Abstract
The human immunodeficiency virus type 1 envelope glycoprotein (Env) complex is the principal focus of neutralizing antibody-based vaccines. The functional Env complex is a trimer consisting of six individual subunits: three gp120 molecules and three gp41 molecules. The individual subunits have proven unsuccessful as vaccines presumably because they do not resemble the functional Env complex. Variable domains and carbohydrates shield vulnerable neutralization epitopes on the functional Env complex. The deletion of variable loops has been shown to improve gp120's immunogenicity; however, problems have been encountered when introducing such modifications in stabilized Env trimer constructs. To address these issues, we have created a set of V1/V2 and V3 loop deletion variants in the context of complete virus to allow optimization by forced virus evolution. Compensatory second-site substitutions included the addition and/or removal of specific carbohydrates, changes in the disulfide-bonded architecture of the V1/V2 stem, the replacement of hydrophobic residues by hydrophilic and charged residues, and changes in distal parts of gp120 and gp41. These viruses displayed increased sensitivity to neutralizing antibodies, demonstrating the improved exposure of conserved domains. The results show that we can select for functionally improved Env variants with loop deletions through forced virus evolution. Selected evolved Env variants were transferred to stabilized Env trimer constructs and were shown to improve trimer expression and secretion. Based on these findings, we can make recommendations on how to delete the V1/V2 domain from recombinant Env trimers for vaccine and X-ray crystallography studies. In general, virus evolution may provide a powerful tool to optimize Env vaccine antigens.
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein complex (Env) is the principal target of vaccine research aimed at raising an antiviral humoral immune response. The isolation of a small number of broadly active neutralizing antibodies from HIV-infected individuals serves as a rationale for the search for vaccines that elicit such antibodies. Although many Env-based HIV-1 vaccines have been tested in preclinical and clinical studies, so far none has raised broadly reactive antibodies that could neutralize diverse primary virus isolates. Thus, relatively straightforward vaccine strategies that worked for other pathogens—for example, the use of unmodified surface antigens—have been explored without satisfactory results for HIV-1, emphasizing the necessity for more-sophisticated vaccine design.
The functional HIV-1 Env complex, which mediates viral entry into CD4+ host cells, is a trimer consisting of six individual subunits: three gp120 molecules and three gp41 molecules. The attachment of HIV-1 to a target cell is followed by the fusion of the viral and cellular membranes. First, gp120 binds to the CD4 receptor, a process that induces conformational changes to create and expose the coreceptor binding site (71, 76). The conformational changes in gp120 involve the movement of the first, second, and third variable loops (V1, V2, and V3 loops) that normally shield the coreceptor binding site (68, 69, 78). Additional conformational changes in the trimeric complex lead to the exposure of hydrophobic fusion peptides at the N termini of the gp41 subunits, culminating in the fusion of the viral and cellular membranes (13, 16, 31, 36, 75).
The individual subunits have proven unsuccessful as vaccines, presumably because they do not resemble the functional Env complex present on infectious virus particles. We and others have generated stabilized trimeric Env, yielding incremental to modest improvements of Env immunogenicity (4, 4-6, 21, 23, 62, 65, 80, 82). These soluble trimeric gp140 molecules provide useful scaffolds for further improvement of its immunogenicity, for example, by uncovering masked neutralization targets.
HIV-1 has evolved several strategies to limit the generation of neutralizing antibodies and to minimize their effect on its replication cycle. A prominent strategy is the shielding of conserved protein domains, for example, the (co)receptor binding sites, by flexible variable loops that can easily be changed by the virus to escape from antibodies. Considerable efforts have therefore been made to generate and characterize forms of Env with loop deletions. Initial functional studies have shown that the deletion of V3 or V4 abrogated Env function and viral infectivity (15, 52, 70, 79). Several constructs have been described lacking V1 or V2 that were compatible with minimal Env function and viral replication, indicating that they are not absolutely required for function (35, 66, 78). However, most constructs with combined V1/V2 deletions were nonfunctional or severely impaired in Env function (78, 79).
The removal of variable domains from recombinant monomeric gp120 has resulted in incremental improvement of immunogenicity (3, 27, 34, 44, 45, 81). We have previously incorporated such deletions into disulfide-stabilized gp140 constructs (SOS and SOSIP) (60, 64; unpublished data), but we encountered difficulties with the protein expression and purification of these recombinant constructs. Moreover, Center and coworkers have found that the deletion of the V1/V2 region from uncleaved JR-FL strain gp140 trimers promotes aggregation (12). Thus, the deletion of variable loops can cause problems in complex recombinant Env constructs that are not apparent in the context of monomeric gp120.
Although the adverse effects of V1/V2 deletions on recombinant trimers are ill-defined, we hypothesized that one could overcome these effects by employing forced virus evolution to select improved Env deletion variants that would have been difficult to obtain through molecular design. In addition to solving the problem, it may assist in better defining the actual problem. We have created a set of different V1/V2 and V3 loop deletion variants in the context of a complete virus to allow optimization by virus evolution. The results described here indicate that it is indeed possible to obtain functionally improved Env variants lacking the entire V1/V2 domain. Compensatory changes were identified that improve the folding and secretion of stable Env trimers with loop deletions and should benefit the generation of recombinant Env trimers for vaccine and structural studies.
MATERIALS AND METHODS
Construction of loop deletion mutants.
Deletion variants 1, 2, 8 to 12, and 14 to 17 were created by splice-overlap extension PCR. The downstream and upstream sequences of the sequences to be deleted were amplified in separate PCRs (PCRs 1 and 2). The complete fragment was amplified in a third PCR using the combined products of the first two PCRs as a template and using primers A (5′-CAGATGCTAAAGCATATGATAC-3′) and B (5′-TTGTTCTCTTAATTTGCTAGCTATC −3′). The restriction sites for NdeI and NheI are underlined. The products of the third PCRs were cloned into pRS1 (57), using NdeI and NheI or NdeI and StuI.
The following primers were used for PCRs 1 and 2 (the nucleotides coding for the linker residues replacing the deleted sequences are in italics): for deletion variant 1, primers A plus C (5′-GGATACCTTTGGTGCTGCTGGCTTTAGGCT-3′) and B plus D (5′-CCAAAGGTATCCTTTGAGC-3′); for variant 2, primers A plus E (5′-GACTGAGGTGTTTGCTGCGAGTGGGGTTAA-3′) and B plus F (5′-AACACCTCAGTCATTACAC-3′); for variant 8, primers A plus G (5′-GGTGTTACAACTGCCGCGGCCAACACAGAGTGG-3′) and B plus H (5′-AGTTGTAACACCTCAGTC-3′); for variants 9 to 12, primers A plus I (5′-GGTGTTACAACTGCCGKYGYCAACACAGAGTGG-3′) and B plus H; for variant 14, primers A plus J (5′-CTTTGGACAGGCGCCTGCGCCTACACATGGCTTTAGG-3′) and B plus K (5′-GCCTGTCCAAAGGTATCC-3′); for variant 15, primers A plus L (5′-CCCTGGTCCCCTTGTATTGTTGTT-3′) and B plus M (5′-AGGGGACCAGGGAGAGCATTTGT-3′); for variant 16, primers A plus N (5′-GTTACAATGTGCTGTAACAAATGC-3′) and B plus O (5′-GCACATTGTAACATTAGTAGAGCA-3′); and for variant 17, primers A plus P (5′-ATGTTACAATGTGCTGTAACAAATGCTCTCCCTGGTCCCCTTGTATTGTTGTTGGG]-3′) and B plus O.
V1/V2 deletion variants 3 and 4 were created previously (and designated ΔV1/V2′ and ΔV1/V2*, respectively [60]) and cloned into pRS1 using the NdeI and StuI sites. Note that these mutants were originally created in a JR-FL SOS gp140 background. The subcloning of these variants in LAI Env using NdeI and StuI resulted in the presence of a few JR-FL-derived residues flanking the V1/V2 region: V87E, N92H, D99N, H105Q, S128T, K130N, T132K, and N197D for mutant 3 and V87E, N92H, D99N, H105Q, and N197D for mutant 4. The numbering of the amino acids is based on the HXB2 sequence according to convention.
Deletion variants 5 and 6 were created by a single PCR amplification using an HXB2 Env template with primers Q (5′-ACTTGTGGGTCACAGTCTATTATGGGGTACC-3′ and R (5′-TCATTCTAGGCCTCAGTGCACTTTAAACTAAC-3′) and Q and S (5′-TTCTTTCTAGGCCTTACCTCTTATGCTTGTGCTG-3′), respectively. The resulting PCR fragments which exclude the V1/V2 or V2 sequences, respectively, were cloned into pRS1 using NdeI and StuI (the restriction sites are underlined). The use of HXB2 as a template for the PCR resulted in a few residues that differ from the LAI sequences (Fig. 1).
FIG. 1.
