin. Interestingly, the virulence region of the CDT-1Φ genome contains, in addition to cdt genes, a truncated cycle-inhibiting factor gene (cif) and a type 3 secreted effector protein gene. Although several other EPEC strains belonging to serogroups O86, O142, and O127 were positive for the presence of cdt genes and produced biologically active CDT, only four of seven CDT-I strains investigated produced infectious CDT-1Φ particles. Southern hybridization analysis revealed the genetic diversity of CDT-I prophages carried by the different CDT-I-producing strains (4).
The regions encoding head and tail proteins and the lysis region in CDT-1Φ have homology to the regions in lambdoid Stx phages, but the locations of virulence regions in CDT-1Φ and Stx phages are significantly different. The stx genes are associated with the late regulatory region in the Stx phages, while the cdt-I and cdt-IV genes are not associated with the late regulatory genes.
Similar to intestinal pathogens, phages could play a fundamental role in the evolution of ExPEC strains. The O acetylation of the capsular polysaccharide is a phenomenon that has been known for a long time (33). Recently, an E. coli K1-specific virulent bacteriophage (CUS-3) carrying an O acetyltransferase gene, neuO, was described. Investigation of CUS-3 revealed that it is a 47,021-bp lambdoid phage, and the neuO gene is uniformly present in typically “high-virulence” E. coli O18 and O45 strains, while it is absent or rarely present in low-virulence strains or in strains belonging to less commonly isolated serotypes (20). If functional K1 and CDT-I phages are produced from the lysogenic ExPEC strains, other sensitive strains may be lysogenized by transduction. Lysogenization could lead to novel pathogenic strains harboring neuO and/or cdt genes (4, 20).
The locations of toxin genes in our sequenced cdt-I and cdt-IV alleles are substantially related to the location of the cdt gene cluster in CDT-1Φ. Although we do not have sequence information for a whole CDT-1Φ-like genome in ExPEC strains IHE3034 and 28C, our sequence data suggest that the ExPEC cdt-I and cdt-IV genes could be also parts of prophages. The overall G+C content of the cdt-I allele in IHE3034 is 51.51%, which is similar to that in E. coli (50%). The G+C content of the cdt-I genes is only 41.77%. The overall G+C content of the cdt-IV allele in 28C is 44.6%, and the G+C content of the cdt-IV genes is only 42.3%. These values indicate that both the cdt-I and cdt-IV genes might have been acquired recently by a perhaps a previously existing E. coli phage through multiple gene transfer events and that these genes evolved differently in their new bacterial hosts. Additionally, both of our sequenced cdt alleles contain several homologous structural genes upstream of the cdt genes, like those in the CDT-1Φ genome. Sequence comparisons indicated that in the cdt-I allele of IHE3034, similar to CDT-I, phage is also located downstream of the head and tail region. One of the differences is that the virulence region in IHE3034 contains only the cdt genes, while in the CDT-1Φ virulence region there are other virulence genes, including a truncated cif gene and a non-locus-of-enterocyte-effacement-encoded type 3 effector gene, as well as (downstream of the cdtC gene) two other ORFs which encode putative hypothetical proteins (Fig. 3.). It is interesting that in the cdt-I allele of IHE3034 there is, upstream of cdtA, a 2.5-kb region which is missing from the genome of CDT-1Φ but is more or less present at the related position in the cdt-IV allele in 28C. In our sequenced cdt-I allele, the cdtC gene is associated with a P4-like integrase gene, while CDT-1Φ carries a different integrase gene downstream of the virulence region. The cdtC-IV genes in ExPEC strains 28C and APEC O1 are framed downstream by two prophage protease genes. The detailed mosaic structure observed for our cdt-I and cdt-IV alleles in ExPEC strains could also be the result of extensive genetic exchanges among different phages, as suggested by Asakura et al. for CDT-I phages carried by EPEC (4). These and similar recombination events might even occur in the human or animal intestine.
In summary, our results indicate that the cdt-I and cdt-IV genes might have been acquired by phage transduction from a common ancestor and that evolution of the CDT-encoding phages in different bacterial hosts generated differences in the cdt genes and their flanking DNA contents. More exact definitions of the differences in flanking regions would require further investigation. Similarly, the fact that the cdt-II, cdt-III, and cdt-V genes show significant homology despite their genetic location raises the possibility that the cdt-II, cdt-III, and cdt-V gene clusters might also have evolved from another ancestor and might have spread with mobile elements (perhaps plasmids) and evolved differently in the different bacterial hosts. The fact that cdt-III genes are located on a large conjugative virulence plasmid (pVir) in prototype strain S5 (36) could suggest the mode of acquisition from a common ancestor, but without sequence data it is impossible to predict the original donor of the cdt-II, cdt-III, and cdt-V genes. Further studies are required to answer all these questions.
Acknowledgments
This study was supported by the ERA-NET project “Deciphering the intersection of extraintestinal pathogenic and commensal Escherichia coli,” by the Hungarian Research Fund OTKA (grant T 37890), and a by bilateral program between INRA (France) and the Hungarian Academy of Sciences.
