Abstract
The antigen-binding fragment of the broadly neutralizing human immunodeficiency virus type 1 (HIV-1) antibody 2G12 has an unusual three-dimensional (3D) domain-swapped structure with two aligned combining sites that facilitates recognition of its carbohydrate epitope on gp120. When expressed as an intact immunoglobulin G (IgG), 2G12 formed typical IgG monomers containing two combining sites and a small fraction of a higher-molecular-weight species, which showed a significant increase in neutralization potency (50- to 80-fold compared to 2G12 monomer) across a range of clade A and B strains of HIV-1. Here we show that the higher-molecular-weight species corresponds to a 2G12 dimer containing four combining sites and present a model for how intermolecular 3D domain swapping could create a 2G12 dimer. Based on the structural model for a 3D domain-swapped 2G12 dimer, we designed and tested a series of 2G12 mutants predicted to increase the ratio of 2G12 dimer to monomer. We report a mutation that effectively increases the 2G12 dimer/monomer ratio without decreasing the expression yield. Increasing the proportion of 2G12 dimer compared to monomer could lead to a more potent reagent for gene therapy or passive immunization.
Broadly neutralizing antibodies against human immunodeficiency virus type 1 (HIV-1) have attracted attention not only for the lessons they provide for designing vaccine antigens to induce a more robust immunological response (2) but also as potential therapeutic reagents. Although HIV infection leads to a vigorous antibody response, most antibodies fail to control the virus due to targeting of non-neutralizing epitopes or the ability of escape mutants to quickly develop against neutralizing antibodies (23). Correlating with the ability of the virus to elude antibodies, the majority of neutralizing antibodies are highly strain specific. Nevertheless, a small set of broadly neutralizing antibodies has been isolated from the blood of HIV-infected individuals, and these reagents have been extensively studied (2). Clinical trials using a cocktail of three such antibodies—2G12, 4E10, and 2F5—have demonstrated a partial ability to suppress viral replication (13, 20, 21).
The 2G12 antibody has an unusual structure that facilitates recognition of its carbohydrate epitope on gp120 (4). Whereas typical immunoglobulin G (IgG) antibodies contain two flexibly attached antigen-binding fragments (Fabs), resulting in two antigen-binding sites separated by distances ranging from 120 to 150 Å in structures of intact IgGs (6, 7, 17), the Fab arms of 2G12 are entwined in such a way as to create a single antigen-binding region with two rigidly arranged antigen-binding sites separated by ∼35 Å (4) (Fig. 1A and B). The entwined structure of the 2G12 Fabs results from three-dimensional (3D) domain swapping (1) in which each 2G12 light chain associates with both heavy chains: the light-chain variable domain (VL) is paired with the variable domain of one heavy chain (VH), while the light constant domain (CL) is paired with constant domain 1 (CH1) of the partner heavy chain (Fig. 1B). This domain-swapped arrangement prevents the Fab arms from having the normal flexibility observed in other antibodies but, by possessing a double-sized antigen-combining site, the 2G12 Fab2 unit is able to recognize clusters of mannose-rich carbohydrates that occur on gp120 (18). Normally, these carbohydrates create a glycan shield on the HIV envelope glycoprotein (Env) spike that helps the virus evade the host antibody response (23).
FIG. 1.
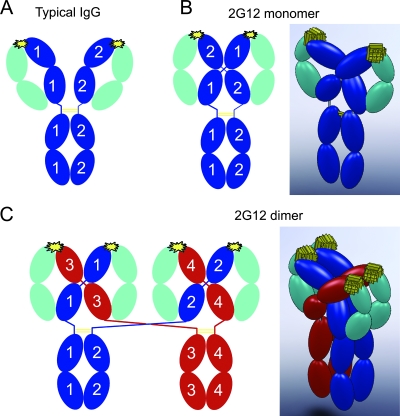
Schematic structures of a typical IgG and 2G12. Heavy chains are blue in panels A and B and blue or red in panel C, light chains are cyan, disulfide bonds are yellow lines, and the antigen combining sites are yellow starbursts. (A) Schematic diagram showing the domain arrangement of a typical IgG, which contains two identical heavy chains and two identical light chains. (B) Schematic diagram (left) and a corresponding 3D model (right) illustrating chain pairing in monomeric 2G12 (based on structural data from reference 4). As a result of intramolecular 3D domain swapping, each heavy chain forms part of both Fab units to create a rigidly arranged Fab2 unit. To distinguish the two heavy chains, they are labeled 1 or 2 in the schematic diagram. (C) Schematic diagram (left) and corresponding 3D model (right) illustrating chain pairing in dimeric 2G12. The proposed dimer structure resulting from intermolecular 3D domains swapping has the same domain-swapped Fab2 unit as the monomer, but the connectivity to the Fc domains is altered. To distinguish the four heavy chains, they are labeled 1, 2, 3, or 4 in the schematic diagram and are red in one of the IgG monomer precursors.
