Abstract
Francisella tularensis, the highly virulent etiologic agent of tularemia, is a low-dose intracellular pathogen that is able to escape from the phagosome and replicate in the cytosol. Although there has been progress in identifying loci involved in the pathogenicity of this organism, analysis of the genome sequence has revealed few obvious virulence factors. We previously reported isolation of an F. tularensis subsp. tularensis strain Schu S4 transposon insertion mutant with a mutation in a predicted hypothetical lipoprotein, FTT1103, that was deficient in intracellular replication in HepG2 cells. In this study, a mutant with a defined nonpolar deletion in FTT1103 was created, and its phenotype, virulence, and vaccine potential were characterized. A phagosomal integrity assay and lysosome-associated membrane protein 1 colocalization revealed that ΔFTT1103 mutant bacteria were defective in phagosomal escape. FTT1103 mutant bacteria were maximally attenuated in the mouse model; mice survived, without visible signs of illness, challenge by more than 1010 CFU when the intranasal route was used and challenge by 106 CFU when the intraperitoneal, subcutaneous, or intravenous route was used. The FTT1103 mutant bacteria exhibited dissemination defects. Mice that were infected by the intranasal route had low levels of bacteria in their livers and spleens, and these bacteria were cleared by 3 days postinfection. Mutant bacteria inoculated by the subcutaneous route failed to disseminate to the lungs. BALB/c or C57BL/6 mice that were intranasally vaccinated with 108 CFU of FTT1103 mutant bacteria were protected against subsequent challenge with wild-type strain Schu S4. These experiments identified the FTT1103 protein as an essential virulence factor and also demonstrated the feasibility of creating defined attenuated vaccines based on a type A strain.
Francisella tularensis is an endemic zoonotic gram-negative bacterium that is found throughout the United States, Europe, and Asia. There are two main subspecies, F. tularensis subsp. tularensis and F. tularensis subsp. holarctica (also known as type A and type B, respectively), that are responsible for the majority of infections and disease. Another related species (possibly a subspecies), Francisella novicida, is highly virulent in mice but is avirulent in immunocompetent humans. Although primarily a vector-borne disease, F. tularensis infection can also occur through inhalation of contaminated materials. Infection with a type A strain can be particularly severe, and this subspecies has been targeted as a potential agent of biological warfare. A live vaccine strain (LVS), which is an attenuated type B strain, has been safely used as a tularemia vaccine for over 50 years in Europe and Asia, but a number of regulatory issues have prevented licensing and approval by the FDA (for a review, see reference 9).
F. tularensis is a facultative intracellular bacterium that can invade a variety of cell types, including macrophages, endothelial cells, and hepatocytes (1, 8, 16). A few factors that regulate or facilitate phagosome escape and intracellular survival of F. tularensis have been identified (22, 31, 40, 43, 49). So far, many of the factors identified are encoded on a 30-kb Francisella pathogenicity island (FPI) that has at least 17 open reading frames (40). Two proteins encoded on the FPI, IglA and IglB, are interacting cytoplasmic proteins that have similarity to a recently described type VI secretion system (12). Most of the work with FPI genes has been done with F. novicida because a single copy of FPI is present in F. novicida, whereas duplicate copies are present in LVS and Schu S4. In part because analysis of the genome sequence revealed few obvious virulence factors, several laboratories have created site-specific mutations or used transposon mutagenesis of LVS or F. novicida as a means to identify virulence-associated loci (20, 33, 36, 42, 44, 51, 53, 58). The mutations have included mutations in purine (42, 44) and lipopolysaccharide (LPS) biosynthetic genes (33), as well as hypothetical genes (53). To identify virulence genes and attenuating mutations that were directly relevant to type A strains, we created a transposon insertion library in Schu S4 and then screened this library for attenuated mutants to identify potential new live vaccine candidates for use against tularemia (43). One of the candidate mutants had a transposon inserted into the FTT1103 locus, which was predicted to encode a hypothetical lipoprotein. This hypothetical protein shares some similarity with DsbA proteins, which are proteins that catalyze disulfide bond formation (6). Whether this protein functions in this manner in F. tularensis still needs to be clarified. The FTT1103 mutant strain was found to be defective in intracellular growth, and in J774A.1 cells there was a decreased ability to escape from phagosomes. This strain was avirulent in mice with all routes of infection that were tested, and importantly, immunization with this strain provided protection against challenge with the wild-type strain. These results indicated that despite the high virulence of type A strains, it was possible to attenuate this subspecies to obtain a low level of reactogenicity and still achieve robust protective immunity.
MATERIALS AND METHODS
Bacterial strains, primers, plasmids, and culture.
All bacterial strains and plasmids used in the experiments in this study are listed in Table 1. The primers used are listed in Table 2. Escherichia coli strains were grown in Luria-Bertani broth or on Luria-Bertani agar plates with kanamycin (50 μg/ml) or ampicillin (100 μg/ml) when required. F. tularensis subsp. tularensis (type A) strain Schu S4 and the F. tularensis subsp. holarctica (type B) live vaccine strain (LVS) were subcultured on cysteine-supplemented Muller-Hinton agar (MHAC). For liquid cultures bacteria were grown in Trypticase soy broth supplemented with cysteine (TSBC). Growth comparison experiments were performed using TSBC and Chamberlain's defined medium as described elsewhere (43). When appropriate, 25 μg/ml rifampin or 15 μg/ml kanamycin was added. All studies involving Schu S4 and derivatives of this strain were carried out in an approved biosafety level 3 laboratory.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source and/or referencea |
|---|---|---|
| Francisella strains | ||
| LVS | F. tularensis subsp. holarctica | K. Elkins |
| Schu S4 | F. tularensis subsp. tularensis | CDC |
| BJM1028 | Schu S4 EZ-Tn5 <rpsL-arr-2>::FTT1103 | A. Qin (43) |
| BJM1028c | Schu S4 EZ-Tn5 <rpsL-arr-2>::FTT1103 containing pAQ021 | |
| BJM1031 | Schu S4 ΔFTT1103 | |
| BJM1033 | BJM1031 containing pAQ036 | |
| BJM1034 | BJM1031 containing pFNLTP-6-groE-gfp | |
| BJM1035 | BJM1031 containing pAQ038 | |
| Plasmids | ||
| pCR2.1-TOPO | Invitrogen | |
| pGIR463 | sacB suicide vector | G. Ramakrishnan |
| pFNLTP-6-groE-gfp | T. C. Zahrt (37) | |
| pFNLTP-6-gfp | Deleted groE promoter | |
| pAQ020 | FTT1103 pCR2.1-TOPO (BM063/BM066) | |
| pAQ021 | FTT1103 pFNLTP-6-groE-gfp (BM063/BM066) | |
| pAQ031 | 5′ and 3′ flanking regions of FTT1103 pGIR463 | |
| pAQ036 | FTT1103 pFNLTP-6-groE-gfp (BM064/BM066) | |
| pAQ038 | FTT1103 pFNLTP-6-gfp (BM064/BM066) |
This study unless indicated otherwise.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Use | Restriction enzyme site |
|---|---|---|---|
| BM063 | TCCATATGCAAGAAATGGCTGCTC | Forward primer for FTT1103 in transcomplementation | NdeI |
| BM064 | GCGGCCGCTATAAGAAGGATAGGC | Forward primer for FTT1103 in transcomplementation | NotI |
| BM065 | TTCTCGAGTATCATCATCTTGGCTGAGC | Reverse primer for FTT1103 in transcomplementation | XhoI |
| BM066 | GGATCCTATCATCATCTTGGCTGAGC | Reverse primer for FTT1103 in transcomplementation | BamHI |
| BM082 | TCCGGATGAGTTAGTCCCTGGTACC | 5′ flanking forward primer for ΔFTT1103 | BspEI |
| BM083 | CATATGGTTGCAATAGCAGCAACAGC | 5′ flanking reverse primer for ΔFTT1103 | NdeI |
| BM084 | CATATGAGTGACAGCTGAAGCTCAAGC | 3′ flanking forward primer for ΔFTT1103 | NdeI |
| BM085 | CTCGAGATTACAGCATTACCAGCTGC | 3′ flanking reverse primer for ΔFTT1103 | XhoI |
Restriction enzyme sites are underlined.
