Abstract
Microbial survival in a host is usually dependent on the ability of a pathogen to undergo changes that promote escape from host defense mechanisms. The human-pathogenic fungus Cryptococcus neoformans undergoes phenotypic switching in vivo that promotes persistence in tissue. By microarray and real-time PCR analyses, the allergen 1 gene (ALL1) was found to be downregulated in the hypervirulent mucoid switch variant, both during logarithmic growth and during intracellular growth in macrophages. The ALL1 gene encodes a small cytoplasmic protein that is involved in capsule formation. Growth of an all1Δ gene deletion mutant was normal. Similar to cells of the mucoid switch variant, all1Δ cells produced a larger polysaccharide capsule than cells of the smooth parent and the complemented strain produced, and the enlarged capsule inhibited macrophage phagocytosis. The mutant exhibited a modest defect in capsule induction compared to all of the other variants. In animal models the phenotype of the all1Δ mutant mimicked the hypervirulent phenotype of the mucoid switch variant, which is characterized by decreased host survival and elevated intracranial pressure. Decreased survival is likely the result of both an ineffective cell-mediated immune response and impaired phagocytosis by macrophages. Consequently, we concluded that, unlike loss of most virulence-associated genes, where loss of gene function results in attenuated virulence, loss of the ALL1 gene enhances virulence by altering the host-pathogen interaction and thereby impairing clearance. Our data identified the first cryptococcal gene associated with elevated intracranial pressure and support the hypothesis that an environmental opportunistic pathogen has modified its virulence in vivo by epigenetic downregulation of gene function.
Cryptococcus neoformans is a major fungal pathogen in patients with AIDS or other diseases that result in impaired T-cell immunity (11, 44). In patients with AIDS, cryptococcosis usually presents as a chronic meningoencephalitis with elevated intracranial pressure (ICP), which is associated with high morbidity and mortality. Despite antifungal therapy, immunocompromised patients often develop persistent disease or relapse (23). Several studies have supported the concept that this yeast changes during chronic infection, which can affect its virulence and allow it to persist in the human host (5, 6, 8).
Pathoadaptation is not unique to C. neoformans and is a common strategy shared by many human pathogens that allows them to survive in their host. Phenotypic phase variation of bacteria and antigenic variation of eukaryotic pathogens change the microbial surface in vivo (9), and these processes can control invasion of mucosal barriers or permit evasion of the host's immune response. Phenotypic switching of colony morphology has been described for several fungal species and includes the white-opaque switching in Candida albicans (45, 46) that is essential for high-efficiency mating (34, 39) and is also associated with changes in host-pathogen interactions (29). Achieving enhanced microbial fitness in the host is a complex phenomenon that involves differential regulation of many genes. Historically, pathogenesis research has focused on characterization of virulence-enhancing factors, which are often upregulated in vivo. Studies that identified so-called “antivirulence genes” (14, 37), whose loss or absence enhances virulence, have been rare. The paucity of results is explained in part by the challenge of assigning augmented virulence to one specific gene when many genes are simultaneously differentially regulated in the process of pathoadaptation. The mechanisms that regulate virulence factors and generate phenotypic variability within a pathogen population are variable and can involve genetic mutations that are inheritable, as well as epigenetic regulation of gene expression that also is often unstable and typically is either not inheritable or inherited in a non-Mendelian pattern (10).
C. neoformans is an excellent model system to study the impact of phenotypic switching with respect to pathoadaptation during chronic infection. In C. neoformans phenotypic switching is observed in strains of all three serotypes (22, 27), including strain RC-2 (21), which undergoes switching from a parent smooth (SM) colony phenotype to a hypervirulent mucoid (MC) colony phenotype. Similar to the white-opaque system in C. albicans, the SM-to-MC switch in C. neoformans involves simultaneous changes in many cellular characteristics (46), and the phenotypic switch variants are hypothesized to be epigenetic states of genetically identical cells. Switching to the MC phenotype can be observed in chronic murine infection models, where it is associated with a poor outcome (21). The MC variant elicits a damage-prone, ineffective inflammatory response in the host, which promotes selection, a process that can be further enhanced in a treatment setting (18). Analogous to poor outcomes observed for human cryptococcosis, infection with the hypervirulent MC variant also leads to high ICP and rapid death in rat models (20).
The aim of this study was to investigate the molecular mechanisms of enhanced virulence as a result of phenotypic switching from the SM phenotype to the MC phenotype. Below we present data for a virulence-modifying gene, ALL1, which is downregulated in the phenotypic switching setting and confers the hypervirulent phenotype of the MC state. Loss of function of this gene changes the polysaccharide capsule, inhibits phagocytosis, and affects the cell-mediated immune response (CMI). This leads to elevated ICP and decreased survival in animal models.
MATERIALS AND METHODS
C. neoformans strains and plasmids.
RC-2 is a variant of strain ATCC 24067 (serotype D) that undergoes phenotypic switching, as previously described (21). Strains were either grown and passaged on Sabouraud's dextrose agar and yeast potato dextrose (YPD) solid medium or cultured in Sabouraud's dextrose broth (SD) or minimal medium (0.6% yeast nitrogen base, 0.2% dropout medium, 2% glucose) overnight. Plasmids pJAF1, pJAF13, and pUC19 were grown in Escherichia coli on standard Luria-Bertani (LB) agar plates with ampicillin and have been described elsewhere (16).
Microarray analysis and real-time PCR (RT-PCR).
For RNA extraction starter cultures of SM, MC, and all1Δ yeast cells were grown overnight in SD at 37°C with agitation (150 rpm). The yeast cells were subcultured in fresh SD and grown for another 12 h to mid-logarithmic phase, as monitored by absorbance at 600 nm. Three independent sets of RNA samples from independent experiments were prepared using a Qiagen RNAeasy kit according to the manufacturer's protocol. Microarray analysis was performed and the results were analyzed by the Genome Sequencing Center, School of Medicine, Washington University, St. Louis, MO, with Genechips. Briefly, slides were scanned immediately after hybridization with a ScanArray Express HT scanner (PerkinElmer) to detect Cy3 and Cy5 fluorescence. The laser power was kept constant, and photomultiplier tube (PMT) values were set for optimal intensity with a minimal background (high-PMT scan). An additional scan was done for each slide with the PMT set so that <1% of the elements were saturated (low-PMT scan) to characterize spots which were saturated in the high-PMT scan. Gridding and analysis of images were performed with the ScanArray Express software (version 3.0; PerkinElmer). The intensity values were imported into GeneSpring 7.3 software (Agilent, Redwood City, CA). The local background intensity was subtracted from individual spot intensities. The mean signal and control intensities of the on-slide duplicate spots were calculated. A Lowess curve was fitted to the plot of log intensity versus log ratio, and 20.0% of the data was used to calculate the Lowess fit at each point. The curve was used to adjust the control value for each measurement. If the control channel value was less than 10 relative fluorescence units (RFU), then 10 RFU was used instead. Mean ratios of the signal to the Lowess-adjusted control were calculated. The cross-chip averages were derived from the antilog of the mean of the natural log ratios across the replicate microarrays. The data were then filtered in the following manner. Oligonucleotide elements that received a “present” call (an intensity of >200 RFU or a local signal-to-background ratio of >2) with the ScanArray software for two of the four high-PMT scans with either Cy3 or Cy5 were identified for each of the samples, and all other elements were excluded from the analysis. The means were calculated using all replicates and were filtered for genes with a P value of <0.05 in the high-PMT scan. DNA sequences were annotated based on the C. neoformans JEC21 genomic database.
