Abstract
We previously identified Rbf as an activator for biofilm formation on polystyrene surfaces in Staphylococcus aureus strain 8325-4. However, strain 8325-4 contains genetic mutations that may affect biofilm formation. To extend the observation to other strains, we used strain Newman, a weak biofilm producer, and strain UAMS-1, an osteomyelitis clinical strain, in this study. We found that mutations in the chromosomal rbf gene did not affect biofilm formation on polystyrene surfaces in these strains, but transformants of these strains carrying a multiple-copy plasmid containing the rbf gene formed stronger biofilms than the wild-type strains and the mutant strains. Using the flow cell method, we found that the chromosomal mutation in the rbf gene delayed biofilm formation, whereas strains with a plasmid containing the rbf gene accelerated biofilm formation in strains Newman and UAMS-1. These results led us to conclude that rbf is an activator of biofilm formation in different strains of S. aureus, although the degree of activation varies among strains. In a murine model of foreign body infection, the rbf mutations in strain Newman, but not in strain UAMS-1, reduced the bacterial survival rate in catheter lumen. However, UAMS-1 carrying multiple copies of rbf in a plasmid increased the bacterial survival rate. The animal studies therefore suggest that Rbf has a role in S. aureus virulence.
Staphylococcus aureus causes a wide range of acute and chronic human infections. The organism is capable of attaching to solid surfaces of native tissues or artificial devices, thereby forming a biofilm matrix that encases the bacteria. The bacteria within the biofilm matrix are protected from the host immune system and from antibiotic attack. As a result, biofilm formation is a major virulence determinant for chronic infections caused by S. aureus. The mechanism of biofilm formation appears to involve the initial attachment of bacteria to solid surfaces, followed by the accumulation of multilayered cell clusters in a slime matrix (11, 16). Poly-N-acetyl-glucosamine, also known as polysaccharide intracellular adhesin (PIA), has been found to be the main component of biofilm in the closely related species Staphylococcus epidermidis, in which the PIA was discovered (16). In S. aureus, the role of PIA in biofilm formation is not universal since PIA-independent biofilm formation mechanisms have been reported in several strains (3, 12, 29, 33-35). The genetics of PIA biosynthesis have been well studied. Four genes, icaADBC in an operon, are required for the biosynthesis of PIA (17, 25). Recently, DNA and some proteins have been implicated as components of the biofilm matrix in certain strains, some of which do not produce detectable PIA (18, 31-33, 35).
Recent studies on biofilm regulation have suggested a complex regulatory mechanism. In S. aureus, the ica genes are negatively regulated by icaR, tcaR, codY, and spx but positively regulated by sarA (2, 10, 19, 26, 30, 36, 38). Several other regulatory loci that regulate biofilm formation have also been reported. These include arlRS (34), clp proteases (9, 14), agr (2, 5, 39, 40), and rbf (23). In addition, mgrA and ssrA have been identified by transposon mutagenesis as potential regulators affecting biofilm formation (37). How these regulators affect biofilm formation has not been elucidated. Previously, we reported the identification of the rbf regulatory locus that positively affects biofilm formation in response to sodium chloride and glucose in laboratory strain 8325-4 (23). However, whether Rbf affects biofilm formation in other strains is not known. In addition, whether Rbf plays a role in S. aureus virulence has not been investigated. In the present study, we found that Rbf was involved in biofilm formation in strains Newman and UAMS-1. We also showed that Rbf contributed to bacterial survival in a murine foreign body infection model.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. aureus strains and plasmids used in the present study are listed in Table 1. Bacterial strains were routinely cultivated in Trypticase soy medium (Difco Laboratories, Detroit, MI) with appropriate antibiotic selection when necessary. Antibiotics used for selection were tetracycline at 3 μg/ml, chloramphenicol at 10 μg/ml, and erythromycin at 10 μg/ml. S. aureus RN4220 was used as a recipient for electroporation by the procedure of Kraemer and Iandolo (20). Phage 52A or 80α was used for plasmid and chromosomal DNA transduction between S. aureus strains.