DNA manipulation and transformation.
DNA was prepared and purified using commercial kits (Qiagen, Valencia, CA). Oligonucleotides were synthesized by Integrated DNA Technologies Inc. Restriction endonucleases and ligase were purchased from New England Biolabs. HotStart Taq (Qiagen) was used for routine PCR. The FastStart high-fidelity PCR system (Roche, Indianapolis, IN) was used for complementation and suicide vector construction. DNA sequencing was performed at the University of Virginia Biomolecular Research Facility. DNA and protein sequences were compared and aligned with CLUSTAL W (55). DNA transformation of Schu S4 was performed as described previously (43).
Construction of nonpolar deletion mutant and complementation plasmids.
DNA fragments corresponding to 1,336 bp upstream of FTT1103 (BM082 and BM083), including the start codon, and to 1,379 bp downstream of FTT1103 (BM084 and BM085), including the stop codon, were amplified by PCR and cloned into the sacB suicide vector pGir463 (a kind gift from Girija Ramakrishnan). The suicide plasmid pAQ031 was sequenced to verify the constructs before transformation. The plasmid was introduced by electroporation into competent strain Schu S4 as described previously (43). Colonies were grown on MHAC plates containing kanamycin and then streaked on an MHAC plate supplemented with 5% sucrose. Chromosomal DNAs were isolated from sucrose-resistant, kanamycin-sensitive colonies. Specific primers (BM064 and BM065) that corresponded to DNA flanking FTT1103 were used in PCR to confirm the deletion. The PCR product was sequenced to verify the nonpolarity of the deletion. The plasmid used for complementation of FTT1103 was constructed by inserting a full-length PCR product (obtained with BM064 and BM065), including 534 bp upstream of FTT1103, into the pFNLTP6-groE-gfp vector (37). The groE promoter was then deleted from this plasmid by digesting it with PacI, repairing the ends with an End-It DNA end repair kit (Epicentre, Madison, WI), and then self-ligating the DNA to obtain pAQ038. The cloned fragment was sequenced to verify the construct.
Invasion and intracellular replication assay.
Human hepatocellular carcinoma HepG2 (ATCC HB-8065) and murine macrophage J774A.1 (ATCC TIB-67) cells were propagated in high- and low-glucose Dulbecco modified Eagle medium, respectively, supplemented with 10% fetal bovine serum. Infection was carried out by adding bacteria to cells in a tissue culture well in a plate and then centrifuging the plate at 1,000 × g for 8 min. The plate was then incubated for 1 h at 37°C. After 1 h fresh medium containing 50 μg/ml gentamicin was added. At the indicated time points the cells were lysed by addition of 0.1% sodium deoxycholate in phosphate-buffered saline (PBS) for 5 min or in water for 10 min. The number of bacteria was determined by plating serial dilutions of the lysates on MHAC plates. CFU were counted 48 h later. These experiments were repeated at least three times. The Student two-tailed t test was used for statistical analysis.
Phagosomal integrity assay.
The phagosomal integrity assay was conducted as previously described (3). Briefly, infected J774A.1 cells (multiplicity of infection [MOI], 50:1) were washed three times with KHM buffer (110 mM potassium acetate, 20 mM HEPES, 2 mM MgCl2; pH 7.3), and this was followed by permeabilization of the plasma membrane with 50 μg/ml digitonin in KHM buffer for 1 min at room temperature. Cells were then washed with KHM buffer and incubated with rabbit anti-Francisella antibody (1:5,000 in KHM buffer; BD Sciences, San Jose, CA) for 12 min at 37°C. Cells were then immediately washed with PBS and fixed with paraformaldehyde. Next, cells were permeabilized with 0.1% saponin-10% goat serum in PBS and incubated with mouse anti-Schu S4 antiserum (1:5,000) and rat anti-lysosome-associated membrane protein 1 (anti-LAMP-1) (1:250; University of Iowa Hybridoma Bank, Iowa City) in 0.1% saponin-10% goat serum in PBS for 1 h at room temperature. Mouse anti-Schu S4 serum was generated by immunizing mice with a Sarkosyl-insoluble membrane fraction of Schu S4. Following washes with PBS, cells were incubated with goat anti-rat Alexa 488, goat anti-rabbit Alexa 546, and goat anti-mouse Alexa 633 (all at a 1:1,000 dilution in 0.1% saponin-10% goat serum in PBS) for 1 h at room temperature. Samples were viewed with a Zeiss LSM 510 laser scanning confocal microscope using a ×63 oil immersion lens and either LSM Image Browser or Image J software. With this procedure, bacteria that were within phagosomes were stained blue, while cytoplasmic bacteria were stained both red and blue. A minimum of 100 bacteria per sample were counted, and each experiment was performed in triplicate. Heat-killed bacteria (15 min at 70°C) were included in each experiment as a control. The Student two-tailed t test was used for statistical analysis.
Mouse virulence and immunization studies.