RT-PCR was used to quantify the relative expression of ALL1 during intracellular growth of SM and MC yeasts in macrophagelike cell line J774.16. Briefly, J774.16 cells were grown in petri dishes, and C. neoformans was added at a fungus/macrophage ratio of 1:5 in the presence of capsule-specific monoclonal antibody (MAb) 18B7. At 2 and 8 h macrophage monolayers were rinsed to wash off extracellular yeast cells. Two separate macrophage experiments were performed. The remaining intracellular C. neoformans cells in attached macrophages were scraped off the wells and suspended in RLT buffer (RNAeasy Miniprep kit; Qiagen). The macrophages were then lysed, and RNA was isolated using standard methods. Total RNA was treated with DNase (Genehunter), and cDNA synthesis was performed using the SuperScript II enzyme system (Invitrogen). The various cDNAs were subjected to a SYBR green-based RT-PCR with ALL1-specific primers. The actin (ACT1) and glyceraldehyde-3-phosphate dehydrogenase (GPD) housekeeping genes were used as controls (Table 1). All experiments were performed in duplicate, and the data were normalized to the expression of the control genes.
TABLE 1.
Primers used in this study
| Primer | Use | Sequence (5′-3′) |
|---|---|---|
| Primers used for RT-PCR | ||
| GAPDH-F | RT-PCR GAPDH control | GGAATCAACGGTTTCGGTCG |
| GAPDH-R | CAATAAAAGGGTCGTTGA | |
| ACT1-F | RT-PCR ACTIN control | AACATTGTCATGTCTGGTGGT |
| ACT1-R | CGGTGATCTCCTTTTGCATAC | |
| ALL1-RT-for | RT-PCR of ALL1 | CGTTACCCAAGGTGTCAAGGA |
| ALL1-RT-rev | CTTGAGGCCTGATATGCTCCT | |
| Primers used for plasmid construction for homologous recombination | ||
| Neo-F | Amplification of neomycin cassette | CCATATGTTGGTAA AACGACGGCCAGTGAATTGTA |
| Neo-R | CCATGAATTGGCAGGAAACAGCTATGACCATGATT | |
| ALL1-R-For | Amplification of ALL1 upstream nucleotides | CCATTTCTTGGTAACCTGTTGTTCAACGCGAGACTG |
| ALL1-R-Rev | CCATAAATTGGCTTCGACTTCCTTCCGGCTATATTCC | |
| ALL1-L-For | Amplification of ALL1 downstream nucleotides | CCATAGATTGGAGGTTTGGGGTTGATTTCGAAGCT |
| ALL1-L-Rev | CCATCATTTGGCGAAAGTAATGTGATGGACCCAAGA | |
| pUC19-F | Amplification of origin of replication and ampicillin resistance gene | CCATTTTTTGGGAAAGGGCCTCGTGATACGCCT |
| pUC19-R | CCATTCTTTGGGCTTTCCAGTCGGGAAACCTGT | |
| ALL1-F | Complementation of ALL1 | CTCGAGACGGGACCTACAGGTACATTCA |
| ALL1-R | TCTAGAAGAGCGTTTGGAAACACCAC | |
| ALL1p-F | Recombinant ALL1 protein | ATGTCCGGCGTTACCCAAGGT |
| ALL1p-R | TTAAAGAGCCTGGGTCTTGCT |
Disruption of the ALL1 gene.
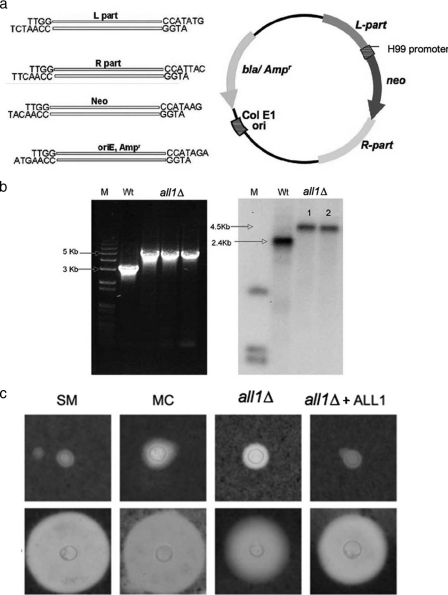
The complete open reading frame of ALL1 was replaced by a neomycin cassette in the SM parent by homologous transformation. Biolistic transformation was performed with a PDS-1000/He hepta system (Bio-Rad) using 5 μg of the purified linear DNA construct. The plasmid construct contained a neomycin cassette under control of the H99 ACT1 promoter and a TRP1 terminator in addition to 1,000 bp up- and downstream of the target open reading frame genomic sequence (see Fig. 2). These elements were amplified from the SM parent genomic template using the appropriate primers (Table 1). The neomycin resistance gene was amplified from plasmid pJAF1 using primers Neo-F and Neo-R. The ampicillin resistance gene with its origin of replication was amplified from the pUC19 plasmid using pUC19-F and pUC19-R. All primers contained a Van91I restriction site to permit one-step directional cloning (Table 1; see Fig. 2). Amplified products were restricted with Van91I and ligated using T4 DNA ligase (New England Biolabs, United States). Clones were selected on LB agar plates containing ampicillin and were confirmed by single digestion with Van91I. For transformation, the plasmid construct was PCR amplified using primers ALL1-L-for and ALL1-R-rev to obtain a linearized product that contained the 5′ and 3′ regions of the target gene flanking the neomycin cassette. Transformants were screened on YPD plates containing 200 μg/ml neomycin, and colonies were screened further by PCR and confirmed by Southern blot analysis.
FIG. 2.
Confirmation and characterization of the all1Δ mutant. (a) Plasmid construct used in this work to generate homologous recombinants. Van91I restriction sites on the individual primers permitted rapid directional cloning. (b) PCR amplification and Southern blotting demonstrated that there was correct homologous recombination in the mutant compared to the SM parent variant (Wt). (c) India ink preparations (magnification, ×1,000) demonstrating differences in capsule size in vitro (upper panel) and in vivo after induction (cells derived from lungs of mice on day 7 after i.t. infection) (lower panel) between the SM, MC, all1Δ, and all1Δ+ALL1 cells.
Complementation of the all1Δ mutant.
The wild-type ALL1 gene was amplified with the native promoter from the SM genomic template with primers ALL1(2)-F and ALL1(2)-R containing XhoI and XbaI restriction sites and was cloned into plasmid pJAF13. The plasmid was linearized using SmaI and randomly inserted into all1Δ cells by biolistic transformation. The all1Δ- and ALL1-positive clones were selected on YPD plates containing 100 μg/ml nourseothricin (Werner Bioagents, Germany). Gene complementation was confirmed by PCR.
Phenotypic characterization.
For measurement of the capsule size, yeast cells grown in SD overnight were suspended in India ink (Becton Dickinson, New Jersey) and visualized at a magnification of ×1,000 with an Olympus AX70 microscope. Images were captured with a QImaging Retiga 1300 digital camera using the QCapture Suite V2.46 software (QImaging, Burnaby, BC, Canada). Capsule measurements were obtained for 25 randomly chosen cells of each strain using Adobe Photoshop 7.0 for Windows, and capsule thickness was calculated using a conversion factor of 45 pixels per μm, as described previously (27, 53). Yeast cells were also stained with MAb 18B7 to the capsular polysaccharide glucuronoxylomannan and visualized with fluorescein isothiocyanate-labeled goat antibody to mouse immunoglobulin G (IgG). The capsular staining patterns were categorized as annular and punctuate (42). Cell wall stability, switching frequencies, doubling times, melanization, and MICs of fluconazole and amphotericin B were determined as previously described (18, 21).
Animal studies.
Female BALB/c mice that were 6 to 8 weeks old and male Fisher rats that weighed 200 to 250 g were obtained from the National Cancer Institute (Bethesda, MD). Mice (10 mice per group) were infected intratracheally (i.t.), intravenously (i.v.), and intraperitoneally (i.p.) as described previously (27) with 104 to 5 × 106 cells. The fungal burden was determined on day 10 and at the time of death by plating homogenized organ suspensions onto Sabouraud's dextrose agar plates. For standard histology and immunohistochemistry, mice were sacrificed and lung tissue was fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS). Tissue sections were stained with hematoxylin and eosin or mucicarmine. To study changes in ICP, Fisher rats were anesthetized and inoculated in the cisterna magna with 100 cells in 100 μl of sterile PBS. ICP was measured on days 7 and 21 by inserting a 23-gauge cannula that was connected to PE-50 tubing and a transducer amplifier. Pressure was recorded as described previously (20).