During expression of 2G12 in mammalian cells, we observed the production of IgG monomers (i.e., two heavy chains, two light chains, and thus two Fabs), as typically formed by other IgGs, and a higher-molecular-weight fraction that exhibited a significantly increased neutralization potency. Here, we show that the higher-molecular-weight fraction corresponded to a 2G12 dimer with four heavy chains, four light chains, and four Fabs and present a model for how 3D domain swapping could create a 2G12 dimer. We used the model of the 2G12 dimer to design mutations predicted to increase the fraction of dimer being expressed and report a mutation that effectively increases the 2G12 dimer/monomer ratio without decreasing the expression yield.
MATERIALS AND METHODS
Materials.
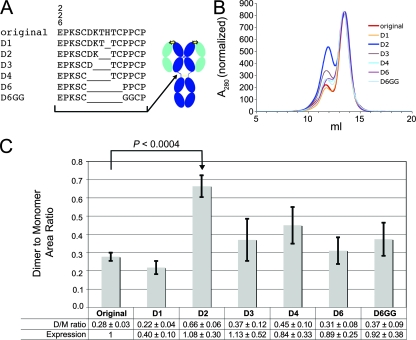
Sequences encoding the 2G12 VH-CH1 and VL-CL domains in the pComb3H expression vector (a gift from Dennis Burton, The Scripps Research Institute) were subcloned into the bicistronic baculovirus vector pAc-κ-Fc (PROGEN Biotechnik), which contains the gene for the Fc region of human IgG1 (G1m marker). The heavy-chain gene, now including the hinge and Fc regions (residues 236 to 446, numbered according to the method of Kabat et al. [11]), and light-chain gene were each subcloned into the mammalian expression vector pTT5 (NRC Biotechnology Research Institute) for expression in HEK293-6E cells. Mutations were introduced in the 2G12 heavy-chain gene by using a QuikChange mutagenesis kit (Stratagene). Seven different forms of the 2G12 heavy-chain gene were constructed: our original 2G12 sequence, which differed from previously reported 2G12 and IgG1 Fc sequences (4) by two substitutions (V5L and H237S), and six hinge deletion mutants constructed from our original 2G12 sequence. The mutants were D1 (deletion of residue 237), D2 (deletion of residues 236 to 237), D3 (deletion of residues 235 to 237), D4 (deletion of residues 232 to 237), D6 (deletion of residues 232 to 239), and D6GG (deletion of residues 232 to 239 and two proline-to-glycine substitutions [P240G and P241G]) (see Fig. 3A). In addition, we produced forms of the original 2G12 sequence and of the D2 mutant that reverted the V5L and H237S substitutions to the previously reported 2G12 and IgG1 Fc sequences.
FIG. 3.
2G12 deletion constructs. (A) Amino acid sequences of the hinge region of the original 2G12 and the six deletion mutants (left) and approximate location of the hinge deletions on a schematic structure of a normal IgG (right). (B) Size exclusion chromatography profiles of the original 2G12 and the deletion mutants. Absorbance traces are shown for one expression trial, normalized on the monomer peak height. (C) Dimer/monomer ratios for the original 2G12 and the deletion mutants. The data are presented as the means and standard deviations for ratios determined from four independent expression trials, each of which involved the expression of all seven constructs. Also shown is the mean total expression relative to the original 2G12 construct. The P value comparing the original 2G12 and the D2 mutant was computed by using the two-sided independent sample Student t test.
Expression of 2G12.
