Abstract
Host-derived plasmin plays a critical role in mammalian infection by Borrelia burgdorferi. The Lyme disease spirochete expresses several plasminogen-binding proteins. Bound plasminogen is converted to the serine protease plasmin and thereby may facilitate the bacterium's dissemination throughout the host by degrading extracellular matrix. In this work, we demonstrate plasminogen binding by three highly similar borrelial outer surface proteins, ErpP, ErpA, and ErpC, all of which are expressed during mammalian infection. Extensive characterization of ErpP demonstrated that this protein bound in a dose-dependent manner to lysine binding site I of plasminogen. Removal of three lysine residues from the carboxy terminus of ErpP significantly reduced binding of plasminogen, and the presence of a lysine analog, ɛ-aminocaproic acid, inhibited the ErpP-plasminogen interaction, thus strongly pointing to a primary role for lysine residues in plasminogen binding. Ionic interactions are not required in ErpP binding of plasminogen, as addition of excess NaCl or the polyanion heparin did not have any significant effect on binding. Plasminogen bound to ErpP could be converted to the active enzyme, plasmin. The three plasminogen-binding Erp proteins can also bind the host complement regulator factor H. Plasminogen and factor H bound simultaneously and did not compete for binding to ErpP, indicating separate binding sites for both host ligands and the ability of the borrelial surface proteins to bind both host proteins.
Lyme disease is the most commonly reported arthropod-borne disease in the United States (8). Borrelia burgdorferi, the causative agent of Lyme disease, is transmitted to its hosts through the bites of infected Ixodes ticks. In the earliest stage of Lyme disease, a bull's-eye-shaped rash, erythema migrans, occurs as the spirochete spreads outward from the site of the tick bite. If left untreated, serious clinical outcomes can occur, including arthritis, neuropathies, and carditis (48).
The bacterium disseminates from the bite site to other host tissues. B. burgdorferi can traverse the epithelium and invade vascular walls but is rarely abundant in blood (1). In addition, B. burgdorferi can pass through the blood-brain barrier to enter the central nervous system (58). The spirochete, unlike many invasive pathogens, lacks surface protease activities (12, 26). Therefore, binding of host proteases to the surface of the bacterium may aid in the spirochete's dissemination. Indeed, B. burgdorferi binds plasminogen, a component of the host's fibrinolytic system (12, 19). Plasminogen circulates in the plasma as an inactive proenzyme and is activated by tissue-type plasminogen activator and urokinase-type plasminogen activator (uPA) to plasmin (55). Plasminogen binding is an important virulence factor for invasive pathogens such as group A streptococci and Staphylococcus, as well as Borrelia species (10, 43, 55). The binding of plasminogen to bacteria and its subsequent activation allow bacteria to degrade the host's extracellular matrix and basement membranes either through the direct protease activity of plasmin or by plasmin's activation of host matrix metalloproteases (MMPs). B. burgdorferi has previously been shown to bind plasminogen, which is rapidly converted to active plasmin in the presence of host plasminogen activator (11). In vitro, plasmin-coated B. burgdorferi is able to penetrate endothelial cell monolayers (12). Surface-associated plasmin on B. burgdorferi can directly degrade fibronectin, a major component of the extracellular matrix, as well as laminin and vitronectin (11, 19). B. burgdorferi induces the release of MMP-9 (gelatinase) and MMP-1 (collagenase) from human cells, and plasmin-coated B. burgdorferi activates pro-MMP-9 (20), initiating a cascade that leads to degradation of basement membranes. Plasminogen has previously been shown to be important in B. burgdorferi pathogenesis. Although not strictly required for infection, plasminogen was required for efficient dissemination in ticks, and its absence decreased spirochetemia in plasminogen-deficient mice (10).
Plasminogen-binding proteins of B. burgdorferi have previously been identified, including the outer-surface lipoprotein OspA (19). A role for OspC in plasminogen binding has also been suggested (31). However, OspA is generally not expressed during human infection, and OspC production ceases within the first few days of mammalian infection (13, 24, 25, 34, 42). Other, unidentified plasminogen-binding proteins have been observed in B. burgdorferi, including a protein(s) with an approximate molecular mass of 20 kDa, which is close to the size of several Erp proteins (12, 19). The members of the Erp family of outer-surface lipoproteins are expressed at high levels during mammalian infection (15, 23, 38-41).
Lyme disease spirochetes contain numerous DNA elements, including the main chromosome as well as linear and circular plasmids (6). Infectious isolates carry several distinct yet homologous elements called cp32s, circular prophages of approximately 32 kb (54). All cp32 elements encode one or two Erp proteins, which can vary widely in amino acid sequence (50). However, all erp loci are preceded by nearly identical promoter regions (36, 53). Hence, most of the erp genes analyzed follow the same pattern of expression, being repressed in the tick vector but synthesized during mammalian infection (15, 21, 23, 35, 37-41). Roles for most of the Erp proteins have yet to be defined. ErpX has been demonstrated to bind host laminin (our unpublished results and reference 3). Three Erp proteins bind the host complement regulator factor H and factor H-related protein 1: ErpP, ErpC, and ErpA (22, 28, 29). Some factor H binding proteins of other human pathogens have been demonstrated to bind multiple ligands, including plasminogen (30, 47). These data, and the presence of unidentified plasminogen-binding proteins in B. burgdorferi, prompted us to examine if Erp proteins are able to bind plasminogen.