All mouse studies were approved by the University of Virginia's Animal Care and Use Committee. For intranasal inoculation or subcutaneous injection, 6- to 8-week-old C57BL/6 or BALB/c mice (Jackson Laboratory) were anesthetized with ketamine-HCl-xylazine. Twenty microliters of bacteria or PBS was inoculated into the nares, or 100 μl was injected. In protection assays mice were intranasally or subcutaneously immunized with 108 CFU of BJM1028 (transposon insertion in FTT1103) or BJM1031 (deletion of FTT1103) and then intranasally challenged with wild-type strain Schu S4 20 to 40 days later. The intended challenge dose was 100 organisms. The actual inoculation, immunization, and challenge doses were confirmed by viable plate counting. Four mice were used for each immunization group. To have a 95% chance of detecting infection in a situation where it is assumed that only 75% of the mice are infected, three mice would be required for each group (13). The infection and mortality rates for the PBS-immunized control groups were 100%, so four mice in each group were sufficient for these analyses. Overall, more than 40 mice were challenged with either the transposon or deletion mutant, and there were six separate vaccine trials. The mice were monitored daily. Mice showing signs of irreversible mortality were humanely euthanized. Death was considered to occur within 24 h.
Bacterial dissemination studies.
Eight-week-old C57BL/6 mice were anesthetized prior to subcutaneous injection or intranasal inoculation of either 200 CFU of Schu S4 or 5.8 × 1011 CFU of BJM1031 as described above. Three mice from each group were euthanized on the indicated days. The lungs, livers, and spleens were harvested, homogenized in 2 ml of PBS with a tissue grinder, and serially diluted, and then the dilutions were plated on MHAC plates. The plates were incubated at 37°C for 48 h, and then the numbers of colonies were counted. For presentation purposes a log10 transformation was performed on the data.
Antibody titers.
At 28 days postimmunization sera, fecal extracts, and lung homogenates were collected from mice immunized with PBS and BJM1031 (1 × 1011 CFU) intranasally to determine the antibody response using enzyme-linked immunosorbent assays. Mouse fecal extracts were prepared by resuspending three pellets in 1 ml of PBS with protease inhibitor cocktail (catalog no. P2714; Sigma, St. Louis, MO), vortexing the preparation vigorously, and then centrifuging it at 16,000 × g for 10 min. Supernatants were stored at −80οC (27). Lung homogenates were prepared by homogenizing lung tissue with a tissue grinder in 2 ml PBS and then pelleting the debris by centrifugation at 16,000 × g for 10 min. Supernatants were kept at −80οC until they were used. Microtiter plates were coated with 100 μl of a 1-mg/ml Schu S4 lysate and incubated overnight at 4°C. The plates were then washed six times with wash buffer (PBS-0.1% Tween 20), blocked at 37°C for 1 h with PBS-1% bovine serum albumin (BSA), and then washed six times with wash buffer. Samples were diluted in PBS-0.1% Tween 20-1% BSA to obtain dilutions ranging from 1:125 to 1:78,125 for sera, 1:1 to 1:125 for fecal extracts, and 1:5 to 1:3,125 for lung homogenates. Each dilution was added (100 μl/well) in duplicate to precoated wells and incubated at 37°C for 1 h. The plates were washed six times with wash buffer, and then 100 μl of horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G1 (IgG1), IgG2a, or IgA (Southern Biotech, Birmingham, AL) (diluted 1:5,000 in PBS-0.1% Tween 20-1% BSA) was added to each well and incubated at 37°C for 1 h. The plates were washed six times with wash buffer, and then 100 μl of 3,3′,5,5′-tetramethylbenzidine single solution substrate (Zymed/Invitrogen, Carlsbad, CA) was added to each well and incubated at 37°C for 15 min, which was followed by addition of 100 μl of 1 N HCl. The optical density at 450 nm (OD450) was determined with a Bio-Rad 680 microplate reader (Bio-Rad, Hercules, CA). Endpoint antibody titers were calculated for each antibody isotype by linear regression analysis and were expressed as log10 of the reciprocal of the highest antiserum dilution that gave an OD450 greater than the cutoff value, which was defined as the average OD450 for the PBS-immunized mice plus two standard deviations. Three mice were included in each group. The assays were performed in duplicate.
Immunoblot analysis.
Whole-cell lysates of Schu S4 were boiled in Laemmli buffer for 10 min, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and then transferred to polyvinylidene difluoride membranes. Pooled preimmune or 45-day postimmune sera were incubated with membranes at a 1:1,000 dilution. HRP-conjugated anti-mouse IgG (Sigma, St. Louis, MO) and HRP-conjugated anti-mouse IgA (Southern Biotech Birmingham, AL) were used as secondary antibodies. Blots were visualized with a Pierce ECL Western blot detection kit (Thermo Scientific, Rockford, IL).
RESULTS
FTT1103 was required for intracellular survival.
We previously identified a Schu S4 mutant with a transposon insertion in the FTT1103 locus that was unable to grow in HepG2 cells (43). FTT1103 is predicted to encode a 365-amino-acid hypothetical lipoprotein protein. To further explore the intracellular survival defect in HepG2 cells, infection with BJM1028 (Tn::FTT1103) was followed over time (Fig. 1). The uptake of BJM1028 was similar to that of wild-type strain Schu S4; however, it was apparent that BJM1028 did not replicate intracellularly within HepG2 cells and was rapidly eliminated, so this strain was undetectable by 48 h. FTT1103 protein was not required for growth in culture because BJM1028 and BJM1031 had growth curves similar to that of Schu S4 in TSBC or Chamberlain's defined medium (data not shown).
FIG. 1.
BJM1028 (Tn::FTT1103) was defective in intracellular survival in HepG2 cells. Bacteria were incubated with HepG2 cells at an MOI of 100:1. After 1 h fresh medium that contained 50 μg/ml of gentamicin was added the wells. At the indicated time points the HepG2 cells were lysed and dilutions of the lysate were plated on MHAC plates.
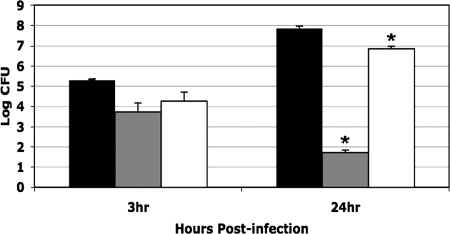
Macrophages are thought to be some of the primary host cells for F. tularensis in vivo (18). To examine survival within macrophages, macrophagelike J774A.1 cells were incubated with Schu S4, BJM1031 (ΔFTT1103), or complemented strain BJM1035. BJM1031 was severely deficient for growth in J774A.1 cells, but growth was restored to nearly wild-type levels by transcomplementation of the locus (Fig. 2).
FIG. 2.
