Abstract
HIV replication occurs throughout the natural course of infection in secondary lymphoid tissues and in particular within the germinal centers (GCs), where follicular dendritic cells (FDCs) are adjacent to CD4+ T cells. Because FDCs provide signaling that increases lymphocyte activation, we postulated that FDCs could increase human immunodeficiency virus (HIV) replication. We cultured HIV-infected CD4+ T cells alone or with FDCs and measured subsequent virus expression using HIV-p24 production and reverse transcription-PCR analyses. When cultured with FDCs, infected CD4+ T cells produced almost fourfold more HIV than when cultured alone, and the rate of virus transcription was doubled. Both FDCs and their supernatant increased HIV transcription and resulted in nuclear translocation of NF-κB and phosphorylated c-Jun in infected cells. FDCs produced soluble tumor necrosis factor alpha (TNF-α) ex vivo, and the addition of a blocking soluble TNF receptor ablated FDC-mediated HIV transcription. Furthermore, TNF-α was found highly expressed within GCs, and ex vivo GC CD4+ T cells supported greater levels of HIV-1 replication than other CD4+ T cells. These data indicated that FDCs increase HIV transcription and production by a soluble TNF-α-mediated mechanism. This FDC-mediated effect may account, at least in part, for the presence of persistent HIV replication in GCs. Therefore, in addition to providing an important reservoir of infectious virus, FDCs increase HIV production, contributing to a tissue microenvironment that is highly conducive to HIV transmission and expression.
The germinal centers (GCs) of secondary lymphoid tissues are unique structures associated with both the immune response and human immunodeficiency virus (HIV) infection (1-4, 9, 14, 23, 27, 35, 36). These sites are composed of highly activated B cells, a specialized form of macrophage (i.e., tingible body macrophage), a population of CD4-positive T cells, and follicular dendritic cells (FDCs). Shortly after infection, immense quantities of HIV particles become trapped on the dendritic processes of FDCs, and virus is found on these cells throughout the natural course of disease (9, 16, 17, 28). FDC-trapped HIV is replication competent and genetically diverse and contains virus collected throughout the disease course, thus creating an archive of infectious virus that can perpetuate infection (22).
We have focused on understanding the contributions of FDCs and the unique microenvironment of the GC to HIV transmission, diversification, and replication. Previous studies by our group found that FDCs maintain HIV in an infectious form for months to years (22, 33). Moreover, when FDCs are present, HIV infection can occur even in the presence of potent neutralizing antibodies (18). FDCs also interact with adjacent GC T cells to increase their expression of the HIV coreceptor CXC chemokine receptor 4 (CXCR4) (11). As a consequence of increased CXCR4, these GC T cells are sensitive to infection with concentrations of X4-tropic HIV that do not appear to infect other CD4 T cells, and analysis of these cells indicates that they frequently carry provirus (13, 20). Collectively, these data indicate potential explanations for why the GC microenvironment, largely contributed by FDCs, is highly conducive to HIV transmission.
The current study seeks to understand how FDCs affect other aspects of the HIV life cycle, specifically those of replication and gene expression. It is known that active HIV replication persists in and around GCs (13), although the reason(s) for this virus activity has not been elucidated. We report here that FDCs contribute to virus replication by increasing HIV transcription both in vitro and in vivo. FDCs therefore not only serve as a dangerous reservoir of replication-competent and genetically diverse HIV that contributes to persisting infection but also increase virus production. These findings explain why both HIV transmission and viral replication occur in GCs.
MATERIALS AND METHODS
Virus preparations.
Viral stocks for infection were prepared by propagating HIV-1IIIB in neoplastic H9 cells. Virus was harvested at the time of peak p24 and/or reverse transcriptase production, pooled, filtered through a 0.45-μm membrane, and stored in aliquots in liquid nitrogen until used. Our HIVIIIB preparations typically contained 1 μg/ml of p24 and 1 × 106 cpm reverse transcriptase activity. Unless otherwise noted, virus infections were performed using the isolate HIV-1IIIB. However, where indicated, the 91US054 (X4) or 92US714 (R5) HIV-1 primary isolate (obtained through the NIH AIDS Research and Reference Reagent Program) was used after propagation in mitogen-stimulated peripheral blood lymphocytes (PBLs) as described below. Primary virus stocks contained 140 to 200 nanograms p24/milliliter and 300,000 to 450,000 cpm reverse transcriptase activity.
HIV infection of PBLs.
PBLs were prepared from whole-blood samples obtained from healthy, HIV-negative donors by using Ficoll-Paque-Plus (Amersham, Piscataway, NJ) as directed. The isolated cells were activated with phytohemagglutinin (5 μg/ml; Sigma, St. Louis, MO) for 3 days in complete tissue culture medium (CM) consisting of RPMI 1640 supplemented with HEPES buffer (20 mM); nonessential-amino-acid solution (1×); l-glutamine (2 mM); 10% heat-inactivated, defined fetal bovine serum (FBS) (all from HyClone Laboratories, Logan, UT); and gentamicin (50 μg/ml; Life Technologies, Gaithersburg, MD), after which they were maintained in CM containing interleukin-2 (20 U/ml; NIH AIDS Reference and Reagent Program; contributed by Hoffmann-La Roche, Inc.). These preparations were >80% CD4+ T cells, with CD8+ T cells as the remainder. In some instances, magnetic-activated cell sorting (MACS) purification of CD4+ T cells was performed using negative selection. These preparations of cells were >95% pure. For infection, the cells were centrifuged and the CM was removed, after which the cells were resuspended in CM (with 20% FBS) containing HIV at a concentration of 50,000 cpm reverse transcriptase activity per 1 × 106 cells. The virus-cell suspension was incubated at 37°C for 2 h, after which unbound virus was removed by washing with fresh RPMI 1640. The resulting infected cells were cultured in CM (20% FBS) with or without FDCs or FDC-supernatant fluid under the conditions described for each experiment. After culture, the contents of each tube were centrifuged (1,000 × g) for 5 min and the supernatant was collected and analyzed for virus particle production using an HIV p24 Ag assay kit with a kinetic standard (Beckman Coulter, Miami, FL) according to the manufacturer's directions.
Isolation of FDCs.
FDCs were isolated from human tonsils as previously described (11, 33). Briefly, tonsils were cut into small pieces and digested with an enzyme cocktail containing Blendzyme (Roche Applied Science; Indianapolis, IN) and DNase I (Sigma, St. Louis, MO). The resulting single-cell suspension was applied to a preformed, 50%-continuous Percoll gradient. The low-density cell fraction (1.050 to 1.060 g/ml) was collected, washed with RPMI 1640 to remove the Percoll, and resuspended in CM. Heat-aggregated mouse immunoglobulin G (IgG; Chrompure, Jackson ImmunoResearch) was added to the cells and incubated on ice for 15 min to minimize nonspecific binding of IgG to FcR. The FDCs were then labeled with mouse IgM, anti-human FDC monoclonal antibody HJ2 (kindly provided by M. Nahm, University of Alabama at Birmingham), and then rat anti-mouse IgM MicroBeads (Miltenyi Biotech, Auburn, CA), and positive selection was performed using an auto-MACS (Miltenyi Biotech). The MACS-enriched cell preparations contained 75 to 90% FDCs, with the remaining cells being roughly equal quantities of T and B lymphocytes. Prior to use, the FDCs preparations were subjected to 3,000 rads of γ irradiation, which does not affect the FDCs but prevents any contaminating CD4+ T cells from supporting HIV replication. In some experiments, FDCs were specifically depleted from isolated lymphoid tissue cells by using HJ2 and magnetic beads (Dynal, Inc., Lake Success, NY). In some experiments, purified FDCs were obtained by cell sorting using a FACSVantage SE equipped with the FACSDiVa option (BD Biosciences, San Jose, CA) after the cells were labeled with HJ2 followed by anti-mouse IgM-fluorescein isothiocyanate (FITC) (Jackson Immuno Research). Fluorescence-activated cell sorter (FACS)-sorted FDCs were routinely >96% pure, with the residual cells being equally distributed between T and B lymphocytes. To obtain FDC supernatant, FACS-isolated FDCs were cultured in CM for 6 days at a concentration of 1 × 106 cells per milliliter. At the end of the culture period, the supernatant was collected, centrifuged (300 × g for 10 min) to remove cell debris, filtered through a 0.2-μm filter, and then used immediately or stored at −80°C. As a control, supernatant was obtained from equal numbers of FACS-sorted HJ2-negative cells. Unless stated otherwise, 1 × 106 infected PBLs were cultured with 1 × 105 FDCs or cells specifically depleted of FDCs in 12- by 75-mm culture tubes containing 0.5 ml CM. In Transwell experiments, the FDCs were cultured on top of the Transwell insert and the infected PBLs on the bottom.
