Abstract
Herpesvirus saimiri (HVS) establishes a persistent infection in squirrel monkeys by maintaining its episome within T lymphocytes. The product of open reading frame 73 (ORF73) plays a key role in episomal maintenance and is the functional homologue of Epstein-Barr virus EBNA1 and Kaposi's sarcoma-associated herpesvirus LANA1 proteins. There is little sequence homology among these proteins, although all contain a central domain of repeating amino acids. The repeat domains of EBNA1 and LANA1 enhance the stability of these proteins and cause a retardation in self-protein synthesis, leading to poor recognition by CD8+ cytotoxic T lymphocytes (CTL). The HVS ORF73 repeat domain is composed of a glutamic acid and glycine repeat linked to a glutamic acid and alanine repeat (EG-EA repeat). Here we show that the EG-EA repeat similarly causes a reduction in the recognition of ORF73 by CD8+ CTL. However, deletion of the EG-EA repeat from HVS ORF73 had no affect on the stability of the protein or its rate of translation. In contrast, the presence of the EG-EA repeat was found to decrease the steady-state levels of ORF73 mRNA. The inhibitory properties of the EG-EA repeat were maintained when transferred to a heterologous protein, and manipulation of the repeat revealed that the motif EEAEEAEEE was sufficient to cause a reduction in recognition of ORF73 by CD8+ CTL. Thus, the EG-EA repeat of HVS ORF73 plays a role in immune evasion but utilizes a mechanism distinct from that of the EBNA1 and LANA1 repeats.
Herpesvirus saimiri (HVS), a member of the gammaherpesvirus subfamily, is the prototypic virus of the Rhadinovirus genus and shares significant homology with Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV) (2, 3, 35). Like all herpesviruses, HVS infects its natural host, the squirrel monkey, as a persistent asymptomatic infection (17). A key feature of this life-long infection is the ability of HVS, and other gammaherpesviruses, to establish and maintain a latent infection in a lymphoid population (17, 30, 33).
Central to this life-long infection is maintenance of the viral genome in latently infected cells. Gammaherpesviruses encode a protein essential for episomal maintenance. The best-characterized of these is the Epstein-Barr nuclear antigen 1 (EBNA1). EBNA1 binds to the EBV genome and tethers the viral episome to host chromosomes, ensuring that the episome is maintained and segregated during cell division (27, 46, 47). EBNA1 is present in all forms of EBV latency, presenting a constant target for the host immune response (33). However, EBNA1 has a unique strategy to evade recognition by CD8+ cytotoxic T lymphocytes (CTL). EBNA1 consists of N- and C-terminal domains, required for episomal maintenance and segregation, separated by an internal repeat domain consisting of glycine and alanine amino acids (GAr domain) (3, 27). Masucci and colleagues showed that the GAr domain contributes to protein stability, inhibits proteasome-mediated degradation, and prevents the generation of CD8+ T-cell peptides from the EBNA1 protein (28, 29). More recently, the GAr domain was shown to inhibit EBNA1 self-protein synthesis (49), and it is thought that this reduction in self-synthesis is the main mechanism for limiting presentation of endogenously processed EBNA1 CD8+ CTL epitopes (26, 39, 40, 45).
Functional homologues of EBNA1 are present in the rhadinoviruses KSHV and HVS. The KSHV protein is known as latency-associated nuclear antigen 1 (LANA1). LANA1 has limited sequence homology to EBNA1 but similarly tethers the KSHV genome to cell chromosomes and ensures segregation of the viral episome (4, 5). These functions also map to the N and C termini (6), which are separated by a large repeat domain. In this case the repeat is highly acidic, consisting of imperfect repeats rich in glutamine, glutamic acid, and aspartic acid (32). The acidic domain has been shown to contribute to LANA1 stability, inhibit self-protein synthesis, and also inhibit the presentation of CD8-restricted antigenic peptides (24, 49).
In HVS the equivalent protein is encoded by open reading frame 73 (ORF73). HVS ORF73 binds to the terminal repeat regions of the viral genome, colocalizes with host mitotic chromosomes and, in isolation, can maintain the viral episome in latently infected cells (9, 10, 43). HVS ORF73 has limited sequence homology to either EBNA1 or LANA1, although again it is comprised of three distinct domains, with a large repeat flanked by small N-terminal and larger C-terminal domains (2). The HVS ORF73 repeat domain is analogous to that of LANA1, being composed of acidic amino acids, although it consists of two distinct elements, a glutamic acid- and glycine-rich region linked to a glutamic acid- and alanine-rich region (EG-EA repeat) (2). No function has yet been attributed to this EG-EA repeat. However, one would predict a similar role to that seen for the repeats of EBNA1 and LANA1. Here we show that the EG-EA repeat domain of HVS ORF73 indeed does function to reduce the presentation of CD8+ peptides. However, this repeat domain acts by reducing the amount of ORF73 mRNA rather than by directly affecting protein translation. These findings imply a common role in immune evasion of CD8+ CTL for these amino acid repeats, yet in the case of HVS ORF73 this is mediated by a distinct mechanism.
MATERIALS AND METHODS
Cell lines.
293 cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (DMEM growth medium). 293 cells transfected with H2-Kb (293-Kb) (42), provided by J. Yewdell (NIH), were maintained in DMEM growth medium containing 0.5 mg/ml G418. The B3Z T-cell hybridoma specific for the H2-Kb-restricted SIINFEKL peptide from ovalbumin (OVA) (23), provided by K. Gould (Imperial College London, United Kingdom), was maintained in RPMI medium supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol.
Plasmids.
p73-GFP and p73Δ-GFP contain the full-length HVS ORF73, or ORF73 with the EG-EA repeat deleted (73Δ), and both are tagged with green fluorescent protein (GFP) at the C terminus (19). p73-E3A-GFP contains a region of the EBV EBNA3A protein in place of the EG-EA repeat and was generated as follows. A 200-amino-acid region of EBNA3A, corresponding to amino acids 71 to 270, was amplified using primers 5′-CCCGAATTCGCCGGTGGGTCAGCTCCAGC-3′ and 5′-CCCGAATTCAGGTGTTAAAACTTCCTCCCAG-3′ (EcoR1 sites underlined) and inserted into the EcoR1 site of p73Δ-GFP.
ORF73 constructs with variable numbers of an EEAEEAEEE repeat motif were generated by annealing the oligonucleotides 5′-AATTAGAAGAAGCTGAAGAAGCTGAAGAAG-3′ and 5′-AATTCTTCTTCAGCTTCTTCAGCTTCTTCT-3′ and insertion into p73Δ-GFP cut with EcoR1, generating p73-1R-GFP, containing one copy of the repeat motif. This plasmid was then used for insertion of a second EEAEEAEEE repeat, generating p73-2R-GFP, and likewise to generate p73-4R-GFP (four EEAEEAEEE repeats) and p73-6R-GFP (six EEAEEAEEE repeats).