We thank Gábor M. Kovács (Budapest, Hungary) for technical help.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 3 November 2008.
REFERENCES
- 1.Acheson, D. W., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 664496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins α, β, γ, and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 4.Asakura, M., A. Hinenoya, M. S. Alam, K. Shima, S. H. Zahid, L. Shi, N. Sugimoto, A. N. Ghosh, T. Ramamurthy, S. M. Faruque, G. B. Nair, and S. Yamasaki. 2007. An inducible lambdoid prophage encoding cytolethal distending toxin (Cdt-I) and a type III effector protein in enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 10414483-14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielaszewska, M., M. Fell, L. Greune, R. Prager, A. Fruth, H. Tschape, M. A. Schmidt, and H. Karch. 2004. Characterization of cytolethal distending toxin genes and expression in Shiga toxin-producing Escherichia coli strains of non-O157 serogroups. Infect. Immun. 721812-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blank, T. E., H. Zhong, A. L. Bell, T. S. Whittam, and M. S. Donnenberg. 2000. Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia coli strains. Infect. Immun. 687028-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, K., J. L. Latimer, D. A. Lewis, and E. J. Hansen. 2001. Investigation of the interaction among the components of the cytolethal distending toxin of Haemophilus ducreyi. Biochem. Biophys. Res. Commun. 285609-615. [DOI] [PubMed] [Google Scholar]
- 9.Dozois, C. M., E. Oswald, N. Gautier, J. P. Serthelon, J. M. Fairbrother, and I. P. Oswald. 1997. A reverse transcription-polymerase chain reaction method to analyze porcine cytokine gene expression. Vet. Immunol. Immunopathol. 58287-300. [DOI] [PubMed] [Google Scholar]
- 10.Elwell, C. A., and L. A. Dreyfus. 2000. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol. Microbiol. 37952-963. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39783-791. [DOI] [PubMed] [Google Scholar]
- 12.Fox, J. G., A. B. Rogers, M. T. Whary, Z. Ge, N. S. Taylor, S. Xu, B. H. Horwitz, and S. E. Erdman. 2004. Gastroenteritis in NF-κB-deficient mice is produced with wild-type Camplyobacter jejuni but not with C. jejuni lacking cytolethal distending toxin despite persistent colonization with both strains. Infect. Immun. 721116-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frisan, T., X. Cortes-Bratti, E. Chaves-Olarte, B. Stenerlow, and M. Thelestam. 2003. The Haemophilus ducreyi cytolethal distending toxin induces DNA double-strand breaks and promotes ATM-dependent activation of RhoA. Cell. Microbiol. 5695-707. [DOI] [PubMed] [Google Scholar]
- 14.Haghjoo, E., and J. E. Galán. 2004. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc. Natl. Acad. Sci. USA 1014614-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassane, D. C., R. B. Lee, and C. L. Pickett. 2003. Campylobacter jejuni cytolethal distending toxin promotes DNA repair responses in normal human cells. Infect. Immun. 71541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2811-22. [DOI] [PubMed] [Google Scholar]
- 17.James, C. E., K. N. Stanley, H. E. Allison, H. J. Flint, C. S. Stewart, R. J. Sharp, J. R. Saunders, and A. J. McCarthy. 2001. Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Appl. Environ. Microbiol. 674335-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janka, A., M. Bielaszewska, U. Dobrindt, L. Greune, M. A. Schmidt, and H. Karch. 2003. The cytolethal distending toxin (cdt) gene cluster in enterohemorrhagic Escherichia coli O157:H− and O157:H7: characterization and evolutionary considerations. Infect. Immun. 713634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 1893228-3236. (Erratum, 189:4554.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King, M. R., R. P. Vimr, S. M. Steenbergen, L. Spanjaard, G. Plunkett III, F. R. Blattner, and E. R. Vimr. 2007. Escherichia coli K1-specific bacteriophage CUS-3 distribution and function in phase-variable capsular polysialic acid O acetylation. J. Bacteriol. 1896447-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korhonen, T. K., M. V. Valtonen, J. Parkkinen, V. Vaisanen-Rhen, J. Finne, F. Ørskov, I. Ørskov, S. B. Svenson, and P. H. Mäkelä. 1985. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150.163. [DOI] [PubMed] [Google Scholar]
- 23.Lara-Tejero, M., and J. E. Galan. 2000. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290354-357. [DOI] [PubMed] [Google Scholar]