Production of All1 recombinant protein.
The ALL1 gene was amplified using C. neoformans serotype D cDNA as a template with primers All1p-F and All1p-R. The resulting fragment was then cloned into the pTrcHis Topo TA expression vector (Invitrogen Life Technologies). This vector contained ALL1 cDNA in frame with Xpress and His tags in the 5′ region of the ALL1 gene. The pTrc-His-ALL1 vector was transformed into E. coli strain TOP10 (Invitrogen Life Technologies). Positive clones were used to induce All1 protein (All1p). Briefly, the clone was grown overnight in LB medium containing 50 μg/ml ampicillin at 37°C with shaking at 250 rpm. The next day an overnight culture that was diluted 1:50 was grown to an optical density at 600 nm of 0.4. Protein production was induced by using isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration, 1 mM), and the cells were grown for 4 h. The cells were centrifuged at 4,000 rpm, and the protein was purified using an Ni-nitrilotriacetic acid fast-start kit (Qiagen). Eluted fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blotting. The protein content was quantified using a DC protein assay kit (Bio-Rad, United States).
Localization and detection of All1 by antibody-based detection assays.
For staining assays, C. neoformans cells were grown in YPD to an optical density of 0.2 to 0.7 and fixed in 0.5 ml of 3.75% formaldehyde in PBS for 30 min at room temperature. Cells were washed with 0.5 ml sorbitol citrate buffer (SCB) to remove the formaldehyde. For intracellular staining, spheroplasts were generated by incubating yeast cells in 0.5 ml SCB with 40 mg/ml lysing enzymes for 6 h at 30°C. Cells were washed with SCB to remove the lysing enzyme and permeabilized in 0.5 ml of a solution containing 0.05% saponin in PBS containing 1% BSA (PBS-BSA) for 20 min. The the cells were washed once with 0.5 ml PBS-BSA to remove the saponin. A polyclonal rabbit serum was generated by Covance Immunology Services (Denver, PA) by inoculating a rabbit with purified All1p. The polyclonal serum titer for All1p was determined by using a standard enzyme-linked immunosorbent assay (ELISA). The high-titer polyclonal serum (titer, >1:51,200) was diluted 1:100 in PBS-BSA, and cells were incubated for 1 h at room temperature. The cells were washed five times and stained with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (1:500; Southern Biotech) in PBS-BSA for 1 h at room temperature. Then the cells were washed five times and resuspended in 50 μl PBS-BSA. Ten microliters of cells was added to 2 μl of Vectashield (Vector Laboratories), and images were collected with a TCS SP2 AOBS confocal microscope (Leica, Mannheim, Germany) with a 63× N.A. 1.4 oil immersion objective. Laser lines at 405 and 488 nm for excitation of 4′-6-diamidino-2-phenylindole (DAPI) and Alexa-488 were provided by a diode and an Ar laser. The scanning was sequential line-by-line scanning to eliminate cross talk between fluorophores. For Western blot analysis cytoplasmic and cell wall proteins were extracted from SM ATCC 24067 and all1Δ cells as described previously (43). Extracted proteins were electrophoresed on a sodium dodecyl sulfate gel at 200 V for 40 min and electroblotted onto nitrocellulose (Hybond-ECL; Amersham Biosciences) using a semidry blot system. Western blotting was performed with rabbit polyclonal sera at a titer of 1:10,000 in 5% milk in PBS, and binding was visualized with horseradish peroxidase-conjugated goat anti-rabbit IgG (Southern Biotech) at a titer of 1:20,000 in PBS, followed by detection by chemiluminescence (Perkin-Elmer Life Sciences). Antibody titers in sera of infected mice were measured on plates coated with 50 μg/ml of purified All1p in PBS by using a standard ELISA protocol.
Immunohistochemistry.
Formalin-fixed and paraffin-embedded lung sections were used for analysis. The Mac-3 anti-mouse MAb (Pharmingen) was used, and staining was performed with an M.O.M immunodetection kit (Vector Laboratories) according to the manufacturer's protocol. For negative controls the experiments were performed without addition of the primary antibody. Stained slides were developed with a DAB substrate kit (Vector Laboratories) and analyzed with a light microscope.
Phagocytosis and T-cell proliferation assays.
For in vitro phagocytosis assays, phagocytosis was performed as described above. The cell layers were washed, fixed, and stained after 2 h, and a phagocytosis index, determined by dividing the number of yeast cells in macrophages by the number of macrophages, was determined. BALB/c, C57/BL6, and CD8 knockout mice were immunized with 20 μg of purified All1p or control protein in an equal volume of complete Freund adjuvant (CFA) in the hind footpad. Popliteal lymph nodes were isolated after 7 days, and a single-cell suspension (2.5 × 106 cells/ml) was plated in 96 well-microtiter plates. Control mice were immunized with only CFA. Cells were incubated with increasing doses of All1p (0.001 to 1 μg) or concanavalin A (positive control) for 48 h, and T-cell proliferation was measured using a Via light bioassay kit according to the manufacturer's protocol (Lonza, Rockland, ME). The assays were done in triplicate. To test if All1p is a protective epitope, BALB/c mice were immunized on day 1 i.p. with 20 μg of purified All1p in an equal volume of CFA or were sham immunized. On day 14 mice were boosted with All1p given i.p. with incomplete Freund adjuvant. On day 21 mice were inoculated via the tail vein with 104 SM, MC, or all1Δ cells. Successful immunization was documented by verifying the antibody titers to All1p in immunized mice prior to infection.
Statistical analysis.
SPSS version 8.0 (SPSS Inc., Chicago, IL) was used to generate Kaplan-Meier survival curves and to perform Student's t test, an analysis of variance, and the Kruskal-Wallis test.
RESULTS
ALL1 is differentially regulated in phenotypic switch variants.
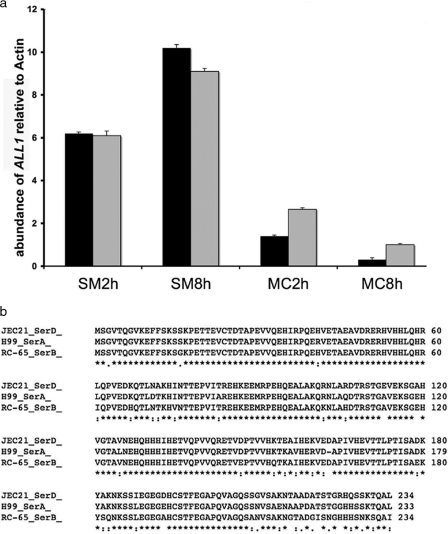
Microarray analysis of SM and MC cells grown at 37°C in SD identified differentially regulated genes that have been described elsewhere (A. Guerrero, N. Jain, E. Cook, A. Casadevall, and B. Fries, presented at the 103rd General Meeting of the American Society for Microbiology, Washington, DC, 2003). Most of these genes encode putative proteins with unknown functions. ALL1 was chosen for further analysis because it was among the most markedly downregulated genes (19.7-fold) in the hypervirulent MC variant compared to SM-phase cells. Differential regulation of ALL1 was also observed during intracellular growth in macrophages by RT-PCR. Following phagocytosis by macrophages, ALL1 was downregulated 3-fold at 2 h and 16.2-fold at 8 h in intracellular MC cells compared to SM cells (Fig. 1a). Homologs of ALL1 with unknown functions are apparent in other fungal genomes.
FIG. 1.