The H2-Kb-restricted SIINFEKL peptide from OVA was inserted into the N- and C-terminal regions of ORF73 and ORF73Δ by using overlapping oligonucleotides. p73-GFP and p73Δ-GFP were digested with either BamH1 or BstX1 and ligated with the following annealed oligonucleotides: for BamH1, sense 5′-GATCCATAATCAACTTTGAAAAACTGG-3′ and antisense 5′-GATCCCAGTTTTTCAAAGTTGATTATG-3′; for BstX1, sense 5′-TATAATCAACTTTGAAAAACTGGCAGAATC-3′ and antisense 5′-CTGCCAGTTTTTCAAAGTTGATTATAGATT-3′, generating p73-S-B and p73Δ-S-B and p73-S-Bx and p73Δ-S-Bx, respectively. The SIINFEKL peptide was inserted into plasmids p73-1R-GFP and p73-2R-GFP at the C-terminal BstX1 site by using the overlapping oligonucleotides described above, generating plasmids p73-1R-S-Bx and p73-2R-S-Bx.
pOVA-GFP was generated by amplification of the OVA open reading frame from pcDNA-OVA (34), with the primers 5′-GGGCTCGAGATGGGCTCCATCGGCGCAGC-3′ (Xho1 site underlined) and 5′-GGGGGATCCCGAGGGGAAACACATCTGCC-3′ (BamH1 site underlined) and insertion into pEGFP-N1. The EG-EA repeat, here termed 73R, was inserted into the Pst1 site of OVA-GFP as follows: the EG-EA repeat spanning amino acids 55 to 247 was amplified with the primers 5′-GGGCTGCAGAAGCAGCACTAACAGAAGAACAACG-3′ and 5′-GGGCTGCAGCACGTGGAGTACTTGGTCC-3′ (Pst1 sites underlined) and cloned into pOVA-GFP, generating pOVA-P73R. pNP-S-GFP contains the influenza virus nuclear protein (NP) linked to SIINFEKL and GFP and was generated by isolating the NP-S-GFP fragment from pSC11-NP-S-GFP (31) and insertion into pcDNA3.
All complementary constructs, with and without the EG-EA repeat, contain the same 5′ and 3′ untranslated regions. All PCR products and annealed oligonucleotide insertions were sequenced to ensure correct reading frames were maintained, and all constructs were verified by protein expression studies.
Immunoblotting.
293 cells (2 × 105 per well) were seeded in a six-well plate, and 24 h later cells were transfected with 2 μg of plasmid using Polyfect reagent (Qiagen). Cells were harvested 48 h posttransfection, and the protein concentration in total cell extracts was measured using a BCA protein assay kit (Pierce). Twenty μg of protein was resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membrane. Blots were probed for GFP-tagged proteins using the mouse monoclonal antibody GFP-3E1 (Cancer Research UK, Research Services) or for actin using a rabbit polyclonal antibody (Sigma). Proteins were visualized using Pierce ECL reagents.
Analysis of protein half-lives.
293 cells (2 × 106) were transfected with 5 μg of either p73-GFP or p73Δ-GFP and incubated at 37°C for 24 h. Cells were washed twice with medium without methionine or cysteine (starvation medium) and incubated for a further 60 min in starvation medium. Transfected cells were then pulsed with starvation medium containing Tran35S-label (MP Biomedical), before being washed twice and addition of fresh complete medium. Labeled transfected cells were harvested and divided into six equal aliquots, one aliquot was harvested as time point zero and the remaining aliquots were harvested at 2, 4, 8, 12, and 24 h. Cells were lysed in radioimmunoprecipitation buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS) containing the protease inhibitors phenylmethylsulfonyl fluoride (50 μg/ml), pepstatin A (1 μg/ml), leupeptin (1 μg/ml), and A-protinin (1 μg/ml). Insoluble material was removed by centrifugation, and supernatants were incubated with normal mouse serum for 60 min at 4°C, followed by addition of protein G-Sepharose with a further 60-min incubation. Cleared supernatant was then incubated overnight at 4°C with a sheep anti-GFP polyclonal antibody (a gift from Ian Prior, University of Liverpool). Protein G-Sepharose beads were then added and the incubation continued for 90 min. Beads were then collected by centrifugation, washed twice with lysis buffer and twice with phosphate-buffered saline, and then resuspended in SDS loading buffer. Immunoprecipitated protein was eluted from beads by incubation at 95°C for 5 min, the beads were pelleted, and supernatant was transferred to a fresh tube. Equal volumes of eluted protein were separated by SDS-PAGE, and the gels were then dried and exposed by autoradiography.
In vitro transcription/translation.
For in vitro transcription, p73-GFP and p73Δ-GFP were linearized by digestion with Nde1. These DNA templates were then used to generated capped mRNA by in vitro transcription with T7 RNA polymerase and an mMESSAGE mMachine high-yield capped RNA transcription kit (Ambion). In vitro-transcribed RNA was purified using RNeasy purification columns (Qiagen) and the concentration determined by UV spectroscopic measurement at 260 nm. Molar equivalents of each RNA were calculated (based on nucleotide length and concentration) for use in uncoupled in vitro translation with a rabbit reticulocyte lysate system (Promega) and [35S]methionine (Perkin-Elmer). Reaction products were resolved by SDS-PAGE, and gels were dried and exposed by autoradiography.
Antigen presentation assays.
A total of 4 × 105 293-Kb cells were transfected with 0.1 to 2 μg of the indicated plasmid and incubated at 37°C for 20 h before being used as targets in antigen presentation assays with the SIINFEKL-specific B3Z T-cell hybridoma. Target cells (2 × 104) were incubated for a further 20 h at 37°C in triplicate with 2 × 104 B3Z cells in a final volume of 200 μl. One hundred microliters of supernatant was then harvested and assayed for interleukin-2 (IL-2) by using a mouse IL-2 DuoSet enzyme-linked immunosorbent assay kit (R&D Systems). Absorbance at 450 nm was measured using a MultiSkan Spectrum microplate reader (ThermoElectron).
Analysis of RNA.
Total RNA was prepared from 293 cells transfected with the indicated plasmid by using RNeasy purification columns (Qiagen) and an extra on-column DNase treatment step to maximize removal of plasmid DNA, and the RNA concentration was determined by UV spectroscopic measurement at 260 nm. RNA integrity was confirmed by determining optical density ratios at 260 and 280 nm (OD260/OD280) (>1.8) and by visualization on agarose gels. Where indicated, transfected cells were treated with 5 μg/ml actinomycin D (ActD; Sigma), cells were harvested at either 90 or 150 min post-ActD treatment, and RNA was isolated as described above. Northern blot analysis was carried out by glyoxalation of RNA samples and electrophoresis as described previously (36). RNA was transferred to nitrocellulose and hybridized with a 32P-labeled probe corresponding to the C terminus of ORF73 (nucleotides 634 to 1170). Duplicate filters were probed for glyceraldehyde-3- phosphate dehydrogenase (GAPDH) as a loading control. Filters were washed with 0.2× SSC-0.1% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and exposed by autoradiography. Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed using an iQ5 real-time PCR detection system (Bio-Rad). Initially, cDNA was synthesized from 1 μg RNA by using a reverse transcription system (Promega). Control cDNA conversions lacking reverse transcriptase were also carried out for each RNA sample to monitor for plasmid DNA contamination. qRT-PCRs were carried out in triplicate with 1 μl of cDNA, 300 nmol primers, and IQ Sybr green supermix. Samples underwent 40 cycles of amplification at 94°C (30 s) and 55°C (30 s), fluorescence was read at 55°C, and melt curves were analyzed. For each sample, the Ct values for test genes were normalized to that for the reference gene β-actin (ACTB), and relative expression is represented as 2ΔCt. Primers used for qRT-PCR were as follows; for ORF73, forward 5′-CAAAATCTTGGTGTTGGGTTG-3′ and reverse 5′-AGCAGTTTGGAATGTTCTGC-3′; for OVA-GFP, forward 5′-CTGTGCAGATGATGTACCAG-3′ and reverse 5′-TTCCTCTCTTCCATAACATTA-3′; for ACTB, forward 5′-CACCTTCTACAATGAGCTGCGTGTG-3′ and reverse 5′-ATAGCACAGCCTGGATAGCAACGTAC-3′.
RESULTS
Deletion of the EG-EA repeat increases steady-state levels of HVS ORF73 protein.