BJM1031 (ΔFTT1103) was defective in intracellular survival in macrophagelike cell line J774A.1. Bacteria were incubated with J774A.1 cells at an MOI of 50:1. After 1 h fresh medium that contained 50 μg/ml of gentamicin was added the wells. At the indicated time points cells were lysed and dilutions of the lysate were plated on MHAC plates. Black bars, Schu S4; gray bars, BJM1031 (ΔFTT1103); open bars, BJM1035 (BJM1031/pAQ38 [FTT1103+]). Asterisks indicate values that are statistically different (P ≤ 0.0004) from the value for Schu S4 based on a two-tailed Student's t test.
The FTT1103 mutant BJM1031 was defective in escape from the phagosome.
After uptake into host cells, Francisella resides in a phagosome that transiently acquires endosomal markers, such as early endosomal antigen, and has limited interaction with LAMP-1 and -2 before it escapes into and replicates in the cytoplasm (4, 21, 30, 45, 46). To determine at what stage of intracellular growth BJM1031 was arrested, J774A.1 cells were infected with LVS, Schu S4, BJM1031, BJM1035, or heat-killed Schu S4 at an MOI of 50:1. At the indicated time points, cytoplasmic bacteria were labeled by perturbing the plasma membrane with digitonin, a selective detergent of cholesterol-containing membranes, and then incubating the preparation with rabbit anti-Francisella LPS antibody. Schu S4-containing vacuoles were negative for staining with cholesterol-associated GM1 (data not shown). Total bacteria were labeled by permeabilizing cell membranes with 0.1% saponin, followed by labeling with mouse anti-Francisella antibody. The cells were then incubated with goat anti-rabbit Alexa 546 and goat anti-mouse Alexa 633. When this procedure was used, bacteria that were within phagosomes appeared to be blue, while cytoplasmic bacteria were stained both red and blue. As shown in Fig. 3A, heat-killed bacteria were retained in phagosomes (at 30 min, 89.2% ± 5.2%; at 60 min, 96.5% ± 3.7%; at 120 min, 93.4% ± 2.4%), while LVS (at 30 min, 42.4% ± 4.0%; at 60 min, 25.5% ± 13.2%; at 120 min, 11.8% ± 2.2%) and Schu S4 (at 30 min, 6.4% ± 4.4%; at 60 min, 8.1% ± 1.8%; at 120 min, 7.1% ± 2.1%) were present in phagosomes at significantly lower levels. In contrast to LVS and Schu S4, BJM1031 was retained in the host cell phagosomes at much higher levels (at 30 min, 70.1% ± 6.1%; at 60 min, 74.6% ± 1.4%; at 120 min, 74.1% ± 6.1%). The ability to escape from the phagosome was restored in the complementation strain, strain BJM1035 (at 30 min, 40.2% ± 3.6%; at 60 min, 46.8% ± 1.7%; at 120 min, 33.6% ± 8.2%).
FIG. 3.
FTT1103 was required for efficient escape from the phagosome. Bacteria were incubated with J774A.1 cells at an MOI of 50:1. A phagosomal integrity assay was performed as described in Materials and Methods and by Checroun et al. (3). Briefly, the plasma membrane was first permeabilized with digitonin, and then cells were incubated with rabbit anti-Francisella antibody. Next, the cells were fixed and then treated with saponin, which permeabilized the entire membrane component of the cells. Finally, the cells were treated with mouse anti-Schu S4 and rat anti-LAMP-1 antisera, washed, and then incubated with goat anti-rat Alexa 488, goat anti-rabbit Alexa 546, and goat anti-mouse Alexa 633. Samples were viewed with a Zeiss LSM 510 laser scanning confocal microscopy. A minimum of 100 bacteria were counted for each sample, and experiments were performed in triplicate. Asterisks indicate values that were statistically different from the value for Schu S4 (P ≤ 0.03) as determined by a two-tailed Student's t test. (A) Percentage of phagosomal bacteria. Bars 1, heat-killed Schu S4; bars 2, LVS; bars 3, Schu S4; bars 4, BJM1031; bars 5, BJM1035. (B) Representative merged confocal images from the phagosomal escape assay. In the merged image cytoplasmic bacteria are red and phagosomal bacteria are blue. HK-Schu S4, heat-killed Schu S4. (C) Percentages of bacteria that colocalized with LAMP-1. Bars 1, heat-killed Schu S4; bars 2, Schu S4; bars 3, BJM1031; bars 4, BJM1035. (D) Colocalization with LAMP-1. Bacteria were detected using mouse anti-Schu S4 and goat anti-mouse Alexa 546 antibodies (green pseudocolor), and LAMP-1 was detected using rat anti-LAMP-1 and goat anti-rat Alexa 488 (red pseudocolor). H-K Schu S4, heat-killed Schu S4.
As a confirmation of the inability of BJM1031 to effectively escape from the host cell phagosome, we next examined the extent of colocalization of LAMP-1 and either heat-killed Schu S4, Schu S4, BJM1031, or BJM1035 by fluorescence microscopy (Fig. 3C and 3D). Heat-killed bacteria extensively colocalized with LAMP-1 (at 30 min, 77.6% ± 4.8%; at 60 min, 88.3% ± 2.9%), while Schu S4 evaded colocalization with LAMP-1 far more effectively (at 30 min, 19.4% ± 6.1%; at 60 min, 15.7% ± 5.1%). Consistent with its inability to escape from the host cell phagosome, BJM1031 colocalized with LAMP-1 at significantly higher levels than Schu S4 (at 30 min, 68.0% ± 4.7%; at 60 min, 66.8% ± 3.5%). Thus, these data suggest that the FTT1103 product is required for efficient escape from the phagosome.
The FTT1103 product was required for in vivo virulence.
Ten or fewer Schu S4 bacteria are lethal to mice when the intranasal route is used (35). To test the virulence of BJM1031, groups of four C57BL/6 mice were inoculated intranasally with the mutant bacteria using amounts in 10-fold increments from 2,000 to 2 × 108 CFU. All mice, including those given the highest dose (2 × 108 CFU), survived for more than 20 days. C57BL/6 mice also survived intranasal challenge by 1.8 × 1010 CFU of BJM1031 without visible signs of illness, while mice challenged with 6 × 104 CFU of complemented strain BJM1035 died 10 days postinfection (Fig. 4). In addition, C57BL/6 mice survived for more than 21 days after infection when 1 × 106 CFU was administered via the intraperitoneal, subcutaneous, or intravenous route.
FIG. 4.