TABLE 1.
S. aureus strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| 8325-4 | Prophage-free laboratory strain | J. Iandolo |
| RN4220 | 8325-4r− | 21 |
| Newman | Wild-type strain | T. Foster |
| UAMS-1 | S. aureus clinical isolate | 15 |
| CYL385 | 8325-4 Δspa rbf::Tn917 | 23 |
| CYL6938 | Newman(pLI50) | This study |
| CYL6932 | Newman(pYL8565) | This study |
| CYL6633 | Newman rbf::Tn917 | This study |
| CYL6962 | Newman Δrbf | This study |
| CYL6969 | Newman Δrbf(pLI50) | This study |
| CYL6975 | Newman Δrbf(pYL8565) | This study |
| CYL6933 | UAMS-1(pYL8565) | This study |
| CYL6965 | UAMS-1 Δrbf | This study |
| CYL6939 | UAMS-1(pLI50) | This study |
| CYL6970 | UAMS-1 Δrbf(pLI50) | This study |
| CYL6976 | UAMS-1 Δrbf(pYL8565) | This study |
| Plasmids | ||
| pGEM-T Easy | Cloning vector | Promega |
| pKOR1 | Vector for allele replacement | 1 |
| pYL8565 | 3.3-kb fragment containing rbf in pLI50 | 23 |
Allele replacement of rbf.
The pKOR1 system (1) was used to construct rbf deletion mutants. A PCR fragment with a 1,323-bp deletion within the rbf gene was first constructed by restriction digestion and ligation of two PCR fragments containing the 5′ region and the 3′ region of the rbf gene, respectively, that had been cloned in the plasmid pGEM-T Easy (Promega, Madison, WI). The fragment containing the rbf 5′ region was amplified by using the primers rbf19 (5′-ACGCTGTAGTGATTGGCTTA-3′) and rbf20 (5′-ATACCGCGGCGCGTTGTCGCATATTCATT-3′), whereas the fragment containing the rbf 3′ region was amplified by using the primers adh15 (5′-GGGCCCAAGCGACTTAAATTCGATTCGT-3′) and adh16 (5′-GTATTCAAAATGTTGCACCCC-3′). An attB-flanked PCR fragment containing the 1,323-bp rbf internal deletion was then amplified by using the primers attB1-rbf19 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACGCTGTAGTGATTGGCTTA-3′) and attB2-adh16 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTATTCAAAATGTTGCACCCC-3′). The PCR fragment containing the deletion was then cloned into the plasmid pKOR1 using the Gateway BP Clonase system (Invitrogen, Carlsbad, CA) and transformed into Escherichia coli DH5α (Invitrogen). The resultant plasmid was purified and electroporated into S. aureus RN4220 and then transduced into strains Newman and UAMS-1. Allele replacement in these strains was carried out as described by Bae and Schneewind (1). The mutations were confirmed by PCR analyses.
Biofilm assay.
Biofilm assays on polystyrene were performed on 96-well microtiter plates as described by Lim et al. (23) with minor modifications. Overnight cultures were diluted by adjusting the optical density at 660 nm to 0.05 with TSB medium enriched with 0.5% glucose and 3% NaCl. A 200-μl diluted sample was applied to each well of the 96-well plate (in quadruplicate), followed by incubation for 24 h at 37°C, and the plates were washed three times with sterile phosphate-buffered saline, air dried, fixed with ethanol, and air dried again. Biofilms were stained with 0.4% crystal violet for 2 min. The plates were then washed 10 times with deionized water to remove unbound crystal violet and then air dried. The biofilm bound crystal violet was extracted with 200 μl of extraction solution containing 40% ethanol and 10% acetic acid for 15 min with shaking. The extracts were diluted 10-fold with deionized water, and the absorbance at 590 nm was recorded with a Molecular Device SpectraMax M2 plate reader using SoftMax Pro Software 4.6.
