Abstract
Although transcription factors of the basic helix-loop-helix family have been shown to regulate enhancers of lymphomagenic gammaretroviruses through E-box motifs, the overlap of an E-box motif (Egre) with the glucocorticoid response element (GRE) has obscured their function in vivo. We report here that Egre, but not the GRE, affects disease induction by the murine T-lymphomagenic SL3-3 virus. Mutating all three copies of Egre prolonged the tumor latency period from 60 to 109 days. Further mutating an E-box motif (Ea/s) outside the enhancer prolonged the latency period to 180 days, suggesting that Ea/s works as a backup site for Egre. While wild-type SL3-3 and GRE and Ea/s mutants exclusively induced T-cell lymphomas with wild-type latencies mainly of the CD4+ CD8− phenotype, Egre as well as the Egre and Ea/s mutants induced B-cell lymphomas and myeloid leukemia in addition to T-cell lymphomas. T-cell lymphomas induced by the two Egre mutants had the same phenotype as those induced by wild-type SL3-3, indicating the incomplete disruption of T-cell lymphomagenesis, which is in contrast to previous findings for a Runx site mutant of SL3-3. Mutating the Egre site or Egre and Ea/s triggered several tumor phenotype-associated secondary enhancer changes encompassing neighboring sites, none of which led to the regeneration of an E-box motif. Taken together, our results demonstrate a role for the E-box but not the GRE in T lymphomagenesis by SL3-3, unveil an inherent broader disease specificity of the virus, and strengthen the notion of selection for more potent enhancer variants of mutated viruses during tumor development.
Murine leukemia viruses (MLVs) are gammaretroviruses with strain-specific patterns of disease induction. The U3 transcriptional enhancers in the long terminal repeat (LTR) of MLVs share a common framework of binding sites for host transcription factors that contribute to the regulation of disease specificity (10, 12, 13, 15, 20, 40, 42, 44, 48, 51). SL3-3 is a potent ecotropic MLV that induces strictly T-cell lymphomas in laboratory mice, with a mean latency of 2 to 4 months depending on the mouse strain (20, 30, 41, 51). The SL3-3 enhancer consists of 2.5 tandem copies of a 72-bp sequence with binding sites for Runx, NF-1, c-Myb, and Ets factors as well as the glucocorticoid receptor (GR) and basic helix-loop-helix (bHLH) factors. Runx and c-Myb binding sites are critical for tumor induction by SL3-3 (20, 30, 51, 60), whereas Ets and NF-1 sites are less important (19, 21, 22, 51, 52, 59, 60). Besides significantly weakening the virus, the mutations of all Runx sites in the SL3-3 transcriptional enhancer (the SL3-3dm mutant) were found to shift disease patterns from exclusively T-cell lymphomas to various hematopoietic malignancies, including B-cell lymphomas and myeloid and erythroid leukemias (57).
The roles of the glucocorticoid response element (GRE) and bHLH binding E-box motifs in tumor induction by SL3-3 have not been investigated; however, the binding of GR and bHLH factors affects SL3-3 transcriptional activity (10, 11, 29, 49, 50, 52). E-box binding proteins belong to the large diverse group of bHLH proteins that are involved in cell cycle control, cell lineage development, and tumorigenesis (1-4, 16-18). The bHLH factors are divided into several classes depending on their dimerization abilities, tissue distribution, and preference of E-box binding motif (the general consensus is NCANNTGN) (35, 46). Class I bHLH proteins, such as E12, E47, HEB, and E2-2 transcription factors, which are involved in lymphocyte development, are the most likely candidates for SL3-3 enhancer binding and activation.
The enhancer of SL3-3 contains three identical GRE-overlapping E-boxes (designated Egre) plus one E-box motif downstream of the tandem repeats (designated Ea/s). The overlapping GRE/Egre site sequence AGAACAGATGGTCCC (the E-box is highlighted) is highly conserved among murine gammaretroviruses (27) but is not found in cellular genes. The glucocorticoid induction of the SL3-3 enhancer is less significant in T cells than in HeLa cells, which are scarce in bHLH factors (10, 11, 29, 52). This indicates that bHLH factors occupy Egre in T cells and, hence, that the GRE does not play a main role for SL3-3 enhancer activation in T cells. In vitro binding studies have shown that human SEF2 (SL3-3 enhancer factor 2, an E2-2 homolog) and murine ALF1 (an HEB homolog), which are expressed in various cell lines, interact with the Egre site and activate the SL3-3 enhancer (14). The bHLH transcription factors that bind to the Ea/s site, which has a slightly different sequence (CCAGATGA), have not been identified (14, 50). The mutation of the Egre sites or the overexpression of bHLH inhibitors, Id proteins, inhibits the ALF1 trans-activation of the SL3-3 LTR-driven expression in T- and B-cell lines, indicating that ALF1 and other bHLH factors bind and drive the activation of the SL3-3 enhancer (14, 49). In this study, we investigate in detail the pathogenic properties and enhancer sequence stability of five SL3-3 E-box and GRE enhancer mutants as well as that of the SL3-3 wild type (wt).
MATERIALS AND METHODS
Viral stocks.
Mutated proviral clones of SL3-3 3mGR, SL3-3 3mGR+mEa/s, SL3-3 mEa/s, SL3-3 3mEgre, and SL3-3 3mEgre+mEa/s were constructed by PCR-based mutagenesis with primers spanning the mutations indicated in Fig. 1. Primer sequences are available upon request. Infectious virus particles, including wt SL3-3, were produced as previously described (20, 30, 43).
FIG. 1.
Structure and pathogenicity of SL3-3 enhancer mutants. (A) The SL3-3 proviral transcriptional enhancer contains 38- and 34-bp repeat elements, represented by white and black boxes, respectively. The nucleotide sequence is given below the diagram, and known binding site sequences for cellular transcription factors are indicated. The GR, Egre, and Ea/s mutations introduced into each repeat in the given enhancer mutant virus are shown at the bottom. The location of PCR primers used for the amplification of tumor DNA is shown. 5′UTR, 5′ untranslated region. (B) Cumulative tumor incidence after the infection of newborn inbred NMRI mice.
Transient expression assays.
The plasmids pN.SL3-3(wt)-Cat, pN.SL3-3(3mEgre)-Cat, and pN.SL3-3(3mGR)-Cat were made by basic cloning techniques (19, 20) and used for the transfection of NIH 3T3 cells by Lipofectamine (Invitrogen) according to the protocol provided by the manufacturer. A luciferase reporter construct was included in the transfections to serve as a reference. Twenty-four hours before transfection, 3 × 104 NIH 3T3 cells were seeded into 10-cm petri dishes, and 24 h after transfection 1 μM dexamethasone (dex) was added to half of the cell transfections. Cells were harvested 24 h later and assayed for chloramphenicol acetyltransferase (CAT) and luciferase activity as described by the manufacturers (CAT enzyme-linked immunosorbent assay and luciferase reporter gene assay; Roche). Transfections and expression assays were done in duplicate and repeated twice.