BJM1031 is avirulent in mice. C57BL/6 mice (three mice per group) were intranasally inoculated with 20 μl of BJM1031 (1.8 × 1010 CFU), BJM1035 (6 × 104 CFU), or Schu S4 (100 CFU). Mice were monitored daily. More than 12 mice survived challenge with >1010 CFU of BJM1031 in three separate experiments.
BJM1031 bacteria exhibit a dissemination defect in vivo.
To examine the ability of BJM1031 to persist and disseminate, C57BL/6 mice were inoculated by the intranasal or subcutaneous route with BJM1031 or Schu S4. Mice were sacrificed on days 1, 3, 5, and 28 after infection. The lungs, livers, and spleens were harvested, and the bacterial burden was determined for each tissue (Fig. 5). When Schu S4 was administered by the intranasal route, it was detected in the lungs, liver, and spleen on day 1, although it was not consistently detected in the spleen. The number of bacteria increased in all organs on days 3 and 5. On day 5 the mice were moribund. The numbers of bacteria recovered from the organs of mice infected intranasally with BJM1031 were similar to the numbers of bacteria recovered from the organs of mice infected intranasally with Schu S4 on day 1, but by day 3 BJM1031 was detectable only in the lungs. No bacteria were recovered from any organ from day 28 mice. When the subcutaneous route of inoculation was used, Schu S4 was detectable in the liver and spleen on day 1 but was not detectable in the lungs until day 3. No subcutaneously injected BJM1031 bacteria were recovered from any organ until day 3, and no bacteria were ever detected in the lungs. Low levels of BJM1031 persisted in the spleen and liver through day 5, and the mice appeared to be healthy when they were euthanized. No bacteria were detected in any organ on day 28. These results suggest that BJM1031 is defective in dissemination. When the intranasal route was used, low levels of bacteria were present in the lungs until at least day 5, but bacteria either did not disseminate to the spleen or liver or were eliminated at these sites. When the subcutaneous injection route was used, the bacteria did not disseminate to the lungs, but low levels did persist in the liver and spleen until at least day 5 in some mice.
FIG. 5.
BJM1031 was defective in dissemination. C57BL/6 mice were intranasally or subcutaneously infected with either 200 CFU of Schu S4 or 5.8 × 1011 CFU of BJM1031. On the indicated days three mice from each group were euthanized. The lungs, livers, and spleens were harvested, homogenized with a tissue grinder, and then serially diluted, and the serial dilutions were plated on MHAC plates. For presentation log10 transformation was performed on the data.
Intranasal immunization with BJM1028 or BJM1031 protected C57BL/6 and BALB/c mice from intranasal challenge with wild-type bacteria.
To test whether prior immunization with the FTT1103 mutants could elicit protective immunity, C57BL/6 mice were intranasally immunized and challenged with intended doses containing 108 mutant bacteria and 100 Schu S4 bacteria, respectively. The actual dose was determined by plating the dose on MHAC plates. In the first trial, mice were intranasally immunized with 1 × 108 CFU of BJM1028 and then challenged 21 days later with 1,000 CFU of wild-type strain Schu S4. Two of the three mice were protected (Table 3). To further test for protective immunity, 12 more mice were immunized with 1.3 × 107 to 2.6 × 107 CFU of BJM1031 and then challenged 30 to 40 days later with 35 to 70 CFU of Schu S4. Ten of the 12 mice survived (Table 3). BALB/c mice were also immunized intranasally with 1 × 108 CFU of BJM1031 and then challenged intranasally with 95 CFU of Schu S4. Three of four mice survived the intranasal challenge with 95 CFU of Schu S4. Intranasally immunized mice had both Schu S4-specific systemic and mucosal antibodies compared to nonimmune mice (Fig. 6). Specific IgA was detected in the serum, fecal homogenates, and lung homogenates. Specific antibodies for both IgG1 and IgG2a isotypes were also detected.
TABLE 3.
Protection of two mouse strains immunized with FTT1103 mutants against intranasal challenge with Schu S4
| Mouse strain | Immunization strain | Intranasal immunization dose (CFU) | Schu S4 challenge dose (CFU) | Day of intranasal challenge | No. of survivors/no. tested | Time to death (days postchallenge)a |
|---|---|---|---|---|---|---|
| C57BL/6 | BJM1028 | 1.6 × 108 | 1,000 | 20 | 2/3 | 7, >21, >21 |
| C57BL/6 | PBS | 1,000 | 20 | 0/3 | 5, 5, 5 | |
| C57BL/6 | BJM1031 | 2.6 × 107 | 68 | 32 | 4/4 | >21, >21, >21, >21 |
| C57BL/6 | BJM1031 | 1.3 × 108 | 37 | 32 | 4/4 | >21, >21, >21, >21 |
| C57BL/6 | BJM1031 | 2.6 × 108 | 68 | 32 | 2/4 | 8, 14, >21, >21 |
| C57BL/6 | PBS | 68 | 32 | 0/4 | 5, 5, 5, 5 | |
| BALB/c | BJM1031 | 1.0 × 108 | 95 | 33 | 3/4 | 10, >21, >21, >21 |
| BALB/c | PBS | 95 | 33 | 0/4 | 5, 5, 5, 5 |
Mice were humanely euthanized when signs of irreversible morbidity were evident. Death was considered to occur in 24 h or less. A value of <21 days indicates that a mouse was healthy upon euthanasia 21 or more days after challenge.
FIG. 6.
Intranasal immunization with BJM1031 elicited specific systemic and mucosal antibody responses. (A) Western blots of whole-cell lysates of Schu S4 were incubated with pooled preimmune sera (lane P) or immune sera (lane I) from mice intranasally immunized with BJM1031 and then with secondary antibodies that were specific for IgA or IgG heavy chains. Reactive proteins were visualized by chemiluminescence. (B) Sera were collected from three mice 28 days after infection with 5.8 × 1011 CFU of BJM1031. Antibody isotype titers were determined by isotype-specific enzyme-linked immunosorbent assays. Endpoint antibody titers were calculated by linear regression analysis and were expressed as log10 of the reciprocal of the highest dilution that gave an OD450 greater than the cutoff value, which was defined as the average OD450 for the PBS-immunized mice plus 2.0 standard deviations. Assays were performed in duplicate.