Biofilm assays in the flow cell system were performed using a disposable three-channel flow cell apparatus (Stovall Life Sciences, Inc., Greensboro, NC) as described by Cassat et al. (8) with some modifications. Briefly, the tubes were clamped, and each channel was injected with 5 × 108 CFU from overnight cultures (grown in the same medium) of the wild-type strain, the rbf deletion mutant, or the complemented strain in either Newman or UAMS-1 background. The flow cells were then incubated at 37°C in an inverted position for 1 h to promote initial attachment. After the flow cells were turned to the upright position, the flow of medium was initiated by adjusting the rate to 1.6 ml per min such that the entire volume of medium in each channel was replaced once every 20 s. The development of biofilm was recorded with a time-lapse camera set at intervals of 30 min.
Murine foreign body infection model.
To assess the role of rbf in virulence, the murine model of foreign body infection described by Beenken et al. (3) was followed closely. Briefly, 6- to 8-week-old male NIH-Swiss mice were obtained from Harlan Sprague-Dawley, Inc. (Indianapolis, Ind.). The mice were anesthetized with 1.25% 2-2-2 tribromoethanol (Avertin; 0.025 ml/g of body weight) and implanted subcutaneously with 1-cm segments of size Fr-10 polyvinyl chloride catheter in the dorsal area. The wound was closed with surgical glue. Infection was initiated 1 day after the implantation procedure by injecting ∼107 CFU of the test strain into the lumen of the catheter. At various time points up to 21 days, mice were euthanized, and the catheters were removed aseptically and washed briefly with phosphate-buffered saline. Catheters were placed in 1 ml of sterile phosphate-buffered saline and sonicated to remove the adherent bacteria. The number of bacteria in the sonicate was determined by plating on tryptic soy agar (TSA). Selected bacterial colonies were tested for lysostaphin sensitivity and antibiotic sensitivity and further confirmed by PCR analyses.
Real-time RT-PCR.
The expression of rbf was assessed by real-time reverse transcription-PCR (RT-PCR) using the primer pair ACGCGTTGCCAAGATGGCATAGTCTT and AGCCTAATTCCGCAAACCAATCGCTA as described previously (24).
Statistics.
Data from animal studies and biofilm assays on polystyrene were analyzed with an unpaired Student t test for comparisons between means. The data from biofilm assays using flow cell were analyzed by using an unpaired t test with Welch's correction. P values of <0.05 were considered statistically significant.
RESULTS
Plasmid-encoded Rbf promotes biofilm formation on polystyrene in strains Newman and UAMS1.
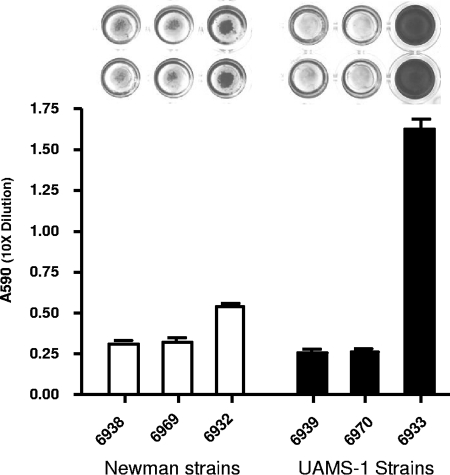
We have preciously used strain 8325-4 to identify Rbf as a regulator for biofilm formation (23). However, strain 8325-4 is sigB deficient and has been maintained as a laboratory strain for a long period of time. To test whether Rbf plays a role in biofilm formation in strains other than 8325-4, we used strain Newman, a sigB-positive strain, and UAMS-1, a recent clinical osteomyelitis strain. To this end, we transduced the rbf::Tn917 mutation from 8325-4 to Newman by phage transduction. In addition, we constructed rbf-null mutants with a 1,323-bp internal segment of the 2,148-bp rbf gene deleted from the chromosomes of Newman and UAMS-1. These strains were tested for biofilm formation in polystyrene microtiter plates with or without precoating with 20% human plasma (precoating with human plasma has been shown to enhance biofilm formation in UAMS-1) (2). The results showed that the chromosomal rbf mutations did not affect biofilm formation in either strain on microtiter plates, whether or not the plates were precoated with plasma (data not shown). However, complementing the mutant strains or the wild-type strains with the plasmid pYL8565 resulted in stronger biofilm formation in both UAMS-1 and Newman backgrounds compared to the wild type or the rbf mutant (P < 0.01 for both backgrounds) (Fig. 1). To confirm that rbf is overly expressed in the complemented strains, we performed real-time RT-PCR experiments. The results showed that the complemented strain produced about 51.7- and 21.52-fold-higher amounts of rbf expression than the wild-type strain in UAMS-1 and Newman backgrounds, respectively. Thus, these results indicate that overexpression of rbf from the multiple-copy pYL8565 plasmid causes Newman, a weak biofilm producer, to form moderate amounts of biofilm and promotes strong biofilm formation in UAMS-1 even without precoating with plasma. Our results therefore suggest that Rbf is also an activator for biofilm formation in these strains.