ALL1 expression in SM and MC variants and high levels of homology of the ALL1 protein in C. neoformans serotypes. (a) Comparison of ALL1 in SM and MC variants during intracellular growth in macrophages by using RT-PCR. In two different macrophage experiments (black bar and gray bar) ALL1 was more highly expressed at 2 and 8 h. Expression was measured relative to actin expression, and similar results were obtained with glyceraldehyde-3-phosphate dehydrogenase. Standard deviations were determined by using triplicate measurements for RT-PCR and are indicated by the error bars. (b) High levels of homology of the putative protein sequences in C. neoformans serotype D, A, and B strains.
The ALL1 gene is highly conserved among C. neoformans strains of different serotypes (Fig. 1b). The ALL1 gene is 1,096 bp long, contains four introns, and encodes a putative protein having a molecular mass of 26 kDa (234 amino acids). ALL1 is located on chromosome 6 just upstream of the APP1 gene, which encodes an antiphagocytic protein (35). Furthermore, searches in the C. neoformans cDNA database (http://www.genome.ou.-edu/cneo.html) showed that the ALL1 cDNA has a 63-bp 5′ untranslated region (UTR) and a 104-bp 3′ UTR. These data were consistent for cDNA sequences of three different strains, strains H99, 184, and B3501. A standard promoter prediction program (http://www.fruitfly.org/seq_tools/promoter.html) identified a promoter sequence 208 to 258 bp upstream of the transcription start site. A search with a 960-bp 5′ UTR upstream genomic sequence using matinspector (http://www.genomatix.de/cgi-bin/matinspector) predicted that the earliest transcription binding site is at −142 to −152 bp upstream of the transcription start site (fungal GATA binding factor). Using PROSITE analysis, no motif resemblance could be determined.
Deletion of the ALL1 gene results in capsule alterations.
The ALL1 gene was disrupted and replaced with a neomycin resistance gene in the SM parent by biolistic transformation (Fig. 2a). Seventy-six transformants were obtained, and successful homologous recombination was confirmed in 8 of 20 representative transformants by PCR and Southern blotting (Fig. 2b). The all1 mutant (all1Δ) was complemented by using standard methods in which the wild-type gene was randomly integrated to generate strain all1Δ+ALL1. The phenotype of the all1Δ mutant was characterized and compared to the phenotypes of the SM parent, the MC switch variant, and the complemented strain. The results are summarized in Table 2 and demonstrate that the all1Δ mutant has a modestly enlarged capsule, similar to the MC switch variant. It is noteworthy that the all1Δ mutant switches to a mucoid colony morphology at a rate comparable to the switching rate of SM and MC cells (0.5 to 1 in 104 cells). The mutation was complemented, and the wild-type capsular morphology was restored by reintroduction of the wild-type gene in the all1Δ+ALL1 complemented strain. Although the capsule of the mutant was larger, the mutant was induced significantly (P < 0.001) less in vitro in response to 10% fetal calf serum and CO2 and in vivo than the SM, MC, and reconstituted strains (Fig. 2c). The capsular staining pattern with anticapsule MAb 18B7 was annular and was the same in SM, MC, and all1Δ cells. Melanin production and susceptibility to antifungal agents were also comparable in these strains (data not shown). The growth of the mutant, as assessed by determining the doubling time, was also comparable to the growth of the wild type in both rich and minimal medium.
TABLE 2.
Characteristics and phenotypes of switch variants and mutants
| Characteristica | SM | MC | all1Δ | all1Δ+ALL1 |
|---|---|---|---|---|
| Colony morphology | Smooth | Mucoid | Smooth/mucoid | Smooth |
| Capsule size (μm) | 0.8 ± 0.4 | 2.5 ± 0.4 | 2.0 ± 0.5 | 0.9 ± 0.2 |
| Capsule vol (μm3) | 6.5 ± 14.1 | 60.1 ± 36.4 | 42.5 ± 34.9 | 4.1 ± 4.6 |
| Induced capsule size (μm) | 3.7 ± 0.9 | 4.8 ± 1.4 | 2.5 ± 1.1 | 3.0 ± 1.3 |
| Induced cell capsule vol (μm3) | 272.8 ± 194 | 597.6 ± 569 | 112.8 ± 154 | 160.6 ± 184 |
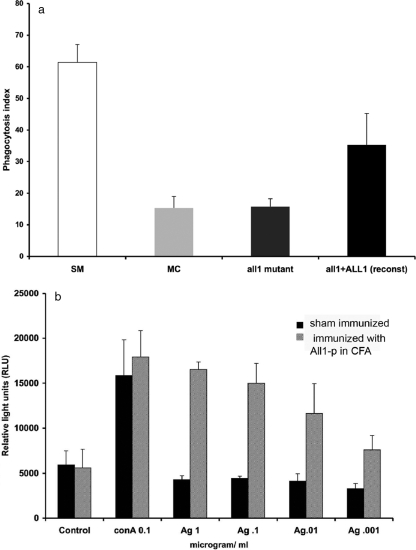
| Phagocytosis index | 61.3 ± 5.6 | 15.3 ± 3.6 | 15.7 ± 2.5 | 35.3 ± 9.8 |
| Phagocytosis index of induced cells | 8.7 ± 1.8 | 4.3 ± 2.8 | 7.1 ± 1.7 | 10.7 ± 5.3 |
| Capsule vol in vivo (μm3) | 2,585 ± 1,744 | 4,071 ± 4,101 | 1,532 ± 1,197 | NDb |
| Capsule induction in vivo (μm) | 8.0 ± 1.8 | 8.6 ± 3.6 | 5.9 ± 1.7 | ND |
| Growth on 3% sodium dodecyl sulfate plates | Normal | Impaired | Normal | Normal |
| Growth on SDA containing 1 M KCl | Normal | Normal | Normal | Normal |
| Growth on SDA containing 1 to 1.8 M NaCl | Normal | Normal | Normal | Normal |
| Growth on YPD containing 3 μg/ml menadione | Normal | Normal | Normal | Normal |
| Growth on YPD containing 10 μM Paraquat | Normal | Normal | Normal | Normal |
| Zone size diam (cm) on YPD containing hydrogen peroxide (3 μl of 30% H2O2) | 2.7 | 2.9 | 2.9 | 2.8 |
| Trichoderma hazaricum lysing enzyme concn (μg/ml) | 48 | 12 | 48 | ND |
| Switching | SM → MC | MC → SM | SM → MC | ND |
| Doubling time in SDA (h) | 2.2 | 2.8 | 2.3 | ND |
| Doubling time in minimal medium (h) | 1.8 ± 0.2 | 3.2 ± 0.8 | 2.7 ± 0.49 | ND |
SDA, Sabouraud's dextrose agar.
ND, not determined.
ALL1 encodes a small cytoplasmic protein.
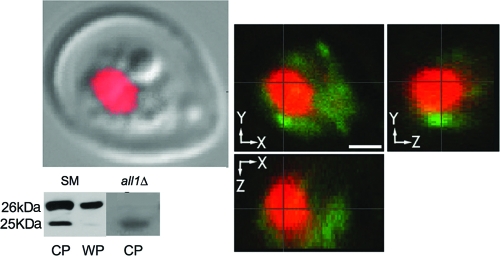
The All1 protein was expressed with a His-tagged system and purified from E. coli. The purified protein was the predicted size, 26 kDa. Purified protein was used to raise polyclonal serum in a rabbit. Both Western blotting and fluorescence staining with polyclonal serum revealed that this protein had a cytoplasmic localization (Fig. 3). Western blot analysis with the polyclonal serum revealed a weakly cross-reacting protein with a lower molecular mass, which is consistent with the predicted 25-kDa protein encoded by a homologous gene in the C. neoformans genome (data not shown). This was confirmed by Western blotting with an antibody specific for this protein. The All1 protein was not shed in the supernatant during in vitro growth, nor cold it be detected by Western blotting in isolated extracellular vesicles. Accordingly, antibody titers in the sera of mice infected by the i.v. or i.t. route were not detected by ELISA, although the protein elicited a strong antibody response in mice and rabbits inoculated with the purified protein (data not shown). Despite the similarities of the All1 protein and the IgE-inducing proteins in Malezzia furfur, we did not detect an IgE response in mice infected with the SM phase variant.