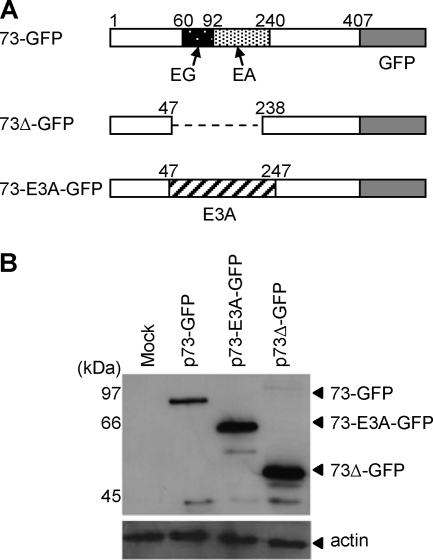
To investigate the role of the HVS ORF73 EG-EA repeat on protein expression, constructs were generated containing either full-length ORF73 (p73-GFP), ORF73 with a deletion of amino acids 47 to 238 spanning the entire EG-EA repeat (p73Δ-GFP), or a chimeric ORF73 where the repeat domain was replaced with a neutral amino acid sequence (p73-E3A-GFP) (Fig. 1A). For all constructs ORF73 was linked at the C terminus to GFP and had the same 5′ and 3′ untranslated regions. Immunoblot analysis of total protein expression 48 h posttransfection of 293 cells revealed that deletion of the EG-EA repeat (73Δ-GFP) caused a significant increase in the steady-state levels of ORF73 protein compared with full-length ORF73 (73-GFP) (Fig. 1B). When the EG-EA repeat was replaced by a nonrepetitive amino acid domain (73-E3A-GFP) containing approximately the same number of amino acids, the level of protein expressed was similar to 73Δ-GFP. Although the predicted molecular weights of 73-GFP and 73-E3A-GFP are approximately equivalent, 73-GFP runs as a higher-molecular-weight protein during SDS-PAGE. This is likely to be a consequence of the acidic nature of the EG-EA repeat domain. Analysis of the transfected cells by flow cytometry revealed comparable transfection efficiencies for each plasmid (73-GFP, 59%; 73-E3A-GFP, 65%; 73Δ-GFP, 55%) and showed the mean fluorescence intensities of 73Δ-GFP and 73-E3A-GFP to be equivalent and significantly higher than 73-GFP (data not shown). This supported the data from immunoblotting and suggests that the observed difference in expression levels was due to the amino acid content of the repeat. This initial experiment implied that the EG-EA repeat was indeed analogous to the internal repeat domains of EBNA1 and LANA1, in that deletion of this repeat caused a significant increase in the steady-sate levels of ORF73 protein.
FIG. 1.
Deletion of the EG-EA repeat from HVS ORF73 leads to an increase in the total amount of ORF73 expressed. (A) Schematic representation of the HVS ORF73 constructs used to investigate the effects of the EG-EA repeat on protein expression. 73-GFP is full-length ORF73, with the EG-EA acidic domain highlighted, tagged with GFP at the C terminus; 73Δ-GFP is ORF73 with the EG-EA repeat deleted and again tagged with GFP; 73-E3A-GFP contains a 200-amino-acid fragment derived from EBV EBNA3A in place of the EG-EA repeat. (B) Immunoblot analysis of expression of 73-GFP, 73Δ-GFP, and 73-E3A-GFP. 293 cells were transfected with the indicated plasmids, and total protein was extracted at 48 h posttransfection. Twenty μg of protein was resolved on 10% SDS-PAGE gels and transferred to nitrocellulose. Duplicate blots were probed with either anti-GFP, to detect the ORF73 constructs, or anti-actin as a loading control.
Presentation of CD8+ T-cell epitopes from HVS ORF73 is inhibited by the EG-EA repeat.
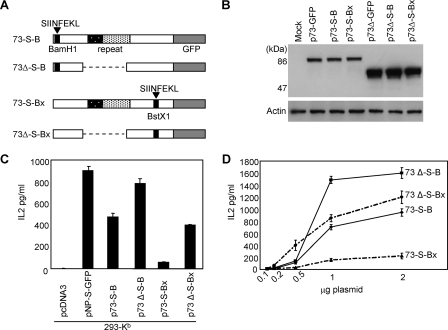
We next investigated if the EG-EA repeat could also inhibit the presentation of CD8+ antigenic peptides. To enable these studies to be carried out we engineered the OVA H2-Kb-restricted peptide SIINFEKL into ORF73, both 5′ (p73-S-B) and 3′ (p73-S-Bx) of the EG-EA repeat, and generated equivalent constructs with the repeat deleted (p73Δ-S-B and p73Δ-S-Bx) (Fig. 2A). Immunoblotting was carried out to confirm that the insertion of the SIINFEKL peptide had no affect on the differential expression of 73 and 73Δ. As seen in Fig. 2B, both 73Δ-S-B and 73Δ-S-Bx were expressed at significantly higher levels than either 73-S-B or 73-S-Bx, indicating that the EG-EA repeat was still effective at reducing ORF73 protein levels in these modified constructs. To investigate presentation of the SIINFEKL peptide, constructs were transfected into 293-Kb cells, which were then used as targets for the SIINFEKL-specific B3Z T-cell hybridoma. Presentation of the SIINFEKL peptide was monitored by assaying release of IL-2 into the supernatant (Fig. 2C). Presentation of the SIINFEKL peptide was inhibited in both constructs containing the EG-EA repeat (73-S-B and 73-S-Bx) compared with the equivalent constructs in which the repeat had been deleted (73Δ-S-B and 73Δ-S-Bx). Importantly, analysis of the target cells by flow cytometry revealed comparable transfection efficiencies for each plasmid (73-S-B, 26%; 73-S-Bx, 30%; 73-S-B, 30%; 73Δ-S-Bx, 32%). A similar result was observed when decreasing amounts of DNA were used to transfect target cells (Fig. 2D). These data indicated that the EG-EA repeat, similar to the GAr domain of EBNA1 and acidic repeat of LANA1, was capable of inhibiting presentation of CD8+ T-cell peptides.
FIG. 2.
The EG-EA repeat of ORF73 inhibits presentation of CD8+ T-cell peptides. (A) Schematic representation of ORF73 with the H2-Kb-restricted SIINFEKL peptide inserted 5′ and 3′ of the EG-EA repeat (73-S-B and 73-S-Bx) and complementary constructs with the EG-EA repeat deleted (73Δ-S-B and 73Δ-S-Bx). All constructs were tagged with GFP at the C terminus. (B) Immunoblot analysis for expression of the ORF73-SIINFEKL chimeras shown in panel A. 293 cells were transfected with the indicated plasmids, and total protein was extracted at 48 h posttransfection. Twenty μg of protein was resolved on 10% SDS-PAGE gels and transferred to nitrocellulose. Duplicate blots were probed as described for Fig. 1. (C) Presentation of the SIINFEKL peptide to B3Z T cells by cells expressing either 73-S-B, 73Δ-S-B, 73-S-Bx, or 73Δ-S-Bx. 293-Kb cells were transfected with p73-S-B, p73Δ-S-B, p73-S-Bx, or p73Δ-S-Bx, and 24 h later these target cells were cocultured with B3Z cells for 18 to 20 h. B3Z T-cell recognition of target cells was monitored by assaying release of IL-2. Control transfections included the empty vector, pcDNA3, or positive control pNP-S-GFP. (D) Presentation of SIINFEKL to B3Z T cells after coculture with 293-Kb cells transfected with various amounts of plasmids p73-S-B, p73Δ-S-B, p73-S-Bx, and p73Δ-S-Bx. Results of T-cell assays represent mean values of IL-2 release into the supernatant measured from triplicate cultures (+ the standard deviation). T-cell data are from one representative experiment out of three.
Deletion of the EG-EA repeat does not affect the stability of ORF73 nor increase the rate of ORF73 mRNA translation.