Pathogenicity studies and tissue analysis.
Two independent series of animal experiments were performed (series A and B) at separate mouse facilities. In both cases, newborn inbred NMRI mice lacking endogenous ecotropic MLVs (39) were injected intraperitoneally with ca. 106 infectious virus particles (0.1 ml). Control mice were injected with 0.1 ml complete medium. All mice were monitored as previously described in details (54). Lymphomatous tissues from lymph nodes, spleen, and liver were dissected, and samples were stored frozen and/or fixed in 4% paraformaldehyde and analyzed by histology and immunohistochemistry as described previously (57).
Three-color flow cytometry.
Cell suspensions of spleen, thymus, and lymph node tissue samples from SL3-3 3mEgre-, SL3-3 3mGR-, and wt SL3-3-induced tumors of series B were prepared and stained as described previously (57). Ten fluorochrome-conjugated rat anti-mouse monoclonal antibodies were used, all obtained from BD Pharmingen: CD11b-fluorescein isothiocyanate (CD11b-FITC), CD3-phycoeryrthrin (CD3-PE), B220-peridinin chlorophyll protein (B220-PerCP), CD8a-PerCP-Cy5.5, CD4-FITC, CD24/heat-stable antigen-FITC (CD24/HSA-FITC), TCRβ-PE-Cy5, Ter119-FITC, Gr-1-PE, and Gr-1-PerCP-Cy5.5. Isotype controls were rat immunoglobulin G2a (IgG2a) and rat IgG2b light-chain monoclonal immunoglobulins conjugated with FITC, PerCP, or PerCP-Cy5.5. Antibodies were used in four combinations: CD3/B220/CD11b, CD3/CD4/CD8a, CD3/T-cell receptor β/HAS (CD3/TCRβ/HSA), and CD11b/Gr-1/Ter119. CD3 is a pan-T marker (26), B220 is a pan-B marker (31), and CD11b (mac-1) was used as a myeloid marker (38). CD3/CD4/CD8a distinguishes between T-cell differentiation stages, as further characterized by CD3/TCRβ/HSA staining. TCRβ is expressed in very early immature thymocytes and upregulated during maturation. HSA (CD24) is present on monocytes, erythroid cells, granulocytes, and lymphocytes and is downregulated during T-cell development. CD11b/Gr-1/Ter119 was used to identify myeloid cells and exclude erythroid cell involvement. Gr-1, which is present on granulocytes, is a marker of myeloid differentiation (23, 34). Ter119 is an erythroid marker (36). Antibody-stained cells were analyzed on a FACSCalibur flow cytometer (BD Bioscience, San Jose, CA) as described earlier (57). Flow cytometry data were represented by two-dimensional dot plots showing all 50,000 events. Quadrants of all blots were defined on the basis of appropriate isotype controls measured together with each stain to determine background fluorescence levels.
Southern hybridizations.
Genomic DNA was purified from frozen tumor tissue samples using a DNeasy tissue kit (Qiagen). Southern blot analysis was done as described earlier (57) with hybridization probes specific for the TCRβ joining regions 1 and 2 (J1 and J2) and the Igκ light-chain gene (43, 57).
PCR amplification of proviral DNA and sequencing of viral enhancer structures.
PCR on genomic tumor DNA used primers 440 and 1112 as described previously (57). Fragments were sequenced with primer 440 and P2(Akv)-rev with a Dyenamic ET terminator cycle sequencing kit (Amersham Pharmacia Biosciences).
RESULTS
Overview of mutants and analyses.
The mutations introduced in the tandem repeat enhancer of SL3-3 are shown in Fig. 1. To impair GRE but not E-box function, all three copies of the GRE core AGAACAGA sequences were changed to ACGACAGA in the 3mGR mutant (the changes are underlined), sites previously shown to abolish GR binding (11). To analyze the effect of the mutations on glucocorticoid inducibility, SL3-3 wt, 3mGR, and 3mEgre LTRs were linked to a CAT reporter gene. Transient expression assays (data not shown) confirmed that both enhancer mutants had wt activities without dex. A fourfold dex induction was observed for the SL3-3 wt but not for the SL3-3 3mGR mutant, thus confirming that the GRE mutations outside the E-box in SL3-3 3mGR abolished the dex inducibility of the enhancer. As expected, the Egre mutation also abolished dex inducibility.
E-boxes were mutated by altering three out of four nucleotides of the core motif NCANNTGN to NACNNCGN, abolishing the binding of bHLH proteins and GR to the site (49, 50). Notably, GR and bHLH factors seem unable to bind Egre simultaneously (14, 50). Finally, an E-box motif downstream of the enhancer repeats (designated Ea/s) was mutated alone or together with 3mGR and 3mEgre. The mutant viruses were analyzed in two separate mouse studies (series A and series B). In short, series A (Fig. 1, Table 1) comprised all five mutants plus SL3-3 wt virus and was used to establish the latency and overall tumor phenotypes (Fig. 1 and 2 and Table 1). Animal series B, done at another mouse facility and used for tumor cell typing by flow cytometry (Fig. 3 and 4 and Table 2), comprised wt SL3-3 and the 3mGR and 3mEgre mutants.
TABLE 1.