Proteins were expressed by using polyethylenimine-mediated transient transfection (5) of suspension HEK293-6E cells (NRC Biotechnology Research Institute). The 2G12 heavy- and light-chain expression vectors were cotransfected at a 1:1 ratio using 25-kDa linear polyethylenimine (Polysciences). Cell culture supernatants were collected at 6 days posttransfection, passed over protein A resin (Pierce Biotechnology), and eluted using a pH 3.0 citrate buffer. Eluates from the protein A column were immediately neutralized and then subjected to size exclusion chromatography in 20 mM Tris (pH 8.0)-150 mM NaCl using a Superdex 200 16/60 or 10/30 column (GE Healthcare). The column profile revealed two major peaks, later identified as 2G12 dimer and 2G12 monomer. Similar results were obtained when the original 2G12 construct and the D2 mutant were purified from cell culture supernatants by passage over a neonatal Fc receptor affinity column in which supernatants were passed over the column at pH 6.0 and eluted at pH 8.0 (8).
Since the sizing column profiles of the monomeric and dimeric forms of 2G12 partially overlapped, samples for neutralization assays were subjected to two size exclusion chromatography purification steps: first, the concentrated protein A eluate was run over a Superdex 200 16/60 column, and then the monomer and dimer fractions were placed over a Superdex 200 10/30 column.
Parallel trials of the original 2G12 construct and six hinge deletion mutants were performed on a 400-ml scale. Five independent expression trials, each involving all seven constructs, were conducted. In each trial, 2G12 proteins were purified on a protein A column as described above and then subjected to size exclusion chromatography. Dimer-to-monomer ratios for each construct (see Fig. 3C) were calculated after integration of the peak areas using UNICORN software. In four of the five trials, the expression of total 2G12 protein in each sample was approximately 2 mg/liter. In the fifth trial, the total expression in each sample was ∼10-fold lower, likely due to suboptimal transfections. The data from this trial were omitted from the calculated means shown in Fig. 3C.
Static light scattering.
The oligomeric states of the two 2G12 peaks isolated by conventional size exclusion chromatography were determined using size exclusion chromatography with in-line static light scattering and refractive index monitoring. Experiments were performed at 25°C using an ÄKTA chromatography system (GE Healthcare) with a Superdex 200 10/30 (GE Healthcare) equipped with a Dawn Helios multi-angle light scattering photometer and an Optilab rEX refractive index detector (Wyatt Technology). Bovine serum albumin was used as a calibration standard. All data were analyzed with ASTRA V software (Wyatt Technology).
In vitro neutralization assays.
We used a previously described pseudovirus neutralization assay, which measures the reduction in luciferase reporter gene expression in the presence of a potential inhibitor following a single round of pseudovirus infection in TZM-bl cells (12, 14). Pseudoviruses were generated by cotransfection of HEK293T cells with an Env expression plasmid and a replication-defective backbone plasmid. Neutralization assays were performed either in-house (data shown only for strain HxBc2) or by the Collaboration for AIDS Vaccine Discovery (CAVD) core neutralization facility (Table 1) . Strains SC422661.8, TRO.11, PVO.4, QH0692.42, 6535.3, and HxBc2 were tested in-house, with results similar to those obtained from the CAVD assays. For in-house assays, each 2G12 sample was tested in triplicate, with 200 infectious viral units per well incubated with a threefold dilution series. After a 1-h incubation at 37°C, 10,000 TZM-bl cells were added to each well, followed by incubation for 2 days. Cells were then lysed and assayed for luciferase expression by using Bright-Glo (Promega) and a Victor3 luminometer (Perkin-Elmer). The percent neutralization was determined by calculating the difference in luminescence between test wells and cell control wells (cell only), dividing this value by the difference between virus control (cell plus virus) and cell control wells, subtracting from 1, and multiplying by 100. Nonlinear regression analysis was used to calculate concentrations at which half-maximal inhibition was observed (IC50s). The average IC50s reported in Table 1 are geometric means calculated by using the formula (Πai)(1/n), where i = 1, 2, …, n. Calculation of geometric means is suitable for data sets covering multiple orders of magnitude (19), as is the case for neutralization data across multiple viral strains.
TABLE 1.