Immunohistochemistry.
Tonsillar tissue obtained from a routine tonsillectomy was placed in Streck's tissue fixative for 1 to 3 days, washed with 80% ethanol, and embedded in paraffin. Immunohistochemistry was performed using a biotin-streptavidin staining method on 5-μm tissue sections mounted on glass slides. Tissues were dewaxed and rehydrated in deionized water. Heat-induced epitope retrieval was performed using a 95°C water bath for 5 min with 1× ReVEAL decloaker reagent (Biocare Medical), followed by cooling to room temperature. Tissues were blocked with 10% Sniper blocking reagent (Biocare Medical) in TNB (0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl, and 0.5% blocking reagent [New England Nuclear]) for 2 h at room temperature. Anti-tumor necrosis factor alpha (anti-TNF-α) antibody (1:100; N-19; Santa Cruz Biotechnology) or anti-TNF-α antibody containing TNF-α blocking peptide (1:10; Santa Cruz Biotechnology) was diluted in 10% Sniper blocking reagent in TNB and incubated overnight at 4°C. The sections were washed in Tris-buffered saline (TBS; pH 7.4) containing 0.05% Tween 20 and endogenous peroxidase blocked with 3% (vol/vol) H2O2 and then incubated with horse anti-goat-biotinylated antibody (Vector Laboratories) for 30 min at room temperature. Tissues were washed in TBS containing 0.05% Tween 20 and incubated with streptavidin-horseradish peroxidase (HRP) reagent (Signet) for 30 min. Tissues were washed again in TBS containing 0.05% Tween 20 and developed with 3,3′-diaminobenzidine (Vector Laboratories, Burlingame, CA). Sections were counterstained with chloramphenicol acetyltransferase hematoxylin (Biocare Medical), mounted in Permount (Fisher Scientific), and examined by light microscopy. The isotype-matched negative control antibody used (data not shown) was goat ChromPure IgG (Jackson Immuno-Research), which uniformly showed no reactivity.
GC T-cell isolation.
CD4+ CD57+ GC T cells were isolated from human tonsillar tissue as previously described (12). Briefly, tonsils were cut into small sections, cells were mechanically separated from tissue by repeat pipetting, and red blood cells were removed by incubation for 5 min at room temperature in red blood cell lysis buffer (155 mM NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA). GC CD4 T lymphocytes were purified by negative selection using a CD4+ T-cell isolation kit (Miltenyi Biotec), followed by positive selection by MACS using anti-CD57 biotin and streptavidin microbeads (Miltenyi Biotec). The resulting CD4+ CD57+ preparations were ≥95% pure as assessed by flow cytometry.
EMSA.
The nuclei of GC T cells (4 × 106 cells) or an equal number of CD4+ T cells isolated from the same tissue were prepared upon isolation from the tissues for electrophoresis, following a standard protocol (6). Prior to the electrophoretic mobility shift assay (EMSA), radiolabeled NF-κB probe (Santa Cruz Biotechnology; Santa Cruz, CA) was prepared by labeling 40 nanograms of probe with [γ32P]ATP (10 μCi) by incubation with T4 polynucleotide kinase for 1 h at 37°C in a total reaction volume of 10 μl. After the labeling, 20 μl of Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) was added to the mixture and the free nucleotides were removed using a DyeEx spin kit (Qiagen, Valencia, CA). After probe preparation, equal quantities of nuclear proteins were preincubated at room temperature for 10 min in gel shift binding buffer (Promega; Madison, WI), after which 900 picograms of labeled probe was added. After 20 min of incubation at room temperature, the samples were applied to 5% polyacrylamide gels (0.5% Tris-buffered EDTA with an acrylamide-to-cross linker ratio of 19:1) and electrophoresed at 200 V. The gel was then dried onto filter paper and exposed to X-ray film (Hyperfilm ECL, GE Healthcare, Piscataway, NJ) for 8 to 72 h.
Reverse transcription-PCR (RT-PCR) analysis.
Total RNA was isolated using RNA-STAT60 (Tel-Test, Friendswood, TX) according to the manufacturer's protocol, except that following precipitation of the RNA with isopropanol, the centrifugation time was increased to 45 min. The resulting RNA preparation was treated with DNase I (DNA-free; Ambion, Inc., Austin, TX) to remove contaminating DNA. To confirm the enzymatic removal of DNA, the samples were analyzed by PCR using β-actin primers (as described below) and subjected, if needed, to additional DNase I treatment. RNA from equal numbers of cells was reverse transcribed using a GeneAmp RNA PCR kit (Applied Biosystems; Foster City, CA) as directed with oligo(dT) or random hexamer primers (Applied Biosystems). PCR amplification was performed in parallel for different numbers of cycles to ensure that comparative analysis occurred during the linear phase of the amplification process. The linear phase of the PCR occurred between cycles 16 and 24 for β-actin and cycles 24 and 30 for HIV. Following amplification, the PCR products were separated by electrophoresis on 1.5% agarose gels, followed by transfer to a nylon membrane (Hybond N+; Amersham Biosciences, Piscataway, NY). Southern blotting was performed using commercially available FITC-labeled probes or probes labeled with FITC by using the ECL 3′-oligolabeling and detection system (Amersham Biosciences) according to the manufacturer's directions. The following primer sets and probes were used: for total HIV RNA, Nef-6 (5′-CCCAGCGGAAAGTCCCTTGTAG-3′) and Nef-7 (5′-CACAAGGCTACTTCCCTGAT T-3′), which recognize a sequence common to spliced and unspliced RNA, yielding a 283-bp amplicon which can be probed with Nefp-3 (5′-TGGATGGTGCTTCAAGCTAGTACCAGTT-3′) (29); for unspliced HIV RNA (gag), SK38 (5′-TAATCCACCTATCCCAGTAGGAGAAAT-3′) and SK39 (5′-TTTGGTCCTTGTCTTATGTCCAGAATGC-3′), yielding a 115-bp amplicon probed by SK19-FITC (5′-FITC-TCCTGGGATTAAATAAAATAGTAAGAATGTATAGCCCTAC-3′) (30); for doubly spliced RNAs (tat, rev, nef), US (5′-TAGTAGCATGCTCTCTCGACGCAGGACTCGGCTTGC-3′) and ART7 (5′-ATGATCTGCAGTTCTATTCCTTCGGGCCTGTCG-3′), yielding amplicons of 402 (Tat), 220 (Rev), and 203 (Nef) bp when probed with S3 (5′-ACCTCGCATGCGAAGAAGCGGAGACAGCGACGAAG-3′) (24); and for β-actin, forward (5′-CATCCTCACCCTGAAGTACC-3′) and reverse (5′-GGTGAGGATCTTCATGAGGT-3′) primers, yielding a 398-bp amplicon (19).