Incidence, latency, and molecular and histological analysis of virus-induced tumors (series A)
| SL3-3 virus | Tumor incidence (no. of mice with tumor development/no. injected) | Mean tumor latency (no. of days ± SD) | Histopathologya | Clonal DNA rearrangementsb (no. of tumors with clonal rearrangements/no. investigated)
|
Complex enhancer changesc (no. of tumors with complex enhancer changes/no. examined) | |
|---|---|---|---|---|---|---|
| Igκe | TCRβ | |||||
| wt | 13/13 | 60 ± 9 | Pre-TLLd | 0/11 | 11/11 | ND |
| mEa/s | 24/24 | 58 ± 8 | ND | 0/11 | 11/11 | 0/24 |
| 3mGR | 21/21 | 57 ± 6 | Pre-TLL | 0/11 | 11/11 | 0/21 |
| 3mGR+mEa/s | 25/25 | 55 ± 9 | ND | 0/11 | 11/11 | 2/24 |
| 3mEgre | 23/23 | 109 ± 44 | Various types | 13/23 | ||
| 14/23 | 105 ± 35 | Pre-TLL | 0/14 | 14/14 | 12/14 | |
| 2/23 | 98 ± 0 | PCT/Pre-TLL | 2/2 | 2/2 | 0/2 | |
| 2/23 | 156 ± 55 | PCT | ND | ND | 0/2 | |
| 2/23 | 125 ± 134 | ML w.M | 0/2 | 0/2 | 0/2 | |
| 2/23 | 87 ± 16 | ML wo.M | 0/2 | 0/2 | 1/2 | |
| 1/23 | 111 | PCT/MKL | 1/1 | ND | 0/1 | |
| 3mEgre+mEa/s | 21/21 | 180 ± 41 | Various types | 12/21 | ||
| 9/21 | 213 ± 15 | PCT/STL | 9/9 | 9/9f | 5/9 | |
| 5/21 | 144 ± 41 | Pre-TLL | 3/5 | 4/5 | 3/5 | |
| 2/21 | 133 ± 18 | PCT/Pre-TLL | 1/2 | 2/2 | 1/1 | |
| 2/21 | 153 ± 49 | PCT | 2/2 | 1/2 | 1/2 | |
| 1/21 | 187 | ML | 1/1 | 1/1 | 0/1 | |
| 1/21 | 187 | PCT/ML wo.M | 1/1 | 0/1 | 1/1 | |
| 1/21 | 206 | PCT/ML | 0/1 | 0/1 | 0/1 | |
Tumor phenotypes were determined by histopathological examination. ML w.M, myeloid leukemia with maturation; ML wo.M, myeloid leukemia without maturation; ML, myeloid leukemia, not otherwise specified; MKL, megakaryoblastic leukemia; PCT, plasmacytomas; pre-TLL, preT-cell lymphoblastic lymphoma; STL, small T-cell lymphoma; and ND, not determined. Some animals developed tumors of mixed phenotypes (data not shown).
Detected by the Southern blot analysis of HindIII-digested genomic tumor DNA with TCRβ (J1 and J2) and Igκ light chain-specific probes.
Complex secondary enhancer changes found in virus-induced tumors by PCR analysis.
Diagnosed for another series of mice injected with wt SL3-3 at the same facility.
All Igκ rearrangements were represented by very weak bands.
T-cell rearrangements observed in four of the nine cases were weak.
FIG. 2.
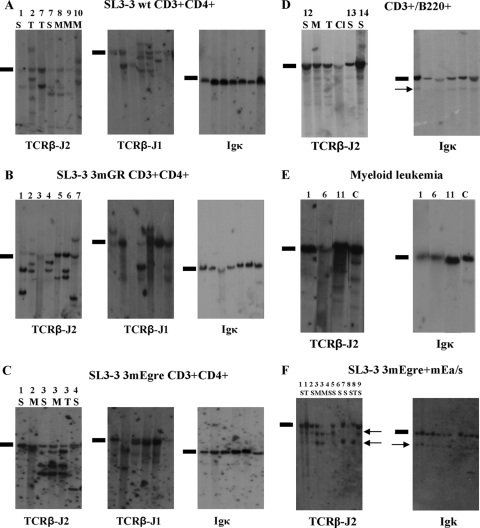
Southern blot analysis of genomic DNA extracted from SL3-3 3mEgre-, SL3-3 3mGR-, and SL3-3 wt-induced tumors of distinct types. (A) DNA from SL3-3 wt-induced T-cell tumors cleaved with HindIII and hybridized with the Igκ, TCR-J1, and TCR-J2 probes. Results are shown for seven tumors from six different mice diagnosed as T-cell lymphomas by flow analysis. (B) Hybridizations with TCRβ-J2, TCRβ-J2, and Igκ of HindIII-digested DNA from seven SL3-3 3mGR-induced tumors. (C) Six SL3-3 3mEgre-induced tumors characterized with the common T-cell lymphoma phenotype from four mice. S, spleen; M, mesenteric lymph node; T, thymus. (D) Tumors from three SL3-3 3mEgre-infected mice diagnosed with the CD3+ B220+ phenotype by flow analysis. Cl, Cervical lymph node. (E) Tumor samples from SL3-3 3mEgre-infected mice characterized with myeloid leukemia (ML) by histopathology. Lane 1, ML with maturation; lanes 6 and 11, ML without maturation. C, control spleen tissue from NRMI mock-infected mice. (F) Results for 10 SL3-3 3mEgre+mEa-injected mice; the arrows mark the weak rearrangements. Note that the rearrangement bands are the same size for all tumor samples, and this consistent pattern was not observed in the other series. The mice are numbered according to latency, with the first terminated mice having the lowest number. Black bars indicate the positions of the unrearranged genomic bands.
FIG. 3.
Characterization of SL3-3 wt- and 3mGR-induced tumors by three-color flow cytometry. Shown are dot plots of data obtained from two different tumor samples from SL3-3 wt-injected (columns A and B) and SL3-3 3mGR-injected (columns C and D) mice with the more frequent (17/23 and 27/30, respectively) (columns A and C) and less frequent (3/23 and 2/30, respectively) phenotypes (columns B and D) stained with fluorochrome-conjugated antibodies to CD3/CD11b, CD3/B220, CD4/CD8a, and TCR/HSA. The region used for the gating of the cells, as defined by forward (FSC) and side (SSC) scatter properties, is shown in all cases. The percentages of cells within each quadrant are indicated in the upper right corner. All CD4-, CD8a-, HAS-, and TCR-positive cells were CD3+.
FIG. 4.
Flow cytometry data from SL3-3 3mEgre-induced tumors with non-T-cell phenotypes. (A) Dot plots showing the staining of CD3/CD11b, CD11b/B220, CD3/B220, and CD11b/Gr-1 from a mouse characterized with a myeloid spleen tumor. (B) A thymic tumor from a mouse characterized with a mixed tumor phenotype of myeloid leukemia and T-cell lymphomas. The following stains are shown: CD3/CD11b, CD11b/B220 CD3/B220, CD11b/Gr-1, CD4/CD3, CD4/CD8a, and CD3/CD8a. (C and D) Spleen and thymus tumors from a mouse characterized with the tumor phenotype of T- and B-cell lymphomas. (C) Dot plots showing the staining of CD3/CD11b, CD11b/B220 CD3/B220, CD11b/Gr-1, CD4/CD8a, HSA/CD3, CD3/TCR, and HSA/TCR from the spleen tumor. (D) Dot plots showing the staining of CD3/CD11b, CD11b/B220 CD3/B220, CD11b/Gr-1, CD4/CD3, CD4/CD8a, and CD3/CD8a from a thymic tumor.
TABLE 2.