Design of the loop deletion variants. (A) Schematic representation of the V1/V2 deletion variants used in this study. The variable loops are indicated in yellow. The deletions are indicated by either a blue line or by blue-colored residues, which replace the deleted sequences. β-strands 2 and 3, components of the conserved bridging sheet, are indicated in green. Cysteines and disulfide bonds are colored in red. Note that the designation of the disulfide bonds is based on studies with the wt protein. We do not know whether the designated disulfide bonds do in fact form in these variants. This is particularly questionable in mutants 5 and 6 where one or two wt cysteine pairs cannot be formed. In variant 5, an alternative and hypothetical disulfide bond between 126 and 131 is drawn. In variant 6, the native C131-C157 bond is drawn and C126 is left unpaired. (B) Assumed 3-D models of selected ΔV1/V2 variants. The top panel provides perspectives on gp120 as seen from CD4 (left) and the coreceptor (right; rotated over the y axis by 90°). The rectangle in the top right panel encloses the V1/V2 stem and the bridging sheet. Colors are the same as described for panel A. The bottom panels represent details of this area for variants 1, 2, and 8 and an overlay of these variants (right bottom panel, variant 1 in red, variant 2 in blue, variant 8 in white, and disulfide bonds in yellow). The four β-strands that compose the bridging sheet and the local disulfide bonds are indicated. The LAI gp120 core and variant cores were modeled by SWISS-MODEL (http://swissmodel.expasy.org//SWISS-MODEL.html) (26) using the HXB2 core (pdb accession code 1G9M [37]) and drawn using Viewerlite (Accelrys, Inc.). The overlay in the bottom right panel was prepared with Deepview/SWISS pdb viewer (http://www.expasy.org/spdbv/) (26) and rendered in Viewerlite. (C) Schematic representation of the V3 deletion variants. Colors are as described for panel A. (D) Rearrangement of the V1/V2 stem in variant 6. The starting situation is as described for panel A. Note that the drawn disulfide bond between residues C131 and C157 is purely speculative. However, in the wt protein, these cysteines do form a disulfide bond. Left panel, a hypothetical situation after the first substitution (C131Y) with a new nonnative disulfide bond between C126 and C157, resulting in the restoration of the V1 to its full length and the formation of a pseudo-V2. Right panel, removal of N156 after prolonged culturing. Note that we observe the removal of the glycosylation site at N156 in two independent cultures in two different substitutions, N156K (as indicated in the figure) in culture 6A and S158F in culture 6B. The sequences were derived from sequencing clones at day 38 (6C) and day 99 (6A).
Cells and transfection.
SupT1 T cells and HEK293T cells were maintained in RPMI 1640 medium and Dulbecco's modified Eagle's medium (DMEM), respectively, supplemented with 10% fetal calf serum (FCS), penicillin (100 U/ml), and streptomycin (100 μg/ml) as previously described (59). HeLa cells (ATCC) and TZM-bl cells were maintained in DMEM containing 10% FCS, MEM nonessential amino acids, and penicillin/streptomycin. SupT1 and HEK293T cells were transfected by electroporation and Ca2(PO4)3 precipitation, respectively, as described elsewhere (18). Peripheral blood mononuclear cells (PBMCs) were isolated from fresh buffy coats (Central Laboratory Blood Bank, Amsterdam, The Netherlands) by standard Ficoll-Hypaque density centrifugation. PBMCs were frozen in multiple vials at a high concentration and, when required, thawed and activated with 5 μg/ml phytohemagglutinin and cultured in RPMI medium containing 10% FCS, penicillin (100 U/ml), streptomycin (100 U/ml), and recombinant interleukin-2 (100 U/ml). On day 4 of the culture, the cells underwent CD4+ enrichment by the depletion of the CD8+ lymphocytes from PBMCs using CD8 immunomagnetic beads.
Viruses and infections.
Virus produced in SupT1 cells and virus from SupT1 evolution cultures were stored at −80°C, and the virus concentration was quantitated by CA-p24 enzyme-linked immunosorbent assay (ELISA) as described previously (32). These values were used to normalize the amount of virus in subsequent infection experiments. The infection experiments were performed with 400 × 103 SupT1 cells or 200 × 103 CD4+ primary lymphocytes and 100 pg CA-p24 or 500 pg CA-p24 of virus, respectively, per well in a 96-well plate. Virus spread was measured for 14 days using CA-p24 ELISA.
Single-cycle infection and neutralization.
The TZM-bl reporter cell line (20, 74) stably expresses high levels of CD4 and the HIV-1 coreceptors CCR5 and CXCR4 and contains the luciferase and β-galactosidase genes under the control of the HIV-1 long-terminal-repeat promoter. The TZM-bl cell line was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (John C. Kappes, Xiaoyun Wu, and Tranzyme, Inc., Durham, NC). One day prior to infection, TZM-bl cells were plated on a 96-well plate in DMEM containing 10% fetal bovine serum, MEM nonessential amino acids, and penicillin/streptomycin (both at 100 U/ml) and incubated at 37°C with 5% CO2. A fixed amount of virus (1.0 ng CA-p24) was preincubated for 30 min at room temperature with increasing concentrations of monoclonal antibodies (MAbs). 2G12 and 4E10 were obtained from Hermann Katinger through the NIH AIDS Research and Reference Reagent Program; b6 and b12 were donated by Dennis Burton (The Scripps Research Institute, La Jolla, CA); 17b was a gift from James Robinson (Tulane University, New Orleans, LA); and CD4-IgG2 was a gift from William Olson (Progenics Pharmaceuticals, Inc., Tarrytown, NY). This mixture was added to TZM-bl cells at 70 to 80% confluence in the presence of 400 nM saquinavir (Roche, Basel, Switzerland) and 40 μg/ml DEAE in a total volume of 200 μl on a 96-well plate. Two days postinfection, the medium was removed, and the cells were washed once with phosphate-buffered saline and lysed in reporter lysis buffer (Promega, Madison, WI). Luciferase activity was measured using a luciferase assay kit (Promega) and a Glomax luminometer (Turner BioSystems, Sunnyvale, CA) according to the manufacturer's instructions. All infections were performed in duplicate, and luciferase measurements were also performed in duplicate. Uninfected cells were used to correct for background luciferase activity. The infectivity of each mutant without inhibitor was set at 100%. Nonlinear regression curves were determined and 50% inhibitory concentrations (IC50) were calculated using Prism software version 5.0c.
Virus evolution.
Evolution experiments were essentially performed as described before (57, 58). A total of 5 × 106 SupT1 cells were transfected with 1, 10, or 40 μg pLAI (cultures A, B, and C/D, respectively), and virus spread was monitored by visual inspection for the appearance of syncytia and by CA-p24 ELISA as indicators of virus replication. Decreasing amounts of supernatant were passaged when the cells were (almost) wasted due to infection by the replicating virus. Viruses were cultured for ∼4½ months and passaged cell free onto uninfected cells when virus replication was apparent. Some cultures were terminated after ∼2½ months because no virus replication was ever observed. Some viruses were lost after cell-free passage. At regular intervals, cells and filtered supernatant were stored at −80°C for subsequent genotypic and phenotypic analysis and virus was quantitated by CA-p24 ELISA. When a putative faster-replicating virus was identified, DNA was extracted from infected cells using the QIAamp DNA mini kit (Qiagen, Valencia, CA), and the complete proviral env sequences were PCR amplified using primers 1 (5′-ATAAGCTTAGCAGAAGACAGTGGCAATG-3′) and 2 (5′-GCAAAATCCTTTCCAAGCCC-3′) and sequenced.
Pulse-chase folding assay.
Subconfluent HeLa cells were infected with recombinant vaccinia virus expressing T7 polymerase to drive the expression of Env mutants under the control of the T7 promoter (25). Thirty minutes postinfection, the cells were transfected with a mixture of 4 μg of mutant or wild-type (wt) pRS1-gp160 and 10 μl poly(ethyleneimine) (Polysciences, Inc., Warrington, PA). Pulse-chase experiments were performed essentially as described previously (8, 42). Five hours after infection, the cells were washed once with Hanks' balanced salt solution (Invitrogen, Carlsbad, CA) and preincubated in starvation medium lacking methionine and cysteine for 15 min at 37°C. The cells were pulse-labeled for 10 min with 50 μCi of Redivue promix [35S] labeling mix (Amersham Biosciences, Sweden) and chased for the indicated times (see Fig. 3) in medium containing an excess of unlabeled cysteine and methionine. The chase was stopped by transferring the cells to ice and washing them with ice-cold Hanks' balanced salt solution containing 20 mM iodoacetamide to alkylate free sulfhydryl groups. The cells were lysed in ice-cold 0.5% Triton X-100 in MNT (20 mM morpholineethanesulfonic acid [MES], 100 mM NaCl, 30 mM Tris-HCl [pH 7.5]) containing 20 mM iodoacetamide and protease inhibitor cocktail (10 μg/ml each of chymostatin, leupeptin, antipain, and pepstatin), 1 mM phenylmethylsulfonyl fluoride, and 1 mM EDTA. The cell lysates were spun for 10 min at 15,000 × g to pellet the nuclei, and postnuclear lysates were immunoprecipitated with a polyclonal antibody that recognizes all forms of HIV-1 Env (42). Cleaved and shed gp120 molecules were immunoprecipitated from culture media at later chase times. Immunoprecipitates were washed twice with a buffer containing 0.05% Triton X-100, 0.05% sodium dodecyl sulfate (SDS), and 300 mM NaCl in 10 mM Tris-HCl (pH 7.4). The washed pellets were resuspended in 0.2% SDS in 100 mM sodium acetate (pH 5.5) and heated to 95°C for 5 min. An equal volume of 100 mM sodium acetate (pH 5.5), which contained 2% Triton X-100 in MNT, protease inhibitor cocktail, and 0.0025 U endo-β-N-acetylglucosaminidase H (Roche, Switzerland), was then added. The samples were incubated for 2 h at 37°C. Concentrated Laemmli sample buffer then was added, and the samples were incubated at 95°C for 5 min. The samples were analyzed by reducing and nonreducing 7.5% SDS-polyacrylamide gel electrophoresis (PAGE). The gels were dried, and signals were detected on Biomax films (Kodak, Rochester, NY).