MATERIALS AND METHODS
Bacteria.
An infectious clone of the sequenced type strain, B. burgdorferi B31-MI-16 (5, 18), which retains all plasmids (40), was grown at 34°C to cell densities of approximately 1 × 107 ml in modified Barbour-Stoenner-Kelly medium as described previously (59). Total DNA (genomic and plasmids) was isolated using the DNeasy blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Recombinant proteins.
Polyhistidine-tagged, full-length ErpA, ErpC, and ErpP have been described previously (17, 49). All recombinant proteins contained amino-terminal tags, with the Erp segment beginning with that protein's first amino acid following the cysteine lipidation site. A recombinant plasmid encoding carboxy-terminally truncated ErpP was produced by PCR of B. burgdorferi B31-MI-16 DNA using primers RERPP-1 (5′-CACCAAAATTCATACTTCATATGATGAG-3′) and RERPP-4 (5′-TTATTAAACCACTTCTAGTGGTATTGCATATTCAG-3′) and insertion into pET200 (Invitrogen, Carlsbad, CA). The resultant plasmid's insert was entirely sequenced on both strands to ensure that no undesired mutations had occurred during PCR or cloning procedures.
Recombinant proteins were expressed in Escherichia coli Rosetta (DE3)pLysS (Novagen, Madison, WI), upon induction with isopropyl-β-d-thiogalactopyranoside (IPTG). Induced E. coli cells were harvested and then lysed by sonication in a mixture of 30 mM imidazole, 0.5 M NaCl, and 20 mM NaPO4 (pH 7.4), and debris was cleared by centrifugation. Recombinant proteins were purified from cleared lysates using MagneHis nickel-conjugated magnetic beads (Promega, Madison, WI) or by His-Trap HP columns and an ÄKTA fast protein liquid chromatograph equipped with a UPC-900 UV absorbance monitor and a Frac920 fraction collector (GE Healthcare, Piscataway, NJ). Proteins were eluted from fast protein liquid chromatography columns by a constantly increasing gradient between the lysis buffer and 0.75 M imidazole, 20 mM NaPO4, and 5 M NaCl (pH 7.4). All recombinant proteins were dialyzed overnight against phosphate-buffered saline (PBS) using 3,500-molecular-weight-cutoff Slide-A-Lyzer cassettes (Pierce, Rockford, IL) at 4°C. Protein purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by staining with Coomassie brilliant blue. Protein concentrations were determined by bicinchoninic acid protein assays (Pierce).
ELISA.
For the enzyme-linked immunosorbent assay (ELISA), Maxisorp 96-well plates (Nalge-Nunc, Rochester, NY) were coated overnight with 10 μg/ml human plasminogen (Sigma-Aldrich, St. Louis, MO), 10 μg/ml recombinant Erp protein, or 10 μg/ml bovine serum albumin (BSA [negative control]) in PBS at 4°C. Plates were brought to room temperature and washed once with PBS supplemented with 0.05% (vol/vol) Tween 20 (PBS-T). Wells were blocked for 2 h at room temperature with PBS with 1% (mass/vol) gelatin and then washed three times with PBS-T. Afterwards, 100 μl/well of either recombinant Erp protein, plasminogen (Sigma-Aldrich), or factor H (Sigma-Aldrich [see text for concentrations]) was added and incubated for 2 h at 37°C. Wells were washed three times with PBS-T and then incubated for 1 h at room temperature with rabbit antiserum raised against B. burgdorferi ErpA (which cross-reacts with the highly similar ErpP and ErpC), diluted 1:500 in PBS, or goat anti-human plasminogen (Novus Biologicals, Littleton, CO) or goat anti-human factor H (Calbiochem, San Diego, CA). Plates were washed three times with PBS-T, and then wells were incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) or horseradish peroxidase-conjugated protein A (GE Healthcare, Little Chalfont, United Kingdom), diluted 1:5,000. Wells were again washed three times with PBS-T, 100 μl/well tetramethylbenzidine substrate was added, and then reactions were stopped by addition of 100 μl/well of 2 N H2SO4. Absorbance was read at 450 nm using a Spectramax plate reader using SoftMax Pro software (Molecular Devices, Sunnyvale, CA).
For experiments examining the role of ionic interactions in ErpP binding to plasminogen, increasing concentrations of NaCl were added to the PBS-based binding buffer with ErpP. For experiments analyzing the role of heparin-binding domains in the ErpP-plasminogen interaction, porcine heparin (final concentrations, 0 to 50 μM [Sigma-Aldrich]) was added to the plasminogen-coated wells for 1 h prior to the addition of tested protein and was also included in the binding buffer along with the tested recombinant protein (51). To determine the role of lysines in plasminogen-ErpP interaction, the lysine analog ɛ-aminocaproic acid (final concentration, 1 mM [Sigma-Aldrich]) was added with plasminogen to ErpP-coated wells. The role of Kringle domains of plasminogen in ErpP binding was assessed using plasminogen lysine-binding site I (LBS I)-coated wells (Sigma-Aldrich).
Protein binding affinities (Kd) were calculated as that concentration of ligand required for half-maximal binding activity.