Incidence, latency, and molecular characterization of SL3-3 3mEgre-induced tumors of different phenotypes (series B)
| Virus injected and tumor phenotypea | No. of mice | Mean latencyf (no. of days ± SD) | DNA rearrangementsb
|
Enhancer change(s) (no. of tumors with changes/no. examined) | |
|---|---|---|---|---|---|
| Igκ | TCR | ||||
| SL3-3 wtc | 13 | 70 ± 7 | |||
| CD4+ | 4 | 67 ± 2 | 0/4 | 4/4 | 0/4 |
| CD4+/CD4+ CD8+, CD8+, or CD4+/CD8+e | 4 | 75 ± 9 | 0/4 | 4/4 | 0/3 |
| SL3-3 3mGRc | 12 | 69 ± 13 | |||
| CD4+ | 8 | 63 ± 8 | 0/8 | 8/8 | 0/8 |
| CD8+ or CD4+/CD8+ | 3 | 77 ± 18 | 0/3 | 3/3 | 0/3 |
| SL3-3 3mEgre | 12 | 135 ± 62 | 6/12 | ||
| CD4+ | 6 | 102 ± 19 | 0/5 | 6/6 | 3/6 |
| CD11b+ GR1+ | 1 | 69 | 0/1 | 0/1 | 0/1 |
| CD4+/CD11b+ Gr-1+ | 1 | 113 | 0/1 | 1/1 | 0/1 |
| CD4+/B220+ or CD4+/CD8+/B220+d | 4 | 226 ± 1 | 4/4 | 4/4 | 3/4 |
Surface expression of SL3-3 3mEgre-induced tumor cells. The antibodies used were CD3, B220, CD11b, CD4, CD8, Gr-1, and Ter119. All CD4+ and CD8+ lymphoma cells were CD3+ (data not shown). The tumor phenotype was determined by flow cytometry.
Rearrangements detected by Southern blot analysis of HindIII-digested genomic tumor DNA with specific TCRβ (J1 and J2) and B-cell receptor Igκ probes.
Not all mice in these series were investigated by flow cytometry and Southern blotting.
All CD3 populations were HSA−, which is in contrast to what was observed in the rest of the study, as determined by the CD3/HSA/TCRβ panel.
Slashes mean that the tumor cells contain several populations with different expression patterns.
The mean latencies were slightly longer than those in series A, possibly reflecting the use of different animal facilities. In all cases the uncommon phenotypes were found together with the CD4+ phenotype, except for one SL3-3-injected mouse, which had only uncommon phenotypes.
SL3-3 wt and GRE and Ea/s mutants induce T-cell lymphomas with similar phenotypes.
As shown in Fig. 1 and Tables 1 and 2, all virus-infected animals developed tumors, but with different latency periods. Among the five mutants, 3mGR, mEa/s, and 3mGR+Ea/s all showed a mean tumor latency periods of approximately 60 days, as did wt SL3-3. Southern blot analysis (Fig. 2) revealed TCRβ, but not Igκ light-chain, rearrangements in 100% of the cases for all three mutants (Tables 1 and 2), as was observed for wt tumors. The histopathological studies of SL3-3 wt- and 3mGR-induced tumors revealed a 99% incidence of pre-T-cell lymphoblastic lymphomas (pre-T LBLs) (Fig. 3, Table 1). Flow cytometry analysis showed that the main T-cell tumor phenotype induced by SL3-3 wt was CD3+ CD4+ CD8− TCRβ+ HSA+ (Fig. 3), confirming our previous results (57). Interestingly, the mutation of GRE did not diverge SL3-3 away from the strict phenotypic CD4+ expression pattern (Fig. 3). Neither gross pathology nor Southern blot analysis indicated that the T-cell lymphomas induced by SL3-3 3mGR+mEa/s and mEa/s differed from SL3-3 wt- and 3mGR-induced tumor phenotypes. These tumors were not analyzed by flow cytometry. We conclude that 3mGR, Ea/s, and 3mGR+Ea/s mutants had pathogenic properties similar to those of the wt, indicating that the GRE and Ea/s site do not play a significant role in T-lymphoma induction by SL3-3.
The Egre mutations increase latency and broaden disease specificity.
The SL3-3 3mEgre mutant induced tumors with an extended latency period of 109 days (Fig. 2). Furthermore, histopathology and Southern blot analysis revealed a shift in the cell type specificity of this mutant virus to the induction of multiple hematopoietic malignancies, including myeloid leukemia and B-cell and T-cell lymphomas (Fig. 2, Table 2). Fourteen of 23 SL3-3 3mEgre-injected mice of series A presented with pre-T LBLs, i.e., the same phenotype as that observed for SL3-3 wt-induced tumors, but the pre-T LBLs appeared after a significantly longer latency period. In support of this, Southern blot analysis revealed clear clonal TCRβ rearrangements in all 14 cases and no clonal Igκ rearrangements (Fig. 2C and Tables 1 and 2). Four mice were diagnosed with myeloid leukemia with or without maturation, none of which showed rearrangements of TCRβ or Igκ (Fig. 2E). The remaining tumors showed more complex or mixed phenotypes (Table 1). Southern hybridization results from these tumor samples (Fig. 2D) supported histopathology data, although most of the rearranged TCRβ and Igκ bands were weak, suggesting tumor oligoclonality or the presence of many nontumor cells in the tissues.
Consistently with results from series A, the flow cytometry analysis of 33 solid tumors from 12 SL3-3 3mEgre-injected mice of series B revealed T-cell lymphomas, myeloid leukemia, and mixed T-cell/B-cell lymphomas (Table 2). The phenotype of the T lymphomas induced by SL3-3 3mEgre (6/12 mice) did not differ from the CD3+ CD4+ HSA+ phenotype seen for wt SL3-3. This is in contrast to Runx site-mutated SL3-3dm, which induces T-cell lymphomas with immunophenotypes different from those of wt SL3-3 (57). Staining with CD11b/Gr-1/Ter119 revealed two cases of CD11b+ tumor cells, indicating a myeloid origin (Fig. 4A and B).
The last four mice in this series, which stayed healthy significantly longer (85 days) than the rest of this group, were diagnosed with mixed T-cell/B-cell lymphomas based on CD3+ and B220+ tumor cell populations (Fig. 5C). Southern blot analysis showed a complex pattern of bands above an intense TCRβ germ line band and very faint Igκ rearrangement bands, suggesting tumor oligoclonality and/or the presence of many nontumor cells (Fig. 2D).
FIG. 5.