FIG. 3.
Oxidative folding of deletion variants. HeLa cells expressing wt gp160 or V1/V2 variants 1, 3, or 4 were pulse-labeled for 10 min and chased for the indicated times. Cells were lysed and Env proteins immunoprecipitated. Immunoprecipitates were deglycosylated and analyzed by reducing (A) and nonreducing (B) 7.5% SDS-PAGE. (C) Shed gp120 was immunoprecipitated from the culture media at later chase times. Folding intermediates (IT), the native form (NT), the reduced state with the signal peptide attached (Ru) or removed (Rc), and shed gp120 (*) are indicated.
SDS-PAGE, BN-PAGE, and Western blotting.
The NdeI and StuI fragments from amplified env genes were cloned into the pPPI4 vector for the expression of recombinant stabilized JR-FL gp140 constructs SOS gp140 and SOSIP gp140 (6, 62). Soluble gp140 was produced in transiently transfected 293T cells in the presence of coexpressed furin as previously described (6, 7, 62). The cell lysates were prepared in lysis buffer: 25 mM Tris-HCl (pH 7.8), 2 mM dithiothreitol, 2 mM trans-1,2-diaminocyclohexane-N,N,N′,N′,-tetraacetic acid (CDTA), 10% glycerol, 1% Triton X-100. The cell extracts containing maturing gp140 and the culture supernatants containing secreted gp140 were subjected to SDS-PAGE, blue native (BN)-PAGE, and Western blot analysis as described before (61, 62), using the JR-FL V3-specific mouse MAb PA-1 (a gift from William Olson, Progenics Pharmaceuticals).
RESULTS
Design of Env with loop deletions.
Env variants with variable loop deletions have been generated by us and others for functional, structural, and vaccine studies, but most of these constructs are slightly different and no comparative studies have been performed. It is a priori hard to predict which deletions are preferable in terms of Env folding and function. We have constructed a set of Env variants with different deletions in the V1/V2 and V3 regions in the context of the CXCR4-using LAI isolate, which is very convenient for evolution experiments. Some of these deletions were of novel design, while others were based on published studies to provide a comparison. Two-dimensional schematics and assumed three-dimensional (3-D) structures of some variants are provided in Fig. 1A to C.
Variants 1 and 2 are of novel design. In contrast to most previously described V1/V2 deletion variants in which the disulfide bonds (between C119 and C205 and/or C126 and C196) are maintained and the V1/V2 region replaced with a Gly-Ala-Gly linker, we replaced the respective cysteines with two adjoining alanines, thus creating a continuous protein backbone (Fig. 1A and B). Variants 3 and 4 were derived and recloned from our previous studies of disulfide-stabilized Env constructs with loop deletions (60). Note that these were derived from the JR-FL strain, and part of the flanking regions between the NdeI and StuI restriction sites that were used for subcloning were also derived from JR-FL (see Materials and Methods). We hypothesized that this sequence variation compared to the other constructs might result in different evolution routes. Variants 5 and 6 were designed to allow for the evolution of an alternative disulfide-bonded architecture of the V1/V2 stem. Variant 5 retained the cysteines at positions 126 and 131, while their counterparts, C157 and C196, were eliminated. We hypothesized that an alternative disulfide bond could be formed between positions 126 and 131 to rearrange the V1/V2 stump. Mutant 6, lacking C196, contained an uneven number of cysteines, which usually is disadvantageous, but we envisaged that it may facilitate new evolution routes by allowing for the addition or removal of a cysteine. In addition, mutant 6 contains an HXB2 V1 region that is 5 amino acids shorter than that of LAI. Some other amino acids in V1 also differ compared to wt LAI (Fig. 1A).
Variant 8 is a copy of the variant described by Wyatt et al. and was used to crystallize the gp120 core (Fig. 1B) (14, 38, 78). After prolonged culturing of this variant in a previous study by Cao et al., an evolved variant was identified with a change in the Gly-Ala-Gly linker region: Asp-Ala-Gly (10). We reproduced this mutant (variant 9). Building on the results of the study by Cao et al., we hypothesized that the Gly-Ala-Gly linker may not be optimal, and we constructed additional variants with changes in the linker region to test their relative functionality and allow for diverse evolution pathways to improve Env function. We generated variants with the following linker residues: variant 10, Gly-Asp-Gly; variant 11, Asp-Asn-Gly; and variant 12, Gly-Thr-Gly. Note that construct 11 contains an extra site for N-linked glycosylation within the linker region. Variant 14 was also taken from the Wyatt studies (78, 79).
Most mutants were constructed such that the bridging sheet between the inner and outer domains, which forms upon CD4 binding and is part of the coreceptor binding domain, remained intact (Fig. 1B). However, variants 1 and 14 lack β2 and β3, while variants 5 and 6 lack β3.
We also constructed three variants with deletions in the V3 loop (variants 15 to 17; Fig. 1C). Variant 15 lacks 7 amino acids in the N-terminal part of the V3 loop, while variant 16 lacks 10 amino acids at the C-terminal end. These deletions are combined in variant 17, in which the conserved tip sequences are retained.
Functional characterization of Env with loop deletions.
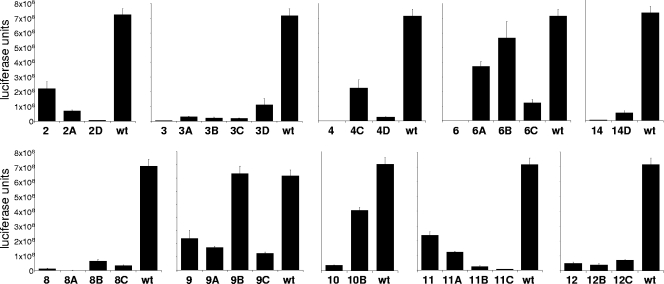
To assess the activity of the Env variants with loop deletions, we performed single-cycle infection assays with the respective mutants. The variant viruses were produced in SupT1 cells and used to infect TZM-bl reporter cells containing the luciferase gene under the control of the long-terminal-repeat promoter. V1/V2 mutants 1, 3 to 6, and 14 and V3 mutants 15 to 17 did not exhibit any activity in these functional assays (Fig. 2 and results not shown). In contrast, V1/V2 variants 2, 9, and 11 remained relatively efficient in infecting the reporter cells, although not as efficient as wt Env. Variants 8, 10, and 12 displayed a low but reproducible level of infection.
FIG. 2.
Functional analysis of the deletion variants. TZM-bl reporter cells (confluence of 70 to 80%) were infected with 1.0 ng of mutant or wt virus in the presence of saquinavir in a 96-well plate, and luciferase activity was measured after 48 h.
Interestingly, V1/V2 variants 8 to 12, which are very similar in design with only minor changes in the linker replacing V1/V2, showed considerably different activities in these infection experiments. Mutants 9 and 11 containing the linkers Asp-Ala-Gly and Asp-Asn-Gly, respectively, were more active than variants 8 (Gly-Ala-Gly), 10 (Gly-Asp-Gly), and 12 (Gly-Thr-Gly). The activity of variant 2 shows that the traditional deletion design—that is, the retention of a disulfide bond linked by a small flexible stretch of amino acids—is not necessary. The Cys126-Cys196 disulfide bond is replaced by two adjoining alanines, thus forming a continuous protein backbone. This Cys126-Cys196 disulfide bond is required for virus replication in the context of the wt virus (73) but apparently not anymore when the V1/V2 domain is deleted.
We also tested these viruses in spreading infections on SupT1 T cells and primary CD4+ T cells (see below). The replication experiments in SupT1 cells mirrored the single-cycle experiments with some qualitative differences. Most of the mutants were not able to replicate in primary CD4+ cells, but some replication was observed for mutants 2, 9, and 11.
Oxidative folding of Env with V1/V2 deleted.
The adverse effects of the V1/V2 deletions on recombinant trimers are ill-defined (11, 60). Since these effects do not become apparent in monomeric gp120, we do expect them to be rather subtle and act primarily on the trimeric complex. To exclude that the deletions impair early Env biosynthesis (i.e., oxidative folding in the endoplasmic reticulum), we examined the oxidative folding of Env mutants with V1/V2 deleted using a previously developed pulse-chase assay (42, 73). Env variants were expressed from plasmids under the control of the T7 promoter, using a recombinant vaccinia virus system (25). The cells were pulse-labeled for 10 min and chased for the indicated times (Fig. 3). The chase was stopped by cooling and the addition of alkylating agent to block free sulfhydryl groups. Lysates were immunoprecipitated and glycans were removed for analysis of the disulfide bond formation (42). The maturation of Env is slow: after 4 h, the majority of wt Env reaches the native state (22, 42, 50). Another striking feature of Env is the completely posttranslational cleavage of the signal peptide. A mutual relationship exists between Env oxidative folding and signal peptide removal: the lack of removal disturbs oxidative folding, whereas the formation of disulfide bonds is necessary to allow cleavage of the signal peptide (42, 43).
Signal peptide cleavage was followed over time by analyzing Env in lysates by reducing SDS-PAGE (Fig. 3A). Directly after the pulse, a single band appeared (Fig. 3A, lane 1, Ru), representing gp160 with its signal peptide still attached. Cleavage started after ∼30 min of chase (Fig. 3A, lane 2, Rc). After 4 h of chase, the majority of the gp160 had lost its signal peptide (Fig. 3A, lane 4). The decrease in signal after a 4-h chase was primarily caused by the cleavage of gp160 into gp120 and gp41 and the subsequent release of gp120 into the culture medium (Fig. 3C, lanes 1 to 2) (47, 67).