The EBNA1 and LANA1 repeats have been shown to contribute to the long half-life of each protein and retard self-protein synthesis, properties predicted to be important for the evasion of CD8+ CTL by these proteins. Thus, we next sought to investigate if the observed decrease in ORF73 protein expression and reduction in presentation of CD8+ T-cell epitopes from ORF73 were mediated by the EG-EA repeat through an effect on protein stability and/or the rate of self-protein synthesis. To investigate if the EG-EA repeat had an effect on the stability of ORF73 we carried out a pulse-chase experiment. 293 cells were transfected with p73-GFP or p73Δ-GFP and then pulsed with 35S-labeled methionine and cysteine. Labeled cells were then chased for up to 24 h, with aliquots of cells harvested at 2, 4, 8, 12, and 24 h for immunoprecipitation of labeled ORF73 proteins. A representative immunoprecipitation is shown in Fig. 3A (top panel). Over the 24-h time period examined there was no significant degradation of either 73-GFP or 73Δ-GFP. When the results of three independent pulse-chase experiments were analyzed by densitometry, it was apparent that deletion of the EG-EA repeat did not destabilize the ORF73 protein (Fig. 3A, lower panel). To investigate if EG-EA exerted its effect at the level of protein translation, capped mRNAs for both 73-GFP and 73Δ-GFP were generated in vitro and equimolar amounts were subjected to in vitro translation by using a rabbit reticulocyte lysate system. Surprisingly, both 73-GFP and 73Δ-GFP mRNAs were translated with similar efficiency (Fig. 3B). These results were reproduced in four independent experiments, and densitometric analysis of the four assays showed that the levels of 73-GFP and 73Δ-GFP translation products were within 10% of one other in each case (data not shown). In a parallel experiment using KSHV LANA1 mRNA, with or without the acidic repeat, we observed the reported retardation of translation due to the presence of the repeat (data not shown). Thus, the EG-EA repeat does not appear to influence the rate at which ORF73 mRNA is translated. These data implied that the EG-EA repeat was exerting its effect on protein synthesis and immune evasion via an alternative mechanism to that of the EBNA1 and LANA1 repeats.
FIG. 3.
The EG-EA repeat has no effect on the stability or translation of ORF73. (A) Deletion of the EG-EA repeat does not destabilize ORF73. 293 cells transfected with either p73-GFP or p73Δ-GFP were pulse-labeled with [35S]methionine-cysteine and chased for the indicated times. ORF73 proteins were immunoprecipitated with anti-GFP antibody, resolved on 10% SDS-PAGE gels, and exposed by autoradiography. Mock-transfected cells (M) were used as a negative control. One representative autograph out of three is shown. The amount of [35S]methionine-cysteine-labeled ORF73 proteins in each immunoprecipitation mixture was quantified by analysis of autoradiographs using Image J software (NIH) (lower panel). The value at time zero is shown as 100%, and the percentage of protein remaining at each time point is indicated. The data shown are the means of three independent experiments (± the standard deviation). (B) 73-GFP and 73Δ-GFP RNAs were translated at the same efficiency in uncoupled in vitro translation assays. Translation reactions were performed using equimolar amounts of T7 polymerase transcribed capped RNA. Luciferase RNA was translated as a positive control, and a reaction was performed without adding RNA as a negative control. The data shown are a representative example of four independent assays.
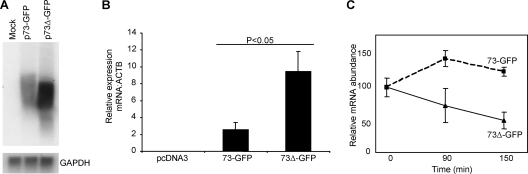
The EG-EA repeat causes a reduction in the steady-state levels of ORF73 mRNA.
An alternative mechanism of action for the EG-EA repeat, which could cause a reduction in the amount of protein expressed, was at the level of ORF73 RNA. Thus, we investigated if deletion of the EG-EA repeat had any effect on ORF73 mRNA levels. Initially we undertook Northern blot analysis of total RNA prepared from 293 cells transfected with either p73-GFP or p73Δ-GFP. As shown in Fig. 4A, deletion of the EG-EA repeat led to a significant increase in ORF73 RNA levels. To be able to accurately quantitate this increase, we investigated the difference in mRNA levels for 73-GFP and 73Δ-GFP by qRT-PCR. mRNA was first converted to cDNA by reverse transcription using an oligo(dT) primer. qRT-PCR was then carried out with primers designed to amplify a region from the C terminus of ORF73. ORF73 mRNA levels were normalized to ACTB, and the results are shown in Fig. 4B. In three separate experiments there was a three- to fivefold increase in 73Δ-GFP mRNA compared with 73-GFP mRNA. qRT-PCR was also carried out on cDNA conversion reaction mixtures in which the reverse transcriptase enzyme had been omitted. There was a greater-than-3,000-fold-lower amplification of 73-GFP or 73Δ-GFP in these reactions than for those containing reverse transcriptase (data not shown), indicating that there was no discernible carryover of plasmid DNA. We next sought to determine if the reduction in steady-state levels of 73-GFP mRNA was due to increased turnover of the ORF73 mRNA. 293 cells were transfected with either p73-GFP or p73Δ-GFP and then 16 h later treated with actinomycin D to inhibit further transcription, and mRNA levels were analyzed over time by qRT-PCR. The results in Fig. 4C demonstrate that, surprisingly, the 73-GFP mRNA was stable over the 150-min time course, whereas 73Δ-GFP mRNA was degraded to approximately 50% of the original level within 150 min. This suggests that the EG-EA repeat has a more complex role in controlling levels of ORF73 mRNA. This is likely to involve a reduction in transcription, but this remains to be determined. Nevertheless, the presence of the EG-EA domain leads to a reduction in the steady-state levels of 73-GFP mRNA, by a mechanism that does not involve increased mRNA turnover, and this is likely responsible for the lower levels of protein expression of 73-GFP compared with 73Δ-GFP.
FIG. 4.
Deletion of the EG-EA repeat increases ORF73 mRNA levels. (A) Northern blot analysis of 73-GFP and 73Δ-GFP RNA in 293 cells transfected with p73-GFP and p73Δ-GFP. Total RNA was prepared 48 h posttransfection, and 15 μg RNA was glyxolated, resolved by agarose gel electrophoresis, and transferred to nitrocellulose. Duplicate filters were probed with 32P-labeled probes corresponding to the 3′ region of ORF73 or, as an internal loading control, GAPDH. (B) qRT-PCR analysis of 73-GFP and 73Δ-GFP mRNA in 293 cells transfected with p73-GFP and p73Δ-GFP. Total RNA was prepared 24 h posttransfection and mRNA converted to cDNA by reverse transcription using an oligo(dT) primer. cDNA was then analyzed by qRT-PCR for 73-GFP or 73Δ-GFP transcripts. Data were normalized to the ACTB reference gene, and 2ΔCt values were calculated to plot relative expression levels. Data shown are the means of a triplicate qRT-PCR analysis (+ the standard deviation) and are one representative example of three independent experiments. (C) Analysis of the effect of the EG-EA repeat on the stability of ORF73 mRNA. 293 cells were transfected with p73-GFP and p73Δ-GFP, followed at 16 h posttransfection by addition of ActD to inhibit transcription. Total RNA was isolated from cells at the indicated times post-ActD treatment and analyzed by qRT-PCR as for panel B. The value at time point zero is shown as 100%, and the percentage of mRNA remaining at each time point is indicated. Data shown are the means of triplicate qRT-PCR analyses (± the standard deviations) and are one representative example of two independent experiments.
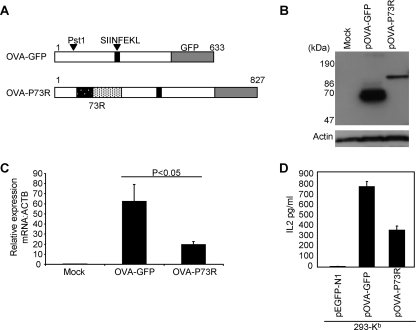
The inhibitory properties of the EG-EA repeat can be transferred to a heterologous protein.