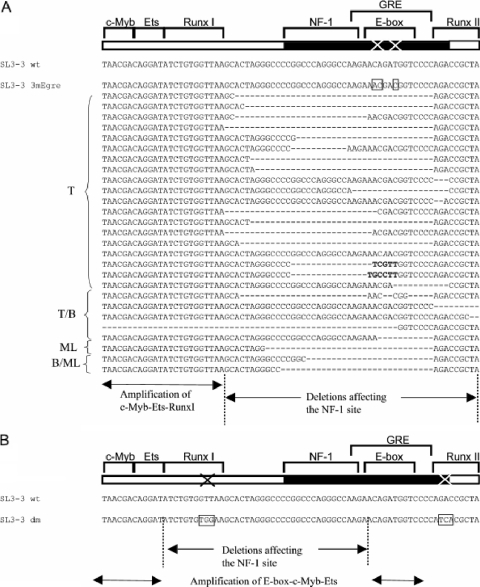
Enhancer repeat deletions found in E-box mutants and the Runx site mutant. (A) The deletions found in SL3-3 3mEgre- and SL3-3 3mEgre+mEa/s-induced tumors. At the bottom are the borders of deletions and amplifications found in the E-box mutant-induced tumors. (B) Borders of deletions and amplifications found in SL3-3 dm (a mutation of RunxI and RunxII sites [57]). The deletions include the outer borders and the amplifications include the inner borders of the enhancer sites. T, T-cell lymphoma; B, B-cell lymphoma; ML, myeloid leukemia; T/B, mixed T-cell/B-cell lymphomas; B/ML, mixed B-cell lymphoma and myeloid leukemia.
Ea/s, the E-box downstream of the tandem repeat, works as a backup site for Egre.
Interestingly, the SL3-3 mEgre+mEa/s virus exhibited a further extended latency period of 180 days (Fig. 1, Table 1), indicating that Ea/s works as a backup site for Egre, since the mutation of Ea/s alone had no effect on viral pathogenicity. Similarly to 3mEgre, the 3mEgre+mEa/s series showed a higher proportion of non-T-cell tumors, including myeloid leukemia and B-cell and T-cell lymphomas, as confirmed by histopathology. Southern blot analysis revealed complex patterns characterized by weak bands of rearranged TCRβ and/or Igκ (Fig. 2E), which were of limited diagnostic value compared to the value of histopathology (Table 1).
In the SL3-3 3mEgre+mEa/s series, only 5/21 mice were diagnosed with the pre-T LBL phenotype that is common to wt SL3-3 and the other mutants analyzed. Three SL3-3 3mEgre+mEa/s-injected mice were diagnosed with myeloid leukemia (Table 1), and the remaining 13 animals had various complex or mixed phenotypes, in all cases including a B-cell component as observed by histopathology (Table 1). Among the latter, nine mice had a mixed plasmacytoma/small T-cell lymphoma (Table 1). In conclusion, the mutation of mEa/s in addition to the Egre sites further extended the latency period as well as broadened the cell type specificity of tumor induction, thereby revealing a backup function of Ea/s for Egre.
Tumor phenotype-associated complex enhancer changes are found in SL3-3 3mEgre- and SL3-3 3mEgre+mEa/s-induced tumors.
The LTR region is known to be dynamic during tumorigenesis, and secondary mutations are found frequently in mice injected with attenuated viruses (19-21, 45, 57). We therefore analyzed the U3 enhancer region in genomic DNA from SL3-3 wt- and mutant-injected mice by PCR amplification and sequencing (Fig. 1). In SL3-3 wt-, 3mGR-, and mEa/s-induced tumors, all introduced mutations were intact, no reversions were found, and only fluctuations in the number of enhancer repeats were observed in almost all cases.
In contrast, several sequences retrieved from SL3-3 3mEgre- and 3mEgre+mEa/s-induced tumors showed numerous complex alterations, including various deletions, point mutations, and insertions in addition to fluctuations in the number of enhancer repeats. Secondary enhancer changes were discovered in 13/23 SL3-3 3mEgre-injected mice and 12/21 SL3-3 3mEgre+mEa/s-injected mice (Tables 1 and 2 and Fig. 5). These enhancer variants are unlikely to be PCR artifacts, since the same structures were found in several independent PCRs.
In the great majority of cases, deletions involved part of or the entire NF1 and/or mutant Egre site, as well as the neighboring Runx site II. The c-Myb-Ets-Runx site I sequence was maintained in almost all cases investigated. Furthermore, in some cases the deletions involved sequences downstream of the enhancer repeat affecting the Ea/s site. The deletions observed in this study were highly similar to those seen in SL3-3dm-injected mice, in which the E-box was left intact in almost all cases, strongly indicating its importance (Fig. 5) (57). Insertions and point mutations were discovered in a few cases, but the origin and possible functional role of these insertions were not clarified. No reversions or gain of E-box motifs were discovered within the U3 region, although several imperfect E-box motifs are present in the U3 region outside the enhancer repeats. Complex secondary enhancer changes were tumor phenotype specific; thus, the mutated SL3-3 3mEgre enhancer seemed to be suboptimal for the induction of T-cell lymphomas and myeloid leukemia without maturation but were sufficient for the induction of myeloid leukemia with maturation and possibly B-cell lymphomas.
DISCUSSION
In this study, we have shown that an E-box (Egre) overlapping a GRE is important for T-lymphoma induction by the gammaretrovirus SL3-3, whereas the GRE itself does not play a significant role. Furthermore, an E-box site (Ea/s) outside the enhancer repeats was shown to work as a backup site for Egre.
In agreement with our earlier work (57), we observed that wt SL3-3 induced strictly T-cell lymphomas in inbred NMRI mice that were characterized by clonal TCRβ rearrangements and a predominant CD3+ CD4+ CD8− HSA+ immunophenotype, similarly to semimature T-helper cells. Such a consistent CD4/CD8 expression pattern on tumor cells from SL3-3-injected mice has not been observed in other strains, including AKR (32, 33) and BALB/c (25). Mutating the GRE in the SL3-3 enhancer did not change the homogenous CD4/CD8 expression pattern in SL3-3-induced tumors in inbred NMRI mice, in agreement with reports of low-level glucocorticoid responses in T cells (11, 29). Thus, our results strongly indicate that the GRE site is dispensable for T-lymphoma induction by SL3-3 and that the E-box is most probably the primary site of activation. A study of the highly related T-lymphomagenic Moloney MLV reported mutations of the GRE to increase the latency of disease induction without changing the cell type specificity (58). However, this early study did not separate the function of GRE and E-box sites or investigate tumors for secondary enhancer changes or the reversion of the two mutated nucleotides.