Plasminogen activation assay.
Maxisorp 96-well plates (Nalge-Nunc) were coated overnight with 10 μg/ml recombinant ErpP or 10 μg/ml BSA in PBS at 4°C. Plates were brought to room temperature and washed once with PBS-T. Wells were blocked for 2 h at room temperature with PBS with 2% (mass/vol) BSA and then washed three times with PBS-T. Afterwards 100 μl/well of 10 μg/ml human plasminogen was added and incubated for 2 h at 37°C. Wells were washed three times with PBS-T, and then 4 ng/well of human uPA (Chemicon, Temecula, CA) was added. Next, the plasmin-specific substrate d-valyl-leucyl-lysine-p-nitroanilide dihydrochloride (Sigma-Aldrich) was added at a final concentration of 0.3 mM in 64 mM Tris, 350 mM NaCl, 0.15% Triton X-100 (pH 7.5). Plates were incubated overnight at 37°C, and absorbance was read at 405 nm.
RESULTS
ErpP, ErpA, and ErpC bind human plasminogen.
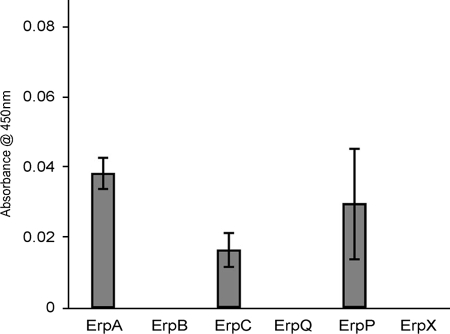
Earlier observations that B. burgdorferi expresses plasminogen-binding surface proteins of approximate masses of 20 kDa (12, 19, 31), similar to some Erp proteins, prompted us to investigate whether Erp proteins were capable of binding human plasminogen. Several Erp proteins from type strain B31 having diverse sequences were chosen for study (5, 49, 52). Recombinant ErpA, ErpB, ErpC, ErpP, ErpQ, and ErpX, and the nonspecific negative control BSA were individually immobilized onto microtiter plates and incubated with human plasminogen, and their abilities to form protein complexes were measured by ELISA. Significant binding of plasminogen was detected only for ErpA, ErpC, and ErpP (Fig. 1). These three proteins are >98% identical and all have molecular masses of 19 to 21 kDa (Fig. 2). Due to those extensive similarities, ErpP was chosen as a representative protein for additional characterization. Analyses using various concentrations of plasminogen demonstrated that ErpP bound plasminogen in a dose-dependent manner, with a Kd of approximately 25 nM (Fig. 3).
FIG. 1.
ErpA, ErpC, and ErpP bind plasminogen. Binding of plasminogen (25 μg/ml) to immobilized Erp proteins (10 μg/ml) was analyzed by ELISA, with bound plasminogen detected using specific antiserum. BSA was used as a negative control for nonspecific binding. Values represent plasminogen binding to each Erp protein minus background absorbance for BSA. Data represent the means and standard errors from two separate experiments with six replicates per Erp protein.
FIG. 2.
Alignment of ErpP, ErpA, and ErpC. Identical amino acid residues found in two or more of the proteins are boxed and shaded. The five amino acids deleted from the truncated ErpP used in this work are indicated by a thick line over the sequence. The black arrowhead indicates the first amino acid following the polyhistidine tag in the recombinant Erp protein.
FIG. 3.
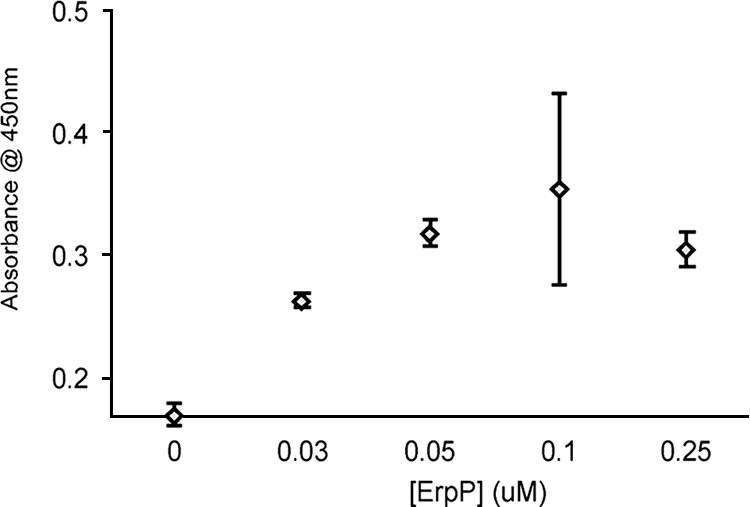
ErpP binds plasminogen in a dose-dependent manner. Binding of ErpP (0 to 1 μM) to immobilized plasminogen (10 μg/ml) was analyzed by ELISA, with bound ErpP detected using polyclonal rabbit antiserum. BSA was used as a negative control for nonspecific binding. Values represent ErpP binding to plasminogen minus background absorbance for BSA. Data represent the means and standard errors from two separate experiments with six replicates per concentration of ErpP.
Role of lysines in ErpP plasminogen binding activity.