IC50s for four forms of 2G12 obtained from in vitro pseudovirus neutralization assays of 2G12-sensitive strainsa
| Env clone | Clade | 2G12 original
|
D2
|
||||
|---|---|---|---|---|---|---|---|
| Monomer IC50 (μg/ml) | Dimer IC50 (μg/ml) | Monomer IC50/dimer IC50 | Monomer IC50 (μg/ml) | Dimer IC50 (μg/ml) | Monomer IC50/dimer IC50 | ||
| 6535.3 | B | 30.1 | 0.34 | 89 | 32.9 | 1.5 | 22 |
| QH0692.42 | B | 9.8 | 0.08 | 123 | 11.1 | 0.1 | 111 |
| SC422661.8 | B | 17.8 | <0.05 | 356 | 19.4 | 0.1 | 194 |
| PVO.4 | B | 4.9 | <0.05 | 98 | 7.1 | <0.05 | 142 |
| TRO.11 | B | 1.6 | <0.05 | 32 | 1.4 | <0.05 | 28 |
| WITO4160.33 | B | 6.8 | <0.05 | 136 | 7.6 | 0.05 | 152 |
| 3988.25 | B | 3.2 | <0.05 | 64 | 2.8 | <0.05 | 56 |
| 7165.18 | B | 6.6 | <0.05 | 132 | 8 | <0.05 | 160 |
| QH0515.1 | B | 0.1 | <0.05 | 2 | 0.1 | <0.05 | 2 |
| 5768.4 | B | 0.1 | <0.05 | 2 | 0.1 | <0.05 | 2 |
| 6101.1 | B | 24 | 0.05 | 480 | 26.5 | 0.06 | 442 |
| TRJO4551.58 | B | >100 | 0.16 | 625 | NDb | ND | ND |
| HxBc2* | B | 0.47 | 0.028 | 17 | ND | ND | ND |
| 0330.v4.c3 | A | 15.3 | <0.05 | 306 | 17.2 | <0.05 | 344 |
| 3415.v1.c1 | A | 8 | <0.05 | 160 | 8.1 | <0.05 | 162 |
| CHO38.12 | CRF07_BC | 0.2 | <0.05 | 4 | 0.3 | <0.05 | 6 |
| 211-9 | CRF02_AG | >100 | 0.4 | 250 | >100 | 1.3 | 77 |
| 235-47 | CRF02_AG | 1.3 | <0.05 | 26 | 1.4 | <0.05 | 28 |
| T280-5 | CRF02_AG | >100 | 1 | 100 | 51 | 4.9 | 10 |
| T250-4 | CRF02_AG | 61.5 | <0.05 | 1230 | 60.7 | 0.4 | 152 |
| T251 | CRF02_AG | 50.8 | 0.1 | 508 | 46.6 | 0.3 | 155 |
| Avg (geometric mean) | 6.3 | 0.08 | 82 | 6.5 | 0.12 | 54 | |
Results from in-house neutralization assays are indicated by an asterisk; all other results were obtained by the CAVD core neutralization facility. In calculating average IC50s, measurements outside of the range of the assay (<0.05 or >100 μg/ml) were assigned to those limiting values, and averages were calculated using only strains for which IC50s were available for all four forms of 2G12. The following clade B and C strains were not neutralized by the original 2G12 monomer or dimer (IC50 values > 100 μg/ml): AC10.0.29, RHPA4259.7, THRO4156.18, REJO4541.67, and CAAN5342.A2 (clade B) and Du156.12, Du172.17, Du422.1, ZM197M.PB7, ZM214M.PL15, ZM233M.PB6, ZM249M.PL1, ZM53M.PB12, ZM109F.PB4, ZM135M.PL10a, CAP45.2.00.G3, and CAP210.2.00.E8 (clade C).
ND, not determined.
Biacore binding studies.
A Biacore T100 biosensor system (GE Healthcare) was used to evaluate binding of the 2G12 proteins to gp120. In this system, a protein is coupled to a gold-dextran layer, and association and dissociation phases for binding to an injected protein are measured in real time in resonance units (RU). The original 2G12 monomer and dimer and the D2 dimer (∼150 RU of each) were captured onto ∼4,000 RU of goat anti-Fc polyclonal antibody (Chromapure; Jackson ImmunoResearch), which was immobilized by primary amine coupling to a CM5 sensor chip as described in the Biacore manual. A concentration series of monomeric gp120 (expressed in baculovirus-infected insect cells; strain HxBc2) was injected at 20 μl/min over the flow cells. After the gp120 dissociation phase, the surface was regenerated by injection of pH 1.5 glycine buffer, and 2G12 proteins were captured again prior to the subsequent gp120 injection.
Structural analysis.
The 2G12 structure was displayed and analyzed by using the program O (10). The 3D model of 2G12 was prepared using SolidWorks (SolidWorks Corp.).
RESULTS
Identification of a dimeric form of 2G12 with increased neutralization potency.