The GRE overlapping the E-box site is conserved in many gammaretroviruses, and it is well established that steroid hormones regulate the replication of a number of retroviruses, including the betaretrovirus mouse mammary tumor virus (53). The T-cell-tropic variant of mouse mammary tumor virus has nonoverlapping GRE and E-box sites in its transcriptional enhancer. Both sites appear to be conserved in proviruses isolated from T lymphomas, suggesting that both are needed for viral T lymphomagenesis (5, 47, 53). Previously, a correlation was made between the presence of a GRE in FIS2 and a significant sex difference in the susceptibility to this virus (6-8). It is conceivable that the conserved GRE motif of SL3-3 also plays a role for infection by means other than the intraperitoneal injection of newborns in which different cell types would be infected.
The mutation of enhancer E-box sites allowed the otherwise strictly T-lymphomagenic SL3-3 to induce a multitude of hematopoietic malignancies, including T-cell lymphomas, myeloid leukemia, plasmacytoma, and mixed T-cell/B-cell lymphomas, clearly demonstrating an important role for the E-box motifs in SL3-3 tumorigenesis. The marked phenotypic heterogeneity suggests that SL3-3 3mEgre and SL3-3 3mEgre+mEa/s infect and transform several different cell types or target an early hematopoietic progenitor. This broad disease specificity is reminiscent of that of natural MLV isolates such as SRS19-6 and Cas-Br-M/Cas-Br-E (9, 24).
Mutating the Ea/s sequence located outside the enhancer framework alone (49, 50) or together with the GRE did not change the pathogenic properties of SL3-3; however, mutating Ea/s along with Egre sites further extended the latency period and increased the induction of B-cell lymphomas. The backup function of Ea/s for Egre parallels the well-established backup function of Runx site II for Runx site I (57) and adds further support to the importance of Egre sites. Ea/s also is present in B-lymphomagenic Akv (41) and erythroleukemic Friend (37) MLVs, while T-lymphomagenic Moloney MLV (55) has an E-box site outside the enhancer repeat with the same core sequence (CAGATG), indicating that other MLVs have exploited this mode of transcription activation.
In contrast to SL3-3dm, a virus with mutations in all Runx sites, which induces T-cell lymphomas of various phenotypes (57), the E-box mutants induced T-cell lymphomas with the same tumor phenotype as that of wt tumors, indicating that the disruption of T-cell lymphomagenesis by E-box mutations was incomplete. Hence, the Runx sites of SL3-3, but not the E-boxes, appear to promote the characteristic CD4 single-positive phenotype of SL3-3-induced tumors in inbred NMRI mice.
In contrast to T-lymphomagenic SL3-3, for which the E-box site and not the GRE site has a primary role in tumorigenic enhancer activity, all E-box/GRE sites appear to play a role for B-lymphomagenic Akv MLV (56), since mutations shift tumor cell specificity to a more differentiated B-cell phenotype. Notably, no secondary enhancer mutations were found in any Akv enhancer mutant-induced tumor, which also is in contrast to the results of the SL3-3 study. It is unclear why the Akv model differs from the SL3-3 model; however, several features of the former point to the importance of an immunostimulatory component in which the overall viral load as well as insertional mutagenesis plays a role (56).
Several tumors from SL3-3 3mEgre and 3mEgre+mEa/s harbored complex secondary enhancer alterations, including various deletions, insertions, and point mutations, which were mainly T-cell tumor phenotype associated. As shown in Fig. 5, the deletions in the SL3-3 E-box and previously studied Runx site mutants (19, 57) share several similarities, but they also share striking differences. In both cases, the deletions of the NF-1 site and the impairment of GRE were observed, and the mutated site often was deleted. NF-1 sites harbor a downregulating function on SL3-3 reporter constructs in T cells (19), and this type of alteration has been shown to partially restore viral enhancer activity and to rescue, to some degree, the pathogenicity of SL3-3 Runx site mutants such as SL3-3dm (19, 20). In SL3-3dm the c-Myb-Ets-E-box sequence was preserved, whereas the c-Myb-Ets-Runx site I sequence was amplified in the E-box mutant. Thus, when Runx site I is mutated the maintenance of the E-box is important, and when the E-box is abolished Runx site I is amplified, strongly supporting the importance of the two sites in T-cell tumor induction by SL3-3. We note that the outer borders of the enhancer deletions observed in tumors with a T-cell lymphoma phenotype coincide precisely with the borders of the respective binding sites, i.e., Runx site I and E-box. The complex enhancer changes were identified primarily in tumors characterized as T-cell lymphomas and in a few cases of myeloid leukemia without maturation, indicating that the E-box mutants are suboptimal for the induction of these phenotypes. Importantly, no reversion or gain of an E-box motif was observed anywhere in sequenced regions, suggesting that T-lymphomas are induced by a virus without the binding of bHLH proteins to the DNA of its transcription control region.
In summary, we have shown that the mutation of the E-box overlapping GRE binding sites in the enhancer of SL3-3 shifts the disease pattern from strictly T lymphomagenicity to a broader variety of hematopoietic malignancies. The GRE itself and an E-box outside the enhancer repeat (Ea/s) appear to be dispensable for the tumor induction process; however, the Ea/s site works as a backup for the Egre sites. While mature myeloid leukemia and plasmacytomas induced by SL3-3 3mEgre and 3mEgre+mEa/s were found to harbor unaltered enhancers, secondary enhancer alterations appeared necessary for the induction of T-cell lymphomas and immature myeloid leukemias by these viruses. Such complex enhancer changes were not observed in the wt or other mutants in this study, suggesting that the E-box mutations induce in vivo the molecular evolution of viral enhancer structures (9, 19, 28), resulting in the redirection of the cell specificity. In conclusion, the E-box sites and their cognate binding factors, the bHLH proteins, play important roles in the potency and specificity of disease induction by SL3-3.
Acknowledgments
The technical assistance of Lone Højgaard, Claudia Klos, Jacqueline Müller, Susan Bensch, Claudia Klos, Jacqueline Müller, and Elenore Samson is gratefully acknowledged, as are the animal caretakers for their dedicated help.
This work was supported by the Danish Cancer Society, the Novo Nordic Foundation, Fabrikant Vilhelm Pedersen og Hustrus Legat, the Danish Medical Research Council, and partially by the National Genome Research Network (NGFN2, 01GR0430). D.E. is a fellow of the Research School in Gene Medicine.
Footnotes
Published ahead of print on 22 October 2008.