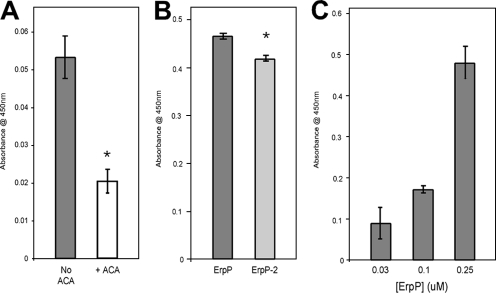
Plasminogen receptors on the surface of host cells bind plasminogen through lysine residues present in Kringle domains, and several bacterial proteins contain lysine residues that are important for the binding of plasminogen (2, 11, 33, 45). ErpP, ErpA, and ErpC are each comprised of approximately 13% lysine residues (24 out of 186 amino acids in ErpP [Fig. 2]). Addition of the synthetic lysine analog ɛ-aminocaproic acid significantly reduced the ErpP-plasminogen interaction, indicating a role for lysines in ErpP binding to the ligand (Fig. 4A).
FIG. 4.
Role of lysines in ErpP plasminogen binding activity. Binding of plasminogen to immobilized ErpP (10 μg/ml) was analyzed by ELISA. (A) Plasminogen (25 μg/ml) was added to ErpP-coated wells in the presence or absence of 1 mM ɛ-aminocaproic acid (ACA). Bound plasminogen was detected using a specific antiserum. BSA was used as a negative control. Data represent the means and standard errors from two separate experiments with 12 replicates per condition. *, P < 0.001, Student's t test assuming unequal variances. (B) Binding of plasminogen to immobilized ErpP and the carboxy-terminal truncation mutant (rErpP2) was analyzed by ELISA. Plasminogen (25 μg/ml) was added to ErpP-coated wells. Bound plasminogen was detected using a specific antiserum. BSA was used as a negative control. Data represent the means and standard errors from two separate experiments with 12 replicates per condition. *, P < 0.001, Student's t test assuming unequal variances. (C) Binding of ErpP (0 to 250 nM) to immobilized LBS I of plasminogen analyzed by ELISA. Bound ErpP was detected using a specific antiserum. BSA was used as a negative control. Data represent the means and standard errors from two separate experiments with six replicates per concentration of ErpP.
Previous studies found that the three carboxy-terminal lysine residues of ErpP are essential for that protein to interact with human complement regulator factor H (29). In addition, the carboxy-terminal lysine residues of some bacterial plasminogen-binding proteins are necessary for plasminogen binding (4, 44). To ascertain the importance of the carboxy-terminal lysines in ErpP binding to plasminogen, a truncated ErpP protein lacking five amino acids at the carboxy terminus was engineered (Fig. 2). Deletion of the carboxy-terminal lysines significantly reduced, but did not completely eliminate, binding of plasminogen to ErpP. Thus, the carboxy terminus of ErpP is involved in plasminogen binding, but apparently so are some or all of ErpP's other 21 lysine residues (Fig. 4B).
Next, the ability of ErpP to interact with the lysine-binding sites of plasminogen was tested. A fragment of plasminogen which contains the first three triple-loop structures in the plasmin A chain (Kringle 1 + 2 + 3), plasminogen LBS I, was used as ligand. ErpP bound LBS I in a dose-dependent manner, again pointing to a role for the lysines of ErpP in the binding of plasminogen (Fig. 4C).
Role of ionic interactions in ErpP binding to plasminogen.
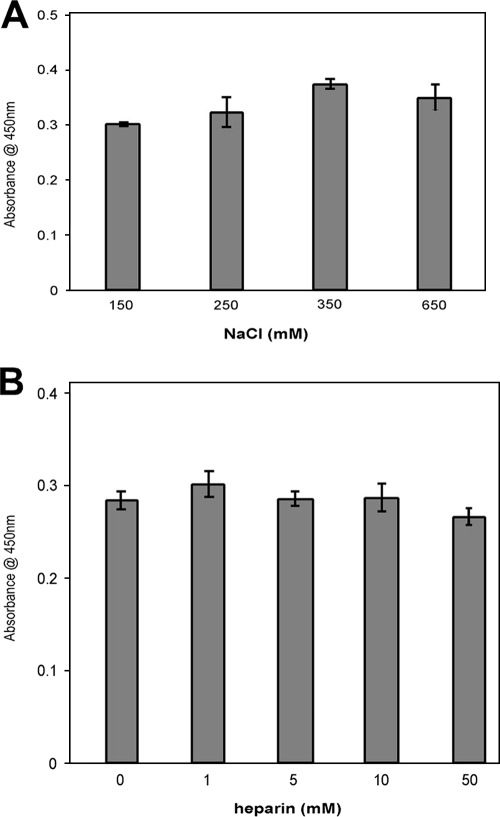
ErpP contains a large number of charged amino acids (Fig. 2), with a theoretical pI of 8.36. To assess the role of ionic interactions in ErpP plasminogen binding, binding assays were performed in the presence of various concentrations of NaCl or the polyanion heparin. NaCl, at up to four times the physiological concentration, did not have any significant effects on plasminogen binding (Fig. 5A). Likewise, heparin had no appreciable effect on ErpP-plasminogen interactions (Fig. 5B). These data suggest that ionic interactions are dispensable for plasminogen-ErpP interactions.