We subjected the V1/V2 variants to the same assay. Since all variants behaved roughly similarly in these pulse-chase assays, we only show the results obtained with variants 1, 3, and 4, which are representative for all variants. Variants 1, 3, and 4 showed similar kinetics in the cleavage of the signal peptide, notwithstanding the mobility differences caused by loop deletions (Fig. 3A). The ratio between uncleaved and cleaved Env after 30 min of chase is similar to that of the wt (Fig. 3A; compare lanes 2, 4, 10, and 14).
To monitor the oxidative folding more directly, we followed the disulfide bond formation by analyzing the samples with nonreducing SDS-PAGE. The formation of disulfide bonds during folding increases the compactness of gp160 and, as a consequence, its electrophoretic mobility (8, 42). Immediately after the pulse, wt gp160 appeared as a fuzzy band that represents gp160 with different sets of disulfide bonds representing folding intermediates (Fig. 3B, lane 1). With time, the folding intermediates gradually disappeared (Fig. 3B, lanes 2 to 4) in favor of a fast migrating sharp band, which represents native, folded gp160 containing all its disulfide bonds. The mutants showed a similar oxidation pattern, as after 30 min of chase, some molecules reached their native state (Fig. 3B, lanes 6, 10, and 14). Folding intermediates did appear somewhat more diffuse and heterogeneous, but the mobility differences between the mutants and the wt preclude any conclusions. The intensity of the native bands increased over the first hour and then disappeared from the lysate because of the cleavage of gp160 into gp120 and gp41 and the subsequent shedding of gp120 into the medium (Fig. 3C).
Major defects thus did not appear in the early biosynthesis of the Env variants with V1/V2 deletions, in contrast to the Env variants lacking disulfide bonds essential for oxidative folding (73). Even the more-drastic deletion variants, 1, 5, and 14, did not show significant defects in oxidative folding. We argue that the consequences of V1/V2 deletion only become apparent later, for example, during trimer assembly or maintenance of the trimeric spike.
Evolution of Env with V1/V2 deleted.
Regardless of the causes underlying the adverse effects of V1/V2 deletion, we examined whether the function of the Env constructs with loop deletions could be rescued. For this purpose, we used spontaneous virus evolution, which is likely to select for compensatory second-site changes. SupT1 cultures transfected with each of the molecular clones (three or four independent cultures per variant) were maintained for prolonged times (4½ months) as described in Materials and Methods and monitored for the appearance of faster-replicating variants by visual inspection for the appearance of syncytia and by CA-p24 ELISA. The initial CA-p24 production after the bulk transfection of SupT1 cells roughly mirrored the single-cycle infection experiments described above (data not shown). Viruses were passaged cell free onto uninfected cells when signs of active virus spread were apparent.
Cultures of the V1/V2 mutants 1 and 5 and all of the V3 mutants never produced replicating virus. These deletions, hence, are incompatible with residual Env function, which is needed to allow virus evolution. The V3 viruses were therefore excluded from further experiments. Virus spread was measured in 4/4 cultures of variant 3, 4/4 of variant 4 (of which two were lost during subsequent cell-free passage [see below]), 3/4 of variant 6, and 1/4 of variant 14, suggesting that although the deletions quite severely affected Env function, the virus was able to rescue this function by evolution.
In some cases, we lost replicating viruses during cell-free passage, indicating that although Env function was sufficient for cell-cell spread, some of these variants were defective in virus-cell infection (some cultures of variants 2, 4, 10, and 12). The virus variants differed in their ability to form syncytia and the morphology of syncytia (data not shown). These different properties, which also changed over the course of the evolution experiment, could not be correlated with CA-p24 production and replication kinetics, pointing at mechanistic differences in Env function.
Functional analysis of revertant Envs with V1/V2 deleted.
To confirm that the evolved viruses indeed gained replication capacity, the mutant and adapted viruses were directly compared in an infection experiment. SupT1 cells were infected with equal amounts of virus, and virus spread was monitored by CA-p24 ELISA (Fig. 4A). Mutant 2 replicated quite efficiently already, and the evolved variants 2A and 2D therefore did not readily display further improvements. Mutants 8 to 12 replicated efficiently as well, but evolution in some cases did improve replication (e.g., 10B, 12B, and 12C). In contrast, variants 8B and 11B lost replication capacity. Mutant 3 was a poorly replicating virus, and the viruses from three out of four cultures (3B to D but not 3A) clearly showed an improvement. Mutant 4 was worse than mutant 3, and both evolved variants were greatly improved, 4C being the best. Mutant 6 was as poor as mutant 4, but all three evolved variants (6A to C) displayed wt-like virus spread. Mutant 14 appeared completely replication defective, but a single evolved variant, 14B, did appear, although it replicated with a 4-day delay compared to that of the wt.
FIG. 4.
Replication of mutant and adapted viruses. (A) 400 × 103 SupT1 cells were infected with 100 pg virus, and replication was monitored for 18 days by CA-p24 ELISA. (B) 200 × 103 primary CD4+ T cells were infected with 500 pg virus, and replication was monitored. The results are representative of three independent experiments using cells from different donors for each experiment.
We also tested the V1/V2 mutants and evolved variants for replication in primary blood cells (Fig. 4B). Most mutants were unable to replicate in primary CD4+ cells, but variants 2, 9, and 11 displayed a low level of virus spread. We observed improved replication for some of the evolved variants (variants 6B, 9B, 10B, and 12C), although virus spread was still very poor compared to that of the wt virus. Some variants lost the capacity to replicate in primary T cells (2A, 2D, and 11C), which may indicate specific adaptation to the SupT1 T-cell line with which the evolution experiment was performed. To establish that the evolved viruses had improved their Env function, we also performed single-cycle infection assays with TZM-bl cells (Fig. 5). Most variants showed improved infectivity in TZM-bl cells upon evolution (3A to D, 4C, 4D, 6A to C, 8B, 8C, 9B, 10B, and 14D), but some variants became less infectious (2A, 2D, 8A, 9A, 9C, and 11A to C), pointing at SupT1-specific adaptations. In summary, using virus evolution, we obtained several fully replication-competent Env variants with large V1/V2 deletions.
FIG. 5.
Functional analysis of adapted Envs. TZM-bl reporter cells were infected with 1.0 ng of mutant or wt virus in the presence of saquanavir, and luciferase activity was measured after 48 h.
Genotypic analysis of revertant Envs with V1/V2 deleted.
We next investigated the unlikely scenario that the evolved virus variants had reintroduced sequences at the location of the V1/V2 deletion. The V1/V2 domain of provirus in revertant cultures after 4½ months of evolution was PCR amplified and subjected to gel electrophoresis, and it showed no change in size compared to that of the input mutant viruses (Fig. 6). Apparently, V1/V2 sequences cannot be easily rebuilt.
FIG. 6.
No restoration of V1/V2 sequences. The V1/V2 domain and surrounding sequences from proviral DNA in evolution cultures were PCR amplified and analyzed by gel electrophoresis. The lengths of the amplified fragments are 460 bp (wt), 253 bp (variant 2), 304 bp (variant 3), 265 bp (variant 4), 364 bp (variant 6), 268 bp (variants 8 to 12), and 220 bp (variant 14).
To identify determinants that are responsible for the improvement of Env function, the complete env genes were PCR amplified and sequenced. The resulting sequences represent the predominant variants in the viral quasispecies. The identified substitutions are given in Table 1. All of the cultures contained changes to the input mutant sequence except for culture 14B, suggesting that mutations elsewhere in the genome underlie the improved replication capacity (for example, elevated transcription or alternative RNA splicing, resulting in more Env mRNA [54]). Several interesting observations can be made based on the sequence analysis (see Discussion). A selected set of mutant and adapted Env variants were pursued in follow-up experiments.
TABLE 1.
Observed amino acid changes in evolved ΔV1/V2 virus variantsa
Amino acid changes that occur in independent cultures containing the same original virus are indicated in red. Substitutions that occur in multiple cultures containing different original viruses are represented in blue. Identical changes that occur in different original viruses in the 8 to 12 cluster are indicated in green. We used the same color code for different variants that result in the loss of the same glycosylation site (N156K and S158F, T303A/K, N295Y and T297I, and N234D and T236I). Mutations that result in the elimination of glycosylation sites are underlined, and mutations that result in the acquisition of a glycosylation site are in italics. Mixed sequences and silent mutations were excluded. The dark gray shading indicates cultures in which we did not find adapted viruses. The medium gray shading indicates cultures in which replicating virus was observed at some point during the 4½ months of culturing but which were lost during cell-free passage.
The D197N mutation was observed in early sequences (data not shown), but the virus was subsequently lost during cell-free passage.
Neutralization sensitivity of variants with V1/V2 deleted.
To assess the neutralization sensitivity of mutants and revertants with V1/V2 deleted, we performed neutralization assays with a set of MAbs to gp120 and gp41. We chose to perform these experiments with the wt and the 2, 4C, 6B, 9, 9B, 10B, 11, and 11A variants because these viruses efficiently infected TZM-bl cells used in the neutralization experiments and because most of these viruses also infected primary CD4+ T cells. We included the mutant-revertant pairs 9 to 9B and 11 to 11A to investigate whether the neutralization sensitivity changed upon evolution. The inhibition curves are shown in Fig. 7, and the respective IC50 are given in Table 2.
FIG. 7.