REFERENCES
- 1.Bain, G., I. Engel, E. C. Robanus Maandag, H. P. te Riele, J. R. Voland, L. L. Sharp, J. Chun, B. Huey, D. Pinkel, and C. Murre. 1997. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 174782-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain, G., S. Gruenwald, and C. Murre. 1993. E2A and E2-2 are subunits of B-cell-specific E2-box DNA-binding proteins. Mol. Cell. Biol. 133522-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain, G., and C. Murre. 1998. The role of E-proteins in B- and T-lymphocyte development. Semin. Immunol. 10143-153. [DOI] [PubMed] [Google Scholar]
- 4.Bain, G., M. W. Quong, R. S. Soloff, S. M. Hedrick, and C. Murre. 1999. Thymocyte maturation is regulated by the activity of the helix-loop-helix protein, E47. J. Exp. Med. 1901605-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhadra, S., M. M. Lozano, and J. P. Dudley. 2005. Conversion of mouse mammary tumor virus to a lymphomagenic virus. J. Virol. 7912592-12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruland, T., H. Y. Dai, L. A. Lavik, L. I. Kristiansen, and A. Dalen. 2001. Gender-related differences in susceptibility, early virus dissemination and immunosuppression in mice infected with Friend murine leukaemia virus variant FIS-2. J. Gen. Virol. 821821-1827. [DOI] [PubMed] [Google Scholar]
- 7.Bruland, T., L. A. Lavik, H. Y. Dai, and A. Dalen. 2003. A glucocorticoid response element in the LTR U3 region of Friend murine leukaemia virus variant FIS-2 enhances virus production in vitro and is a major determinant for sex differences in susceptibility to FIS-2 infection in vivo. J. Gen. Virol. 84907-916. [DOI] [PubMed] [Google Scholar]
- 8.Bruland, T., L. A. Lavik, H. Y. Dai, and A. Dalen. 2003. Identification of Friend murine retrovirus-infected immune cells and studies of the effects of sex and steroid hormones in the early phase of infection. APMIS 111878-890. [DOI] [PubMed] [Google Scholar]
- 9.Bundy, L. M., M. Ru, B. F. Zheng, L. Cheng, P. K. Pattengale, J. L. Portis, and H. Fan. 1995. Biological characterization and molecular cloning of murine C-type retroviruses derived from the TSZ complex from mainland China. Virology 212367-382. [DOI] [PubMed] [Google Scholar]
- 10.Celander, D., and W. A. Haseltine. 1987. Glucocorticoid regulation of murine leukemia virus transcription elements is specified by determinants within the viral enhancer region. J. Virol. 61269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celander, D., B. L. Hsu, and W. A. Haseltine. 1988. Regulatory elements within the murine leukemia virus enhancer regions mediate glucocorticoid responsiveness. J. Virol. 621314-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatis, P. A., C. A. Holland, J. W. Hartley, W. P. Rowe, and N. Hopkins. 1983. Role for the 3′ end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc. Natl. Acad. Sci. USA 804408-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatis, P. A., C. A. Holland, J. E. Silver, T. N. Frederickson, N. Hopkins, and J. W. Hartley. 1984. A 3′ end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J. Virol. 52248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corneliussen, B., A. Thornell, B. Hallberg, and T. Grundstrom. 1991. Helix-loop-helix transcriptional activators bind to a sequence in glucocorticoid response elements of retrovirus enhancers. J. Virol. 656084-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DesGroseillers, L., and P. Jolicoeur. 1984. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J. Virol. 52945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engel, I., C. Johns, G. Bain, R. R. Rivera, and C. Murre. 2001. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 194733-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel, I., and C. Murre. 2004. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 23202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engel, I., and C. Murre. 1999. Ectopic expression of E47 or E12 promotes the death of E2A-deficient lymphomas. Proc. Natl. Acad. Sci. USA 96996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ethelberg, S., B. Hallberg, J. Lovmand, J. Schmidt, A. Luz, T. Grundstrom, and F. S. Pedersen. 1997. Second-site proviral enhancer alterations in lymphomas induced by enhancer mutants of SL3-3 murine leukemia virus: negative effect of nuclear factor 1 binding site. J. Virol. 711196-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ethelberg, S., J. Lovmand, J. Schmidt, A. Luz, and F. S. Pedersen. 1997. Increased lymphomagenicity and restored disease specificity of AML1 site (core) mutant SL3-3 murine leukemia virus by a second-site enhancer variant evolved in vivo. J. Virol. 717273-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ethelberg, S., A. B. Sorensen, J. Schmidt, A. Luz, and F. S. Pedersen. 1997. An SL3-3 murine leukemia virus enhancer variant more pathogenic than the wild type obtained by assisted molecular evolution in vivo. J. Virol. 719796-9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ethelberg, S., B. D. Tzschaschel, A. Luz, S. J. Diaz-Cano, F. S. Pedersen, and J. Schmidt. 1999. Increased induction of osteopetrosis, but unaltered lymphomagenicity, by murine leukemia virus SL3-3 after mutation of a nuclear factor 1 site in the enhancer. J. Virol. 7310406-10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleming, T. J., M. L. Fleming, and T. R. Malek. 1993. Selective expression of Ly-6G on myeloid lineage cells in mouse bone marrow. RB6-8C5 mAb to granulocyte-differentiation antigen (Gr-1) detects members of the Ly-6 family. J. Immunol. 1512399-2408. [PubMed] [Google Scholar]
- 24.Fredrickson, T. N., W. Y. Langdon, P. M. Hoffman, J. W. Hartley, and H. C. Morse III. 1984. Histologic and cell surface antigen studies of hematopoietic tumors induced by Cas-Br-M murine leukemia virus. J. Natl. Cancer Inst. 72447-454. [PubMed] [Google Scholar]
- 25.Glud, S. Z., A. B. Sorensen, M. Andrulis, B. Wang, E. Kondo, R. Jessen, L. Krenacs, E. Stelkovics, M. Wabl, E. Serfling, A. Palmetshofer, and F. S. Pedersen. 2005. A tumor-suppressor function for NFATc3 in T-cell lymphomagenesis by murine leukemia virus. Blood 1063546-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldrath, A. W., and M. J. Bevan. 1999. Selecting and maintaining a diverse T-cell repertoire. Nature 402255-262. [DOI] [PubMed] [Google Scholar]