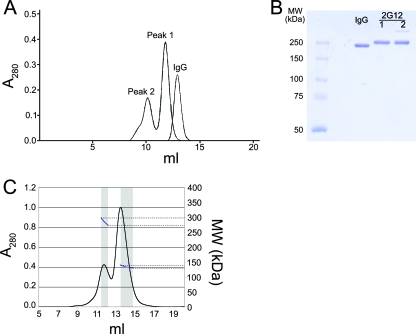
During purification of 2G12 IgG expressed in mammalian cells, we noticed that the protein eluted as two peaks from a size exclusion chromatography column (peak 1, eluting at ∼12 ml, and peak 2, eluting at ∼10 ml) (Fig. 2A). Peak 1 migrated as if it were a somewhat larger protein than a typical IgG, which eluted at ∼13 ml, suggesting that it corresponded to a 2G12 monomer, which has a more elongated structure than other IgG monomers due to the 3D domain-swapped structure of its Fabs (4) (Fig. 1B). Peak 2 appeared to correspond to a higher-molecular-weight form of 2G12. Samples from both peaks migrated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis under nonreducing conditions, as expected for an IgG (Fig. 2B), indicating that covalent modification(s) could not account for the altered migration of peak 2. Once isolated, both peaks retained their size exclusion profile over a period of weeks to months, indicating that there was little tendency for conversion between the two forms.
FIG. 2.
Size exclusion chromatography profile of 2G12. (A) Comparison of the elution profiles of protein A-purified 2G12 with a typical human IgG. The more elongated structure of the 2G12 IgG monomer (peak 1) caused it to elute slightly earlier than typical IgGs. (B) Sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis of a typical human IgG and the 2G12 proteins in peaks 1 and 2. Samples were run under nonreducing conditions. (C) Multi-angle light scattering data obtained by size exclusion chromatography with in-line light scattering and refractive index monitoring. UV absorbance at 280 nm is shown in black, and the calculated molecular weight based on multi-angle light scattering data is indicated in blue with units shown on the right axis. The windows used for calculating the molecular masses of peak 1 and peak 2 are shown in gray.
In-line static multi-angle light scattering was used to determine the absolute molecular masses, and therefore the oligomeric states, of both peaks. For this analysis, we used the major portion of peak 2 and did not include the shoulder slightly ahead of the peak. The experimentally determined molecular masses derived from these data were 135 kDa for peak 1 and 285 kDa for peak 2. By comparing these values with the molecular mass predicted from the 2G12 sequence (145 kDa), we identified peak 1 as 2G12 monomer and peak 2 as 2G12 dimer.
We next compared the potencies of purified 2G12 monomer and dimer to neutralize a range of HIV-1 pseudoviruses by using an in vitro neutralization assay (12, 14). The 2G12 monomer neutralized a range of clade A and B, but not clade C, strains (Table 1). Over the range of strains that were sensitive to 2G12 neutralization, the 2G12 dimer was generally 1 to 2 orders of magnitude more potent than the 2G12 monomer, with an average increased potency of 82-fold (Table 1). Three of the twenty strains that were not neutralized by monomeric 2G12 (IC50 > 100 μg/ml) were neutralized by dimeric 2G12: strains TRJO4551.58 (clade B) and strains 211-9 and T280-5 (circulating recombinant forms) (Table 1). The clade C strains remained resistant to 2G12 dimer (Table 1 legend).
Design and expression of 2G12 mutants.
Given that the 2G12 dimer was not formed by covalent joining of two monomers (Fig. 2B), we reasoned that the dimeric form could arise from intermolecular 3D domain swapping in which each light chain was paired with heavy chains derived from two IgGs (Fig. 1C). In contrast, the intramolecular domain swapping observed in monomeric 2G12 involves the pairing of each light chain with both heavy chains from a single IgG (4) (Fig. 1B).
To increase the dimeric fraction of 2G12, we designed mutants predicted to favor intermolecular domain swapping instead of the intramolecular swapping that led to the monomer. Compared to a typical antibody, the hinge regions of the domain-swapped 2G12 are forced into a relatively extended conformation. In other words, the C-terminal ends of the 2G12 CH1 domains (defined by residue K228) are held fixed at a large distance: the K228-K228 distance is 40.6 Å in the 2G12 Fab2 structure (pdb code 1OP3) (4) versus 25.0 Å for the corresponding distance in an unswapped antibody structure (b12 IgG; pdb code 1HZH) (17). Intramolecular domain swapping, as occurs in monomeric 2G12, may be disfavored by a shorter hinge region, which could instead accommodate an intermolecular domain swap resulting in an intertwined IgG dimer instead of an intertwined IgG monomer (Fig. 1B and C). Based on this structural hypothesis we designed forms of 2G12 with shorter hinge regions.