FIG. 1.
(Top) Biofilm formation on polystyrene microtiter plates. Only two wells of each strain are shown. (Bottom) Quantitative determination of biofilm formation by CYL6938, Newman(pLI50); CYL6969, Newman Δrbf(pLI50); CYL6932, Newman(pYL8565); CYL6939, UAMS-1(pLI50); CYL6970, UAMS-1 Δrbf(pLI50); CYL6933, and UAMS-1(pYL8565). The dye in each well of the microtiter plate was eluted and quantitated by spectrophotometry. Error bars indicate standard deviations from four independent measurements.
Rbf enhances biofilm formation in flow cells.
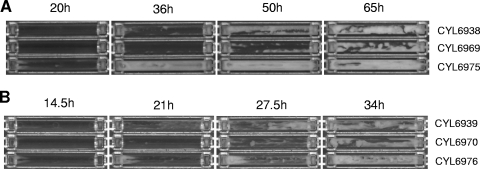
Although the polystyrene microtiter plate assay is a simple means for testing biofilm formation, it measures biofilm formation under static conditions but not in a hydrodynamic environment. To demonstrate further the role of Rbf on biofilm formation in UAMS-1 and Newman under a flow condition, we used the flow cell method as described in Materials and Methods. Two independent experiments were performed, and the results at selected time points in one of the experiments are shown in Fig. 2. Under our experimental condition, on average, strain Newman began to form biofilm at about 22.5 ± 0.5 h, and the biofilm peaked at about 46 ± 3.0 h. In contrast, the Newman rbf mutant began to form biofilm at about 29 ± 0 h and took 59.5 ± 5.5 h to reach the peak. The complemented strain began to form biofilm at about 10.25 ± 1.25 h and reached the peak at 33 ± 3.0 h (Fig. 2A). There were significant differences in the length of time for biofilm initiation when the wild type was compared to the mutant (P = 0.0244) and with the complemented stain (P = 0.0348), but there was no significant difference on the length of time to reach the peak among these strains. However, at the peak, we found that the complemented strain filled up the flow chamber almost completely, while the mutant strain filled up the least among the three strains, indicating that the complemented strain produced the most amounts, followed by the wild type, and the mutant produced the least. In UAMS-1, the average times to initiate biofilm from two independent experiments for the wild type, the mutant, and the complemented strain were 10.75 ± 0.25 h, 14.5 ± 0.5 h, and 6.0 ± 0 h, respectively; to reach the peak, the respective times were 32 ± 0.5 h, 34 ± 2.0 h, and 30.25 ± 0.75 h (Fig. 2B). The length of time for biofilm initiation in the wild type was significantly different from that of the mutant (P = 0.0471) and from that of the complemented stain (P = 0.0168), whereas the times to reach the peak were not significantly different among these strains. As in Newman, the complemented strain produced the most biofilm, and the mutant produced the least at the peak. Thus, these results are similar between the UAMS-1 and the Newman backgrounds. However, the time to initiate biofilm formation and to reach maximal quantity was generally shorter in UAMS-1 genetic background. In addition, the differences between strains, especially the time to reach the peak, were greater in the Newman background than in the UAMS-1 background. Taken together, these data showed that the rbf mutation resulted in not only a delay in forming biofilm, especially the time to initiate biofilm formation, but also a reduction in biofilm quantity, indicating that rbf is involved in the regulation of biofilm formation in the two strains. In addition, we also noted that the biofilms of the complemented strains of both Newman and UAMS-1 took a much longer time to detach than those formed by the wild type or the mutant, suggesting that Rbf may enhance attachment or strengthen biofilm structure.