Neutralization sensitivity of mutant and adapted viruses. TZM-bl cells were infected with 1.0 ng virus as described in the legend to Fig. 2 and in Materials and Methods. Virus was preincubated with the indicated amount of MAb for 30 min at room temperature prior to infection of the reporter cells. The luciferase activity in the absence of antibody was set at 100%.
TABLE 2.
Neutralization sensitivity of the ΔV1/V2 variantsa
| Variant | IC50 (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| gp41 (4E10) | Glycan (2G12) | CD4BS
|
CD4i (17b) | |||
| CD4 | b12 | b6 | ||||
| wt | >10 | 6.1 | 0.58 | 0.77 | >10 | >30 |
| 2 | 2.8 | 0.12 | 0.0090 | 0.018 | 0.39 | 1.7 |
| 9 | 2.7 | 0.13 | 0.0062 | 0.010 | 0.072 | 0.39 |
| 11 | 2.1 | 0.11 | 0.0055 | 0.017 | 0.14 | 0.24 |
| 4C | 1.3 | 0.080 | 0.0070 | 0.0066 | 0.13 | 3.5 |
| 6B | 2.6 | 0.22 | 0.034 | 0.37 | 2.13 | >30 |
| 9B | 1.9 | 0.081 | 0.16 | 0.013 | 0.16 | >30 |
| 10B | 1.2 | >10 | 0.0029 | NDb | 0.054 | 25.8 |
| 11A | 1.7 | 0.16 | 0.015 | 0.0077 | 0.083 | >30 |
IC50 were derived from the experiment for which the results were depicted in Fig. 7 as described in Materials and Methods. CD4-IgG2 was used as a surrogate for CD4.
ND, not determined.
We first tested the neutralization sensitivity to the 4E10 MAb directed against the membrane proximal region in gp41. We hypothesized that the exposure of the 4E10 epitope would not be affected significantly by the truncation of the V1/V2 domain. However, we found that all variants were more sensitive than the wt virus (IC50 ranging from 1.2 to 2.8 μg/ml versus ∼10 μg/ml for the wt). The deletion of the V1/V2 domain in gp120 apparently increased the accessibility of the membrane proximal domain in gp41. Mutant viruses and evolved variants did not show major differences (compare 9 to 9B and 11 to 11A).
The epitope for the glycan-dependent 2G12 MAb is located on the outer face of gp120 (61, 63, 72). Most viruses exhibited increased sensitivity to 2G12 (IC50 ranging from 0.080 to 0.22 μg/ml versus 6.1 μg/ml for the wt), suggesting that the exposure of the epitope is increased upon the deletion of the V1/V2 domain. As for 4E10, no major differences were apparent for the two mutant-revertant pairs. Revertant 10B was resistant to 2G12 neutralization, consistent with the loss of the 295 glycan which is part of the 2G12 epitope as a result of the T297I substitution (Table 1) (61, 63, 72). Resistance was not complete, since even low 2G12 concentrations resulted in an infection inhibition of 25%. Possibly a 2G12-sensitive subpopulation was present within the viral quasispecies that had not acquired the T297I substitution yet.
Mutants 2, 9, and 11 were highly sensitive to neutralization by CD4-IgG2 used as a surrogate for CD4 (IC50 of 0.0055 to 0.0090 μg/ml) compared to the wt virus (IC50 of 0.58 μg/ml), confirming that the V1/V2 domain is involved in limiting the accessibility of CD4BS. Of the adapted viruses, the 6B variant, which contains most of the V1 sequences, was more resistant to CD4 than the variants with V1/V2 deleted (IC50 of 0.034 μg/ml) but more sensitive than the wt virus. These results suggest that on the native trimer, both V1 and V2 are involved in the shielding of CD4BS. The evolved 9B and 11A variants (IC50 of 0.16 and 0.015 μg/ml, respectively) were more resistant than the original mutants 9 and 11. Their affinities for CD4 may be lower, caused by the substitutions located in or near CD4BS (G458D, F382L, and M434T). Residue 458 is a contact residue for CD4 through main chain interactions (38, 83), and a G458A substitution indeed causes a dramatic decrease in CD4 binding (51).
We next tested the sensitivity to the broadly neutralizing antibody b12 directed to CD4BS (9). All mutants were highly sensitive to b12 neutralization (IC50 of 0.010 to 0.018 μg/ml for the mutants and 0.77 μg/ml for the wt), but interestingly, complete neutralization was not achieved even at high b12 concentrations (∼35% residual infectivity). We have repeatedly observed this phenomenon but do not have an adequate explanation. Most adapted viruses were also highly sensitive to b12 (IC50 of 0.0066 to 0.013 μg/ml) but could be inhibited completely. An exception was the 6B variant, which was similarly sensitive to b12 as to the wt (IC50 of 0.37 μg/ml).
MAb b6 is derived from the same phage library as b12 and also targets CD4BS (56), but it does not neutralize the wt virus, presumably because its angle of binding to gp120 is incompatible with binding to the Env trimer (51). Indeed, wt LAI was resistant to b6 neutralization. In contrast, we found that the mutant viruses with V1/V2 deleted were sensitive to b6 neutralization, although b6 was less potent than b12 against these viruses (IC50 of 0.072 to 0.39 μg/ml). The adapted viruses were equally sensitive to b6 neutralization (IC50 of 0.054 to 0.14 μg/ml), except 6B, which was quite resistant (IC50, 2.13 μg/ml), although not as resistant as the wt virus, suggesting that both V1 and V2 play a role in covering the b6 epitope on trimeric Env.
The 17b MAb is directed to an epitope overlapping the coreceptor binding site. The creation and exposure of the epitope are induced by CD4 binding. The neutralizing potency of 17b is therefore limited (41). While the wt virus was resistant to 17b neutralization, the mutant viruses were sensitive to 17b (IC50 of 0.24 to 1.7 μg/ml), indicating that V1/V2 deletion increases the exposure of the epitope (78). Consistent with this, variant 6B, containing most of V1, was resistant to 17b neutralization. The evolved variants 9B, 10B, and 11A were also resistant to 17b neutralization, most likely because the acquisition of the V120E, P417T, and M434T changes located in β2, β19, and β21 of the four-stranded bridging sheet damaged the 17b epitope (38, 55).
Improvement of the synthesis and secretion of soluble stabilized Env.
Our goal is to use substitutions selected during evolution experiments to improve the folding and secretion of soluble stabilized Env immunogens. To provide a proof of principle that this is possible, we cloned selected deletion variants into an expression vector for SOS gp140 based on the JR-FL isolate (6). The PCR fragments from different time points during the evolution of the ΔV1/V2.6 variant (Fig. 6) were cloned into an SOS gp140 vector and sequenced. Most clones contained the mutations identified in the population sequence at the respective time points (indicated in the labels of Fig. 8), but one clone of culture 6A contained an additional substitution (M104I) (Fig. 8). The resulting SOS gp140 constructs were transiently expressed in 293T cells in the presence of abundant furin (6). The cell lysates and supernatants were subjected to SDS-PAGE and Western blotting to analyze the intracellular and secreted Env (Fig. 8). Full-length SOS gp140 was expressed efficiently as reported previously (6), but the expression of the SOS ΔV1/V2.6 variant was reduced. In contrast, all four evolved variants were efficiently expressed and secreted in the culture supernatant, with some quantitative differences. These results indicate that compensatory changes were selected in the virus evolution experiments that improved the biosynthesis and secretion of stabilized gp140.
FIG. 8.
Substitutions improve the folding and secretion of stabilized gp140 constructs. The ΔV1/V2.6 deletion and compensatory changes identified in various evolution cultures were introduced in an expression vector for SOS gp140. The variants were expressed in 293T cells, and intracellular and secreted Env was analyzed by SDS-PAGE, BN-PAGE, and Western blotting. d, day.
Since we wish to employ evolved variants in recombinant trimer constructs, we also analyzed the oligomerization of the SOS gp140 constructs using native electrophoresis (BN-PAGE; Fig. 8, bottom panel). The wt gp140 forms a mixture of predominantly dimers and trimers, as does SOS gp140, although the oligomerization of SOS gp140 varies considerably between experiments and can be modulated by additional stabilization (62). The evolved SOS ΔV1/V2.6 variants showed a similar pattern, indicating that the deletion and compensatory substitutions are compatible with efficient oligomerization.
DISCUSSION
The deletion of variable loops from recombinant HIV-1 Env trimer constructs for vaccine and structural studies has proven difficult. We decided to pursue a practical virological approach to address this problem. By means of spontaneous virus evolution, we aimed to select for variants with improved Env biosynthesis, trimer formation, and function. We focused on the combined deletion of the V1 and V2 loops for several reasons. Of the five variable loops, V4 and V5 locate to the outer domain of gp120, relatively distant from the receptor binding sites, the most obvious targets for neutralizing antibodies on gp120. The deletion of V4 causes major biosynthesis defects (53). We did attempt to delete V3, but V3 participates in coreceptor binding (28), and its deletion can cause biosynthesis defects (15, 60, 70). As a consequence, we did not obtain replicating virus from V3 deletion variants, and recent studies confirmed that only relatively subtle changes in V3 are allowed (40, 49).
In contrast to V3, the V1/V2 domain does not appear to be critical for function. It represents the largest chunk of variable gp120 and a large number of carbohydrates are anchored in the V1/V2 domain (seven sites in LAI). We considered the construction of V1 or V2 single-deletion variants, but this may increase the exposure and antigenicity of the reciprocal loop. Indeed, the deletion of V2 has been shown to induce V1-specific antibodies (19, 65). We hence focused on Env constructs in which both V1 and V2 were deleted.