To determine if the HVS ORF73 acidic repeat could function as an independent element we inserted the EG-EA repeat into OVA-GFP, 5′ of the SIINFEKL peptide, generating the construct pOVA-P73R (Fig. 5A). We then investigated the effect of the repeat on OVA-GFP expression, RNA levels, and presentation of the native SIINFEKL peptide to B3Z cells. Insertion of the EG-EA repeat into OVA lead to a decrease in steady-state levels of the chimeric protein (OVA-P73R) when compared with OVA-GFP (Fig. 5B). Aliquots of the same transfected cells analyzed by flow cytometry showed that the transfection efficiency was comparable for each plasmid. By analogy to the observation for ORF73, this was likely to be due to an EG-EA-induced decrease in OVA-P73R mRNA levels. To confirm that this was the case, qRT-PCR was carried out to compare the mRNA levels of OVA-GFP and OVA-P73R in 293 cells transfected with the relevant plasmids. In two separate experiments the presence of the EG-EA repeat reduced OVA mRNA by up to fivefold (Fig. 5C), similar to the reduction seen in the natural ORF73 setting. To determine if this also correlated with a reduction in presentation of CD8+ T-cell epitopes, we investigated the effect of the EG-EA repeat on the native SIINFEKL peptide. Using 293-Kb cells transfected with either pOVA-GFP or pOVA-P73R as targets and coculturing these cells with B3Z T cells, presentation of SIINFEKL was assayed by monitoring IL-2 release. The SIINFEKL peptide was presented efficiently from pOVA-GFP, whereas the insertion of the EG-EA repeat (pOVA-P73R) led to a significant reduction in IL-2 secretion (Fig. 5D). Thus, in this chimeric construct the EG-EA repeat inhibits presentation of SIINFEKL. Taken together these data show that the EG-EA repeat can be transferred to a heterologous protein and on its own is sufficient to reduce the steady-state levels of mRNA and hence protein of the chimeric construct, leading to inhibition of endogenous presentation of CD8+ T-cell peptides.
FIG. 5.
Insertion of the EG-EA repeat into ovalbumin reduces protein expression and mRNA production and inhibits presentation of SIINFEKL. (A) Schematic representation of OVA tagged with GFP at the C terminus (OVA-GFP) and OVA-GFP with the EG-EA repeat (73R) inserted into the Pst1 site (OVA-P73R). (B) Immunoblot analysis of expression of OVA-GFP and OVA-P73R. 293 cells were transfected with either pOVA-GFP or pOVA-P73R or mock transfected as a control, and total protein was extracted at 48 h posttransfection. Twenty μg of protein was resolved on 10% SDS-PAGE gels and transferred to nitrocellulose. Duplicate blots were probed with either an anti-GFP monoclonal antibody, to detect OVA constructs, or an anti-actin polyclonal antibody as a loading control. (C) qRT-PCR analysis of OVA-GFP and OVA-P73R mRNA in 293 cells transfected with pOVA-GFP and pOVA-P73R. Total RNA was prepared 24 h posttransfection and mRNA converted to cDNA by reverse transcription using an oligo(dT) primer. cDNA was then analyzed by qRT-PCR for OVA-GFP or OVA-P73R transcripts. Data were normalized to the ACTB reference gene, and 2ΔCt values were calculated to plot relative expression levels. Data shown are the means of triplicate qRT-PCR analyses (+ the standard deviation) and are one representative example of two independent experiments. (D) Presentation of SIINFEKL peptide to B3Z T cells by cells expressing either OVA-GFP or OVA-P73R. 293-Kb cells were transfected with pOVA-GFP or pOVA-P73R, and 24 h later these target cells were cocultured with B3Z cells for 18 to 20 h. B3Z T-cell recognition of target cells was monitored by assaying release of IL-2. Cells transfected with pEGFP-N1 acted as a negative control. Results represent the mean values of IL-2 released into the supernatant measured from triplicate cultures (+ the standard deviation) and are a representative example from three independent experiments.
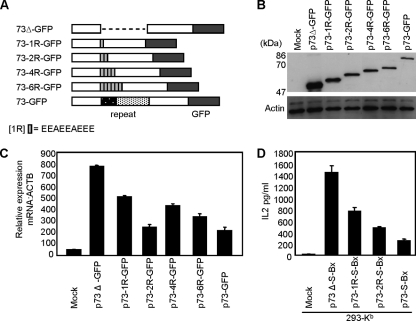
The short motif of EEAEEAEEE is sufficient to inhibit presentation of CD8+ T-cell epitopes.
In an effort to characterize the immune evasion property of the EG-EA repeat further, we sought to determine the minimal sequence required to inhibit presentation of T-cell peptides. We analyzed the amino acid content of the EG-EA domain for the presence of distinct motifs in the two strains of HVS for which ORF73 sequence data were available, A11 used in this study and C488. For strain A11 the EG-EA repeat is composed of a 31-amino-acid EG region and a 149-amino-acid EA region, and C488 is composed of a 139-amino-acid EG region and 132-amino-acid EA region (2, 14). We focused on the more-conserved EA-rich region and determined that the sequence EEA[E]1-3A[E]2-4 occurred multiple times within this region in the A11 strain. Thus, we chose to use as our model repeat motif the sequence EEAEEAEEE and inserted multimers of this into p73Δ-GFP in place of the normal EG-EA repeat. Constructs were generated containing one, two, four, or six copies of the repeat motif (p73-1R-GFP, p73-2R-GFP, p73-4R-GFP, and p73-6R-GFP, respectively) (Fig. 6A). The effects of these multiple short motifs on steady-state protein levels were initially investigated. 293 cells were transfected with the above constructs, along with both 73-GFP and 73Δ-GFP, and immunoblot analysis was carried out. As seen in Fig. 6B, a single motif (73-1R-GFP) was sufficient to reduce the steady-state levels of ORF73 protein. Although not directly quantitative, inclusion of an additional motif (73-2R-GFP) reduced expression further, with little obvious reduction upon addition of further motifs (73-4R-GFP and 73-6R-GFP). When similarly transfected cells were analyzed by flow cytometry for GFP fluorescence, there was a reduction in mean fluorescence intensity compared to 73Δ-GFP of 42% for 73-1R-GFP, 66% for 73-2R-GFP, 68% for 73-4R-GFP, 70% for 73-6R-GFP, and 66% for 73-GFP (data not shown). We then determined if these short motifs also caused a reduction in the steady-state levels of ORF73 mRNA. qRT-PCR was carried out on total RNA prepared from 293 cells transfected with these constructs and compared to 73-GFP and 73Δ-GFP. When compared to 73Δ, mRNA levels were reduced by 35% when one copy of the EEAEEAEEE motif was present (73-1R-GFP), 68% for 73-2R-GFP, 46% for 73-4R-GFP, 57% for 73-6R-GFP, and 62% for 73-GFP (Fig. 6C). This was in general agreement with the protein data and clearly indicated that the EEAEEAEEE motif led to a reduction in steady-state levels of ORF73 mRNA. Based on these data we chose the constructs containing one and two copies of the EEAEEAEEE motif to investigate the effect of this sequence on presentation of CD8+ T-cell epitopes. In experiments similar to those described earlier we inserted the SIINFEKL peptide into the BstX1 site of both 73-1R-GFP and 73-2R-GFP, generating the constructs p73-1R-S-Bx and p73-2R-S-Bx. Analysis of steady-state protein levels confirmed that insertion of the SIINFEKL peptide did not alter the expression profiles of these chimeric ORF73 proteins (data not shown). To investigate presentation of the SIINFEKL peptide, 293-Kb cells were transfected with these constructs along with p73-S-Bx and p73Δ-S-Bx and used as targets for the SIINFEKL-specific B3Z T-cell hybridoma. Both one and two EEAEEAEEE motifs reduced presentation of SIINFEKL to B3Z cells (Fig. 6C). In the representative assay shown, the levels of IL-2 release were reduced by 47% for 73-1R-S-Bx and 67% for 73-2R-S-Bx, in comparison to 82% for the full-length repeat, 73-S-Bx. Thus, it is clear that one EEAEEAEEE motif is sufficient to decrease antigen presentation.