FIG. 2.
Biofilm development in flow cells. (A) Newman and derivatives: CYL6938, Newman(pLI50); CYL6969, Newman Δrbf(pLI50); CYL6975, Newman Δrbf(pYL8565). (B) UAMS1 and derivatives: CYL6939, UAMS-1(pLI50); CYL6970, UAMS-1 Δrbf(pLI50); CYL6976, UAMS-1 Δrbf(pYL8565). Selected pictures taken every 30 min are shown with the time point indicated above each image. The medium flow was from left to right.
Rbf promotes virulence of strain Newman in a murine foreign body infection model.
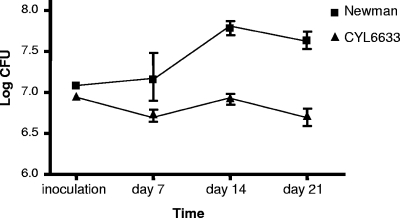
To determine whether Rbf affects the virulence of S. aureus, we performed animal studies using a murine model of foreign body infection. We first compared 8325-4 and 8325-4 rbf::Tn917. One day postimplantation, 20 mice were divided into two groups; one group was inoculated with the wild type, and the other was inoculated with the mutant into the lumen of the implanted catheters. Mice were sacrificed at 3 and 10 days after infection for bacterial enumeration. We found that these strains were resolved quickly in mice (more than a 2-log reduction at day 3 for both wild type and mutant), and there were very few live bacteria left at day 10, resulting in no significant difference between the wild type and the mutant. These results indicate that 8325-4 strain does not persist in this animal model, suggesting that it may not be suitable for in vivo biofilm study. We then used strain Newman in our animal study since we showed above that Rbf was involved in biofilm formation in this strain. Furthermore, Newman has been shown to form biofilm in vivo (13). To this end, we first performed a pilot study with six mice in each group using the murine foreign body infection model. The mice were sacrificed for bacterial counts in the catheter lumen at 5 and 12 days after infection. We found that at day 12, there was a significant difference in bacterial persistence between the wild type and the mutant (data not shown). Based on these data, we performed a power analysis and used a larger number of mice. Thirty-two mice were each implanted as described above. One day postimplantation, the mice were divided into two groups of 16; one group was inoculated with wild-type Newman, and the other was inoculated with Newman rbf::Tn917. At days 7, 14, and 21, at least five mice from each group were sacrificed, and bacterial counts were taken from the explanted catheters. As shown in Fig. 3, the wild type persisted in the catheter better than the mutant at days 14 and 21 (P = 0.0079 and 0.0043, respectively). These data suggest that Rbf is promoting Newman survival in vivo, most likely due to biofilm formation in the implanted catheters.
FIG. 3.
Survival of Newman and CYL6633 (Newman rbf::Tn917) in catheters implanted in mice. Error bars indicate standard deviations.
Overproduction of Rbf in UAMS-1 promotes virulence in a murine foreign body infection model.