Surprisingly, none of the viruses with V1/V2 deleted replicated very efficiently in primary CD4+ T cells. One explanation may be the absence of the recognition motif for integrin α4β7 located within the V2 loop, which facilitates virus spread by the upregulation of adhesion molecules such as LFA-1 (2). Alternatively, V1/V2 deletions may alter interactions with CD4 and/or CXCR4. This may not be apparent in SupT1 cells that express relatively high levels of CD4 and CXCR4 (33; R. E. Jeeninga, personal communication) but may become an obstacle for replication in primary cells where receptor levels are lower. This speculation may be supported by the fact that the V1/V2 domain modulates coreceptor usage (48) and by the hypothesis that V1/V2 deletion can reduce the affinity for CCR5 in a CCR5-using virus strain (40). Thus, the V1/V2 domain may harbor determinants that are involved in the interactions with cellular receptors, and this may account for the poor replication of virus with V1/V2 deleted in primary cells.
We have provided evidence that compensatory changes can improve the expression, folding, and secretion of stabilized gp140 constructs with V1/V2 deleted. Whether gp140 variants based on this design will be better immunogens remains to be established and is the topic of ongoing studies. A concern with the modification of Env for vaccine purposes is the creation of neoepitopes that elicit antibodies that do not recognize wt Env on virus and hence are nonneutralizing. This is particularly relevant for major modifications such as the ∼40- to 80-amino-acid deletions described in this study. Env constructs with V1/V2 deleted may elicit increased numbers of CD4i antibodies and nonneutralizing CD4BS antibodies. One hint that this may occur can be found in the neutralization studies with the antibodies 17b (CD4i) and b6 (nonneutralizing CD4BS), which generally neutralized the viruses with V1/V2 deleted more efficiently than the wt virus. V1/V2 deletion may also enhance the elicitation of antibodies directed against V3. Thus, we need to identify constructs with the optimal exposure and presentation of neutralizing epitopes, without the efficient expression of neoepitopes and/or nonneutralizing epitopes. When immunizing with Env variants with loop deletions, the viruses generated in this study can be used as tools to monitor neutralization mediated by neoepitopes.
Interestingly, Env with V1/V2 deleted appears to be highly sensitive to 2G12 neutralization. One could imagine that the deletion of the entire V1/V2 domain could affect the accessibility and modification of the carbohydrates that form the 2G12 epitope such that they would become complex sugars, resulting in resistance to 2G12. This is not what we observed. On the contrary, we observed an increased sensitivity to 2G12, implying that the 2G12 oligomannose chains on Env with V1/V2 deleted are not more accessible to the modifying enzymes in the Golgi, while they are more accessible to 2G12.
Env with V1/V2 deleted was neutralized efficiently by b6. It is thought that b6 can only bind monomeric gp120, not trimeric Env, and hence is nonneutralizing. However, since b6 can neutralize Env with V1/V2 deleted, it is able to bind Env trimers with V1/V2 deleted. Two possible explanations come to mind. First, the V1/V2 domain on trimeric Env is oriented differently than that on monomeric gp120. Alternatively, the V1/V2 domain from one subunit in the trimer shields the b6 epitope on a neighboring subunit.
Molecular determinants for functional Env with V1/V2 deleted.
The replication and evolution capacities of the V1/V2 deletion variants teach us about the minimal sequence requirements for Env function. Deletion variants 1 and 5 never gained replication competence. Essential sequences must be missing in these viruses. Variant 1 lacks β-strands 2 and 3, which, combined with β20 and β21, form the bridging sheet upon CD4 binding (Fig. 1B). Residues of strands β2 and β3 are involved in the interaction with CD4 and the coreceptor, and the removal of these two strands may cause replication defects. Surprisingly, mutant 6, which lacks β3, was defective but could regain replication capacity, suggesting that a triple-stranded bridging sheet is sufficient for Env function. Mutant 5 lacks β3 but also glycan 197 (see below) and two conserved cysteines (157 and 196), leaving their counterparts (126 and 131) either unpaired or involved in a nonnative disulfide bond with each other or with other cysteines. The combination of these problems caused defects that were beyond repair.
Among the mutant viruses, variant 2 represented the best-replicating virus (Fig. 2). The minimal sequences of the bridging sheet (β2 and β3) are present in this mutant. The fact that this mutant is functionally superior to the other variants indicates that the traditional design of loop deletions (maintaining the cysteine pair and replacing the deleted sequences by a Gly-Ala-Gly linker) may not be optimal. The design of mutant 8 was similar to a previously described construct, which was crystallized successfully (14, 38, 78). Building on the previous observation of a change in the Gly-Ala-Gly linker region after prolonged culturing of this variant (10), we reconstructed this evolved variant (mutant 9 [Asp-Ala-Gly]), which indeed was more infectious than mutant 8. Variants 9 and 11 were more infectious than variants 8, 10, and 12, suggesting that especially the first glycine in the linker is not tolerated very well in Env with V1/V2 deleted. Interestingly, in prolonged cultures of variant 4 beyond the 4½ months described in this study, we also found a substitution of the first glycine (to serine) resulting in a Ser-Ala-Gly linker (data not shown), further strengthening the notion that the standard Gly-Ala-Gly linker is suboptimal.
Several interesting phenomena can be distilled from the evolution paths.
(i) Loss of N-linked glycosylation sites.
N-linked carbohydrates were lost frequently, perhaps because they are important only in the presence of the V1/V2 domain or in vivo for protection from the humoral immune response and hence may be lost in vitro upon culturing. On the other hand, this loss of glycans likely relates to their importance for protein folding. Glycans recruit the calnexin/calreticulin chaperone machinery in the endoplasmic reticulum, and loss of a glycan may change the assistance this protein domain receives during folding (29). Carbohydrates also affect protein conformation directly by adding bulk during synthesis. Especially in positions close to cysteines, glycosylation and disulfide bond formation are known to compete, as disulfide bond formation may preclude the use of a glycosylation site, and glycosylation may sterically hinder two cysteines from reaching each other for disulfide bond formation (1, 30). This implies that glycosylation and disulfide bond formation mutually influence each other.
(ii) Changes in or around the V3 domain.
Many of the lost glycosylation sites resided in V3, although the primary defect was a V1/V2 deletion. For mutant 3, three of four cultures showed loss of the glycosylation site at position 301 (3A, 3B, and 3D). The V3 base glycan (295) was eliminated in variants 10B and 11C, and glycosylation site 339 at the C-terminal side of V3 was eliminated in culture 8C. Other changes in or around V3 included S306N (11C), A329V (9A), R335K (3B), and A340D (found twice [10A and 10B]). It is unclear if and why these are compensatory changes, but they may underline the intimate interactions between the V1/V2 region and the V3 region (48).
(iii) Rearrangement of the V1/V2 stem in mutant 6.
Mutant 6 showed a similar replication/evolution pattern as variant 4. It was replication incompetent but quickly reverted to regain replication capacity. The mutation responsible for this initial restoration was the C131Y change, based on early sequences (data not shown; Fig. 1D). The elimination of a cysteine may not come as a surprise since variant 6 harbors an odd number of cysteines, but in contrast to the expected removal of C126, which would leave the native 131-157 disulfide bond intact, C131 was eliminated. We speculate that this may result in the formation of a nonnative alternative 126-157 disulfide bond. The alternative evolution route to add a cysteine did not occur in our cultures. Perhaps not a coincidence, the formation of a 126-157 disulfide bond would result in a V1 loop closer to the core but with exactly the number of residues of the LAI V1 loop, because it now includes residues 127 to 131 (Fig. 1D). The remaining V2 sequences then form a rudimentary V2 loop. Of note is that variant 6 misses β3, the outer strand of the four-stranded bridging sheet. Possibly β2 can still form a triple-stranded sheet with β20 and β21. The spacing between the native 119-205 and the nonnative 126-157 disulfide bonds may be important for the proper formation of the bridging sheet.
The changes in the disulfide-bonded architecture of the V1/V2 stem in variant 6 are followed by changes in glycosylation: the removal of the glycan attached to N156, either through the N156K or the S158F change (Fig. 1D). Interestingly, Wu et al. (77) suggested that the glycan at position 156, which is the only absolutely conserved glycosylation site in the V1/V2 domain, is important for the correct pairing of C157 with C131 and that the lack of the 156 glycan results in aberrant disulfide bond formation and misfolding. The loss of glycan 156 in our variant 6 may thus be advantageous once C131 is eliminated to facilitate the formation of a nonnative disulfide bond between C157 and C126.
(iv) Introduction of hydrophilic residues at the V1/V2 stump and bridging sheet.
Hydrophilic and often charged residues were introduced at the V1/V2 stump and/or in the bridging sheet (V120E, V127D, F423S, I424K, E429K, and M434T). Moreover, the D197N substitution results in the addition of a polar carbohydrate. The deletion of the V1/V2 domain likely exposes hydrophobic domains, which can trigger protein misfolding and/or protein aggregation (17, 46), and these amino acid replacements may reduce or shield the hydrophobic surface.