FIG. 6.
The amino acid motif EEAEEAEEE functions to inhibit presentation of CD8+ T-cell peptides. (A) Schematic representation of ORF73 containing one, two, four, or six copies of the EEAEEAEEE repeat motif (p73-1R-GFP, p73-2R-GFP, p73-4R-GFP, and p73-6R-GFP) in place of the full-length EG-EA acidic repeat. (B) Immunoblot analysis of expression of ORF73 containing the short EEAEEAEEE motif. 293 cells were transfected with p73-GFP, p73Δ-GFP, p73-1R-GFP, p73-2R-GFP, p73-4R-GFP, or p73-6R-GFP, and total protein was extracted at 48 h posttransfection. Twenty μg of protein was resolved on 10% SDS-PAGE gels and transferred to nitrocellulose. Duplicate blots were probed as described for Fig. 1. (C) qRT-PCR analysis of ORF73 mRNA in 293 cells transfected with p73-GFP, p73Δ-GFP, p73-1R-GFP, p73-2R-GFP, p73-4R-GFP, or p73-6R-GFP. Total RNA was prepared 24 h posttransfection, and mRNA was converted to cDNA by reverse transcription using an oligo(dT) primer. cDNA was then analyzed by qRT-PCR for ORF73 transcripts. Data were normalized to the ACTB reference gene, and 2ΔCt values were calculated to plot relative expression levels. Data shown are the means of triplicate qRT-PCR analyses (+ the standard deviation) and are one representative example of two independent experiments. (D) Presentation of SIINFEKL peptide to B3Z T cells by cells expressing either 73-S-Bx, 7 3Δ-S-Bx, 73-1R-S-Bx, or 73-2R-S-Bx. 293-Kb cells were transfected with p73-S-Bx, p73Δ-S-Bx, p73-1R-S-Bx, or p73-2R-S-Bx, and 24 h later these target cells were cocultured with B3Z cells for 18 to 20 h. B3Z T-cell recognition of target cells was monitored by assaying release of IL-2. Cells transfected with pcDNA3 acted as a negative control. Results of T-cell assays represent the mean values of IL-2 release into the supernatant measured from triplicate cultures (+ the standard deviation). T-cell data are from one representative experiment of two.
DISCUSSION
Gammaherpesviruses are extremely efficient at colonizing and persisting within their host species and utilize a number of methods for evasion of host immune responses. The primary strategy during long-term latency is the reduction in expression of viral proteins to only those required to establish and maintain latent infection. In an in vitro model of HVS latency, three ORFs (71 to 73) are expressed (20). ORF71 and ORF72 encode proteins involved in preventing apoptosis (v-FLIP) and cell transformation (v-cyclin D), respectively (11, 41). The third protein, ORF73, plays an essential role in maintaining the viral genome within latently infected cells (10) and is likely to be continually present during HVS latency. Thus, ORF73 will present a constant target for CD8+ CTL. The functional homologues of HVS ORF73 in the human gammaherpesviruses EBV and KSHV have been shown to have a strategy for evasion of CD8+ CTL that relies on the presence of a central domain of repeating amino acids. The GAr domain in EBNA1 and an acidic amino acid domain in LANA1 enhance the stability of these proteins and retard self-protein synthesis, leading to the inhibition of presentation of antigenic peptides to CD8+ CTL (24, 28, 49, 50). HVS ORF73 contains an analogous repeat domain, the EG-EA repeat, and in this study we have shown that it also inhibits presentation of CD8+ T-cell epitopes present in ORF73. However, we have shown that its mechanism is distinct from that of the EBNA1 and LANA1 repeats.
When analyzed at the level of protein expression, deletion of the EG-EA repeat from HVS ORF73 led to an increase in the steady-state levels of the ORF73 protein, which correlated with an increase in the recognition of ORF73 by CD8+ T cells. This was directly comparable with the EBNA1 GAr domain and LANA1 acidic repeat. However, in contrast to the EBV and KSHV proteins, the EG-EA repeat was found to act at the level of RNA rather than protein. HVS ORF73 was found to be a stable protein but, surprisingly, deletion of the EG-EA repeat had no affect on its stability over a 24-h period. We cannot rule out that over a longer chase period an effect due to deletion of the EG-EA repeat would be observed. Interestingly, it should be noted that deletion of the GAr domain from EBNA1 was originally reported to result in a half-life of approximately 20 h (28). However, a recent study suggested that the GAr domain plays no role in preventing EBNA1 degradation and implied that the GAr does not interfere with the proteolytic action of the proteasome (13). These contradictory data remain to be resolved. However, more strikingly, deletion of the acidic repeat from LANA1 resulted in a half-life of approximately 2 h (50). We have not tested whether the EG-EA repeat can inhibit proteasomal degradation; however, under the conditions used in this study it would seem that the EG-EA domain plays no role in stabilizing the ORF73 protein. This lack of turnover of HVS ORF73 can be related to its role in the virus life cycle. Although not formally proven, it seems likely that HVS will have a form of latency equivalent to that reported for EBV (22), where no viral antigens are expressed but ORF73 is present to ensure the viral episome is tethered to the cell chromosome. Therefore, a stable ORF73 protein would be crucial for maintenance of the viral episome in cells that are not actively expressing viral antigens.
The EBNA1 GAr domain and the LANA1 acidic repeat have both been shown to retard self-protein synthesis in in vitro translation assays (24, 49). CD8+ T-cell epitopes are primarily generated from newly synthesized proteins and the turnover of rapidly degraded polypeptides. Thus, a key property of retardation of self-protein synthesis is the reduction in the substrates for the generation of antigenic peptides. However, in this study ORF73 mRNA was translated very efficiently and we were surprised to find that deletion of the EG-EA repeat had no noticeable effect on the rate of translation. This is an important observation, because although the EG-EA repeat does not regulate self-protein synthesis it does have a role in protecting ORF73 from recognition by CD8+ CTL. These data implied that control of ORF73 protein levels, and subsequent generation of CD8+ T-cell peptides, was not at the level of protein stability or translation but involved a mechanism unique to HVS ORF73. We have shown here that the EG-EA repeat exerts its effect on the steady-state levels of mRNA. Analysis of the levels of mRNA in cells transfected with either 73-GFP or 73Δ-GFP showed that the presence of the EG-EA repeat caused up to a fivefold reduction in ORF73 mRNA. It is assumed that the reduction in mRNA levels due to the presence of the EG-EA repeat influences the generation of defective ribosomal products (DRiPs). DRiPs, aberrant translation products generated from newly synthesized proteins, are now accepted as the main source of antigenic peptides (48). Thus, the reduction in ORF73 mRNA levels due to the EG-EA repeat (compared to 73Δ) will lead to a reduction in the amount of protein translated (rather than the rate of translation), and this will reduce the generation of ORF73 DRiPs and subsequently of antigenic peptide. This remains to be formally proven; however, if this is indeed the case, it is analogous to the repeats of EBNA1 and LANA1, although by a novel mechanism. Importantly, when transferred to the model protein OVA, the EG-EA repeat alone was sufficient to reduce the steady-state levels of OVA mRNA and protein and was capable of inhibiting the presentation of the native SIINFEKL peptide. Thus, at least for this in vitro model no other region of the ORF73 is required.