Quantitative RT-PCR assay for HIV RNA production from CD4+ T cells.
Supernatant fluid from cultures was collected and the HIV RNA isolated using a QIAamp viral RNA minikit (Qiagen) as directed. The isolated viral RNA was then reverse transcribed into cDNA by using random primers (Invitrogen) and SuperScript III reverse transcriptase (Invitrogen). The cDNA template was then used for quantitative PCR (TaqMan) with the following primers (RT) and probe: forward primer (nucleotide 3696 in HXB2), 5′-TGGGTACCAGCACACAAAGG-3′; reverse primer (nucleotide 3850), 5′-ATCACTAGCCATTGCTCTCCAAT-3′; and probe, ATTGGAGGAAATGAAC-MBG (6-carboxyfluorescein labeled). The quantitative PCR conditions were as follows: 1 cycle at 50°C for 2 min and 1 cycle at 95°C for 10 min, followed by 60 cycles at 95°C for 15 s and 60°C for 1 min. The primers were used at 900 nM each, and the probe was used at 250 nM. The cell equivalents are based on β-actin amplification.
ELISA for HIV p24.
The supernatant fluid from T-cell or coculture cultures was collected, and the amount of p24 per milliliter was assessed using an HIV p24 antigen enzyme-linked immunosorbent assay (ELISA) kit (ZeptoMetrix Corporation), following the manufacturer's directions. Briefly, particle-associated p24 was obtained by treating the samples with lysis buffer, and the resulting product was added to a 96-well plate coated with p24-specific antibody. After incubation for 2 h at 37°C, the plate was washed to remove unbound material. HIV p24 detector antibody was added to each well and incubated as before for 1 h. The plate was then washed to remove unbound antibody and incubated with a secondary antibody conjugated to HRP. After a 30-min incubation, substrate was added and reacted for 30 min, after which the reaction was stopped and the absorbance was measured at 450 nm with an ELISA plate reader. The absorbance of the samples was compared to a standard curve to determine the number of picograms per milliliter p24 present.
Run-on transcription analysis.
Nuclear lysates were prepared as previously described from cocultured cells (6). The cells were centrifuged at 15,000 rpm for 10 min in an Eppendorf 5417R refrigerated microcentrifuge, after which the supernatant was removed and the cells were resuspended in cold (4°C) phosphate-buffered saline (PBS). After two wash steps using cold PBS, the cells were suspended in 5 ml of cold NP-40 lysis buffer (10 mM Tris-Cl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) and incubated on ice for 5 min, followed by centrifugation at 1,000 rpm for 10 min at 4°C. The cell pellet was suspended in 500 μl of nuclear freezing buffer (50 mM Tris-Cl [pH 8.3], 40% [vol/vol] glycerol, 5 mM MgCl2, 0.1 mM EDTA) and stored at −80°C until tested. Radiolabeling of the nascent transcripts was initiated by thawing the isolated nuclei on ice and adding 225 μl to 60 μl of 5× run-on buffer (25 mM Tris-Cl [pH 8.0], 12.5 mM MgCl2, 750 mM KCl, 1.5 mM each of ATP, GTP, CTP). Radiolabeled UTP (15 μl) was then added (150 μCi of 3,000-Ci/mmol [10-mCi/ml] [α-32P]UTP; Perkin Elmer), and the mixture was incubated at 37°C for 15 min, followed by addition of 20 μl of calcium chloride (10 mM). Genomic DNA was digested with RNase-free DNase I (10 μl of a 1-mg/ml solution) for 5 min at 30°C. After digestion, 35 μl of proteinase K buffer (10% sodium dodecyl sulfate [SDS], 50 mM EDTA, 10 mM Tris-Cl [pH 7.4], with freshly added proteinase K [3 mg/ml]) was added and the mixture was incubated at 37°C for 45 min. The RNA was isolated by extracting it twice with 400 μl phenol-chloroform and once with chloroform. The RNA was then precipitated by mixing it with an equal volume of ammonium acetate (5 M) followed by an equal volume of isopropanol, followed by incubation at −20°C for 30 min. The precipitated RNA was collected by centrifugation and was suspended in 100 μl Tris-EDTA buffer. To further purify the RNA, an additional DNase treatment and NaOH degradation were performed by adding 100 μl magnesium-calcium buffer (magnesium chloride [19 mM] plus calcium chloride [5 mM]) and 10 μl RNase-free DNase (1 mg/ml). After incubation at 37°C for 5 min, the reaction mixture was placed on ice for 5 min, after which 50 μl sodium hydroxide (1 N) was added and the mixture was incubated on ice for an additional 2 min. For neutralization, 77 μl of HEPES (1 M) was added and the RNA was precipitated with 340 μl of ammonium acetate (5 M) plus 700 μl isopropanol, followed by incubation at −20°C for 1 h. The RNA was centrifuged and the supernatant removed, after which the nucleic acid was air dried and suspended in 100 μl Tris-EDTA. One microliter of each sample was spotted onto filter paper and the radioactivity quantitated using a scintillation counter. For detection of HIV-specific transcripts, the plasmid pNL4-3, containing the full-length HIV genome (obtained through the NIH AIDS Research and Reference Reagent Repository Program), was used. The parental plasmid pUC18 (Invitrogen, Carlsbad, CA) was used to control for nonspecific binding and binding to non-HIV DNA. The capture and control plasmids were linearized using EcoRI, after which the DNA was extracted twice with phenol-chloroform, followed by precipitation with ethanol and sodium acetate. The plasmid DNA (10 μg) was then diluted in 400 μl of Tris-EDTA buffer (pH 8.0) and denatured for 30 min at room temperature using 40 μl of 3 M NaOH. The samples were neutralized by the addition of 300 μl of 2.5 M ammonium acetate (pH 7.0), after which they were applied to Hybond N+ (Amersham) by using a slot blot apparatus. The nylon membrane was washed with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and the DNA was immobilized by irradiation in a UV Stratalinker (Stratagene), using the auto cross-link option. Immediately prior to use, the membrane was washed with 2× SSC. For prehybridization, the membrane containing the DNA was washed with 2× SSC buffer and incubated at 65°C for 30 min in Perfect Hyb Plus solution (Sigma), after which hybridization was performed at 65°C overnight using 1 ml of Perfect Hyb Plus solution containing the radiolabeled transcripts. Following hybridization, the membrane was subjected to two washes at 65°C for 1 h in 2× SSC, after which it was exposed to film.
Image analysis.
Developed X-ray films from Southern blotting and nuclear run-on transcription assays were subjected to densitometric analysis using ImageJ software developed by Wayne Rasbad, National Institutes of Health (available at http://rsb.info.nih.gov/ij/).
Analysis of TNF-α production by FDCs.
The cell-free supernatant fluid from cultures of FACS-purified FDCs from a number of different individuals was assayed for TNF-α by using ELISA (R&D Systems, Minneapolis, MN) as instructed. Values were expressed in picograms TNF-α per milliliter culture fluid on replicate samples. Data points represent the means ± standard deviations.
Isolation of nuclear and cytoplasmic fractions of cultured CD4+ T cells.