- 24.Lee, R. B., D. C. Hassane, D. L. Cottle, and C. L. Pickett. 2003. Interactions of Campylobacter jejuni cytolethal distending toxin subunits CdtA and CdtC with HeLa cells. Infect. Immun. 714883-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, D. A., M. K. Stevens, J. L. Latimer, C. K. Ward, K. Deng, R. Blick, S. R. Lumbley, C. A. Ison, and E. J. Hansen. 2001. Characterization of Haemophilus ducreyi cdtA, cdtB, and cdtC mutants in in vitro and in vivo systems. Infect. Immun. 695626-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., A. Sharipo, E. Chaves-Olarte, M. G. Masucci, V. Levitsky, M. Thelestam, and T. Frisan. 2002. The Haemophilus ducreyi cytolethal distending toxin activates sensors of DNA damage and repair complexes in proliferating and non-proliferating cells. Cell. Microbiol. 487-99. [DOI] [PubMed] [Google Scholar]
- 27.McSweeney, L. A., and L. A. Dreyfus. 2004. Nuclear localization of the Escherichia coli cytolethal distending toxin CdtB subunit. Cell. Microbiol. 6447-458. [DOI] [PubMed] [Google Scholar]
- 28.Nesic, D., Y. Hsu, and C. E. Stebbins. 2004. Assembly and function of a bacterial genotoxin. Nature 429429-433. [DOI] [PubMed] [Google Scholar]
- 29.Nishikubo, S., M. Ohara, Y. Ueno, M. Ikura, H. Kurihara, H. Komatsuzawa, E. Oswald, and M. Sugai. 2003. An N-terminal segment of the active component of the bacterial genotoxin cytolethal distending toxin B (CDTB) directs CDTB into the nucleus. J. Biol. Chem. 27850671-50681. [DOI] [PubMed] [Google Scholar]
- 30.Nougayrede, J. P., S. Homburg, F. Taieb, M. Boury, E. Brzuszkiewicz, G. Gottschalk, C. Buchrieser, J. Hacker, U. Dobrindt, and E. Oswald. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313848-851. [DOI] [PubMed] [Google Scholar]
- 31.Nougayrède, J. P., F. Taieb, J. De Rycke, and E. Oswald. 2005. Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 13103-110. [DOI] [PubMed] [Google Scholar]
- 32.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9481-485. [DOI] [PubMed] [Google Scholar]
- 33.Orskov, F., I. Orskov, A. Sutton, R. Schneerson, W. Lin, W. Egan, G. E. Hoff, and J. B. Robbins. 1979. Form variation in Escherichia coli K1: determined by O-acetylation of the capsular polysaccharide. J. Exp. Med. 149669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oswald, E., J. P. Nougayrède, F. Taieb, and M. Sugai. 2005. Bacterial toxins that modulate host cell-cycle progression. Curr. Opin. Microbiol. 883-91. [DOI] [PubMed] [Google Scholar]
- 35.Oswald, E., M. Sugai, A. Labigne, H. C. Wu, C. Fiorentini, P. Boquet, and A. D. O'Brien. 1994. Cytotoxic necrotizing factor type 2 produced by virulent Escherichia coli modifies the small GTP-binding proteins Rho involved in assembly of actin stress fibers. Proc. Natl. Acad. Sci. USA 913814-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peres, S. P., O. Marches, F. Daigle, JP. Nougayrède, F. Herault, C. Tasca, J. De Rycke, and E. Oswald. 1997. A new cytolethal distending toxin (CDT) from Escherichia coli producing CNF2 blocks HeLa cell division in G2/M phase. Mol. Microbiol. 241095-1107. [DOI] [PubMed] [Google Scholar]
- 37.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409529-533. [DOI] [PubMed] [Google Scholar]
- 38.Pickett, C. L., D. L. Cottle, E. C. Pesci, and G. Bikah. 1994. Cloning, sequencing, and expression of the Escherichia coli cytolethal distending toxin genes. Infect. Immun. 621046-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pratt, J. S., K. L. Sachen, H. D. Wood, K. A. Eaton, and V. B. Young. 2006. Modulation of host immune responses by the cytolethal distending toxin of Helicobacter hepaticus. Infect. Immun. 744496-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purdy, D., C. M. Buswell, A. E. Hodgson, K. McAlpine, I. Henderson, and S. A. Leach. 2000. Characterisation of cytolethal distending toxin (CDT) mutants of Campylobacter jejuni. J. Med. Microbiol. 49473-479. [DOI] [PubMed] [Google Scholar]
- 41.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 653855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott, D. A., and J. B. Kaper. 1994. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 62244-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tóth, I., F. Herault, L. Beutin, and E. Oswald. 2003. Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: establishment of the existence of a new cdt variant (type IV). J. Clin. Microbiol. 414285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tóth, I., H. Schmidt, M. Dow, A. Malik, E. Oswald, and B. Nagy. 2003. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a Shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 697242-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Bost, S. Roels, E. Oswald, and J. Mainil. 2003. Putative roles of the CNF2 and CDTIII toxins in experimental infections with necrotoxigenic Escherichia coli type 2 (NTEC2) strains in calves. Microbes Infect. 51189-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young, R. S., K. R. Fortney, V. Gelfanova, C. L. Phillips, B. P. Katz, A. F. Hood, J. L. Latimer, R. S. Munson, Jr., E. J. Hansen, and S. M. Spinola. 2001. Expression of cytolethal distending toxin and hemolysin is not required for pustule formation by Haemophilus ducreyi in human volunteers. Infect. Immun. 691938-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]