The mechanism by which the EG-EA repeat causes a reduction in the steady-state levels of ORF73 mRNA was predicted to be via either destabilizing the mRNA or inhibiting ORF73 transcription. Surprisingly, analysis of the effect of the repeat on mRNA stability revealed that ORF73 mRNA was stable over a period of 150 min, whereas ORF73Δ mRNA levels decayed by approximately 50% over this period. Thus, this implies that the EG-EA repeat actually stabilizes ORF73 mRNA, suggesting a complex mechanism for controlling steady-state levels of ORF73 mRNA. This may include a reduction in transcription rates; however, this remains to be determined. While it will be of interest in the future to fully characterize the mechanism of action, the data reported here are important observations toward elucidating how HVS ORF73 evades recognition of CD8+ CTL. Of particular relevance to this study is a recent report from Tellam and colleagues (38). Based on an earlier observation that the EBNA1 GAr domain utilizes codons which are heavily biased toward purines at position 3, as opposed to pyrimidines for other EBV genes (12), Tellam et al. showed that this purine loading of the GAr domain influences the mRNA structure. When they changed the codon usage to a more humanized pyrimidine bias, they reported a significant increase in the secondary structure of the GAr RNA, which enhanced the translation of EBNA1 and subsequently the generation of CD8+ antigenic peptides. How do these data impact on HVS ORF73? First, the purine bias of codons in the GAr domain did not affect the steady-state levels of EBNA1 mRNA but rather influenced the rate of self-protein translation. In our study the EG-EA repeat clearly had no effect on translation but did reduce the steady-state levels of ORF73 mRNA. Second, the overall codon usage within the ORF73 of HVS strain A11 used in this study, although not using the most frequent human codons, is less biased to either purines or pyrmidines. Within the EG-EA repeat, for alanine there is a bias for pyrimidine codons (95%), but this is offset by the glutamic acid and glycine codons, which are almost exclusively purine based (99%). This supports the experimental data showing that the EG-EA repeat has a different mechanism of action compared to the EBNA1 GAr.
We mapped the inhibitory function of the EG-EA repeat to a short motif, EEAEEAEEE, stemming from the larger EA repeat of HVS strain A11. We have shown that one copy of this motif is sufficient to reduce steady-state levels of ORF73 protein and mRNA, and although a greater degree of inhibition was seen when two motifs were combined this was not enhanced by addition of further copies. Significantly, there was also a reduction in presentation of the SIINFEKL peptide due to the presence of one EEAEEAEEE motif, and the addition of a second copy of the motif reduced presentation levels further. We did not look at the effect of addition of further copies, and it may be that an increase in inhibition would be seen. However, it is clear that the EEAEEAEEE sequence maintains the ability to reduce steady-state levels of ORF73 protein and mRNA and, importantly, inhibit antigen presentation. These data are consistent with the reports for EBNA1 and LANA1 showing that the inhibitory properties of their repeats can be mapped to short elements from within the larger repeats (24, 49). This is intriguing, given the different amino acid contents and proposed mechanisms of action of these distinct repeats. Moreover, large amino acid repeat domains are present in the two HVS strains sequenced (2, 14) and in all EBV and KSHV virus isolates studied to date (16, 18), perhaps suggesting there is a requirement for larger repeats to be effective in vivo or indeed indicating additional functionality yet to be mapped to the large repeat domains.
The conservation of amino acid repeat domains and the associated immune evasion functions within these proteins clearly suggest a vital role for this function for long-term latency of these viruses. ORF73 homologues have been found in the majority of gammaherpesviruses characterized, and internal amino acid repeat domains have been identified in many of these proteins (1, 8, 15, 21). However, no homologue for ORF73 has been found in equine herpesvirus 2 (37), while in bovine herpesvirus 4 and murine gammaherpesvirus 68 (MHV-68) the ORF73 proteins lack a large internal repeat domain (25, 44). Although, by a mechanism analogous to EBNA1 and LANA1, MHV68 ORF73 is also protected from recognition by CD8+ CTL due to a retardation of self-protein synthesis (7). It is worthy of comment that many viral proteins contain regions of repeating amino acids, and although the exact sequence may vary and different mechanisms may be utilized, this may be a common immune evasion strategy that has evolved for unrelated proteins. Thus, the presence of such repeating amino acid sequences prevents the rapid expression of viral antigens and thus delays the production of substrate for processing to generate antigenic peptides, creating an opportunity for the virus to replicate before an efficient immune response targets the infected cell. In the future it may be that by manipulation of such sequences it will be possible to induce or enhance an antiviral T-cell response to help prevent infection.
Acknowledgments
We thank K. Gould, J. Yewdell, I. Prior, and C. Rush for providing various cell lines, antibodies, and plasmids. We acknowledge T. Astill for help generating plasmids p73-S-B and p73Δ-S-B.
This work was supported by the Biotechnology and Biological Sciences Research Council (grant number S16586).
Footnotes
Published ahead of print on 22 October 2008.
REFERENCES
- 1.Albrecht, J.-C. 2000. Primary structure of the herpesvirus ateles genome. J. Virol. 741033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J.-C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittman, M. A. Craxton, H. Coleman, B. Fleckenstein, and R. W. Honess. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 665047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tufnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310207-211. [DOI] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates epsiome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 753250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284641-644. [DOI] [PubMed] [Google Scholar]
- 6.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking system for Kaposi's sarcoma herpesvirus LANA. Science 311856-861. [DOI] [PubMed] [Google Scholar]
- 7.Bennett, N. J., J. S. May, and P. Stevenson. 2005. Gamma-herpesvirus latency requires T cell evasion during episome maintenance. PLoS Biol. 3638-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnside, K. L., J. T. Ryan, H. Bielefeldt-Ohmann, A. G. Bruce, M. E. Thouless, C.-C. Tsai, and T. M. Rose. 2006. RFHVMn ORF73 is structurally related to the KSHV ORF73 latency-associated nuclear antigen (LANA) and is expressed retroperitoneal fibromatosis (RF) tumor cells. Virology 354103-115. [DOI] [PubMed] [Google Scholar]
- 9.Calderwood, M. A., K. T. Hall, D. A. Matthews, and A. Whitehouse. 2004. The herpesvirus saimiri ORF73 gene product interacts with host-cell mitotic chromosomes and self-associates via its C terminus. J. Gen. Virol. 85147-153. [DOI] [PubMed] [Google Scholar]
- 10.Calderwood, M., R. E. White, R. A. Griffiths, and A. Whitehouse. 2005. Open reading frame 73 is required for herpesvirus saimiri A11-S4 episomal persistence. J. Gen. Virol. 862703-2708. [DOI] [PubMed] [Google Scholar]
- 11.Chang, Y. P., P. S. Moore, S. J. Talbot, C. H. Boshoff, T. Zarkowska, K. Godden, H. Paterson, R. A. Weiss, and S. Mittnacht. 1996. Cyclin encoded by KS herpesvirus. Nature 382410. [DOI] [PubMed] [Google Scholar]
- 12.Cristillo, A. D., J. R. Mortimer, I. H. Barratte, T. P. Lillicrap, and D. R. Forsdyke. 2001. Double-stranded RNA as a not-self alarm signal: to evade, most viruses purine-load their RNAs but some (HTLV-1, Epstein-Barr) pyrmidine-load. J. Theor. Biol. 208475-491. [DOI] [PubMed] [Google Scholar]