FIG. 3.
Cytoplasmic location of All1 protein. The Western blot is a Western blot of cytoplasmic protein (CP) extracts from SM and all1Δ cells (WP, whole-cell proteins). Membranes were stained with polyclonal serum to All1. Note that there is a smaller 25-kDa cross-reacting band. This is consistent with detection of the second homologous protein encoded by CNM02200. Examination of single optical sections by confocal microscopy showed that red staining and green staining occurred in different compartments of the cell. Green, intracellular staining of All1; red, nuclear staining with DAPI.
By using microarray analysis, we compared the transcriptomes of SM parent cells and all1Δ mutant cells that were grown to log phase at 37°C in SD. Analysis of two microarrays probed in two separate experiments revealed five genes that were differentially regulated (range, 2.0- to 2.8-fold) in the mutant in addition to the ALL1 gene (see the table in the supplemental material). When RT-PCR was used, only one of these genes was confirmed, and it was found to be upregulated 18.8-fold in all1Δ mutant cells compared to SM phase cells. This gene (CNC03730) encodes a small putative protein (300 amino acids) that has high levels of homology with several short-chain dehydrogenases from other fungal species. In summary, ALL1 encodes a small cytoplasmic 26-kDa protein with an unknown function that is not secreted and that does not induce an IgE response.
Loss of ALL1 function alters virulence.
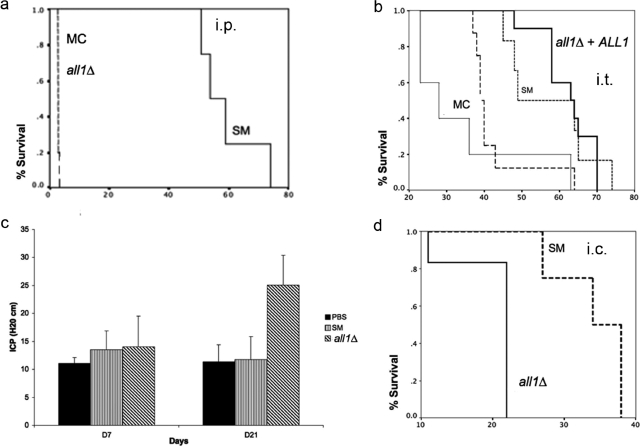
Next, we compared the virulence of the the all1Δ, SM, MC, and all1Δ+ALL1 strains in BALB/c mice using i.p., i.t., and i.v. infection models. The parameters used to measure virulence were organ fungal burden and survival. Similar to the MC variant, the all1Δ mutant was significantly more virulent in i.t., i.p., and cisterna magna infection models. In the i.p. infection model, the median survival time for SM cells was 56 days, compared with 2 days for MC and all1Δ cells (P = 0.003) (Fig. 4a). Also, in the i.t. infection model, the all1Δ mutant was more virulent than the SM parent variant and slightly less virulent than the MC switch variant, as the median survival times were 57, 37, and 30 days for the SM, all1Δ, and MC strains, respectively (P = 0.013) (Fig. 4b). There were no differences in fungal burden at day 10, but there was a significantly lower fungal burden for SM cells at the time of death (log 4.8 ± 0.7 and log 7 ± 0.4 for SM and all1Δ cells, respectively [P < 0.01]). Virulence was enhanced by loss of ALL1 function, and the hypervirulence was alleviated in the all1Δ+ALL1 complemented strain, which exhibited the wild-type virulence level. Hypervirulence was also documented in i.t. infections in another mouse strain (C57/BL6). No difference in virulence was observed in i.v. infection models (data not shown).
FIG. 4.
Virulence of the all1 mutant is enhanced. (a and b) Differences in survival of the SM variant-, all1Δ mutant-, all1Δ+ALL1 mutant-, and MC variant-infected mice in an i.p. and i.t. infection model. (c) Increased ICP in all1Δ mutant-infected rats compared to sham- and SM variant-infected rats. (d) Different ICP results are consistent with premature mortality in all1Δ mutant-infected rats.
Loss of ALL1 function increases ICP.
Next, we examined if the presence of ALL1 affected the development of ICP, because this is a predominant clinical sign associated with poor outcomes in C. neoformans-infected humans and MC variant-infected mice. In addition, previous work in our laboratory documented that the MC variant causes increased ICP in a rat model of cryptococcal meningitis (20). ICP was measured on days 7, 21, and 34 for SM variant- and all1Δ mutant-infected rats. The ICP of all1Δ mutant- and SM variant-infected rats were comparable to the ICP of sham-infected rats on day 7 (14.0 cm H2O ± 5.5 and 13.5 cm H2O ± 3.4 [P = 0.8]), whereas on day 21 the ICP of all1Δ mutant-infected rats were significantly higher than those of SM variant-infected rats (25.0 cm H2O ± 5.3 versus 11.8 cm H2O ± 4.0 [P = <0.001]) (Fig. 4c). Consistent with the 11-cm H2O augmentation of ICP, all1Δ mutant-infected rats died significantly faster than SM variant-infected rats, and the median survival times were 22 and 34 days, respectively (Fig. 4d). Despite the differences in ICP, the brain fungal burdens on day 21 (6.1 ± 0.1 and 6.3 ± 0.4 for SM variant- and all1Δ mutant-infected rats) were comparable. Taken together, the results of the virulence studies with diverse animal models demonstrated that loss of ALL1 function in C. neoformans is associated with significant enhancement of virulence and even leads to increases in ICP.
ALL1 modifies two major components of the host response.
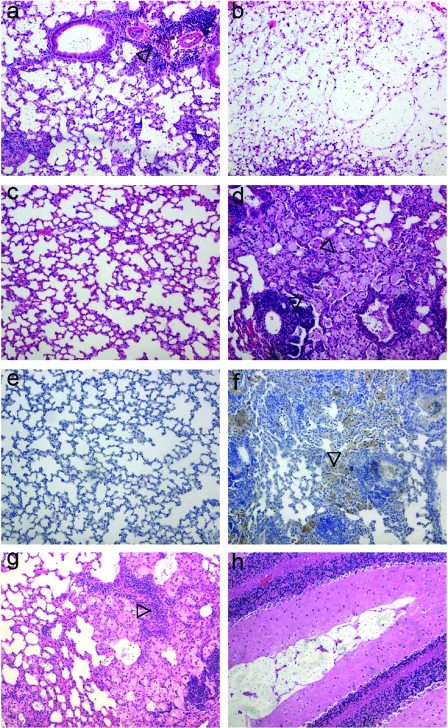
We also examined the inflammatory responses in the lungs and brains of all1Δ mutant-infected mice. Consistent with the absence of a major growth defect, the fungal lung burdens on day 10 were comparable for SM variant- and all1Δ mutant-infected mice (log 4.7 ± 0.4 and log 4.8 ± 0.7, respectively). Therefore, we surmised that differences in virulence reflected an altered host-pathogen interaction that failed to clear the mutant and led to increased damage to the host. This interpretation was supported by a histopathological analysis of infected tissues (Fig. 5). Lung sections obtained on day 10 postinfection exhibited early signs of an organized inflammatory response in SM variant-infected mice (Fig. 5a), whereas all1Δ mutant-infected mice lacked signs of an organized inflammatory response and instead had abundant extracellular C. neoformans in the alveolar spaces (Fig. 5b). By the time of death (day 46 for low-dose infections), all1Δ mutant-infected mice exhibited a disorganized excessive inflammatory response, including robust macrophage recruitment (Fig. 5d). In contrast, SM variant-infected mice cleared the infection from the lungs and exhibited minimal inflammatory changes and intact alveolar microarchitecture by day 46 (Fig. 5c). Immunohistochemistry demonstrated that there was more recruitment of macrophages in all1Δ mutant-infected lungs (Fig. 5f) than in SM variant-infected lungs (Fig. 5e). Interestingly, an SM variant-infected mouse whose infecting strain switched in vivo to a predominantly MC phenotype (as shown by the recovery of mucoid colonies from tissue) exhibited an intense inflammatory response that was similar to the inflammatory response elicited by the all1Δ mutant (Fig. 5g). An inability to clear the all1Δ mutant from tissue was also observed in the rat central nervous system (CNS) infection model, where accumulation of yeast cells in the meningeal spaces was observed only in all1Δ mutant-infected rats (Fig. 5h).