A series of deletions was made in the 2G12 heavy chain between residue C230, which normally participates in a disulfide bond with the light chain, and residues C239 and C242, which form disulfide bonds between the heavy chains (Fig. 3A). The mutants were named according to how many residues were deleted, with D1 referring to a single residue deletion, up to D6, which represented a six-residue deletion. An additional mutant, D6GG, in which two prolines were replaced with glycines to allow increased flexibility, was also prepared.
The original 2G12 construct was expressed, along with the suite of mutants in multiple expression trials. During each trial, the expressed IgGs were purified from transfected cell supernatants on a protein A column, followed by size exclusion chromatography to separate 2G12 monomer and dimer. The amounts of dimer and monomer produced by each construct were determined by integrating the area under the relevant size exclusion chromatography peak (Fig. 3B). As shown in Fig. 3C, the D2 mutant had a significantly higher dimer/monomer ratio (0.66 ± 0.06) than the original 2G12 (0.28 ± 0.03). The other mutants showed only slightly higher dimer/monomer ratios (D3 to D6, D6GG) or a lower ratio (D1) than the original 2G12. A comparison of the total expression levels (monomer plus dimer) of the original and D2 constructs showed that the D2 mutation did not impair overall expression (Fig. 3C).
2G12 protein has been reported to aggregate under strongly acidic conditions (16). To address whether the low pH used during elution from the protein A column affected the ability of 2G12 to dimerize, we purified 2G12 from transfected cell supernatants by using a neonatal Fc receptor affinity column in which samples were loaded at pH 6.0 and eluted at pH 8.0 (8). Size exclusion chromatography profiles of the original 2G12 and the D2 mutant were similar for both purification schemes, indicating that dimer formation was not affected by the protein A elution conditions. In addition, we verified that our results were not influenced by two substitutions in our 2G12 construct compared to the published sequences of 2G12 Fab2 and human IgG1 Fc that were introduced during cloning (see Materials and Methods). To ascertain the effects of the substitutions, we produced versions of the original 2G12 construct and the D2 mutant in which the substitutions were reverted and found no significant differences in the total yields of 2G12 protein and the dimer/monomer ratios for the reverted constructs compared to the constructs containing the two substitutions: the average total yields for both reverted constructs were within 8% of the unreverted constructs, and the dimer/monomer ratios for the reverted 2G12 and the D2 mutant constructs were 0.34 ± 0.03 (reverted 2G12) and 0.76 ± 0.06 (reverted D2 mutant).
To verify that the substitutions in the D2 mutant did not affect its neutralization activity, we determined the neutralization potencies of monomeric and dimeric D2 for the strains that were tested for the original 2G12 monomer and dimer. As seen for the original 2G12 dimer, the D2 dimer was more potent in neutralization of 2G12-sensitive strains than the corresponding monomer, and two of the strains that were resistant to the original 2G12 monomers were sensitive to D2 2G12 dimers (Table 1). The average IC50 for the D2 dimer (0.12 μg/ml) was slightly higher than the average value for the original dimer (0.08 μg/ml), and the overall monomer IC50/dimer IC50 ratio for the D2 mutant (54-fold) was lower than the corresponding ratio for the original 2G12 dimer (82-fold). However, the differences in neutralization potencies of the D2 and original 2G12 dimers are not necessarily significant because the dimer IC50 averages that were used to derive the ratios were calculated using strains with IC50s that were below the sensitivity limit (0.05 μg/ml) of our assay; these are reported as <0.05 μg/ml in Table 1 and input as 0.05 μg/ml in the calculations of averages.
To address whether dimeric 2G12 exhibits higher neutralization potencies than monomeric 2G12 because of avidity effects (i.e., higher apparent affinity resulting from multivalent binding), we compared the binding of gp120 to the two forms of 2G12. In these experiments, the original 2G12 monomer and dimer and the D2 dimer were coupled to a biosensor chip, and the binding of injected monomeric HxBc2 gp120 was evaluated. No significant differences were observed for the interactions with gp120 of the dimers compared to the monomer: the equilibrium dissociation constants (KDs) were ∼1 nM for the binding of gp120 to the original monomer, the original dimer, and the D2 dimer (data not shown). These results do not rule out avidity effects as a mechanism for increased neutralization potency, however, since the gp120 monomers would not contain the same arrangement of carbohydrate epitopes as a trimeric gp120/gp41 envelope spike on the surface of a virus.