FIG. 5.
Role of ionic interactions in ErpP binding of plasminogen. Binding of plasminogen to immobilized ErpP (10 μg/ml) was analyzed by ELISA. (A) Plasminogen (25 μg/ml) was incubated in the presence of increasing concentrations of NaCl. Bound plasminogen was detected using a specific antiserum. BSA was used as a negative control. Data represent the means and standard errors from two experiments with six replicates per concentration of NaCl. (B) Heparin (0 to 50 μM) was added to immobilized ErpP for 1 h prior to the addition of plasminogen. After a single wash with PBS-T, plasminogen (25 μg/ml) was incubated in the presence of heparin (0 to 50 μM). Bound plasminogen was detected using a specific antiserum. BSA was used as a negative control. Data represent the means and standard errors from two experiments with six replicates per concentration of heparin.
Factor H and plasminogen do not compete for binding to ErpP.
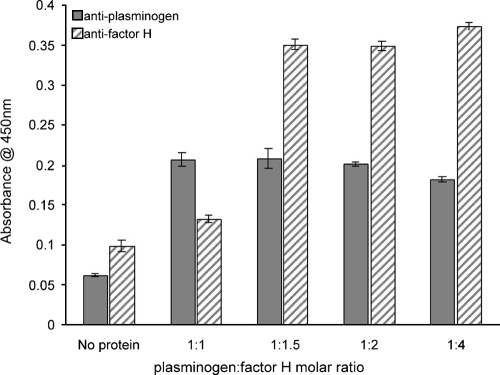
Purified ErpP, ErpA, and ErpC can also bind the human complement regulator factor H. The deletion of nine residues of the carboxy terminus of ErpP, which contains three lysines, abrogates factor H binding (29). As described above, deletion of the same three lysine residues significantly impaired ErpP binding of plasminogen. To assess if factor H and plasminogen share identical or overlapping binding sites on ErpP, binding of plasminogen by full-length ErpP was assayed in the presence of increasing concentrations of factor H. In normal human sera, the molar ratio of plasminogen to factor H is 1:1.4. At experimental ratios of up to 1:4, no competition between plasminogen and factor H for ErpP binding was observed (Fig. 6). These data suggest that plasminogen and factor H bind to separate sites of ErpP and demonstrate that the two host proteins can adhere to ErpP simultaneously.
FIG. 6.
Factor H and plasminogen do not compete for binding to ErpP. Plasminogen (200 nM) binding to immobilized ErpP (10 μg/ml) was assayed in the presence of increasing concentrations of factor H. Both serum proteins were detected by individual, specific antisera. In normal human serum, the molar ratio of plasminogen to factor H is 1:1.4. Data represent the means and standard errors from two different experiments with six replicates per condition.
Plasminogen bound to ErpP can be converted to plasmin.
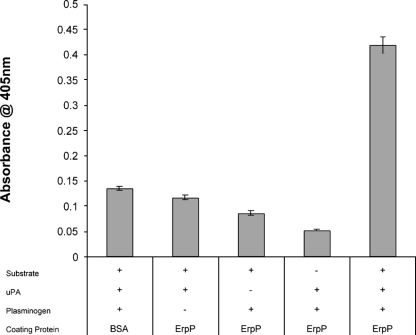
Plasminogen bound to the surface of B. burgdorferi can be converted to plasmin (19). To determine if ErpP-bound plasminogen can similarly be activated, multiwell plates were coated with ErpP, blocked, and then incubated with plasminogen. uPA was added, and the proteolytic activity was quantified using a plasmin-specific chromogenic substrate. As shown in Fig. 7, ErpP-bound plasminogen was converted to plasmin. These data demonstrate that Erp-bound plasminogen is accessible to activators and can be converted into the active enzyme.
FIG. 7.
ErpP-bound plasminogen is converted into plasmin. ErpP-coated wells of microtiter plates were incubated with plasminogen, urokinase (uPA), and/or a plasmin-specific chromogenic substrate. Proteolytic activity was measured by absorbance at 450 nM. Data represent the means and standard errors from two different experiments with six replicates per condition.
DISCUSSION
Binding of plasminogen to the surface of bacteria is often important for virulence, both by providing a means of initial anchoring to endothelium via host plasminogen receptors and by adding a potent protease activity to the bacterial surface. Activation of plasmin on the surface of pathogens can therefore facilitate extracellular matrix degradation, allowing the pathogen to disseminate throughout the host. Plasminogen has previously been shown to be important for B. burgdorferi dissemination in both vector ticks and mammalian hosts (10). Plasminogen is also important in kidney and brain invasion by a related spirochete, Borrelia crocidurae (43). Multiple plasminogen-binding proteins are present in Lyme disease spirochetes, as demonstrated by plasminogen overlay assays, but only a few have been identified and characterized. Of these, OspA is generally not produced during human infection, and OspC is only transiently expressed during transmission from ticks to mammals (13, 24, 25, 35, 42, 56). In the present study, we identified three members of the polymorphic Erp protein family, ErpP, ErpA, and ErpC, as being plasminogen-binding proteins. Those Erp proteins are known to be produced through all stages of mammalian infection (15, 35, 37-41), and therefore acquisition of plasminogen by ErpP, ErpA, and ErpC may play a role(s) in long-term infection.