FIG. 5.
Host response is modified in the absence of ALL1. (a and b) Hematoxylin- and eosin-stained and mucicarmine-stained lung tissue of SM variant-infected (a) and all1Δ mutant-infected (b) mice on day 10 after the mice were given 106 cells i.t. Note the beginning of granuloma formation in the SM variant-infected mouse (a) and the extracellular accumulation of yeast cells in the alveolar spaces of the all1Δ mutant-infected mouse (b). At day 46 hematoxylin and eosin staining of lung tissue revealed an excessive but ineffective host response in the all1Δ mutant-infected mice (d), whereas the SM variant-infected mice (c) successfully cleared the infection. (e and f) Immunohistochemistry with a macrophage-specific MAb, MAC3, revealed significantly more macrophages in all1Δ mutant-infected lungs (f) than in SM variant-infected lungs (e). (g) The inflammatory response in an SM variant-infected mouse whose strain switched to a MC phenotype is similar to the inflammatory response in all1Δ mutant-infected mice. (h) Accumulation of yeast cells in the subarachnoid space in an all1Δ mutant-infected rat.
Consistent with extracellular accumulation of the all1Δ mutant in infected tissue, despite enhanced recruitment of macrophages in vitro phagocytosis assays revealed that the all1Δ mutant exhibited reduced phagocytosis, as shown by a lower phagocytosis index (15.7 ± 2.5) compared to the SM variant (61.3 ± 5.6) (Fig. 6a). Hind footpad immunization experiments with purified All1 protein revealed a dose-dependent T-cell response in popliteal lymph nodes, which demonstrates that this protein contains a T-cell epitope (Fig. 6b). Similar All1-induced T-cell proliferation was also seen in CD8 knockout mice (data not shown), suggesting that ALL1 is a CD4 epitope. However, experiments comparing the survival data for mice that were immunized with All1 in CFA and mice that were sham immunized or immunized with control protein and then infected i.v. with SM, MC, and all1Δ cells did not demonstrate that immunization provided protection (data not shown). Thus, we concluded that although ALL1 may be a relevant T-cell epitope, it does not protect animals in this model. Based on these findings, we concluded that loss of all1Δ confers the hypervirulent phenotype of the MC variant. Our data demonstrate that two key components of an effective host response in chronic cryptococcosis are modified, namely, macrophage-mediated phagocytosis and T-cell-mediated immune responses.
FIG. 6.
Phagocytosis and T-cell proliferation are affected by loss of ALL1. (a) The phagocytosis index is significantly (P < 0.05) lower in macrophages incubated with the all1Δ mutant and the MC variant than in macrophages incubated with the SM variant and the all1Δ+ALL1 mutant. The standard deviations for phagocytosis in three wells were determined by Student's t test and are indicated by the error bars. (b) Dose-dependent T-cell proliferation in lymphocytes incubated with All1. The test was done in triplicate, and the error bars indicate the standard deviations. The lymphocytes were isolated from popliteal lymph nodes of mice on day 7 after immunization with CFA and All1. The T-cell proliferation was comparable to concanavalin A (conA)-induced proliferation and was significantly greater (P < 0.05, as determined by Student's t test) than the proliferation of lymphocytes of control antigen (Ag) (protein from empty vector bacterial lysate)-immunized mice.
DISCUSSION
Phenotypic switching occurs during chronic cryptococcal infection and generates hypervirulent MC variants that are selected in the host. Hence, phenotypic switching is a form of pathoadaptation. We identified a virulence-modifying gene, ALL1, which is downregulated in the setting of phenotypic switching. We observed that there was enhancement of virulence as a consequence of loss of function of this gene, which sets this gene apart from the majority of virulence genes that enhance virulence if they are expressed. The loss of ALL1 function mimics the hypervirulent phenotype of the MC switch variant, where the gene is downregulated. Analogous to infection with the MC variant, infection with the all1Δ mutant was associated with differences in the immune response, suggesting a mechanism of enhanced virulence through immunoregulation whereby a disorganized granulomatous response is correlated with reduced survival. All1 is an intracellular protein, and its effect on the immune system appears to occur in part through regulation of C. neoformans capsule production. We present evidence here that the mechanism of immunoregulation involves modification of macrophage-mediated phagocytosis and possibly the presence of a T-cell epitope in this protein that is recognized during the immune response. Failure to clear the pathogen also leads to augmented ICP, which in human cryptococcosis is the major predictor of morbidity and mortality (23).
ALL1 is downregulated when C. neoformans strain RC-2 switches from an SM phenotype to a hypervirulent MC phenotype. This gene is not essential for survival of this yeast in the host niche and, more importantly, is not a regulator of switching as it does not affect the switching rate. Although the precise function of All1 is not known, the cytoplasmic location and capsular morphology of this protein suggest that it is part of a metabolic pathway related to capsule formation. Interestingly, although the polysaccharide capsule of the all1Δ mutant is larger at baseline, it exhibits impaired capsule induction in vivo or in vitro. In other mutants impaired capsule induction has been associated with hypovirulence rather than hypervirulence (4). Interestingly, known capsule genes were not differentially regulated in the all1Δ mutant and the SM parent. Instead, one gene, encoding a short-chain alcohol dehydrogenase homolog, was upregulated 18-fold in the all1Δ mutant. This class of enzymes is heterogeneous in eukaryotes, but most of the enzymes are small proteins (<300 amino acids), like the proteins encoded by ALL1 and CN03730 (303 amino acids). They can play roles in a broad spectrum of metabolic processes. It is conceivable that CN03730 has a similar function and therefore compensates for the loss of ALL1 in the all1Δ mutant.
Survival studies with mice and rats demonstrated that enhanced virulence was the result of loss of ALL1. Downregulation of this gene in the parent strain significantly augmented the already virulent phenotype (SM) of this strain (infection of mice results in death within 60 to 70 days when the dose is 104 cells). The cause of death in mice is only partially understood and depends on the infection model. In humans elevated ICP is associated with organic brain syndrome, blindness, deafness, and acute mortality in patients with advanced AIDS (23). Previous studies demonstrated that MC variant-infected rats, but not SM variant-infected rats, developed high ICP (20), implying that pathogen-related characteristics were responsible for the development of ICP. We demonstrate here that loss of ALL1 function affects ICP and decreases survival. Also, in the pulmonary model, dissemination to the CNS occurs, and mice die with hunched backs and gait disturbances, both of which are clinical symptoms of meningitis and increased ICP. Despite a comparable number of lung CFU at day 10, histological analysis of lung tissue demonstrated that there were already signs of a disturbed host response with disorganized granuloma formation and extracellular accumulation of yeast. Consistent with this ineffective immune response, the number of lung CFU in the all1Δ mutant-infected animals progressively increased, whereas the CFU were cleared in SM variant-infected mice.
Based on human autopsy studies and murine models of infection, the alveolar macrophage is a primary phagocytic cell in pulmonary cryptococcosis (13). Alveolar macrophages are able to kill yeasts inoculated into the bronchial alveolar system. Phagocytosis is required for epitope presentation and links the innate immune response to CMI. In this respect proliferation of lymphocytes in response to C. neoformans correlates with the magnitude of phagocytosis (48, 49). The all1Δ mutant has an enlarged capsule, which impairs phagocytosis. As reported for the MC variant (38), other aspects of the capsular polysaccharide, such as the distribution of charged residues, could also be altered and affect phagocytosis. In addition, our immunization studies indicate that All1p can elicit T-cell proliferation in a dose-dependent matter. Although CMI is crucial for an effective host response to C. neoformans, the antigen specificity of the T-cell response to C. neoformans appears to be diverse (33) and derived mostly from culture supernatants (1, 26, 32, 36, 40, 41, 47). All1 is not shed into the extracellular space, but after phagocytosis, macrophages could present this epitope to T cells. Although our immunization studies failed to show that All1 is protective, T-cell assays have suggested that a T-cell epitope is present, and more detailed studies are required to determine the precise role of All1 with respect to CMI.