DISCUSSION
Unlike other known broadly neutralizing anti-HIV-1 antibodies, 2G12 recognizes a carbohydrate, rather than protein, epitope on gp120 (18, 22). The unusual domain-swapped structure of its Fab2 unit facilitates recognition of adjacent oligomannose units on the same envelope spike (3). Multimerization of 2G12 has been reported to increase the neutralization potency of 2G12; an engineered IgM form of 2G12, which contained 10 Fabs (as compared to 2 in a monomeric IgG), was shown to neutralize up to 28-fold better than monomeric 2G12 IgG (24). Previous investigators suggested that 2G12 IgG might form dimers with higher neutralization potency; for example, the severalfold higher neutralization potency for 2G12 produced in transgenic maize as opposed to mammalian cells was ascribed to a higher dimer or aggregate content (15), and 2G12 IgG dimers are mentioned but not characterized or directly compared to 2G12 IgM or 2G12 monomers by Wolbank et al. (24). Here we report a higher-molecular-weight form of 2G12 IgG that is naturally produced during expression in mammalian cells, and we present biophysical evidence that it represents a dimeric IgG.
We compared the potencies of isolated 2G12 dimer and monomer for neutralization of a range of clade A and clade B HIV-1 strains, and report that the 2G12 dimer has a >50-fold average increased potency compared to the monomer. The high neutralization potency of 2G12 dimers suggests that the dimeric fraction of 2G12 would be a better candidate for passive immunotherapy to treat HIV-1 infection than the monomeric fraction, which appears to have been used in previous studies of the efficacy of passive immunization using anti-HIV antibodies (13, 21). Another form of 2G12 that could be considered for passive immunizations is an engineered IgM form, which was reported to exhibit up to an ∼28-fold increase in neutralization potency compared to 2G12 IgG (24). Because the previous study used a different neutralization assay and different viral strains, the average neutralization potency increases cannot be directly compared, but their similarity in magnitude suggests that the extra Fab2 units in the pentameric IgM form compared to the dimeric form do not contribute significantly to more effective neutralization. In any case, the choice of an optimal therapeutic reagent depends on other factors in addition to neutralization potency, including in vivo half-life, tissue distribution, antibody-dependent cellular cytotoxicity activity, and expression yield. Given the longer serum half-lives of IgG versus IgM antibodies (9), the activity of IgG antibodies in antibody-dependent cellular cytotoxicity (9) and the more complex assembly and heterogeneity of recombinant J chain-containing IgM and IgA, the dimeric IgG form of 2G12 is likely to have several advantages compared to the IgM form.
Based on the propensity of 2G12 monomers to form an intramolecular domain-swapped Fab2 structure, we present a structural model for the organization of the 2G12 dimer that involves intermolecular 3D domain swapping (Fig. 1C). We used the structural model to design and express 2G12 mutants predicted to have a higher fraction of the more potent dimeric form. One of these mutants, D2, had a significantly higher dimer content and equivalent overall expression. Due to the much higher neutralization potencies of the original and the D2 mutant dimers, a modest fraction of dimer can dominate the overall potency of a 2G12 preparation with a mixture of forms. Different 2G12 dimer fractions may occur in different expression contexts, perhaps relating to the density of assembling 2G12 in the endoplasmic reticulum, which could influence the rates of intra- versus intermolecular domain swapping. Further experiments will be required to investigate the extent to which the D2 mutant increases dimer fraction in other expression systems. Although the mechanism by which the D2 mutant causes a higher dimer content is not known for certain, this mutant would be a potentially useful therapeutic reagent in any context in which unfractionated 2G12 is utilized. This includes gene therapy applications, topical (microbicide) use, and use as an injectable reagent.
Acknowledgments
We thank Adrian Rice for assistance with light scattering experiments, Lili Yang and David Baltimore for advice concerning the neutralization assay, David Stolzer for making the 3D model figures, Marta Murphy for assistance with figures, and Dennis Burton of the Scripps Research Institute for cDNAs. We also thank the CAVD Neutralizing Antibody Core Laboratories for performing in vitro neutralization assays.