Plasminogen interacts with its substrates, inhibitors, and effectors through five Kringle domains (7). These approximately 80-amino-acid loop structures are linked by triple-disulfide bonds and undergo conformational change upon binding to lysine-containing sequences. This conformational change allows plasminogen to achieve a form that is easily activated and protected from plasmin inhibitors (32). A number of bacterial proteins contain lysine residues that are crucial for the binding of plasminogen (2, 11, 33). In the present study, we demonstrated that lysine residues in ErpP are directly involved in plasminogen binding, as the lysine analog ɛ-aminocaproic acid significantly inhibited binding of ErpP to plasminogen. Deletion of the three carboxy-terminal lysine residues also decreased ligand binding. In addition, ErpP bound the LBS I (Kringle domains 1 to 3) of plasminogen in a dose-dependent manner. Most of the Erp proteins contain high percentages of lysine residues, yet only ErpP, ErpA, and ErpC bind plasminogen. Intriguingly, addition of anions (Cl−, heparin) did not affect ErpP-plasminogen interactions, suggesting that noncharged amino acids and/or protein conformations are also important for binding. Indeed, internal binding motifs flanked by hydrophobic residues are involved in plasminogen binding by enolases from Streptococcus pneumoniae and Neisseria meningitidis (16, 27).
Beyond its role in degradation of basement membranes and the facilitation of dissemination to diverse tissues in the host, plasminogen binding by B. burgdorferi might be involved in immune avoidance. The acquired proteolytic activity of plasmin may degrade specific antibodies and components of the complement system. For example, staphylokinase of Staphycoccus aureus exhibits antiopsonic activity by binding plasminogen. The subsequent protease activity of plasmin degrades the opsonins C3b and IgG (46). Some plasminogen-binding proteins are also immunosuppressive, such as enolase of Streptococcus sobrinus, which increases interleukin-10 production and dampens immune responses in mice (57).
ErpP also binds factor H, the key regulator of the alternative pathway of complement. ErpP appears to bind factor H and plasminogen through discrete domains, as factor H did not compete with plasminogen for binding to ErpP. This echoes the dual function of the unrelated CRASP-1Bh/FhbA protein of Borrelia hermsii, which likewise binds factor H and plasminogen through separate regions (47). Another example of a dual-function protein that binds multiple serum components is Tuf of Pseudomonas aeruginosa. However, unlike ErpP, Tuf shares similar or overlapping binding sites for factor H and plasminogen (30). The carboxy-terminal lysine residues of ErpP are essential for binding to factor H and are important for binding to plasminogen. Yet the absence of competition for ErpP suggests that the two ligands use multiple contact points for binding of factor H and/or plasminogen. While the significance of binding multiple serum components remains to be defined, many bacterial adhesins have multiple binding partners (9, 14). An obvious advantage to the spirochete would be that fewer surface lipoproteins need to be expressed in order to disseminate and evade host immune responses, conserving energy and allowing B. burgdorferi to safely and swiftly transition from blood to tissues. The combination of plasminogen and factor H binding to the same proteins suggests a rather sophisticated escape mechanism of B. burgdorferi for dissemination and persistence in the mammalian host.
Acknowledgments
This work was supported by NIAID grant AI44254 to B. Stevenson and Deutsche Forschungsgemeinschaft grant Kr3383/1 to P. Kraiczy.
We thank Logan Burns, Sean Riley, Ashutosh Verma, Erin Welsh, and Michael Woodman for helpful comments and technical assistance.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 10 November 2008.
REFERENCES
- 1.Barbour, A. G., and S. F. Hayes. 1986. Biology of Borrelia species. Microbiol. Rev. 50381-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle, M. D., and R. Lottenberg. 1997. Plasminogen activation by invasive human pathogens. Thromb. Haemost. 771-10. [PubMed] [Google Scholar]
- 3.Brissette, C. A., A. E. Cooley, L. H. Burns, S. P. Riley, A. Verma, M. E. Woodman, T. Bykowski, and B. Stevenson. 2008. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int. J. Med. Microbiol. 298(Suppl. 1)257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broder, C. C., R. Lottenberg, G. O. von Mering, K. H. Johnston, and M. D. Boyle. 1991. Isolation of a prokaryotic plasmin receptor. Relationship to a plasminogen activator produced by the same micro-organism. J. Biol. Chem. 2664922-4928. [PubMed] [Google Scholar]
- 5.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35490-516. [DOI] [PubMed] [Google Scholar]
- 6.Casjens, S. R., W. M. Huang, E. B. Gilcrease, W. Qiu, W. D. McCaig, B. J. Luft, S. E. Schutzer, and C. M. Fraser. 2006. Comparative genomics of Borrelia burgdorferi, p. 79-95. In F. C. Cabello, D. Hulinska, and H. P. Godfrey (ed.), Molecular biology of spirochetes. IOS Press, Amsterdam, The Netherlands.