CD4+ T cells (107 cells/sample) were collected and washed three times with ice-cold PBS. The resulting cell pellet was then suspended in 150 μl buffer A (10 mM Tris-HCl [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 2 mM dithiothreitol, 0.1% Triton X-100, 2.5 mM NaH2PO4, 1 mM Na3VO4, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Roche]) and inverted gently. This cell buffer mixture was incubated on ice for 15 min and centrifuged at 250 × g for 5 min at 4°C, after which the supernatant fluid was discarded. One hundred microliters of buffer A was then added to the cell pellet, and the mixture was passed five times through a syringe with a small gauge needle. Sequentially, the mixture was centrifuged at 8,000 × g for 20 min at 4°C. The supernatant fluid was collected and represented the cytoplasmic protein. To extract the nuclear protein, the remaining nuclear fraction was resuspended in 80 μl buffer B (10 mM Tris-HCl [pH 7.9], 1.5 mM MgCl2, 420 mM KCl, 0.2 mM EDTA, 2 mM dithiothreitol, 1% Igepal CA-630, 25% [vol/vol] glycerol, 2.5 mM NaH2PO4, 1 mM Na3VO4, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Roche]) and the nuclei were disrupted by passing this mixture through a syringe as described above. The mixture was then agitated gently at 4°C for 50 min and centrifuged at 16,000 × g for 5 min, after which the supernatant was collected to represent the nuclear protein. Both the cytoplasmic and the nuclear proteins were then stored at −80°C until used.
Western blot assay for activated NF-κB P65, P50, and phosphorylated c-Jun in CD4+ T cells.
The nuclear and cytoplasm protein fractions were each mixed with 4× loading buffer (250 mM Tris-HCl [pH 8.8], 4% [wt/vol] SDS, 4% [vol/vol] β-mercaptoethanol, 40% [vol/vol] glycerol, 0.01% [wt/vol] bromophenol blue) and incubated for 20 min at room temperature. After the initial incubation, the mixture was then incubated at 100°C for 5 min. The proteins from each fraction were separated by 12% SDS-polyacrylamide gel electrophoresis and transferred to a Hybond C membrane (Amersham Life Science). The membrane was then incubated in TBS with 5% nonfat milk for 2 h at room temperature to block its binding ability and then incubated with the a primary antibody to detect NF-κB p50 or p65 (from the NF-κB family transcription factor assay kit; Chemicon International) or phosphorylated c-Jun (Ser73) (Millipore). The antibodies were diluted in TBS with 5% nonfat milk protein and incubated for 2 h at room temperature. The membrane was then washed three times in TBS with 0.1% Tween 20 and 5% nonfat milk to remove unbound antibody. After being washed, the membrane was incubated with HRP-conjugated secondary antibody (anti-rabbit IgG-HRP from the NF-κB family transcription factor assay kit; Chemicon International) diluted in TBS with 5% nonfat milk. Finally, the membrane was washed in TBS with 0.1% Tween 20 and 5% nonfat milk and developed using chemiluminescence. Semiquantitation was performed with a phosphorimager.
ELISA for NF-κB binding activity in CD4+ T cells.
The binding activity of NF-κB was detected using an NF-κB family transcription factor assay kit (Chemicon International), following the manufacturer's instructions. Briefly, the nuclear or cytoplasmic protein was added to a plate coated with NF-κB binding sequence (5′-GGGACTTTCC-3′) and incubated for 1 h at room temperature. After the plate was washed three times, the primary antibody (rabbit, anti-human antibody) was added and incubated for another hour as before. The plate was again washed three times and incubated with secondary antibody (anti-rabbit IgG-HRP) for 30 min at room temperature. The plate was again washed three times as above, and binding activity was detected with an ELISA plate reader by measuring absorbance at 450 nm.
Statistical analysis.
Analysis of data was performed using a one-tailed Student t test. P values of ≤0.05 were considered significant. Unless specifically stated, the error bars indicate the standard errors of the means (SEM).
RESULTS
FDCs and HIV transcription.
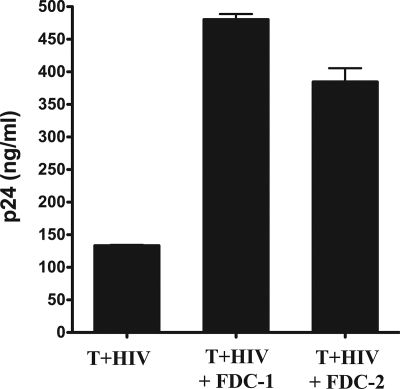
To determine whether FDCs contributed to HIV replication and gene expression, we first cocultured FDCs with in vitro-infected CD4+ lymphocytes and measured the resulting p24 production (Fig. 1). We observed a two- to threefold increase in HIV p24 production when FDCs were cocultured with infected T cells, even though the FDCs do not become infected themselves (31, 32). In independent experiments, we saw an average increase of 4.3-fold in viral p24 production, with the data ranging between 2.5- and 5.5-fold. FDCs increased both regulatory (multiply spliced) and structural (unspliced) RNAs, and we confirmed the general nature of the FDC-mediated effect using both a primary R5 and an X4 isolate of HIV (not shown).
FIG. 1.
FDCs cocultured with HIV-infected CD4+ T cells increased virus p24 production. In vitro-infected CD4+ T cells (T+HIV) were cultured for 6 days with or without FDCs (10 T cells per FDC). The tissue culture supernatant was then harvested and virus particle production measured by p24 ELISA. FDC-1 and FDC-2 refer to FDCs obtained from two separate, unrelated donors. All assays were performed in duplicate, and the error bars represent the SEM. Statistical significance (* signifies P values of <0.05) was determined by Student's t test. These data are representative of three independent experiments using different sources of primary CD4+ T cells and FDCs.
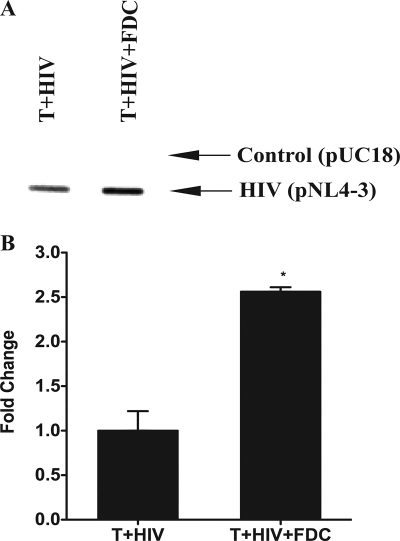
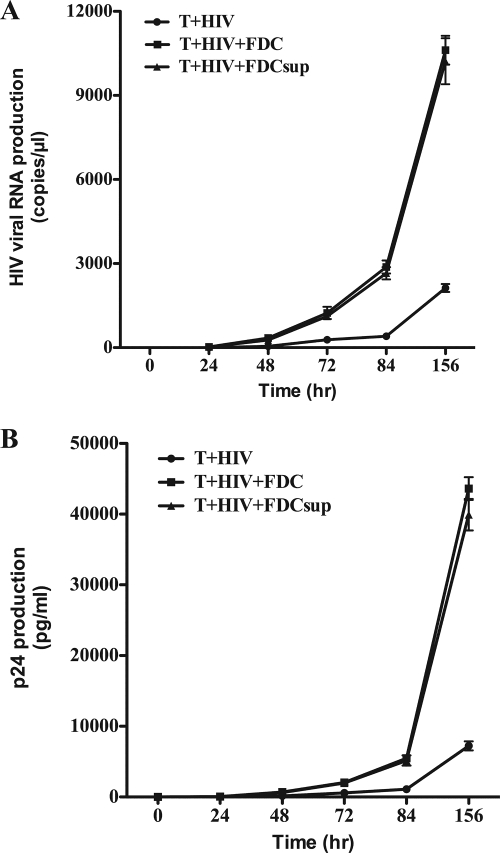
Because we observed increases in both p24 production and viral RNA levels when FDCs and infected CD4+ T cells were cocultured, we examined the level of HIV transcription in these cultures by using nuclear run-on analysis (Fig. 2A). In the presence of FDCs, the rate of HIV transcription was more than doubled in a single round of virus replication (Fig. 2B). To determine whether the FDC signaling that increased HIV transcription was soluble or membrane bound, we incubated infected T cells with FDCs or a cell-free supernatant from FDC cultures (Fig. 3). Consistent with the presence of a soluble mediator, the FDC supernatant recapitulated the effect of intact FDCs, as measured by both quantitative viral RNA levels (Fig. 3A) and p24 protein production (Fig. 3B). FDCs also increased HIV transcription when they were physically separated from the infected CD4+ T cells by a semipermeable membrane (Transwell), thereby confirming the soluble nature of the FDC signal (data not shown).