This study was supported by a grant from the Bill and Melinda Gates Foundation through the Grand Challenges in Global Health Initiative.
Footnotes
Published ahead of print on 22 October 2008.
REFERENCES
- 1.Bennett, M. J., and D. Eisenberg. 2004. The evolving role of 3D domain swapping in proteins. Structure 121339-1341. [DOI] [PubMed] [Google Scholar]
- 2.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody versus HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA 10214943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calarese, D. A., H. K. Lee, C. Y. Huang, M. D. Best, R. D. Astronomo, R. L. Stanfield, H. Katinger, D. R. Burton, C. H. Wong, and I. A. Wilson. 2005. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. USA 10213372-13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 3002065-2071. [DOI] [PubMed] [Google Scholar]
- 5.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris, L. J., S. B. Larson, K. W. Hasel, and A. McPherson. 1997. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 361581-1597. [DOI] [PubMed] [Google Scholar]
- 7.Harris, L. J., E. Skaletsky, and A. McPherson. 1998. Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 275861-872. [DOI] [PubMed] [Google Scholar]
- 8.Huber, A. H., R. F. Kelley, L. N. Gastinel, and P. J. Bjorkman. 1993. Crystallization and stoichiometry of binding of a complex between a rat intestinal Fc receptor and Fc. J. Mol. Biol. 2301077-1083. [DOI] [PubMed] [Google Scholar]
- 9.Janeway, C. A., P. Travers, M. Walport, and M. J. Schlomchik. 2005. Immunobiology, 6th ed. Garland Science Publishing, New York, NY.
- 10.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt. 2)110-119. [DOI] [PubMed] [Google Scholar]
- 11.Kabat, E. A., T. T. Wu, H. M. Perry, K. S. Gottesman, and C. Foeller. 1991. Sequences of proteins of immunological interest. Department of Health and Human Services, Washington, DC.
- 12.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 7910108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehandru, S., B. Vcelar, T. Wrin, G. Stiegler, B. Joos, H. Mohri, D. Boden, J. Galovich, K. Tenner-Racz, P. Racz, M. Carrington, C. Petropoulos, H. Katinger, and M. Markowitz. 2007. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J. Virol. 8111016-11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montefiori, D. C. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12:Unit 12.11. [DOI] [PubMed]
- 15.Ramessar, K., T. Rademacher, M. Sack, J. Stadlmann, D. Platis, G. Stiegler, N. Labrou, F. Altmann, J. Ma, E. Stoger, T. Capell, and P. Christou. 2008. Cost-effective production of a vaginal protein microbicide to prevent HIV transmission. Proc. Natl. Acad. Sci. USA 1053727-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roux, K. H., P. Zhu, M. Seavy, H. Katinger, R. Kunert, and V. Seamon. 2004. Electron microscopic and immunochemical analysis of the broadly neutralizing HIV-1-specific, anti-carbohydrate antibody, 2G12. Mol. Immunol. 411001-1011. [DOI] [PubMed] [Google Scholar]
- 17.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 2931155-1159. [DOI] [PubMed] [Google Scholar]
- 18.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 767306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheskin, D. 2004. Handbook of parametric and nonparametric statistical procedures, 3rd ed. Chapman & Hall/CRC, Boca Raton, FL.
- 20.Stiegler, G., C. Armbruster, B. Vcelar, H. Stoiber, R. Kunert, N. L. Michael, L. L. Jagodzinski, C. Ammann, W. Jager, J. Jacobson, N. Vetter, and H. Katinger. 2002. Antiviral activity of the neutralizing antibodies 2F5 and 2G12 in asymptomatic HIV-1-infected humans: a phase I evaluation. AIDS 162019-2025. [DOI] [PubMed] [Google Scholar]
- 21.Trkola, A., H. Kuster, P. Rusert, B. Joos, M. Fischer, C. Leemann, A. Manrique, M. Huber, M. Rehr, A. Oxenius, R. Weber, G. Stiegler, B. Vcelar, H. Katinger, L. Aceto, and H. F. Gunthard. 2005. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat. Med. 11615-622. [DOI] [PubMed] [Google Scholar]
- 22.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 701100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 24.Wolbank, S., R. Kunert, G. Stiegler, and H. Katinger. 2003. Characterization of human class-switched polymeric (immunoglobulin M [IgM] and IgA) anti-human immunodeficiency virus type 1 antibodies 2F5 and 2G12. J. Virol. 774095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]