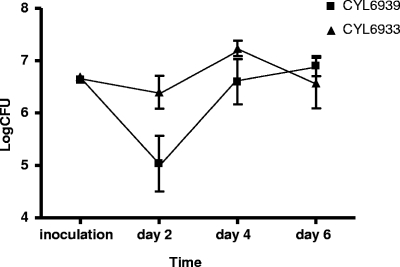
To confirm that Rbf plays an important role in virulence in a clinical strain, we used UAMS-1 in the murine model of foreign body infection using a total of 32 mice in the experiment. However, we found no significant difference between the wild-type UAMS-1 and its rbf mutant at all three time points (days 7, 14, and 21). This was surprising since the Rbf mutation in UAMS-1 affected biofilm phenotype similar to that of the Newman strain in flow cells. To further evaluate whether Rbf plays a role in virulence in UAMS-1, we tested whether overproduction of Rbf from a plasmid would promote virulence. To this end, we compared UAMS-1(pYL8565) and UAMS-1(pLI50) in the foreign body infection model using 32 mice as described above. Since these plasmids are unstable without antibiotic selection, mice were sacrificed at earlier time points, as shown in Fig. 4. The numbers of bacteria from the catheter lumen were determined by plating on TSA without selection. At day 2 postinfection, we found that UAMS-1(pYL8565) had significant higher (P = 0.0303) bacterial counts than the wild-type strain containing only the vector. However, at 4 days postinfection, there was no significant difference (P = 0.2037) between the two strains, although UAMS-1(pYL8565) was consistently higher than UAMS-1(pLI50). At day 6 postinfection, no difference between the two strains was detected. To determine the plasmid retention rates, about 150 colonies were replica plated from TSA plates onto chloramphenicol selection plates (pLI50 carries a chloramphenicol resistance gene). We found plasmid retention rates of 27.5 and 36.8% at day 2 postinfection and 26.6 and 33.8% at day 4 postinfection for UAMS-1(pYL8565) and UAMS-1(pLI50), respectively. At day 6 postinfection, the rates for UAMS-1(pYL8565) and UAMS-1(pLI50) dropped considerably to 8.4 and 17.3%, respectively. The low rate of plasmid retention probably is the reason that significant differences in bacterial counts were observed only at an early time point.
FIG. 4.
Survival of CYL6939 [UAMS-1(pLI50)] and CYL6933 [UAMS-1(pYL8565)] in catheters implanted in mice. Error bars indicate standard deviations.
DISCUSSION
Rbf was identified as a transcriptional regulator affecting biofilm formation in strain 8325-4 (23). In that study, we used a microtiter plate method to demonstrate the Rbf effect on biofilm. However, in the present study, using the same method, we were unable to demonstrate any difference in biofilm formation between the wild type and the rbf mutant in both the Newman and the UAMS-1 genetic backgrounds. However, introduction of the rbf gene carried in a multiple-copy plasmid vector enabled these strains to form detectable biofilms (Fig. 1). On the other hand, by using the flow cell method, we found that the rbf mutation delayed biofilm formation, whereas rbf overexpression accelerated biofilm development in Newman and UAMS-1 (Fig. 2). Based on these data, we speculate that Rbf may be produced in much lower quantity in Newman and UAMS-1 than in 8325-4 such that there are only very small differences in biofilm production between the wild type and the rbf mutant in the Newman and UAMS-1 strains. These small differences may not be detected using the microtiter plate method but are detectable by the more sensitive flow cell method. However, our real-time RT-PCR experiments showed that rbf is expressed at a very low level in all three strains (not shown), suggesting that the quantity of Rbf may not contribute to the in vitro biofilm differences among these strains. Alternatively, the composition of biofilms and/or biofilm regulatory networks among these strains may differ such that the effect of regulation may impact biofilm detection using different methods. Variation in gene regulation among different S. aureus strains is rather common (4, 6, 7). Although it is not currently known what differences in biofilm composition there are among different S. aureus strains, at least two genes have been found defective in 8325-4 but not in UAMS-1 or Newman. One of these is the tcaR gene that has been shown to be involved in repression of biofilm formation (27). Strain 8325-4 is also deficient in SigB due to an 11-bp deletion in the rsbU gene of the sigB operon (22). However, the role of SigB in biofilm regulation in S. aureus is still inconclusive, although it is clearly important in S. epidermidis (reviewed in reference 28). It is likely that the differences in these two regulators and perhaps other factors result in differences in biofilm formation in these strains.