FIG. 2.
FDCs increased the rate of HIV transcription in HIV-infected T cells. (A) In vitro-infected CD4+ T cells were cultured with FDCs for 18 h. The nuclei were then isolated and subjected to nuclear run-on transcription analysis. A plasmid containing the full-length HIV genome (pNL4-3) was used to capture HIV-specific RNA while the parental plasmid without the HIV genome was used as a control for nonspecific binding. The Southern blot presented is representative of two independent experiments. (B) The density of the Southern blot was determined using ImageJ software. The differences between the control and pNL4-3 densities are plotted and represent the means for two independent experiments. The error bars represent the SEM. Statistical significance (* signifies P values of <0.05) was determined by Student's t test.
FIG. 3.
FDC augmentation of virus transcription was mediated by a soluble factor. In vitro-infected CD4+ T cells were cultured with FDCs or supernatant (FDCsup; 10%, vol/vol) obtained from 6-day cultures of FACS-sorted FDCs. (A) Viral RNA was harvested from the culture supernatants at the times indicated and examined using quantitative RT-PCR. (B) The supernatant fluid from the same cultures was harvested and examined for HIV p24 production at the times indicated. Error bars represent the standard deviations for replicate cultures. These data are representative of three independent experiments using different sources of primary CD4+ T cells and FDCs.
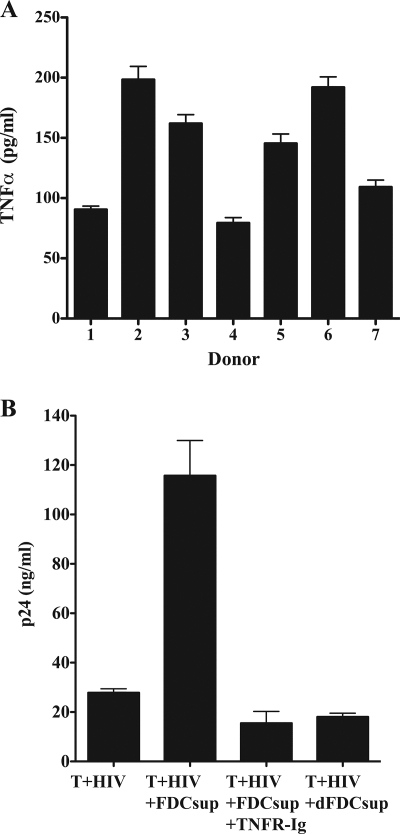
Because TNF-α is known to be a major activator of HIV transcription (8, 10) as well as being important in the development and maintenance of FDCs (15, 24, 29), we examined the supernatant from FDC cultures to determine if these cells produced soluble TNF-α (Fig. 4A). Tonsillar FDCs produced between 79 and 198 picograms TNF-α per milliliter culture fluid after 6 days. We also found that the addition of a similar amount (i.e., 167 nanograms) of recombinant TNF-α per milliliter to infected CD4+ T cells recapitulated the effect of FDCs or their supernatant in viral RNA generation (i.e., 24 ± 3 versus 27 ± 3 or 27 ± 12 copies/μl at 24 h, 372 ± 21 versus 342 ± 51 or 267 ± 60 copies/μl at 48 h, and 1,137 ± 118 versus 1,237 ± 218 or 1,116 ± 93 copies/μl with TNF-α versus FDCs or FDC supernatant, respectively) and p24 production (i.e., 54 ± 9 versus 51 ± 12 or 49 ± 13 ng/ml at 24 h, 572 ± 36 versus 692 ± 71 or 632 ± 61 ng/ml at 48 h, and 1,737 ± 106 versus 2,037 ± 317 or 1,975 ± 117 ng/ml at 72 h with TNF-α versus FDCs or FDC supernatant, respectively). To confirm that FDC-produced TNF-α resulted in the observed increase in HIV production, we cultured infected cells with FDC supernatant and soluble TNF receptor fusion protein (TNFR-Ig) to block endogenously produced TNF-α (Fig. 4B). The addition of TNFR-Ig completely inhibited the signal present in FDC supernatant, confirming the role of FDC-produced TNF-α in increasing HIV transcription and virus production. As a further control, we also tested supernatant obtained from culturing of secondary lymphoid tissue cells that were specifically depleted of FDCs and found that in the absence of the FDCs, no increase in HIV p24 production was observed. In a separate experiment, we also confirmed the specificity of TNFR-Ig by comparing it to purified mouse immunoglobulin and found that the control antibody did not substantively affect the production of p24 (i.e., FDCs cultured with infected T cells produced 1,758 ± 119, 1,882 ± 173, and 530 ± 184 picograms p24/ml after 72 h in the presence of medium alone, medium plus control antibody, and medium plus TNFR-Ig, respectively, while infected T cells without FDCs produced 541 ± 39 picograms p24/ml).
FIG. 4.
FDCs produce TNF-α that increases HIV replication. (A) FACS-sorted FDCs (1 × 105/ml) from seven unrelated donors were cultured for 6 days, after which the culture supernatant fluid was examined for TNF-α production using ELISA. Data are expressed in picograms TNF-α per milliliter of culture fluid and represent the means ± standard deviations for triplicate wells. (B) In vitro-infected CD4+ T cells were cultured with supernatant (FDCsup) obtained from cultured FDCs with or without TNFR-Ig (40 μg/ml). As a negative control, the same cells were cultured with supernatant from secondary lymphoid tissue cells that had been specifically depleted of FDCs (dFDCsup). After 6 days of culture, the supernatants were assayed for virus production by p24 ELISA. The data are representative of two independent experiments using different donors of FDCs and T cells and represent the means for replicate cultures. The error bars represent the SEM.
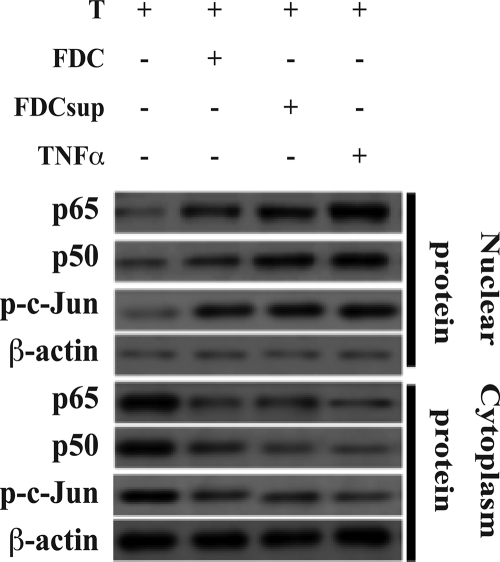
The U3 region of the HIV long terminal repeat contains a number of different binding sites for many cellular transcription factors, including NF-κB (21, 26). We reasoned that if FDC-produced TNF-α were responsible for the observed increased HIV transcription, then an increased level of activated (i.e., nuclear) NF-κB p50 and p65 would be present in the infected T cells incubated with FDCs or their supernatant. We therefore isolated the nuclear and cytoplasmic fractions of the HIV-infected T cells cultured with FDCs, FDC supernatant, or recombinant TNF-α (Fig. 5). In support of our hypothesis, we saw increased nuclear localizations of both p50 and p65 and corresponding decreases of the cytoplasmic concentrations of these NF-κB factors. Quantitative ELISA confirmed these increases and indicated that nuclear p50 and p65 increased 1.6- and 2.4-fold, respectively, by FDCs and 2.1 and 3-fold, respectively, by FDC supernatant (not shown). We also observed increased amounts of nuclear phosphorylated c-Jun, consistent with activation of the transcription factor AP-1, another factor activated through TNFR1 (Fig. 5).