Compared with other hypervirulent mutants of C. neoformans, such as cas1Δ, pkr1Δ, sch9Δ, crg1Δ, and app1Δ mutants (12, 28, 35, 50, 51), the all1Δ mutant has a distinct phenotype. The PKR1 gene encodes the regulatory subunit of protein kinase A and is part of the Gα protein cyclic AMP-protein kinase A signaling pathway, which regulates several virulence traits (25). The size of the polysaccharide capsule of both the sch9Δ and pkr1Δ mutants dramatically increases in vivo, and mortality is associated with large differences in the number of CFU in the brain. In contrast, the capsule size of the all1Δ mutant is well within the variation observed in clinical strains, and the microarray data indicate that ALL1 does not regulate other virulence-associated genes. The crg1Δ mutant also has only a mildly enlarged capsule; however, both melanization and mating are affected in this mutant (51). The cas1Δ hypervirulent mutant exhibits changes in the binding of MAbs to the capsule, whereas the all1Δ mutant does not. Finally, the app1Δ mutant exhibits enhanced phagocytosis and is hypervirulent only in mice deficient for T cells and NK cells (Tgɛ26). The ALL1 and APP1 genes are closely linked in the genome, but loss of ALL1 does not alter APP1 expression, suggesting that the hypervirulent mutant phenotypes are attributable to distinct mechanisms.
Although the immunocompromised state of the host contributes to treatment failure, it is also evident that delayed clearance by a weakened host promotes pathoadaptation of the fungus in the host niche. This can result in virulence differences in mice, which have been demonstrated for passaged strains and for serial patient isolates (2, 5, 6, 8, 15, 17, 19). Although the abilities of mouse models of cryptococcosis to predict strain-related virulence in humans have not been validated, a recent study elegantly demonstrated that a high level of CTR4 expression was correlated with dissemination to the CNS of humans, whereas a low level of expression was not correlated with dissemination (52). In addition, the inflammatory response in patients with cryptococcal meningoencephalitis and comparable levels of immunosuppression is variable, suggesting that both host- and pathogen-related factors contribute to the pathogenesis and differences in outcome (31).
Chronic infectious diseases exhibit complex pathogenesis in part because the host-pathogen interaction is dynamic. The RC-2 C. neoformans switching strain allowed us to identify virulence determinants in an isogenic background that changed during chronic infection and enhanced fitness in the host niche. Dynamic virulence factors that are regulated during chronic infection are likely to be highly relevant. Because such factors are often epigenetically regulated, they may not be detected in classic genetic screens. In the white-opaque switching strain of C. albicans more than 350 genes are differentially upregulated and downregulated in the two switch variants (30), and no single gene can confer the white or opaque phenotype. In contrast, loss of ALL1 can result in a phenotype that mimics the hypervirulent phenotype to a large extent.
Relatively few loci whose loss confers increased virulence have been described for pathogens, and the concept of antivirulence genes remains controversial (3, 14, 37). One example involves mutations in the CsrR/CsrS two-component regulatory system in Streptococcus pneumoniae, which represses synthesis of multiple virulence factors, including the capsule. Mutants are more virulent and spontaneously arise in vivo because they are selected (24). Also, in Leishmania major downregulation of the pteridine reductase (PTR1) reduces H4B levels, which is responsible for elevated metacyclogenesis and virulence. It was hypothesized that expression of the PTR1gene, which limits virulence, was maintained because the parasite needs the host to survive in order to complete its complex life cycle (7).
In C. albicans the white and opaque phenotypes are maintained over generations via complex interlocking transcriptional feedback loops (54, 55). The selective force behind maintaining the complex regulation of these epigenetic phenotypes is that they are required for mating of C. albicans. The cost of the loss of ALL1 function is presently not known, but in the host niche there seems to be no impairment of fitness. The epigenetic state of loss (or downregulation) of ALL1 specifically in the MC variant has a biological advantage in the host niche and is selected. For C. neoformans, the human host is not required for survival; hence, it is still unclear why epigenetic downregulation of this gene was maintained in evolution. Virulence in C. neoformans is probably a serendipitous consequence of an intrinsic ability of this environmental microbe to adapt and survive in certain animal hosts. To better understand the virulence strategy of this pathogen, future studies will focus on understanding the epigenetic regulation of phenotypic variability.
Supplementary Material
Acknowledgments
We thank Jim Kronstad for help with sequence analysis of C. neoformans var. gattii, Micheal Cammer for assistance with staining of intracellular proteins, and A. Casadevall for critical reading of the manuscript.
This work was supported by grant RO1 AI059681-04.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 27 October 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Biondo, C., C. Beninati, D. Delfino, M. Oggioni, G. Mancuso, A. Midiri, M. Bombaci, G. Tomaselli, and G. Teti. 2002. Identification and cloning of a cryptococcal deacetylase that produces protective immune responses. Infect. Immun. 702383-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackstock, R., K. L. Buchanan, A. M. Adesina, and J. W. Murphy. 1999. Differential regulation of immune responses by highly and weakly virulent Cryptococcus neoformans isolates. Infect. Immun. 673601-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall, A., and L. A. Pirofski. 2003. “Anti-virulence” genes—further muddling the lexicon? Response from Arturo Casadevall and Liise-anne Pirofski. Trends Microbiol. 11413-414. [DOI] [PubMed] [Google Scholar]
- 4.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 641977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L. C., and A. Casadevall. 1999. Variants of a Cryptococcus neoformans strain elicit different inflammatory responses in mice. Clin. Diagn. Lab. Immunol. 6266-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherniak, R., L. C. Morris, T. Belay, E. D. Spitzer, and A. Casadevall. 1995. Variation in the structure of glucuronoxylomannan in isolates from patients with recurrent cryptococcal meningitis. Infect. Immun. 631899-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham, M. L., R. G. Titus, S. J. Turco, and S. M. Beverley. 2001. Regulation of differentiation to the infective stage of the protozoan parasite Leishmania major by tetrahydrobiopterin. Science 292285-287. [DOI] [PubMed] [Google Scholar]
- 8.Currie, B., H. Sanati, A. S. Ibrahim, J. E. Edwards, Jr., A. Casadevall, and M. A. Ghannoum. 1995. Sterol compositions and susceptibilities to amphotericin B of environmental Cryptococcus neoformans isolates are changed by murine passage. Antimicrob. Agents Chemother. 391934-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deitsch, K. W., E. R. Moxon, and T. E. Wellems. 1997. Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiol. Mol. Biol. Rev. 61281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domergue, R., I. Castano, A. De Las Penas, M. Zupancic, V. Lockatell, J. R. Hebel, D. Johnson, and B. P. Cormack. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308866-870. [DOI] [PubMed] [Google Scholar]
- 11.Dromer, F., S. Mathoulin-Pelissier, O. Launay, and O. Lortholary. 2007. Determinants of disease presentation and outcome during cryptococcosis: the CryptoA/D study. PLoS Med. 4e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 213179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldmesser, M., Y. Kress, P. Novikoff, and A. Casadevall. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 684225-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foreman-Wykert, A. K., and J. F. Miller. 2003. Hypervirulence and pathogen fitness. Trends Microbiol. 11105-108. [DOI] [PubMed] [Google Scholar]