DISCUSSION
We identified a novel locus, FTT1103, in the Schu S4 strain that is required for F. tularensis subsp. tularensis virulence. Mutation of this locus rendered this strain avirulent, but the strain was still able to elicit a protective immune response when it was introduced by the intranasal route. FTT1103 is predicted to encode a hypothetical lipoprotein. However, this protein also has a region with similarity (e value, 1.4e-06) to the conserved DsbA Pfam domain (PFO1323). DsbA proteins are periplasmic oxidoreductase enzymes that catalyze disulfide bond formation in nascent proteins as they enter or transit through the periplasmic space (6). Several pathogenic bacteria with mutations in dsbA have attenuated phenotypes. The activity of DsbA has been shown to be important for the formation of a number of virulence factors, including the biogenesis of type IV pili in bacteria such as enteropathogenic E. coli and Pseudomonas aeruginosa (23, 60), efflux pumps in E. coli and Burkholderia (24, 52), the assembly and function of type III secretion systems in Salmonella enterica serovar Typhimurium and Shigella flexneri (14, 39, 61), and toxins such as cholera and pertussis toxins (61). DsbA is also important for the intracellular survival of S. flexneri, S. enterica serovar Typhimurium, and P. aeruginosa (14, 23, 39, 61). It remains to be determined whether FTT1103 encodes a DsbA ortholog in Schu S4. Although the FTT1103 protein contains the conserved DsbA active site, this protein is only 67% similar in a 12-amino-acid overlapping region to E. coli DsbA (Bestfit, GCG version 11.1; Accelrys Inc., San Diego, CA). If FTT1103 encodes a DsbA ortholog, this suggests that the virulence defect of the FTT1103 mutant is indirect and could be due to the inactivation of multiple virulence factors. The virulence factors encoded by the FPI are some of the logical candidates. The genome of Schu S4 also contains all the genes necessary to assembly type IV pili, although the presence of pili on type A strains has not been demonstrated (17). Clearly, these putative FTT1103-dependent factors would have to have roles in phagosomal escape and/or intracellular survival since the FTT1103 mutant was rapidly cleared from J774A.1 cells and at 3 h postinfection there was more than a 1-log difference in the number of bacteria recovered. A phagosomal integrity assay and LAMP-1 colocalization data also demonstrated that the FTT1103 mutant was impaired in the ability to escape from phagosomes. FTT1103 lipoprotein has been shown to be capable of inducing Toll-like receptor 2 (TLR2)-dependent proinflammatory chemokines, which suggests that it has a role in innate immunity, although Francisella has multiple TLR2 agonists (54). Innate immune responses to LVS are mediated primarily through TLR2 (5, 25, 28, 32).
Despite the high level of attenuation, the FTT1103 mutant was able to induce protective immunity against intranasal challenge with almost 10 times the reported lethal dose (<10 organisms) in two different mouse genetic backgrounds. Thus, although the FTT1103 protein is required for virulence, it is not an essential protective antigen. We initially chose a challenge dose of 100 bacteria to guard against dilution errors, which were significant. Although we did not specifically test the limits of protection, the fact that a few mice did not survive a challenge with 68 CFU suggests that protective immunity can be overwhelmed by higher challenge doses.
The specific growth characteristics of an attenuated Francisella vaccine candidate, particularly a vaccine candidate for use against challenges with type A strains, that are required to elicit a protective immune response remain unclear. iglC mutants of LVS or F. novicida have been shown to be severely impaired in phagosomal escape (34, 47). Less than 1% of LVS and 10 to 20% of F. novicida iglC mutants are able to escape into the cytoplasm (34, 47). However, intradermal immunization with a Schu S4 iglC mutant does not protect against an intradermal challenge with wild-type strain Schu S4, despite the ability of the mutant to persist for at least 4 days in mouse skin, livers, and spleens (56). Although the FTT1103 mutant was defective in phagosomal escape, approximately 25% of the bacteria appeared to be able to escape into the cytoplasm. Perhaps, as suggested by other workers (9), this low level of escape facilitates better antigen presentation and thus a more robust adaptive immune response. Dissemination or at least persistence in the liver was not required because BJM1031 was not detectable in the liver or spleen on day 3 but did persist in the lungs. Intranasal immunization with BJM1031 elicited both systemic and mucosal antibody responses. Both IgG1 and IgG2b isotypes were detected, suggesting that both Th-1 and Th-2 cytokine responses occur. Immune sera have been shown to protect against challenge with LVS (29, 41, 50), but both antibody and cell-mediated responses are necessary to control a type A infection (10, 11, 19). We noticed that preimmune sera recognized several Schu S4 proteins. It seems unlikely that this reactivity represented prior exposure to Francisella because of the high degree of lethality and low lethal dose of Francisella (35). Other investigators have observed Francisella proteins that cross-react with preimmune sera, including a ribosomal protein and GroE (15, 57).
Human and animal studies with LVS have demonstrated that a live tularemia vaccine can be safe and efficacious (for a review, see reference 9). Optimal protection against aerosol or intranasal challenge requires immunization by the same route (7, 59). One of the limitations of LVS is that is it more reactogenic when it is given by the aerosol or intranasal route (9, 26). One approach to overcome reactogenicity would be to further attenuate LVS by creating defined mutants. One attempt at this was an LVS strain with a mutation in LPS biosynthesis; this strain protected against intranasal challenge with wild-type type B strains but not against intranasal challenge with type A strains (48). However, an attenuated LVS mutant with a mutation in iron superoxide dismutase has shown increased protection against a respiratory challenge with Schu S4 compared to the protection provided by LVS (2).
The large difference between the virulence of LVS, which has an intranasal 50% lethal dose of ∼1,000 bacteria (38), and the virulence of Schu S4 (essentially <10 bacteria given intranasally can kill 100% of the mice tested) might suggest that it is not possible to adequately or effectively attenuate a type A strain for use as a live vaccine. However, a spontaneous attenuated mutant of Schu S4, FSC043, provides good protection against intranasal challenge with wild-type bacteria (56). It was discovered that FSC043 lacks a 58-kDa protein (FTT918). Twine et al. created a defined ΔFTT918 mutant. This mutant also provides protection against wild-type challenge but is too virulent to be used as an attenuated vaccine. In contrast, we found that large doses of the FTT1103 mutant, given by every route tested, did not produce any overt symptoms of disease, and intranasal immunization was able to elicit an immune response that protected against a challenge that was at least 10 times the lethal dose. It is also noteworthy that this vaccine was efficacious in both the BALB/c and C57BL/6 mouse backgrounds. Wu et al. found that intranasal vaccination with LVS protected BALB/c mice but not C57BL/6 mice against intranasal challenge with a type A strain (59). Our work demonstrates that it is possible to create a broadly protective attenuated or avirulent type A live vaccine with just a single defined mutation.
Acknowledgments
This work was supported by grant U54 AI057168 from the NIH/NIAID Middle Atlantic Regional Center of Excellence for Biodefense and Emerging Infectious Diseases.
The LAMP-1 monoclonal antibody developed by J. Thomas August was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the Department of Biological Sciences, University of Iowa, Iowa City.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 3 November 2008.