FIG. 5.
FDCs induce activated NF-κB in in vitro-infected CD4+ T cells. FDCs (1 × 105 cells/ml), supernatant fluid from FDC cultures (FDCsup; 10%, vol/vol), or recombinant TNF-α (166 picograms/ml) was cultured with in vitro-infected CD4+ T cells (1 × 106 cells/ml) for 24 h, after which the cells were examined for cytoplasmic and nuclear NF-κB p50, p65, and phosphorylated c-Jun (p-c-Jun) by Western blotting as described in Materials and methods. β-Actin was also immunoblotted and served as a loading control. These data are representative of two independent experiments using different sources of primary CD4+ T cells and FDCs.
TNF-α production in vivo and HIV transcription in ex vivo GC T cells.
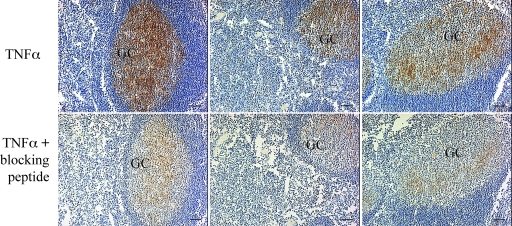
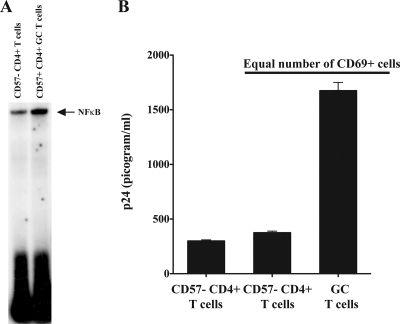
To ensure that FDC production of TNF-α was not just an in vitro phenomenon, we performed immunohistochemical analysis on tonsil sections from three different donors (Fig. 6). Examination revealed the presence of TNF-α in GCs. The reticular labeling pattern is characteristic of the FDC network and suggests that the TNF-α is associated with this network. Control sections labeled with the same antibody in the presence of a neutralizing peptide confirmed that the observed labeling was specific for TNF-α. We reasoned that if FDC-produced TNF-α affected adjacent CD4+ T cells, then isolated GC T cells would possess higher levels of activated NF-κB than non-GC T cells. Consistent with our hypothesis, GC T cells had higher levels of nuclear NF-κB than did non-GC T cells obtained from the same tissues (Fig. 7A).
FIG. 6.
TNF-α is present in GCs. Tonsillar sections from three unrelated individuals were examined for the presence of TNF-α as described in Materials and Methods. Original magnification, ×100. The scale bar represents 50 μm.
FIG. 7.
Ex vivo GC CD4+ T cells are like CD4+ T cells cultured with FDCs in vitro. GC CD4+ T cells were isolated from the tonsils of healthy, uninfected donors as described in Materials and Methods. (A) GC T cells (CD4+ CD57+) express more activated NF-κB than non-GC CD4+ T cells obtained from the same tissue. Immediately after isolation, the nuclei were obtained from equal numbers of GC T cells (4 × 106) and non-GC T cells and prepared and analyzed by an EMSA for NF-κB as described in Materials and Methods. (B) GC T cells infected in vitro produce more virus than other CD4+ T cells from the same tissue. Equal numbers (5 × 105) of GC T cells and other CD4+ T cells from the same tissue were infected with HIV-1 92US714 (R5-tropic; 0.025 cpm reverse transcriptase activity) as described in Materials and Methods. After 6 days, the culture supernatant was harvested and the HIV produced assessed by p24 ELISA. These data are representative of three independent experiments. Because GC T cells express three- to fourfold more CXCR4 than other CD4+ T cells, we used an R5 primary isolate of virus for these studies to avoid potential differences due to virus binding and entry.
We next determined whether GC T cells with their increased levels of activated NF-κB and proximity to FDCs in vivo would produce greater amounts of HIV than other, non-GC T cells from the same donor. Because we previously found that GC T cells possess higher levels of CXCR4, thereby making them more susceptible to X4 HIV binding and entry than other CD4+ T cells (11), we infected GC T cells immediately upon isolation with HIV-1 92US714, an R5-tropic, primary isolate of virus (Fig. 6B). We observed that the GC T cells generated a threefold increase in p24 production compared to the level for non-GC T cells from the same tissue. Furthermore, because the majority of GC T cells are CD69+, indicating an activated state, we also infected an equal number of CD69+ non-GC CD4+ T cells to ensure that the activation state of the cells alone was not responsible for the observed results. Once again, we found that the GC T cells produced a much higher level of HIV than other activated CD4+ T cells. In an independent experiment, we also confirmed that FDCs increased HIV production by non-GC T cells, indicating that these cells were susceptible to FDC-mediated augmentation of virus production and not somehow deficient in their ability to support HIV replication (not shown). Collectively, these data indicate that not only do FDCs increase HIV replication and virus production in vitro but also GC T cells that reside adjacent to FDCs in vivo have higher levels of nuclear NF-κB and produce substantively more virus than other CD4+ T cells. These findings suggest a high level of activation of CD4 T cells residing adjacent to the FDC. In fact, the level of activation appears to be comparable to that seen with CD4+ T cells activated in vitro (not shown). These data add to a growing list of complex interactions between FDCs, HIV, and the natural targets of viral infection, thereby providing new information about how FDCs promote HIV replication and progeny virus production.
DISCUSSION
FDCs are a large reservoir of infectious and genetically diverse HIV that can perpetuate infection (22). These cells are located in the GCs of secondary lymphoid tissues, where they interact intimately with surrounding lymphocytes, including CD4+ GC T cells. In addition to providing a continual source of infectious HIV to surrounding CD4+ cells (i.e., both T cells and macrophages) throughout infection (22), FDCs also increase HIV transcription and virus production in CD4+ T cells. This increased generation of viral RNA was a general effect and applied to both spliced and unspliced viral RNAs and to laboratory-adapted and primary isolates of virus. FDCs mediated the increased virus transcription and production by the secretion of TNF-α, which in turn resulted in the nuclear translocation of both NF-κB p50 and p65 (c-Rel) in infected CD4+ T cells.
In vivo, we observed the presence of TNF-α in GCs, and the reticular immunohistochemical labeling pattern suggested that the TNF-α was associated with FDCs, although other cells could also be involved. The ability to increase HIV replication is not unique to FDCs, as dendritic cells have also been shown to increase HIV production by CD4+ T cells (30, 34). We reason that in vivo, other TNF-α-producing cells, including dendritic cells, macrophages, and T cells, could act in concert with the FDCs to create a microenvironment that is especially suited to HIV replication. It should be noted, however, that FDCs alone produced on average 178 picograms TNF-α per milliliter in culture, and we found that adding a similar quantity of recombinant TNF-α to cultures of infected T cells recapitulated the presence of FDCs or their supernatant. We also found that adding this amount of TNF-α to cultures resulted in activation of NF-κB in the same manner observed when FDCs were present. Thus, FDC-produced TNF-α appears sufficient to account for the observed increases in HIV replication. It is also likely that because FDCs “embrace” adjacent cells, the net FDC-TNF-α signaling may be far more efficient and concentrated than observed in the in vitro setting where soluble signal could diffuse throughout the CM. Additionally, FDC “contacts” with GC T cells would likely transmit HIV, while providing TNF-α-mediated activation signals to facilitate productive infection of the target cell. Collectively, the overall contribution of FDC-T-cell signaling may play an even more significant role in vivo than what we observed in vitro.