To investigate this in more detail, we analyzed the gp120 surface of variants 8, 9, and 9B. These variants were chosen because variant 9 contains a G128D substitution in the Gly-Ala-Gly linker replacing V1/V2, while variant 9B incorporated two additional changes from hydrophobic to hydrophilic/charged, V120E and G458D (Fig. 9A), and these variants showed a progressive improvement of Env function (Fig. 5 and 9B). Analysis of the surface associated with hydrophobic residues (Fig. 9C) revealed a large relatively hydrophobic patch at the base of the V1/V2 stem and the bridging sheet (Fig. 9C, top panels) and a highly hydrophobic backside (Fig. 9C, bottom panels). All three substitutions contributed to a decrease in the hydrophobic surface. We next analyzed the electrostatic potential of the gp120 surface (Fig. 9D). Within the predominantly basic surface facing the target cell (39) and in agreement with its hydrophobic character, the V1/V2 stem stands out as an electrostatically neutral surface. The three substitutions caused a progressive and dramatic change in the negatively charged electrostatic surface potential (visualized by the increase in red surface in Fig. 9D). The negative charge per se is not crucial because basic residues were introduced as well (e.g., I424K and E429K), and the prolonged culturing of mutant 9 beyond the 4½ months yielded a V120K substitution (data not shown). The improvement of Env variants with V1/V2 deleted by increasing the hydrophilic surface resembles strategies that are used in biotechnology to enhance the stability and solubility of catalytic enzymes and therapeutic proteins (for an example, see reference 24).
FIG. 9.
Surface analysis of variants 8, 9, and 9B. (A) Locations of the mutations and substitutions (residues 120, 128, and 458) on the 3-D structure of gp120 (same view as in Fig. 1B, top left panel). (B) Relative Env function of variants 8, 9, and 9B compared to that of the wt (based on the data depicted in Fig. 5). (C) Analysis of surface hydrophobicity. The top panels present a view similar to that shown in Fig. 1B (top right panel). The lower panels show gp120 rotated over the z axis by 180°. These perspectives provide good views on both sides of the V1/V2 stump. The surface associated with nonpolar residues is indicated in gray, and the surface associated with polar residues is in cyan. Mutations and substitutions are indicated. The LAI gp120 core and variant cores were modeled by SWISS-MODEL and drawn using Viewerlite. (D) Analysis of electrostatic surface potential. Electrostatic surface potentials were calculated and rendered using Deepview (red, acidic; blue, basic). The mutations and substitutions that are responsible for the changes in the electrostatic surface potential are indicated. Note that we underestimated the polar surface of gp120, since we only considered the surface associated with protein, not with carbohydrate.
(v) Changes in or around the CD4BS.
Of the many changes we found close to or in the CD4BS (N234D, V254I, V255G, A340D, S375N, G458D, and D474G), the latter three form direct contacts with CD4, and several of the residues have been shown to affect CD4 binding (38, 51). Several of the revertants displayed a decreased sensitivity to CD4 compared to the original mutants, suggesting that an extremely high affinity for CD4 may not be beneficial for Env function. This may, for example, interfere with conformational changes following coreceptor binding that are needed for fusion.
(vi) Changes in gp41.
A number of changes emerged in gp41 (four in the ectodomain [L544S, A607T, L619R, and N625S] and three in the intracellular tail [V701I, A792V, and R841H]). This finding and the increased neutralization by 4E10 underscore the influence of the V1/V2 deletion on gp41. Indeed, Johnson et al. studied an evolved simian immunodeficiency virus mac239 with V1/V2 deleted and found that the crucial compensatory changes were located in the gp41 ectodomain (35).
Recommendations for the design of Env with V1/V2 deleted.
Our results provide a number of recommendations for the deletion of the V1/V2 domain for vaccine and/or structural studies. (i) The frequently used replacement of V1/V2 sequences by Gly-Ala-Gly, leaving a few residue disulfide bonded loops, is not optimal. The replacement of the first glycine with, for example, aspartic acid already provides a major improvement. (ii) Retention of the 126 to 196 disulfide bond is not necessary: it may be replaced by an Ala-Ala linker as in mutant 2. (iii) It is desirable to have an N-linked glycosylation site at position 197. Although most virus isolates in fact contain a glycosylation site at this position, some, including the often-used JR-FL isolate, do not. If such isolates are used, the introduction of this site may be beneficial. The carbohydrate may mask the hydrophobic surface or may otherwise assist protein folding. (iv) The hydrophobic surface at the V1/V2 stem and base should be reduced by substitutions in the V1/V2 stem and bridging sheet. For example, the valines at positions 120 and 127 can be replaced by glutamic or aspartic acids. (v) Several distal changes may be helpful, but their exact contribution needs to be investigated in more detail. Needless to say, these recommendations will have different value with different Env backgrounds.
In conclusion, we have performed an extensive set of evolution experiments with Env variants with V1/V2 deleted. These studies provide a proof of principle that compensatory changes identified in forced evolution experiments can be useful for the production of stabilized Env trimer constructs for vaccine and structural studies.
Acknowledgments
R.W.S. is a recipient of an amFAR Mathilde Krim fellowship, an Anton Meelmeijer fellowship, and a VENI fellowship from The Netherlands Organization for Scientific Research (NWO)—Chemical Sciences. B.B. is supported by an NWO-VICI grant and by the Dutch AIDS fund (grant #2005021), and I.B. and A.L. are supported by an ECHO grant from NWO—Chemical Sciences.
We acknowledge William Olson, James Robinson, James Binley, and Dennis Burton for the kind gift of reagents. We thank Stephan Heynen for technical assistance.
Footnotes
Published ahead of print on 15 October 2008.
REFERENCES
- 1.Allen, S., H. Y. Naim, and N. J. Bulleid. 1995. Intracellular folding of tissue-type plasminogen activator. Effects of disulfide bond formation on N-linked glycosylation and secretion. J. Biol. Chem. 2704797-4804. [DOI] [PubMed] [Google Scholar]
- 2.Arthos, J., C. Cicala, E. Martinelli, K. Macleod, D. Van Ryk, D. Wei, Z. Xiao, T. D. Veenstra, T. P. Conrad, R. A. Lempicki, S. McLaughlin, M. Pascuccio, R. Gopaul, J. McNally, C. C. Cruz, N. Censoplano, E. Chung, K. N. Reitano, S. Kottilil, D. J. Goode, and A. S. Fauci. 2008. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat. Immunol. 9301-309. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 755526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beddows, S., M. Franti, A. K. Dey, M. Kirschner, S. P. Iyer, D. C. Fisch, T. Ketas, E. Yuste, R. C. Desrosiers, P. J. Klasse, P. J. Maddon, W. C. Olson, and J. P. Moore. 2006. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 360329-340. [DOI] [PubMed] [Google Scholar]
- 5.Beddows, S., N. Schulke, M. Kirschner, K. Barnes, M. Franti, E. Michael, T. Ketas, R. W. Sanders, P. J. Maddon, W. C. Olson, and J. P. Moore. 2005. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 798812-8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant HIV-1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binley, J. M., R. W. Sanders, A. Master, C. S. Cayanan, C. L. Wiley, L. Schiffner, B. Travis, S. Kuhmann, D. R. Burton, S.-L. Hu, W. C. Olson, and J. P. Moore. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 762606-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braakman, I., H. Hoover-Litty, K. R. Wagner, and A. Helenius. 1991. Folding of influenza hemagglutinin in the endoplasmic reticulum. J. Cell Biol. 114401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, and P. L. Nara. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 2661024-1027. [DOI] [PubMed] [Google Scholar]
- 10.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 719808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Center, R. J., J. Lebowitz, R. D. Leapman, and B. Moss. 2004. Promoting trimerization of soluble human immunodeficiency virus type 1 (HIV-1) Env through the use of HIV-1/simian immunodeficiency virus chimeras. J. Virol. 782265-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Center, R. J., P. Schuck, R. D. Leapman, L. O. Arthur, P. L. Earl, B. Moss, and J. Lebowitz. 2001. Oligomeric structure of virion-associated and soluble forms of the simian immunodeficiency virus envelope protein in the prefusion activated conformation. Proc. Natl. Acad. Sci. USA 9814877-14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89263-273. [DOI] [PubMed] [Google Scholar]
- 14.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433834-841. [DOI] [PubMed] [Google Scholar]
- 15.Chiou, S. H., E. O. Freed, A. T. Panganiban, and W. R. Kenealy. 1992. Studies on the role of the V3 loop in human immunodeficiency virus type 1 envelope glycoprotein function. AIDS Res. Hum. Retrovir. 81611-1618. [DOI] [PubMed] [Google Scholar]
- 16.Cladera, J., I. Martin, J. M. Ruysschaert, and P. O'Shea. 1999. Characterization of the sequence of interactions of the fusion domain of the simian immunodeficiency virus with membranes. Role of the membrane dipole potential. J. Biol. Chem. 27429951-29959. [DOI] [PubMed] [Google Scholar]
- 17.Cromwell, M. E., E. Hilario, and F. Jacobson. 2006. Protein aggregation and bioprocessing. AAPS J. 8E572-E579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das, A. T., B. Klaver, and B. Berkhout. 1999. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 7381-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derby, N. R., Z. Kraft, E. Kan, E. T. Crooks, S. W. Barnett, I. K. Srivastava, J. M. Binley, and L. Stamatatos. 2006. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J. Virol. 808745-8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 748358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earl, P. L., C. C. Broder, D. Long, S. A. Lee, J. Peterson, S. Chakrabarti, R. W. Doms, and B. Moss. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 683015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earl, P. L., B. Moss, and R. W. Doms. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 652047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eijsink, V. G., A. Bjork, S. Gaseidnes, R. Sirevag, B. Synstad, B. van den Burg, and G. Vriend. 2004. Rational engineering of enzyme stability. J. Biotechnol. 113105-120. [DOI] [PubMed] [Google Scholar]