- 7.Castellino, F. J., and S. G. McCance. 1997. The kringle domains of human plasminogen. Ciba Found. Symp. 21246-65. [DOI] [PubMed] [Google Scholar]
- 8.CDC. 2007. Lyme disease—United States, 2003-2005. MMWR Morb. Mortal. Wkly. Rep. 56573-576. [PubMed] [Google Scholar]
- 9.Coburn, J., J. R. Fischer, and J. M. Leong. 2005. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol. Microbiol. 571182-1195. [DOI] [PubMed] [Google Scholar]
- 10.Coleman, J. L., J. A. Gebbia, J. Piesman, J. L. Degen, T. H. Bugge, and J. L. Benach. 1997. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell 891111-1119. [DOI] [PubMed] [Google Scholar]
- 11.Coleman, J. L., E. J. Roemer, and J. L. Benach. 1999. Plasmin-coated Borrelia burgdorferi degrades soluble and insoluble components of the mammalian extracellular matrix. Infect. Immun. 673929-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman, J. L., T. J. Sellati, J. E. Testa, R. R. Kew, M. B. Furie, and J. L. Benach. 1995. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect. Immun. 632478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crother, T. R., C. I. Champion, J. P. Whitelegge, R. Aguilera, X.-Y. Wu, D. R. Blanco, J. N. Miller, and M. A. Lovett. 2004. Temporal analysis of the antigenic composition of Borrelia burgdorferi during infection in rabbit skin. Infect. Immun. 725063-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, S., S. W. Barthold, S. Stocker Giles, R. R. Montgomery, S. R. Telford, and E. Fikrig. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Investig. 99987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehinger, S., W. D. Schubert, S. Bergmann, S. Hammerschmidt, and D. W. Heinz. 2004. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 343997-1005. [DOI] [PubMed] [Google Scholar]
- 17.El-Hage, N., K. Babb, J. A. Carroll, N. Lindstrom, E. R. Fischer, J. C. Miller, R. D. Gilmore, Jr., M. L. Mbow, and B. Stevenson. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147821-830. [DOI] [PubMed] [Google Scholar]
- 18.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390580-586. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs, H., R. Wallich, M. M. Simon, and M. D. Kramer. 1994. The outer surface protein A of the spirochete Borrelia burgdorferi is a plasmin(ogen) receptor. Proc. Natl. Acad. Sci. USA 9112594-12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebbia, J. A., J. L. Coleman, and J. L. Benach. 2001. Borrelia spirochetes upregulate release and activation of matrix metalloproteinase gelatinase B (MMP-9) and collagenase 1 (MMP-1) in human cells. Infect. Immun. 69456-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3799-808. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann, K., C. Corvey, C. Skerka, M. Kirschfink, M. Karas, V. Brade, J. C. Miller, B. Stevenson, R. Wallich, P. F. Zipfel, and P. Kraiczy. 2006. Functional characterization of BbCRASP-2, a distinct outer membrane protein of Borrelia burgdorferi that binds host complement regulators factor H and FHL-1. Mol. Microbiol. 611220-1236. [DOI] [PubMed] [Google Scholar]
- 23.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, and D. R. Akins. 2002. Changes in the temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 703468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodzic, E., S. Feng, K. J. Freet, and S. W. Barthold. 2003. Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect. Immun. 715042-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodzic, E., S. Feng, K. J. Freet, D. L. Borjesson, and S. W. Barthold. 2002. Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect. Immun. 703382-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klempner, M. S., R. Noring, M. P. Epstein, B. McCloud, R. Hu, S. A. Limentani, and R. A. Rogers. 1995. Binding of human plasminogen and urokinase-type plasminogen activator to the Lyme disease spirochete, Borrelia burgdorferi. J. Infect. Dis. 1711258-1265. [DOI] [PubMed] [Google Scholar]
- 27.Knaust, A., M. V. R. Weber, S. Hammerschmidt, S. Bergmann, M. Frosch, and O. Kurzai. 2007. Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis. J. Bacteriol. 1893246-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraiczy, P., J. Hellwage, C. Skerka, H. Becker, M. Kirschfink, M. M. Simon, V. Brade, P. F. Zipfel, and R. Wallich. 2004. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 2792421-2429. [DOI] [PubMed] [Google Scholar]
- 29.Kraiczy, P., J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, and R. Wallich. 2003. Immune evasion of Borrelia burgdorferi: mapping of a complement inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 33697-707. [DOI] [PubMed] [Google Scholar]
- 30.Kunert, A., J. Losse, C. Gruszin, M. Huhn, K. Kaendler, S. Mikkat, D. Volke, R. Hoffmann, T. S. Jokiranta, H. Seeberger, U. Moellmann, J. Hellwage, and P. F. Zipfel. 2007. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J. Immunol. 1792979-2988. [DOI] [PubMed] [Google Scholar]
- 31.Lagal, V., D. Portnoi, G. Faure, D. Postic, and G. Baranton. 2006. Borrelia burgdorferi sensu stricto invasiveness is correlated with OspC-plasminogen affinity. Microbes Infect. 8645-652. [DOI] [PubMed] [Google Scholar]
- 32.Lahteenmaki, K., S. Edelman, and T. K. Korhonen. 2005. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 1379-85. [DOI] [PubMed] [Google Scholar]