To address the biological significance of Rbf, we compared the wild type and the isogenic rbf mutant by using a biofilm-relevant murine foreign body infection model. We did not find a significant impact on virulence in 8325-4 using the same model, although we could detect a major difference in biofilm formation in vitro. Perhaps strain 8325-4 is relatively avirulent in this specific animal model. On the other hand, we found that Rbf caused a significant difference in virulence in Newman strain. In UAMS-1, however, no statistical difference was observed between the mutant strain and the wild-type strain. Nonetheless, expression of rbf from a multiple-copy plasmid vector did result in increased virulence in UAMS-1. Thus, Rbf seems to promote virulence more in Newman than in UAMS-1 using this animal model, despite the fact that the rbf mutation in Newman and UAMS-1 caused similar biofilm phenotypes when assayed by in vitro methods. However, by examining the flow cell data more closely, we found that there were subtle differences between the two genetic backgrounds. First, UAMS-1 formed biofilm more quickly than did Newman. Second, the difference between the wild type and the rbf mutant was more profound in Newman than in UAMS-1. It is likely that these differences are the reasons that we could observe a significant difference in bacterial persistence in Newman but not in UAMS-1 when the wild-type strain and the rbf mutant were compared in the animal model used in the present study.
Biofilm formation in S. aureus is a complex genetic trait that involves various factors and regulators, most of which have not been fully characterized with respect to biofilm regulation. Rbf belongs to the AraC/XylS family of regulators that possesses a tandem helix-turn-helix DNA-binding motif (23). It is not known what genes involved in biofilm formation are regulated by Rbf. Using a promoter fusion to the xylE reporter gene, we previously showed that Rbf did not affect transcription of the icaADBC operon nor transcription of icaR, suggesting that it may not regulate biofilm through poly-N-acetyl-glucosamine (23). However, our recent unpublished data suggest that Rbf may partially regulate icaADBC genes. How Rbf regulates biofilm formation is currently under intense study in our laboratory.
In conclusion, we showed here that Rbf is involved in biofilm formation in different strains of S. aureus, although the extent of the effect in each strain was different. More importantly, we showed that Rbf played a role in virulence in the biofilm relevant foreign body infection animal model in strain Newman and UAMS-1. Whether Rbf also has a role in virulence in other animal models merits further studies.
Acknowledgments
This study was supported by grant AI067857 from the National Institute of Allergy and Infectious Diseases.
We thank Mark Smeltzer for valuable advice and consultation on animal studies and David Cue for critical reading and editing of the manuscript and for providing some real-time RT-PCR data.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 27 October 2008.
REFERENCES
- 1.Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 5558-63. [DOI] [PubMed] [Google Scholar]
- 2.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 714206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 1864665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boles, B. R., and A. R. Horswill. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassat, J. E., P. M. Dunman, F. McAleese, E. Murphy, S. J. Projan, and M. S. Smeltzer. 2005. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J. Bacteriol. 187576-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassat, J. E., P. M. Dunman, E. Murphy, S. J. Projan, K. E. Beenken, K. J. Palm, S. J. Yang, K. C. Rice, K. W. Bayles, and M. S. Smeltzer. 2006. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 1523075-3090. [DOI] [PubMed] [Google Scholar]
- 8.Cassat, J. E., C. Y. Lee, and M. S. Smeltzer. 2007. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus, p. 127-144. In Y. Ji (ed.), Methicillin-resistant Staphylococcus aureus (MRSA) protocols. Humana Press, Inc., Totowa, NJ.