REFERENCES
- 1.Anthony, L. D., R. D. Burke, and F. E. Nano. 1991. Growth of Francisella spp. in rodent macrophages. Infect. Immun. 593291-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakshi, C. S., M. Malik, M. Mahawar, G. S. Kirimanjeswara, K. R. Hazlett, L. E. Palmer, M. B. Furie, R. Singh, J. A. Melendez, T. J. Sellati, and D. W. Metzger. 2008. An improved vaccine for prevention of respiratory tularemia caused by Francisella tularensis SchuS4 strain. Vaccine 265276-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Checroun, C., T. D. Wehrly, E. R. Fischer, S. F. Hayes, and J. Celli. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA 10314578-14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 723204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, L. E., A. Santiago, E. Barry, T. J. Kang, K. A. Shirey, Z. J. Roberts, K. L. Elkins, A. S. Cross, and S. N. Vogel. 2008. Macrophage proinflammatory response to Francisella tularensis live vaccine strain requires coordination of multiple signaling pathways. J. Immunol. 1806885-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collet, J. F., and J. C. Bardwell. 2002. Oxidative protein folding in bacteria. Mol. Microbiol. 441-8. [DOI] [PubMed] [Google Scholar]
- 7.Conlan, J. W., W. Chen, H. Shen, A. Webb, and R. KuoLee. 2003. Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb. Pathog. 34239-248. [DOI] [PubMed] [Google Scholar]
- 8.Conlan, J. W., and R. J. North. 1992. Early pathogenesis of infection in the liver with the facultative intracellular bacteria Listeria monocytogenes, Francisella tularensis, and Salmonella typhimurium involves lysis of infected hepatocytes by leukocytes. Infect. Immun. 605164-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlan, J. W., and P. C. Oyston. 2007. Vaccines against Francisella tularensis. Ann. N. Y. Acad. Sci. 1105325-350. [DOI] [PubMed] [Google Scholar]
- 10.Conlan, J. W., H. Shen, R. Kuolee, X. Zhao, and W. Chen. 2005. Aerosol-, but not intradermal-immunization with the live vaccine strain of Francisella tularensis protects mice against subsequent aerosol challenge with a highly virulent type A strain of the pathogen by an alphabeta T cell- and interferon gamma- dependent mechanism. Vaccine 232477-2485. [DOI] [PubMed] [Google Scholar]
- 11.Conlan, J. W., H. Shen, A. Webb, and M. B. Perry. 2002. Mice vaccinated with the O-antigen of Francisella tularensis LVS lipopolysaccharide conjugated to bovine serum albumin develop varying degrees of protective immunity against systemic or aerosol challenge with virulent type A and type B strains of the pathogen. Vaccine 203465-3471. [DOI] [PubMed] [Google Scholar]
- 12.de Bruin, O. M., J. S. Ludu, and F. E. Nano. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dell, R. B., S. Hollaran, and R. Ramakrishnan. 2002. Sample size determination. ILAR J. 43207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellermeier, C. D., and J. M. Slauch. 2004. RtsA coordinately regulates DsbA and the Salmonella pathogenicity island 1 type III secretion system. J. Bacteriol. 18668-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ericsson, M., A. Tarnvik, K. Kuoppa, G. Sandstrom, and A. Sjostedt. 1994. Increased synthesis of DnaK, GroEL, and GroES homologs by Francisella tularensis LVS in response to heat and hydrogen peroxide. Infect. Immun. 62178-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forestal, C. A., J. L. Benach, C. Carbonara, J. K. Italo, T. J. Lisinski, and M. B. Furie. 2003. Francisella tularensis selectively induces proinflammatory changes in endothelial cells. J. Immunol. 1712563-2570. [DOI] [PubMed] [Google Scholar]
- 17.Forslund, A. L., K. Kuoppa, K. Svensson, E. Salomonsson, A. Johansson, M. Bystrom, P. C. Oyston, S. L. Michell, R. W. Titball, L. Noppa, E. Frithz-Lindsten, M. Forsman, and A. Forsberg. 2006. Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 591818-1830. [DOI] [PubMed] [Google Scholar]
- 18.Fortier, A. H., S. J. Green, T. Polsinelli, T. R. Jones, R. M. Crawford, D. A. Leiby, K. L. Elkins, M. S. Meltzer, and C. A. Nacy. 1994. Life and death of an intracellular pathogen: Francisella tularensis and the macrophage. Immunol. Ser. 60349-361. [PubMed] [Google Scholar]
- 19.Fulop, M., P. Mastroeni, M. Green, and R. W. Titball. 2001. Role of antibody to lipopolysaccharide in protection against low- and high-virulence strains of Francisella tularensis. Vaccine 194465-4472. [DOI] [PubMed] [Google Scholar]
- 20.Gallagher, L. A., E. Ramage, M. A. Jacobs, R. Kaul, M. Brittnacher, and C. Manoil. 2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. USA 1041009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjostedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 715940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray, C. G., S. C. Cowley, K. K. Cheung, and F. E. Nano. 2002. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 21553-56. [DOI] [PubMed] [Google Scholar]
- 23.Ha, U. H., Y. Wang, and S. Jin. 2003. DsbA of Pseudomonas aeruginosa is essential for multiple virulence factors. Infect. Immun. 711590-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi, S., M. Abe, M. Kimoto, S. Furukawa, and T. Nakazawa. 2000. The dsbA-dsbB disulfide bond formation system of Burkholderia cepacia is involved in the production of protease and alkaline phosphatase, motility, metal resistance, and multi-drug resistance. Microbiol. Immunol. 4441-50. [DOI] [PubMed] [Google Scholar]
- 25.Hong, K. J., J. R. Wickstrum, H. W. Yeh, and M. J. Parmely. 2007. Toll-like receptor 2 controls the gamma interferon response to Francisella tularensis by mouse liver lymphocytes. Infect. Immun. 755338-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornick, R. B., and H. T. Eigelsbach. 1966. Aerogenic immunization of man with live tularemia vaccine. Bacteriol. Rev. 30532-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houpt, E. R., D. J. Glembocki, T. G. Obrig, C. A. Moskaluk, L. A. Lockhart, R. L. Wright, R. M. Seaner, T. R. Keepers, T. D. Wilkins, and W. A. Petri, Jr. 2002. The mouse model of amebic colitis reveals mouse strain susceptibility to infection and exacerbation of disease by CD4+ T cells. J. Immunol. 1694496-4503. [DOI] [PubMed] [Google Scholar]
- 28.Katz, J., P. Zhang, M. Martin, S. N. Vogel, and S. M. Michalek. 2006. Toll-like receptor 2 is required for inflammatory responses to Francisella tularensis LVS. Infect. Immun. 742809-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirimanjeswara, G. S., J. M. Golden, C. S. Bakshi, and D. W. Metzger. 2007. Prophylactic and therapeutic use of antibodies for protection against respiratory infection with Francisella tularensis. J. Immunol. 179532-539. [DOI] [PubMed] [Google Scholar]