- 27.Golemis, E. A., N. A. Speck, and N. Hopkins. 1990. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J. Virol. 64534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granger, S. W., L. M. Bundy, and H. Fan. 1999. Tandemization of a subregion of the enhancer sequences from SRS 19-6 murine leukemia virus associated with T-lymphoid but not other leukemias. J. Virol. 737175-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallberg, B., and T. Grundstrom. 1988. Tissue specific sequence motifs in the enhancer of the leukaemogenic mouse retrovirus SL3-3. Nucleic Acids Res. 165927-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallberg, B., J. Schmidt, A. Luz, F. S. Pedersen, and T. Grundstrom. 1991. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J. Virol. 654177-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardy, R. R., C. E. Carmack, S. A. Shinton, J. D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 1731213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hays, E. F., and G. Bristol. 1992. Observations on lymphomagenesis and lymphoma in AKR mice. A description of prelymphoma changes in the thymus and phenotypic diversity of lymphomas induced by SL3-3 virus. Thymus 19219-234. [PubMed] [Google Scholar]
- 33.Hays, E. F., G. C. Bristol, S. McDougall, J. L. Klotz, and M. Kronenberg. 1989. Development of lymphoma in the thymus of AKR mice treated with the lymphomagenic virus SL 3-3. Cancer Res. 494225-4230. [PubMed] [Google Scholar]
- 34.Hestdal, K., F. W. Ruscetti, J. N. Ihle, S. E. Jacobsen, C. M. Dubois, W. C. Kopp, D. L. Longo, and J. R. Keller. 1991. Characterization and regulation of RB6-8C5 antigen expression on murine bone marrow cells. J. Immunol. 14722-28. [PubMed] [Google Scholar]
- 35.Jones, S. 2004. An overview of the basic helix-loop-helix proteins. Genome Biol. 5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kina, T., K. Ikuta, E. Takayama, K. Wada, A. S. Majumdar, I. L. Weissman, and Y. Katsura. 2000. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br. J. Haematol. 109280-287. [DOI] [PubMed] [Google Scholar]
- 37.Koch, W., W. Zimmermann, A. Oliff, and R. Friedrich. 1984. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J. Virol. 49828-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagasse, E., and I. L. Weissman. 1996. Flow cytometric identification of murine neutrophils and monocytes. J. Immunol. Methods 197139-150. [DOI] [PubMed] [Google Scholar]
- 39.Leib-Mosch, C., J. Schmidt, M. Etzerodt, F. S. Pedersen, R. Hehlmann, and V. Erfle. 1986. Oncogenic retrovirus from spontaneous murine osteomas. II. Molecular cloning and genomic characterization. Virology 15096-105. [DOI] [PubMed] [Google Scholar]
- 40.Lenz, J., D. Celander, R. L. Crowther, R. Patarca, D. W. Perkins, and W. A. Haseltine. 1984. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. Nature 308467-470. [DOI] [PubMed] [Google Scholar]
- 41.Lenz, J., R. Crowther, S. Klimenko, and W. Haseltine. 1982. Molecular cloning of a highly leukemogenic, ecotropic retrovirus from an AKR mouse. J. Virol. 43943-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis, A. F., T. Stacy, W. R. Green, L. Taddesse-Heath, J. W. Hartley, and N. A. Speck. 1999. Core-binding factor influences the disease specificity of Moloney murine leukemia virus. J. Virol. 735535-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovmand, J., A. B. Sorensen, J. Schmidt, M. Ostergaard, A. Luz, and F. S. Pedersen. 1998. B-cell lymphoma induction by Akv murine leukemia viruses harboring one or both copies of the tandem repeat in the U3 enhancer. J. Virol. 725745-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martiney, M. J., L. S. Levy, and J. Lenz. 1999. Suppressor mutations within the core binding factor (CBF/AML1) binding site of a T-cell lymphomagenic retrovirus. J. Virol. 732143-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martiney, M. J., K. Rulli, R. Beaty, L. S. Levy, and J. Lenz. 1999. Selection of reversions and suppressors of a mutation in the CBF binding site of a lymphomagenic retrovirus. J. Virol. 737599-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mertz, J. A., F. Mustafa, S. Meyers, and J. P. Dudley. 2001. Type B leukemogenic virus has a T-cell-specific enhancer that binds AML-1. J. Virol. 752174-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison, H. L., B. Soni, and J. Lenz. 1995. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J. Virol. 69446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen, A. L., P. L. Norby, F. S. Pedersen, and P. Jorgensen. 1996. Various modes of basic helix-loop-helix protein-mediated regulation of murine leukemia virus transcription in lymphoid cell lines. J. Virol. 705893-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen, A. L., N. Pallisgaard, F. S. Pedersen, and P. Jorgensen. 1994. Basic helix-loop-helix proteins in murine type C retrovirus transcriptional regulation. J. Virol. 685638-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nieves, A., L. S. Levy, and J. Lenz. 1997. Importance of a c-Myb binding site for lymphomagenesis by the retrovirus SL3-3. J. Virol. 711213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nilsson, P., B. Hallberg, A. Thornell, and T. Grundstrom. 1989. Mutant analysis of protein interactions with a nuclear factor I binding site in the SL3-3 virus enhancer. Nucleic Acids Res. 174061-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ringold, G. M., K. R. Yamamoto, G. M. Tomkins, M. Bishop, and H. E. Varmus. 1975. Dexamethasone-mediated induction of mouse mammary tumor virus RNA: a system for studying glucocorticoid action. Cell 6299-305. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt, J., A. Luz, and V. Erfle. 1988. Endogenous murine leukemia viruses: frequency of radiation-activation and novel pathogenic effects of viral isolates. Leuk. Res. 12393-403. [DOI] [PubMed] [Google Scholar]
- 55.Shinnick, T. M., R. A. Lerner, and J. G. Sutcliffe. 1981. Nucleotide sequence of Moloney murine leukaemia virus. Nature 293543-548. [DOI] [PubMed] [Google Scholar]
- 56.Sørensen, K. D., S. Kunder, L. Quintanilla-Martinez, J. Sorensen, J. Schmidt, and F. S. Pedersen. 2007. Enhancer mutations of Akv murine leukemia virus inhibit the induction of mature B-cell lymphomas and shift disease specificity towards the more differentiated plasma cell stage. Virology 362179-191. [DOI] [PubMed] [Google Scholar]
- 57.Sørensen, K. D., L. Quintanilla-Martinez, S. Kunder, J. Schmidt, and F. S. Pedersen. 2004. Mutation of all Runx (AML1/core) sites in the enhancer of T-lymphomagenic SL3-3 murine leukemia virus unmasks a significant potential for myeloid leukemia induction and favors enhancer evolution toward induction of other disease patterns. J. Virol. 7813216-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Speck, N. A., B. Renjifo, E. Golemis, T. N. Fredrickson, J. W. Hartley, and N. Hopkins. 1990. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 4233-242. [DOI] [PubMed] [Google Scholar]
- 59.Zaiman, A. L., and J. Lenz. 1996. Transcriptional activation of a retrovirus enhancer by CBF (AML1) requires a second factor: evidence for cooperativity with c-Myb. J. Virol. 705618-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaiman, A. L., A. Nieves, and J. Lenz. 1998. CBF, Myb, and Ets binding sites are important for activity of the core I element of the murine retrovirus SL3-3 in T lymphocytes. J. Virol. 723129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]