Support for the in vivo role of FDC-produced TNF-α in augmenting virus replication comes from the observation that freshly isolated GC T cells possessed higher levels of activated NF-κB than other CD4+ T cells obtained from the same tissue. Furthermore, upon infection, these GC T cells supported a three- to fourfold increase in HIV replication compared to that seen with other CD4+ T cells. The addition of FDCs to non-GC T cells obtained from the same tissue as the GC T cells also resulted in increased HIV production, and this was ablated when TNFR-Ig was present (not shown). While these data do not unequivocally indicate that FDC signals cause increased HIV transcription in vivo, they are highly suggestive and strongly supportive. Moreover, our data are also in agreement with those of Hufert et al. and Folkvord et al., who found that GC T cells from HIV-infected subjects demonstrated a 10-fold higher frequency of HIV infection than other CD4+ lymphocytes and that these same cells possessed a higher rate of virus expression than other CD4 T cells (13, 20). Although these authors did not postulate why GC T cells were more readily infected and showed higher virus replication, our data provide a plausible explanation for this and confirm the potential importance of the GC environment, in large part contributed by FDCs, to HIV pathogenesis.
Even though it is known that TNF-α increases the transcription of HIV in vitro, our current study is the first report indicating that isolated FDCs produce this cytokine and that it augments HIV transcription and virus production in infected CD4+ lymphocytes. Our observation that FDCs secrete TNF-α is in contrast to the findings of Butch et al., who failed to find TNF-α mRNA in cDNA preparations of FDCs (5). The reason for this difference is not understood but may relate to differences in isolation and testing procedures as well as different tissue samples. In our hands, we found that TNF-α-specific mRNA was present in our FACS-sorted FDC preparations (not shown) and that the different specimens we examined produced levels of TNF-α ranging from a low of 79 to a high of 198 picograms per milliliter. The reason for the differing amounts of TNF-α in our different donors may be related to dynamic differences in the local activation statuses of the tissues from which our FDCs were isolated. We reason that the production of TNF-α by FDCs could also play a role in the persistent hyperimmune activation status seen in many HIV-infected individuals.
The GC is an ideal location for the transmission, perpetuation, and replication of HIV. FDCs trap and retain large amounts of HIV particles, and these are maintained in an infectious form for many months (7, 17, 22, 33). Not only do FDCs serve as a repository of infectious virus, but this virus contains archived quasispecies that are not found elsewhere (22). FDC-trapped virus also escapes the effects of neutralizing antibody (18). Furthermore, FDCs provide CXCL13, which serves as a chemoattractant for GC lymphocytes, and additional FDC signals to GC T cells increase their expression of CXCR4 (11, 12). As a consequence of altered CXCR4 expression, GC T cells are very sensitive to infection by even small amounts of X4-tropic HIV. Additionally, FDC-CD4+ T-cell interactions lead to the generation of two regulators of G-coupled protein signaling, RGS13 and RGS16, which correlate directly with an impaired migration to CXCL12, a signal outside the GC that may help GC T cells to egress this site (12). This inhibited migration capability likely prolongs the time a given CD4+ T-cell remains in the GC adjacent to FDCs, thereby increasing the likelihood of infection by FDC-trapped virus. The net result of these FDC contributions to the GC microenvironment is to create an ideally suited site where infectious virus, susceptible target cells, and activation signals come together, thereby perpetuating the disease state.
The FDC augmentation of virus transcription may be important to viral pathogenesis in helping to ensure that initial contact of GC T cells with FDC-trapped HIV leads to the de novo establishment of infection followed by virus expression. Additionally, activated lymphocytes that were infected at other sites may migrate into GCs, where they too may be affected by FDC signaling, leading to increased virus expression. In light of the current interest in HIV reservoirs, it is interesting to postulate whether circulating, latently infected T cells that come into contact with FDCs could become activated to the extent needed for virus expression. This question has not yet been tested but seems a reasonable consideration in light of the observation that TNF-α induces latently infected cell lines to express virus (25). Thus, both de novo and previously infected T cells may be affected by FDC production of TNF-α and thereby contribute to HIV expression in vivo. These observations provide a mechanism to explain persisting virus expression in GCs surrounding FDCs (9, 28). A better knowledge of FDC contributions to virus transmission and disease progression will certainly increase our understanding of HIV pathogenesis and may aid in the design of better intervention strategies that specifically target the large and dangerous FDC reservoir of HIV.
Acknowledgments
This research complied with all relevant federal and institutional policies. We acknowledge that we have no financial or other relationships that would pose a conflict of interest relating to the research presented in this study.
This work was supported by Public Health Service grant AI-39963 from the National Institute of Allergy and Infectious Diseases and complied with all relevant federal guidelines and institutional policies.
We gratefully acknowledge the assistance of Kipp M. Robins, Randal B. Gibb, the Utah Valley Regional Medical Center, the Health South Provo Surgical Center, and the Central Utah Surgical Center for providing tissues. We also acknowledge the NIH AIDS Reference and Reagent Program for providing reagents.
Footnotes
Published ahead of print on 29 October 2008.
REFERENCES
- 1.Armstrong, J. A. 1991. Ultrastructure and significance of the lymphoid tissue lesions in HIV infection, p. 69-82. In P. Racz, C. D. Dijkstra, and J. C. Gluckman (ed.), Accessory cells in HIV and other retroviral infections. Karger, Basel, Switzerland.