- 25.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 182714-2723. [DOI] [PubMed] [Google Scholar]
- 27.Gzyl, J., E. Bolesta, A. Wierzbicki, D. Kmieciak, T. Naito, M. Honda, K. Komuro, Y. Kaneko, and D. Kozbor. 2004. Effect of partial and complete variable loop deletions of the human immunodeficiency virus type 1 envelope glycoprotein on the breadth of gp160-specific immune responses. Virology 318493-506. [DOI] [PubMed] [Google Scholar]
- 28.Hartley, O., P. J. Klasse, Q. J. Sattentau, and J. P. Moore. 2005. V3: HIV's switch-hitter. AIDS Res. Hum. Retrovir. 21171-189. [DOI] [PubMed] [Google Scholar]
- 29.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 2912364-2369. [DOI] [PubMed] [Google Scholar]
- 30.Holst, B., A. W. Bruun, M. C. Kielland-Brandt, and J. R. Winther. 1996. Competition between folding and glycosylation in the endoplasmic reticulum. EMBO J. 153538-3546. [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs, A., H. Garg, M. Viard, Y. Raviv, A. Puri, and R. Blumenthal. 2008. HIV-1 envelope glycoprotein-mediated fusion and pathogenesis: implications for therapy and vaccine development. Vaccine 263026-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of HIV-1 subtypes A through G. J. Virol. 743740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeeninga, R. E., B. Jan, B. Van der Linden, H. Van den Berg, and B. Berkhout. 2005. Construction of a minimal HIV-1 variant that selectively replicates in leukemic derived T-cell lines: towards a new virotherapy approach. Cancer Res. 653347-3355. [DOI] [PubMed] [Google Scholar]
- 34.Jeffs, S. A., C. Shotton, P. Balfe, and J. A. McKeating. 2002. Truncated gp120 envelope glycoprotein of human immunodeficiency virus 1 elicits a broadly reactive neutralizing immune response. J. Gen. Virol. 832723-2732. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 762075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, P. L., T. Korte, and R. Blumenthal. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273404-409. [DOI] [PubMed] [Google Scholar]
- 37.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 81329-1339. [DOI] [PubMed] [Google Scholar]
- 38.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in a complex with the CD4 receptor and a neutralizing human antibody. Nature 393648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 741961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laakso, M. M., F. H. Lee, B. Haggarty, C. Agrawal, K. M. Nolan, M. Biscone, J. Romano, A. P. Jordan, G. J. Leslie, E. G. Meissner, L. Su, J. A. Hoxie, and R. W. Doms. 2007. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS. Pathog. 3e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 7710557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Land, A., D. Zonneveld, and I. Braakman. 2003. Folding of HIV-1 envelope glycoprotein involves extensive isomerization of disulfide bonds and conformation-dependent leader peptide cleavage. FASEB J. 171058-1067. [DOI] [PubMed] [Google Scholar]
- 43.Li, Y., J. J. Bergeron, L. Luo, W. J. Ou, D. Y. Thomas, and C. Y. Kang. 1996. Effects of inefficient cleavage of the signal sequence of HIV-1 gp120 on its association with calnexin, folding, and intracellular transport. Proc. Natl. Acad. Sci. USA 939606-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu, S., R. Wyatt, J. F. Richmond, F. Mustafa, S. Wang, J. Weng, D. C. Montefiori, J. Sodroski, and H. L. Robinson. 1998. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/2 and V3 regions. AIDS Res. Hum. Retrovir. 14151-155. [DOI] [PubMed] [Google Scholar]
- 45.McKeating, J. A., C. Shotton, S. Jeffs, C. Palmer, A. Hammond, J. Lewis, K. Oliver, J. May, and P. Balfe. 1996. Immunogenicity of full length and truncated forms of the human immunodeficiency virus type I envelope glycoprotein. Immunol. Lett. 51101-105. [DOI] [PubMed] [Google Scholar]
- 46.Moremen, K. W., and M. Molinari. 2006. N-linked glycan recognition and processing: the molecular basis of endoplasmic reticulum quality control. Curr. Opin. Struct. Biol. 16592-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moulard, M., and E. Decroly. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469121-132. [DOI] [PubMed] [Google Scholar]
- 48.Nabatov, A. A., G. Pollakis, T. Linnemann, A. Kliphius, M. I. M. Chalaby, and W. A. Paxton. 2004. Intrapatient alterations in the human immunodeficiency virus type 1 gp120 V1V2 and V3 regions differentially modulate coreceptor usage, virus inhibition by CC/CXC chemokines, soluble CD4, and the b12 and 2G12 monoclonal antibodies. J. Virol. 78524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nolan, K. M., A. P. Jordan, and J. A. Hoxie. 2008. Effects of partial deletions within the human immunodeficiency virus type 1 V3 loop on coreceptor tropism and sensitivity to entry inhibitors. J. Virol. 82664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otteken, A., P. L. Earl, and B. Moss. 1996. Folding, assembly, and intracellular trafficking of the human immunodeficiency virus type 1 envelope glycoprotein analyzed with monoclonal antibodies recognizing maturational intermediates. J. Virol. 703407-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pantophlet, R., S. E. Ollmann, P. Poignard, P. W. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pollard, S. R., W. Meier, P. Chow, J. J. Rosa, and D. C. Wiley. 1991. CD4-binding regions of human immunodeficiency virus envelope glycoprotein gp120 defined by proteolytic digestion. Proc. Natl. Acad. Sci. USA 8811320-11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pollard, S. R., M. D. Rosa, J. J. Rosa, and D. C. Wiley. 1992. Truncated variants of gp120 bind CD4 with high affinity and suggest a minimum CD4 binding region. EMBO J. 11585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purcell, D. F. J., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 676365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 2801949-1953. [DOI] [PubMed] [Google Scholar]
- 56.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 684821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanders, R. W., E. Busser, J. P. Moore, M. Lu, and B. Berkhout. 2004. Evolutionary repair of HIV type 1 gp41 with a kink in the N-terminal helix leads to restoration of the six-helix bundle structure. AIDS Res. Hum. Retrovir. 20742-749. [DOI] [PubMed] [Google Scholar]
- 58.Sanders, R. W., M. M. Dankers, E. Busser, M. Caffrey, J. P. Moore, and B. Berkhout. 2004. Evolution of the HIV-1 envelope glycoproteins with a disulfide bond between gp120 and gp41. Retrovirology 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders, R. W., E. C. de Jong, C. E. Baldwin, J. H. Schuitemaker, M. L. Kapsenberg, and B. Berkhout. 2002. Differential transmission of human immunodeficiency virus type 1 by distinct subsets of effector dendritic cells. J. Virol. 767812-7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 745091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 767293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanders, R. W., M. Vesanen, N. Schuelke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, P. J. Maddon, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 768875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. O. Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 767306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 767760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Srivastava, I. K., L. Stamatatos, E. Kan, M. Vajdy, Y. Lian, S. Hilt, L. Martin, C. Vita, P. Zhu, K. H. Roux, L. Vojtech, C. Montefiori, J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2003. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J. Virol. 7711244-11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stamatatos, L., M. Wiskerchen, and C. Cheng-Mayer. 1998. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV type 1 isolate on viral envelope structure, cell entry, and replication. AIDS Res. Hum. Retrovir. 141129-1139. [DOI] [PubMed] [Google Scholar]
- 67.Stein, B. S., and E. G. Engleman. 1990. Intracellular processing of the gp160 HIV-1 envelope precursor. J. Biol. Chem. 2652640-2649. [PubMed] [Google Scholar]
- 68.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 724694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 673978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Travis, B. M., T. I. Dykers, D. Hewgill, J. Ledbetter, T. T. Tsu, S. L. Hu, and J. B. Lewis. 1992. Functional roles of the V3 hypervariable region of HIV-1 gp160 in the processing of gp160 and in the formation of syncytia in CD4+ cells. Virology 186313-317. [DOI] [PubMed] [Google Scholar]
- 71.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384184-187. [DOI] [PubMed] [Google Scholar]
- 72.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 701100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Anken, E., R. W. Sanders, I. M. Liscaljet, A. Land, I. Bontjer, S. Tillemans, A. A. Nabatov, W. A. Paxton, B. Berkhout, and I. Braakman. 2008. Only five out of ten strictly conserved disulfide bonds are essential for folding and eight for function of the HIV-1 envelope glycoprotein. Mol. Biol. Cell 194298-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387426-430. [DOI] [PubMed] [Google Scholar]
- 76.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384179-183. [DOI] [PubMed] [Google Scholar]
- 77.Wu, Z., S. C. Kayman, W. Honnen, K. Revesz, H. Chen, S. Vijh-Warrier, S. A. Tilley, J. McKeating, C. Shotton, and A. Pinter. 1995. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J. Virol. 692271-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 695723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wyatt, R., N. Sullivan, M. Thali, H. Repke, D. Ho, J. Robinson, M. Posner, and J. Sodroski. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 674557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 751165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang, Z. Y., B. K. Chakrabarti, L. Xu, B. Welcher, W. P. Kong, K. Leung, A. Panet, J. R. Mascola, and G. J. Nabel. 2004. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J. Virol. 784029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, P. F., F. Cham, M. Dong, A. Choudhary, P. Bouma, Z. Zhang, Y. Shao, Y. R. Feng, L. Wang, N. Mathy, G. Voss, C. C. Broder, and G. V. Quinnan, Jr. 2007. Extensively cross-reactive anti-HIV-1 neutralizing antibodies induced by gp140 immunization. Proc. Natl. Acad. Sci. USA 10410193-10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]