- 9.Chatterjee, I., P. Becker, M. Grundmeier, M. Bischoff, G. A. Somerville, G. Peters, B. Sinha, N. Harraghy, R. A. Proctor, and M. Herrmann. 2005. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J. Bacteriol. 1874488-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 1844400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 2841318-1321. [DOI] [PubMed] [Google Scholar]
- 12.Fitzpatrick. F., H. Humphreys, and J. P. O'Gara. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 431973-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flückiger, U., M. Ulrich, A. Steinhuber, G. Döring, D. Mack, R. Landmann, C. Goerke, and C. Wolz. 2005. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 731811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 541445-1462. [DOI] [PubMed] [Google Scholar]
- 15.Gillaspy, A., S. Hickmon, R. Skinner, J. Thomas, C. Nelson, and M. Smeltzer. 1995. Role of the accessory gene regulatory (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 633373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 431367-1378. [DOI] [PubMed] [Google Scholar]
- 17.Heilman, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 201083-1091. [DOI] [PubMed] [Google Scholar]
- 18.Izano, E. A., M. A. Amarante, W. B. Kher, and J. B. Kaplan. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesion locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 1862449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporations. Curr. Microbiol. 21373-376. [Google Scholar]
- 21.Kreiswirth, B., S. Lofdahl, M. Betley, M. O'Reilly, P. Schlievert, M. Bergdoll, and R. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 22.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 1804814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luong, T. T., and C. Y. Lee. 2006. The arl locus regulates Staphylococcus aureus type 5 capsule via an mgr-dependent pathway. Microbiology 1523123-3131. [DOI] [PubMed] [Google Scholar]
- 25.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesion involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majerczyk, C. D., M. R. Sadykov, T. T. Luong, C. Lee, G. A. Somerville, and A. L. Sonenshein. 2008. Staphylococcus aureus CodY represses virulence gene expression. J. Bacteriol. 1902257-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCallum, N., M. Bischoff, H. Maki, A. Wada, and B. Berger-Bächi. 2004. TcaR, a putative MarR-like regulator of sarS expression. J. Bacteriol. 1862966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270179-188. [DOI] [PubMed] [Google Scholar]
- 29.O'Neill, E., C. Pozzi, P. Houston, H. Humphreys, D. A. Robinson, A. Loughman, T. J. Foster, and J. P. O'Gara. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 1903835-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pamp, S., D. Frees, S. Englemann, M. Hecker, and H. Ingmer. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 1884861-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin, Z., Y. Ou, L. Yang, Y. Zhu, T. Tolker-Nielsen, S. Molin, and D. Qu. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 1532083-2092. [DOI] [PubMed] [Google Scholar]
- 32.Rice, K. C., E. E. Mann, J. L. Endres, E. C. Weiss, J. E. Cassat, M. S. Smeltzer, and K. W. Bayles. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 1048113-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohde, H., C. Burdelski, K. Bartscht, M. Hussain, F. Buck, M. A. Horstkotte, J. K. Knobloch, C. Heilmann, M. Herrmann, and D. Mack. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 551883-1895. [DOI] [PubMed] [Google Scholar]
- 34.Toledo-Arana, A., N. Merino, M. Vergara-Irigaray, M. Débarbouillé, J. R. Penadés, and I. Lasa. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 1875318-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tormo, M. A., E. Knecht, F. Gotz, I. Lasa, and J. R. Penades. 2005. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 1512465-2475. [DOI] [PubMed] [Google Scholar]
- 36.Tormo, M. A., M. Martí, J. Valle, A. C. Manna, A. L. Cheung, I. Lasa, and J. R. Penades. 2005. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 1872348-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu Quoc, P., P. Genevaux, M. Pajunen, H. Savilahti, C. Georgopoulos, J. Schrenzel, and W. Kelley. 2007. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immun. 741079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valle, J., A. Toledo-Arana, C. Berasain, J.-M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not SigB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 481075-1087. [DOI] [PubMed] [Google Scholar]
- 39.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 1821688-1693. [DOI] [PubMed] [Google Scholar]
- 40.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 1861838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]