- 33.Lahteenmaki, K., P. Kuusela, and T. K. Korhonen. 2001. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25531-552. [DOI] [PubMed] [Google Scholar]
- 34.Liang, F. T., M. B. Jacobs, L. C. Bowers, and M. T. Philipp. 2002. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 196275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marconi, R. T., S. Y. Sung, C. A. N. Hughes, and J. A. Carlyon. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 1785615-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDowell, J. V., S. Y. Sung, G. Price, and R. T. Marconi. 2001. Demonstration of the genetic stability and temporal expression of select members of the Lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 694831-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. C., K. Narayan, B. Stevenson, and A. R. Pachner. 2005. Expression of Borrelia burgdorferi erp genes during infection of non-human primates. Microb. Pathog. 3927-33. [DOI] [PubMed] [Google Scholar]
- 39.Miller, J. C., and B. Stevenson. 2006. Borrelia burgdorferi erp genes are expressed at different levels within tissues of chronically infected mammalian hosts. Int. J. Med. Microbiol. 296(Suppl. 1)185-194. [DOI] [PubMed] [Google Scholar]
- 40.Miller, J. C., K. von Lackum, K. Babb, J. D. McAlister, and B. Stevenson. 2003. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect. Immun. 716943-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, J. C., K. von Lackum, M. E. Woodman, and B. Stevenson. 2006. Detection of Borrelia burgdorferi gene expression during mammalian infection using transcriptional fusions that produce green fluorescent protein. Microb. Pathog. 4143-47. [DOI] [PubMed] [Google Scholar]
- 42.Montgomery, R. R., S. E. Malawista, K. J. M. Feen, and L. K. Bockenstedt. 1996. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J. Exp. Med. 183261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nordstrand, A., A. Shamaei-Tousi, A. Ny, and S. Bergstrom. 2001. Delayed invasion of the kidney and brain by Borrelia crocidurae in plasminogen-deficient mice. Infect. Immun. 695832-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pancholi, V., and V. A. Fischetti. 1998. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 27314503-14515. [DOI] [PubMed] [Google Scholar]
- 45.Plow, E. F., T. Herren, A. Redlitz, L. A. Miles, and J. L. Hoover-Plow. 1995. The cell biology of the plasminogen system. FASEB J. 9939-945. [DOI] [PubMed] [Google Scholar]
- 46.Rooijakkers, S. H., W. J. van Wamel, M. Ruyken, K. P. van Kessel, and J. A. van Strijp. 2005. Anti-opsonic properties of staphylokinase. Microbes Infect. 7476-484. [DOI] [PubMed] [Google Scholar]
- 47.Rossmann, E., P. Kraiczy, P. Herzberger, C. Skerka, M. Kirschfink, M. M. Simon, P. F. Zipfel, and R. Wallich. 2007. Dual binding specificity of a Borrelia hermsii-associated complement regulator-acquiring surface protein for factor H and plasminogen discloses a putative virulence factor of relapsing fever spirochetes. J. Immunol. 1787292-7301. [DOI] [PubMed] [Google Scholar]
- 48.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345115-125. [DOI] [PubMed] [Google Scholar]
- 49.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 662648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson, B., T. Bykowski, A. E. Cooley, K. Babb, J. C. Miller, M. E. Woodman, K. von Lackum, and S. P. Riley. 2006. The Lyme disease spirochete Erp lipoprotein family: structure, function and regulation of expression, p. 354-372. In F. C. Cabello, H. P. Godfrey, and D. Hulinska (ed.), Molecular biology of spirochetes. IOS Press, Amsterdam, The Netherlands.
- 51.Stevenson, B., H. A. Choy, M. Pinne, M. L. Rotondi, M. C. Miller, E. Demoll, P. Kraiczy, A. E. Cooley, T. P. Creamer, M. A. Suchard, C. A. Brissette, A. Verma, and D. A. Haake. 2007. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS ONE 2e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson, B., and J. C. Miller. 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 57309-324. [DOI] [PubMed] [Google Scholar]
- 53.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 1783508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevenson, B., W. R. Zc̈kert, and D. R. Akins. 2001. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species, p. 87-100. In M. H. Saier and J. Garcia-Lara (ed.), The spirochetes: molecular and cellular biology. Horizon Press, Oxford, United Kingdom. [PubMed]
- 55.Sun, H. 2006. The interaction between pathogens and the host coagulation system. Physiology 21281-288. [DOI] [PubMed] [Google Scholar]
- 56.Tilly, K., J. G. Krum, A. Bestor, M. W. Jewett, D. Grimm, D. Bueschel, R. Byram, D. Dorward, M. J. VanRaden, P. Stewart, and P. Rosa. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 743554-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veiga-Malta, I., M. Duarte, M. Dinis, D. Tavares, A. Videira, and P. Ferreira. 2004. Enolase from Streptococcus sobrinus is an immunosuppressive protein. Cell. Microbiol. 679-88. [DOI] [PubMed] [Google Scholar]
- 58.Wilske, B., V. Fingerle, and U. Schulte-Spechtel. 2007. Microbiological and serological diagnosis of Lyme borreliosis. FEMS Immunol. Med. Microbiol. 4913-21. [DOI] [PubMed] [Google Scholar]
- 59.Zückert, W. R. 2007. Laboratory maintenance of Borrelia burgdorferi, p. 12C.1.1-12C.1.10. In R. T. Coico, T. F. Kowalik, J. Quarles, B. Stevenson, and R. Taylor (ed.), Current protocols in microbiology. J. Wiley and Sons, Hoboken, NJ. [DOI] [PubMed]