- 15.Franzot, S. P., J. Mukherjee, R. Cherniak, L. C. Chen, J. S. Hamdan, and A. Casadevall. 1998. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect. Immun. 6689-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser, J. A., R. L. Subaran, C. B. Nichols, and J. Heitman. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 21036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fries, B. C., and A. Casadevall. 1998. Serial isolates of Cryptococcus neoformans from patients with AIDS differ in virulence for mice. J. Infect. Dis. 1781761-1766. [DOI] [PubMed] [Google Scholar]
- 18.Fries, B. C., E. Cook, X. Wang, and A. Casadevall. 2005. Effects of antifungal interventions on the outcome of experimental infections with phenotypic switch variants of Cryptococcus neoformans. Antimicrob. Agents Chemother. 49350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fries, B. C., D. L. Goldman, and A. Casadevall. 2002. Phenotypic switching in Cryptococcus neoformans. Microbes Infect. 41345-1352. [DOI] [PubMed] [Google Scholar]
- 20.Fries, B. C., S. C. Lee, R. Kennan, W. Zhao, A. Casadevall, and D. L. Goldman. 2005. Phenotypic switching of Cryptococcus neoformans can produce variants that elicit increased intracranial pressure in a rat model of cryptococcal meningoencephalitis. Infect. Immun. 731779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fries, B. C., C. P. Taborda, E. Serfass, and A. Casadevall. 2001. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J. Clin. Investig. 1081639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldman, D. L., B. C. Fries, S. P. Franzot, L. Montella, and A. Casadevall. 1998. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc. Natl. Acad. Sci. USA 9514967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graybill, J. R., J. Sobel, M. Saag, C. van Der Horst, W. Powderly, G. Cloud, L. Riser, R. Hamill, and W. Dismukes. 2000. Diagnosis and management of increased intracranial pressure in patients with AIDS and cryptococcal meningitis. The NIAID Mycoses Study Group and AIDS Cooperative Treatment Groups. Clin. Infect. Dis. 3047-54. [DOI] [PubMed] [Google Scholar]
- 24.Heath, A., A. Miller, V. J. DiRita, and C. N. Engleberg. 2001. Identification of a major, CsrRS-regulated secreted protein of group A streptococcus. Microb. Pathog. 3181-89. [DOI] [PubMed] [Google Scholar]
- 25.Hu, G., B. R. Steen, T. Lian, A. P. Sham, N. Tam, K. L. Tangen, and J. W. Kronstad. 2007. Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog. 3e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, C., S. H. Nong, M. K. Mansour, C. A. Specht, and S. M. Levitz. 2002. Purification and characterization of a second immunoreactive mannoprotein from Cryptococcus neoformans that stimulates T-cell responses. Infect. Immun. 705485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain, N., L. Li, D. C. McFadden, U. Banarjee, X. Wang, E. Cook, and B. C. Fries. 2006. Phenotypic switching in a Cryptococcus neoformans variety gattii strain is associated with changes in virulence and promotes dissemination to the central nervous system. Infect. Immun. 74896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozel, T. R., S. M. Levitz, F. Dromer, M. A. Gates, P. Thorkildson, and G. Janbon. 2003. Antigenic and biological characteristics of mutant strains of Cryptococcus neoformans lacking capsular O acetylation or xylosyl side chains. Infect. Immun. 712868-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lachke, S. A., S. R. Lockhart, K. J. Daniels, and D. R. Soll. 2003. Skin facilitates Candida albicans mating. Infect. Immun. 714970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan, C. Y., G. Newport, L. A. Murillo, T. Jones, S. Scherer, R. W. Davis, and N. Agabian. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 9914907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, S. C., D. W. Dickson, and A. Casadevall. 1996. Pathology of cryptococcal meningoencephalitis: analysis of 27 patients with pathogenetic implications. Hum. Pathol. 27839-847. [DOI] [PubMed] [Google Scholar]
- 32.Levitz, S. M., S. Nong, M. K. Mansour, C. Huang, and C. A. Specht. 2001. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 9810422-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindell, D. M., M. N. Ballinger, R. A. McDonald, G. B. Toews, and G. B. Huffnagle. 2006. Diversity of the T-cell response to pulmonary Cryptococcus neoformans infection. Infect. Immun. 744538-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lockhart, S. R., R. Zhao, K. J. Daniels, and D. R. Soll. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luberto, C., B. Martinez-Marino, D. Taraskiewicz, B. Bolanos, P. Chitano, D. L. Toffaletti, G. M. Cox, J. R. Perfect, Y. A. Hannun, E. Balish, and M. Del Poeta. 2003. Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J. Clin. Investig. 1121080-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansour, M. K., L. E. Yauch, J. B. Rottman, and S. M. Levitz. 2004. Protective efficacy of antigenic fractions in mouse models of cryptococcosis. Infect. Immun. 721746-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurelli, A. T. 2007. Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol. Lett. 2671-8. [DOI] [PubMed] [Google Scholar]
- 38.McFadden, D. C., B. C. Fries, F. Wang, and A. Casadevall. 2007. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot. Cell 61464-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110293-302. [DOI] [PubMed] [Google Scholar]
- 40.Mody, C. H., C. J. Wood, R. M. Syme, and J. C. Spurrell. 1999. The cell wall and membrane of Cryptococcus neoformans possess a mitogen for human T lymphocytes. Infect. Immun. 67936-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy, J. W., and N. Pahlavan. 1979. Cryptococcal culture filtrate antigen for detection of delayed-type hypersensitivity in cryptococcosis. Infect. Immun. 25284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nussbaum, G., W. Cleare, A. Casadevall, M. D. Scharff, and P. Valadon. 1997. Epitope location in the Cryptococcus neoformans capsule is a determinant of antibody efficacy. J. Exp. Med. 185685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson, G. M., D. S. Fox, P. Wang, J. A. Alspaugh, and K. L. Buchanan. 2007. Role of protein O-mannosyltransferase Pmt4 in the morphogenesis and virulence of Cryptococcus neoformans. Eukaryot. Cell 6222-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perfect, J. R., and A. Casadevall. 2002. Cryptococcosis. Infect. Dis. Clin. N. Am. 16837-874, v-vi. [DOI] [PubMed] [Google Scholar]
- 45.Slutsky, B., J. Buffo, and D. R. Soll. 1985. High-frequency switching of colony morphology in Candida albicans. Science 230666-669. [DOI] [PubMed] [Google Scholar]
- 46.Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller, and D. R. Soll. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Specht, C. A., S. Nong, J. M. Dan, C. K. Lee, and S. M. Levitz. 2007. Contribution of glycosylation to T cell responses stimulated by recombinant Cryptococcus neoformans mannoprotein. J. Infect. Dis. 196796-800. [DOI] [PubMed] [Google Scholar]
- 48.Syme, R. M., T. F. Bruno, T. R. Kozel, and C. H. Mody. 1999. The capsule of Cryptococcus neoformans reduces T-lymphocyte proliferation by reducing phagocytosis, which can be restored with anticapsular antibody. Infect. Immun. 674620-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Syme, R. M., J. C. Spurrell, L. L. Ma, F. H. Green, and C. H. Mody. 2000. Phagocytosis and protein processing are required for presentation of Cryptococcus neoformans mitogen to T lymphocytes. Infect. Immun. 686147-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, P., G. M. Cox, and J. Heitman. 2004. A Sch9 protein kinase homologue controlling virulence independently of the cAMP pathway in Cryptococcus neoformans. Curr. Genet. 46247-255. [DOI] [PubMed] [Google Scholar]
- 51.Wang, P., J. Cutler, J. King, and D. Palmer. 2004. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot. Cell 31028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterman, S. R., M. Hacham, G. Hu, X. Zhu, Y. D. Park, S. Shin, J. Panepinto, T. Valyi-Nagy, C. Beam, S. Husain, N. Singh, and P. R. Williamson. 2007. Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J. Clin. Investig. 117794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaragoza, O., B. C. Fries, and A. Casadevall. 2003. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2. Infect. Immun. 716155-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zordan, R. E., D. J. Galgoczy, and A. D. Johnson. 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. USA 10312807-12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zordan, R. E., M. G. Miller, D. J. Galgoczy, B. B. Tuch, and A. D. Johnson. 2007. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 5e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.