- 30.Lai, X. H., I. Golovliov, and A. Sjostedt. 2001. Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 694691-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoeba and intramacrophage survival. Proc. Natl. Acad. Sci. USA 1014246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, H., S. Nookala, X. R. Bina, J. E. Bina, and F. Re. 2006. Innate immune response to Francisella tularensis is mediated by TLR2 and caspase-1 activation. J. Leukoc. Biol. 80766-773. [DOI] [PubMed] [Google Scholar]
- 33.Li, J., C. Ryder, M. Mandal, F. Ahmed, P. Azadi, D. S. Snyder, R. D. Pechous, T. Zahrt, and T. J. Inzana. 2007. Attenuation and protective efficacy of an O-antigen-deficient mutant of Francisella tularensis LVS. Microbiology 1533141-3153. [DOI] [PubMed] [Google Scholar]
- 34.Lindgren, H., I. Golovliov, V. Baranov, R. K. Ernst, M. Telepnev, and A. Sjostedt. 2004. Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53953-958. [DOI] [PubMed] [Google Scholar]
- 35.Lyons, R. C., and T. H. Wu. 2007. Animal models of Francisella tularensis infection. Ann. N. Y. Acad. Sci. 1105238-265. [DOI] [PubMed] [Google Scholar]
- 36.Maier, T. M., M. S. Casey, R. H. Becker, C. W. Dorsey, E. M. Glass, N. Maltsev, T. C. Zahrt, and D. W. Frank. 2007. Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect. Immun. 755376-5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maier, T. M., A. Havig, M. Casey, F. E. Nano, D. W. Frank, and T. C. Zahrt. 2004. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl. Environ. Microbiol. 707511-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metzger, D. W., C. S. Bakshi, and G. Kirimanjeswara. 2007. Mucosal immunopathogenesis of Francisella tularensis. Ann. N. Y. Acad. Sci. 1105266-283. [DOI] [PubMed] [Google Scholar]
- 39.Miki, T., N. Okada, and H. Danbara. 2004. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J. Biol. Chem. 27934631-34642. [DOI] [PubMed] [Google Scholar]
- 40.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 1866430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pammit, M. A., E. K. Raulie, C. M. Lauriano, K. E. Klose, and B. P. Arulanandam. 2006. Intranasal vaccination with a defined attenuated Francisella novicida strain induces gamma interferon-dependent antibody-mediated protection against tularemia. Infect. Immun. 742063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pechous, R., J. Celli, R. Penoske, S. F. Hayes, D. W. Frank, and T. C. Zahrt. 2006. Construction and characterization of an attenuated purine auxotroph in a Francisella tularensis live vaccine strain. Infect. Immun. 744452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin, A., and B. J. Mann. 2006. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quarry, J. E., K. E. Isherwood, S. L. Michell, H. Diaper, R. W. Titball, and P. C. Oyston. 2007. A Francisella tularensis subspecies novicida purF mutant, but not a purA mutant, induces protective immunity to tularemia in mice. Vaccine 252011-2018. [DOI] [PubMed] [Google Scholar]
- 45.Santic, M., M. Molmeret, and Y. Abu Kwaik. 2005. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell. Microbiol. 7957-967. [DOI] [PubMed] [Google Scholar]
- 46.Santic, M., M. Molmeret, K. E. Klose, and Y. Abu Kwaik. 2006. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 1437-44. [DOI] [PubMed] [Google Scholar]
- 47.Santic, M., M. Molmeret, K. E. Klose, S. Jones, and Y. A. Kwaik. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7969-979. [DOI] [PubMed] [Google Scholar]
- 48.Sebastian, S., S. T. Dillon, J. G. Lynch, L. T. Blalock, E. Balon, K. T. Lee, L. E. Comstock, J. W. Conlan, E. J. Rubin, A. O. Tzianabos, and D. L. Kasper. 2007. A defined O-antigen polysaccharide mutant of Francisella tularensis live vaccine strain has attenuated virulence while retaining its protective capacity. Infect. Immun. 752591-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sjostedt, A. 2006. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 8561-567. [DOI] [PubMed] [Google Scholar]
- 50.Stenmark, S., H. Lindgren, A. Tarnvik, and A. Sjostedt. 2003. Specific antibodies contribute to the host protection against strains of Francisella tularensis subspecies holarctica. Microb. Pathog 3573-80. [DOI] [PubMed] [Google Scholar]
- 51.Su, J., J. Yang, D. Zhao, T. H. Kawula, J. A. Banas, and J. R. Zhang. 2007. Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 753089-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takatsuka, Y., and H. Nikaido. 2007. Site-directed disulfide cross-linking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J. Bacteriol. 1898677-8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tempel, R., X. H. Lai, L. Crosa, B. Kozlowicz, and F. Heffron. 2006. Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 745095-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thakran, S., H. Li, C. L. Lavine, M. A. Miller, J. E. Bina, X. R. Bina, and F. Re. 2008. Identification of Francisella tularensis lipoproteins that stimulate the Toll-like receptor (TLR) 2/TLR1 heterodimer. J. Biol. Chem. 2833751-3760. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Twine, S., M. Bystrom, W. Chen, M. Forsman, I. Golovliov, A. Johansson, J. Kelly, H. Lindgren, K. Svensson, C. Zingmark, W. Conlan, and A. Sjostedt. 2005. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 738345-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Twine, S. M., M. Petit, H. Shen, N. C. Mykytczuk, J. F. Kelly, and J. W. Conlan. 2006. Immunoproteomic analysis of the murine antibody response to successful and failed immunization with live anti-Francisella vaccine. Biochem. Biophys. Res. Commun. 347999-1008. [DOI] [PubMed] [Google Scholar]
- 58.Weiss, D. S., A. Brotcke, T. Henry, J. J. Margolis, K. Chan, and D. M. Monack. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. USA 1046037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, T. H., J. A. Hutt, K. A. Garrison, L. S. Berliba, Y. Zhou, and C. R. Lyons. 2005. Intranasal vaccination induces protective immunity against intranasal infection with virulent Francisella tularensis biovar A. Infect. Immun. 732644-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, J. 1998. Inactivation of DsbA, but not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect. Immun. 663909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu, J., and J. S. Kroll. 1999. DsbA: a protein-folding catalyst contributing to bacterial virulence. Microbes Infect. 11221-1228. [DOI] [PubMed] [Google Scholar]