- 2.Biberfeld, P., A. Porwit, G. Biberfield, M. Harper, A. Bodner, and R. Gallo. 1988. Lymphadenopathy in HIV (HTLV-III LAV) infected subjects: the role of virus and follicular dendritic cells. Cancer Detect. Prev. 12217-224. [PubMed] [Google Scholar]
- 3.Burton, G. F., B. F. Keele, J. D. Estes, T. C. Thacker, and S. Gartner. 2002. Follicular dendritic cell contributions to HIV pathogenesis. Semin. Immunol. 14275-284. [DOI] [PubMed] [Google Scholar]
- 4.Burton, G. F., A. Masuda, S. L. Heath, B. A. Smith, J. G. Tew, and A. K. Szakal. 1997. Follicular dendritic cells (FDC) in retroviral infection: host/pathogen perspectives. Immunol. Rev. 156185-197. [DOI] [PubMed] [Google Scholar]
- 5.Butch, A. W., G. H. Chung, J. W. Hoffmann, and M. H. Nahm. 1993. Cytokine expression by germinal center cells. J. Immunol. 15039-47. [PubMed] [Google Scholar]
- 6.Carey, M., and S. Smale (ed.). 2000. Transcriptional regulation in eukaryotes: concepts, strategies, and techniques, p. 87. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 7.Cavert, W., D. W. Notermans, K. Staskus, S. W. Wietgrefe, M. Zupancic, K. Gebhard, K. Henry, Z. Q. Zhang, R. Mills, H. McDade, C. M. Schuwirth, J. Goudsmit, S. A. Danner, and A. T. Haase. 1997. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science 276960-964. [DOI] [PubMed] [Google Scholar]
- 8.Duh, E. J., W. J. Maury, T. M. Folks, A. S. Fauci, and A. B. Rabson. 1989. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc. Natl. Acad. Sci. USA 865974-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Embretson, J., M. Zupancic, J. L. Ribas, A. Burke, P. Racz, K. Tenner-Racz, and A. T. Haase. 1993. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature 362359-362. [DOI] [PubMed] [Google Scholar]
- 10.Emilie, D., R. Fior, B. Jarrousse, A. Marfaing-Koka, D. Merrien, O. Devergne, M. C. Crevon, M. C. Maillot, and P. Galanaud. 1994. Cytokines in HIV infection. Int. J. Immunopharmacol. 16391-396. [DOI] [PubMed] [Google Scholar]
- 11.Estes, J. D., B. F. Keele, K. Tenner-Racz, P. Racz, M. A. Redd, T. C. Thacker, Y. Jiang, M. J. Lloyd, S. Gartner, and G. F. Burton. 2002. Follicular dendritic cell-mediated up-regulation of CXCR4 expression on CD4 T cells and HIV pathogenesis. J. Immunol. 1692313-2322. [DOI] [PubMed] [Google Scholar]
- 12.Estes, J. D., T. C. Thacker, D. L. Hampton, S. A. Kell, B. F. Keele, E. A. Palenske, K. M. Druey, and G. F. Burton. 2004. Follicular dendritic cell regulation of CXCR4-mediated germinal center CD4 T cell migration. J. Immunol. 1736169-6178. [DOI] [PubMed] [Google Scholar]
- 13.Folkvord, J. M., C. Armon, and E. Connick. 2005. Lymphoid follicles are sites of heightened human immunodeficiency virus type 1 (HIV-1) replication and reduced antiretroviral effector mechanisms. AIDS Res. Hum. Retrovir. 21363-370. [DOI] [PubMed] [Google Scholar]
- 14.Fox, C. H., K. Tenner-Racz, P. Racz, A. Firpo, P. A. Rizzo, and A. S. Fauci. 1991. Lymphoid germinal centers are reservoirs of human immunodeficiency virus type 1 RNA. J. Infect. Dis. 1641051-1057. [DOI] [PubMed] [Google Scholar]
- 15.Fu, Y. X., and D. D. Chaplin. 1999. Development and maturation of secondary lymphoid tissues. Annu. Rev. Immunol. 17399-433. [DOI] [PubMed] [Google Scholar]
- 16.Haase, A. T. 1999. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu. Rev. Immunol. 17625-656. [DOI] [PubMed] [Google Scholar]
- 17.Haase, A. T., K. Henry, M. Zupancic, G. Sedgewick, R. A. Faust, H. Melroe, W. Cavert, K. Gebhard, K. Staskus, Z.-Q. Zhang, P. Dailey, H. H. Balfour, Jr., A. Erice, and A. S. Perelson. 1996. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 274985-989. [DOI] [PubMed] [Google Scholar]
- 18.Heath, S. L., J. G. Tew, J. G. Tew, A. K. Szakal, and G. F. Burton. 1995. Follicular dendritic cells and human immunodeficiency virus infectivity. Nature 377740-744. [DOI] [PubMed] [Google Scholar]
- 19.Huang, L., I. Bosch, W. Hofmann, J. Sodroski, and A. B. Pardee. 1998. Tat protein induces human immunodeficiency virus type 1 (HIV-1) coreceptors and promotes infection with both macrophage-tropic and T-lymphotropic HIV-1 strains. J. Virol. 728952-8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hufert, F. T., J. van Lunzen, G. Janossy, S. Bertram, J. Schmitz, O. Haller, P. Racz, and D. von Laer. 1997. Germinal centre CD4+ T cells are an important site of HIV replication in vivo. AIDS 11849-857. [DOI] [PubMed] [Google Scholar]
- 21.Jones, K. A., and B. M. Peterlin. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63717-743. [DOI] [PubMed] [Google Scholar]
- 22.Keele, B. F., L. Tazi, S. Gartner, Y. Liu, T. B. Burgon, J. D. Estes, T. C. Thacker, K. A. Crandall, J. C. McArthur, and G. F. Burton. 2008. Characterization of the follicular dendritic cell reservoir of human immunodeficiency virus type 1. J. Virol. 825548-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klaus, G. G. B., J. H. Humphrey, A. Kunkl, and D. W. Dongworth. 1980. The follicular dendritic cell: its role in antigen presentation in the generation of immunological memory. Immunol. Rev. 533-28. [DOI] [PubMed] [Google Scholar]
- 24.Korner, H., M. Cook, D. S. Riminton, F. A. Lemckert, R. M. Hoek, B. Ledermann, F. Kontgen, B. Fazekas de St. Groth, and J. D. Sedgwick. 1997. Distinct roles for lymphotoxin-alpha and tumor necrosis factor in organogenesis and spatial organization of lymphoid tissue. Eur. J. Immunol. 272600-2609. [DOI] [PubMed] [Google Scholar]
- 25.Kutsch, O., E. N. Benveniste, G. M. Shaw, and D. N. Levy. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 768776-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luciw, P. A. 1996. Human immunodeficiency viruses and their replication, p. 845-916. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott-Raven, Philadelphia, PA.
- 27.Mandel, T. E., R. P. Phipps, A. Abbot, and J. G. Tew. 1980. The follicular dendritic cell: long term antigen retention during immunity. Immunol. Rev. 5329-59. [DOI] [PubMed] [Google Scholar]
- 28.Pantaleo, G., C. Graziosi, J. F. Demarest, L. Butini, M. Montroni, C. H. Fox, J. M. Orenstein, D. P. Kotler, and A. S. Fauci. 1993. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature 362355-358. [DOI] [PubMed] [Google Scholar]
- 29.Pasparakis, M., L. Alexopoulou, V. Episkopou, and G. Kollias. 1996. Immune and inflammatory responses in TNF alpha-deficient mice: a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J. Exp. Med. 1841397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pope, M., N. Gallo, L. Hoffman, S. Gezelter, and R. M. Steinman. 1995. Cutaneous dendritic cell-T cell conjugates are actively infected with HIV-1. J. Cell. Biochem. Suppl. 21B224. [Google Scholar]
- 31.Reinhart, T. A., M. J. Rogan, G. A. Viglianti, D. M. Rausch, L. E. Eiden, and A. T. Haase. 1997. A new approach to investigating the relationship between productive infection and cytopathicity in vivo. Nat. Med. 3218-221. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz, J., J. van Lunzen, K. Tenner-Racz, G. Grossschupff, P. Racz, H. Schmitz, M. Dietrich, and F. T. Hufert. 1994. Follicular dendritic cells retain HIV-1 particles on their plasma membrane, but are not productively infected in asymptomatic patients with follicular hyperplasia. J. Immunol. 1531352-1359. [PubMed] [Google Scholar]
- 33.Smith, B. A., S. Gartner, Y. Liu, A. S. Perelson, N. I. Stilianakis, B. F. Keele, T. M. Kerkering, A. Ferreira-Gonzalez, A. K. Szakal, J. G. Tew, and G. F. Burton. 2001. Persistence of infectious HIV on follicular dendritic cells. J. Immunol. 166690-696. [DOI] [PubMed] [Google Scholar]
- 34.Steinman, R. M., A. Granelli-Piperno, M. Pope, C. Trumpfheller, R. Ignatius, G. Arrode, P. Racz, and K. Tenner-Racz. 2003. The interaction of immunodeficiency viruses with dendritic cells. Curr. Top. Microbiol. Immunol. 2761-30. [DOI] [PubMed] [Google Scholar]
- 35.Tew, J. G., M. H. Kosco, G. F. Burton, and A. K. Szakal. 1990. Follicular dendritic cells as accessory cells. Immunol. Rev. 117185-211. [DOI] [PubMed] [Google Scholar]
- 36.Tew, J. G., M. H. Kosco, and A. K. Szakal. 1989. The alternative antigen pathway. Immunol. Today 10229-231. [DOI] [PubMed] [Google Scholar]