- 13.Daskalogianni, C., S. Apcher, M. M., Candeias, N. Naski, F. Calvo, and R. Fahraeus. 29 August 2008, posting date. GLY-ALA repeats induce position and substrate specific regulation of 26S proteasome-dependent partial processing. J. Biol. Chem. doi: 10.1074/jbc.M803290200. [DOI] [PMC free article] [PubMed]
- 14.Ensser, A., M. Thurau, S. Wittmann, and H. Fickenscher. 2003. The genome of herpesvirus saimiri C488 which is capable of transforming human T cells. Virology 314471-487. [DOI] [PubMed] [Google Scholar]
- 15.Ensser, A., R. Pflanz, and B. Fleckenstein. 1997. Primary structure of the alcelaphine herpesvirus 1 genome. J. Virol. 716517-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falk, K., J. W. Gratama, M. Rowe, J. Z. Zou, F. Khanim, L. S. Young, M. A. Oosterveer, and I. Ernberg. 1995. The role of repetitive DNA sequences in the size variation of Epstein-Barr virus (EBV) nuclear antigens, and the identification of different EBV isolates using RFLP and PCR analysis. J. Gen. Virol. 76779-790. [DOI] [PubMed] [Google Scholar]
- 17.Fickenscher, H., and B. Fleckenstein. 2001. Herpesvirus saimiri. Philos. Trans. R. Soc. Lond. B 356545-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, S.-J., Y.-J. Zhang, J.-H. Deng, C. S. Rabkin, O. Flore, and H. B. Jenson. 1999. Molecular polymorphism of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent nuclear antigen: evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J. Infect. Dis. 1801466-1476. [DOI] [PubMed] [Google Scholar]
- 19.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, A. F. Markham, and A. Whitehouse. 2000. Characterization of the herpesvirus saimiri ORF73 gene. J. Gen. Virol. 812653-2658. [DOI] [PubMed] [Google Scholar]
- 20.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, I. M. Carr, A. J. Stevenson, A. F. Markham, and A. Whitehouse. 2000. Analysis of gene expression in a human cell line stably transduced with herpesvirus saimiri. J. Virol. 747331-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hart, J., M. Ackermann, G. Jayawardane, G. Russell, D. M. Haig, H. Reid, and J. P. Stewart. 2007. Complete sequence and analysis of the ovineherpesvirus 2 genome. J. Gen. Virol. 8828-39. [DOI] [PubMed] [Google Scholar]
- 22.Hochberg, D., J. M. Middeldorp, M. Catalina, J. L. Sullivan, K. Luzuriaga, and D. A. Thorley-Lawson. 2004. Demonstration of the Burkitt's lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo. Proc. Natl. Acad. Sci. USA 101239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karttunen, J., S. Sanderson, and N. Shastri. 1992. Detection of rare antigen-presenting cells by the lacZ T-cell activation assay suggests an expression cloning strategy for T-cell antigens. Proc. Natl. Acad. Sci. USA 896020-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwun, H. J., S. Ramos da Silva, I. Shah, N. Blake, P. S. Moore, and Y. Chang. 2007. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J. Virol. 818225-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomonte, P., M. Bublot, V. van Santen, G. M. Keil, P. Pastoret, and E. Thiry. 1995. Analysis of bovine herpesvirus 4 genomic regions located outside the conserved gammaherpesvirus gene blocks. J. Gen. Virol. 761835-1841. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. P., J. M. Brooks, H. Al-Jarrah, W. A. Thomas, T. A. Haigh, G. S. Taylor, S. Humme, A. Schepers, W. Hammerschmidt, J. L. Yates, A. B. Rickinson, and N. W. Blake. 2004. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J. Exp. Med. 1991409-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leight, E. R., and B. Sugden. 2000. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 1083-100. [DOI] [PubMed] [Google Scholar]
- 28.Levitskaya, J., A. Sharipo, A. Leonchikis, A. Ciechanover, and M. G. Masucci. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. USA 9412616-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levitskay, J., M. Coram, V. Levitsky, S. Imreh, P. M. Steigerwald-Mullen, G. Klein, M. G. Kurilla, and M. G. Masucci. 1995. Inhibition of antigen processing by the internal repeat region of Epstein-Barr virus nuclear antigen-1. Nature 375685-688. [DOI] [PubMed] [Google Scholar]
- 30.Moore, P. S., and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus, p. 2803-2834. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 31.Norbury, C. C., D. Malide, J. S. Gibbs, J. R. Bennink, and J. W. Yewdell. 2002. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat. Immunol. 3265-271. [DOI] [PubMed] [Google Scholar]
- 32.Rainbow, L., G. Platt, G. Simpson, R. Sarid, S. Gao, H. Stoiber, C. Herrington, P. Moore, and T. Schulz. 1997. The 222- to 234-kilodalton latent nuclear antigen (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by ORF73 and is a component of the latency-associated nuclear antigen. J. Virol. 715915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2628. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 34.Rush, C., T. Mitchell, and P. Garside. 2002. Efficient Priming of CD4 and CD8 T cells by DNA vaccination depends on appropriate targeting of sufficient levels of immunologically relevant antigen to appropriate processing pathways. J. Immunol. 1694951-4960. [DOI] [PubMed] [Google Scholar]
- 35.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 9314862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbour, NY.
- 37.Telford, E. A. R., M. S. Watson, H. C. Aird, J. Perry, and A. J. Davison. 1995. The DNA sequence of equine herpesvirus 2. J. Mol. Biol. 249520-528. [DOI] [PubMed] [Google Scholar]
- 38.Tellam, J., C. Smith, M. Rist, N. Webb, L. Cooper, T. Vuocolo, G. Connolly, D. C. Tscharke, M. P. Devoy, and R. Khanna. 2008. Regulation of protein translation through mRNA structure influences MHC class I loading and T cell recognition. Proc. Natl. Acad. Sci. USA 1059319-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tellam, J., G. Connolly, K. J. Green, J. J. Miles, D. J. Moss, S. R. Burrows, and R. Khanna. 2004. Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1. J. Exp. Med. 1991421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tellam, J., M. H. Fogg, M. Rist, G. Connolly, D. Tscharke, N. Webb, L. Heslop, F. Wang, and R. Khanna. 2007. Influence of translation efficiency of homologous viral proteins on the endogenous presentation of CD8+ T cell epitopes. J. Exp. Med. 204525-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, C. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386517-521. [DOI] [PubMed] [Google Scholar]
- 42.Tscharke, D. C., G. Karupiah, J. Zhou, T. Palmore, K. R. Irvine, S. M. Mansour Haeryfar, S. Williams, J. Sidney, A. Sette, J. R. Bennink, and J. W. Yewdell. 2005. Identification of poxvirus CD8 T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 20195-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma, S. C., and E. S. Robertson. 2003. ORF73 of herpesvirus saimiri strain C488 tethers the viral genome to metaphase chromosomes and binds to cis-acting DNA sequences in the terminal repeats. J. Virol. 7712494-12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpevirus 68. J. Virol. 715894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voo, K. S., T. Fu, H. Y. Wang, J. Tellam, H. E. Heslop, M. K. Brenner, C. M. Rooney, and R.-F. Wang. 2004. Evidence for the presentation of major histocompatibility complex class I-restricted Epstein-Barr virus nuclear antigen 1 peptides to CD8+ T lymphocytes. J. Exp. Med. 199459-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yates, J., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313812-815. [DOI] [PubMed] [Google Scholar]
- 47.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 813806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yewdell, J. W., U. Schubert, and J. R. Bennink. 2001. At the crossroads of cell biology and immunology: DRiPs and other sources of peptide ligands for MHC class I molecules. J. Cell Sci. 114845-851. [DOI] [PubMed] [Google Scholar]
- 49.Yin, Y., B. Manoury, and R. Fahraeus. 2003. Self-inhibition of synthesis and antigen presentation by the Epstein-Barr virus-encoded EBNA1. Science 3011371-1374. [DOI] [PubMed] [Google Scholar]
- 50.Zaldumbide, A., M. Ossevoort, E. J. H. J. Wietrz, and R. C. Hoeben. 2007. In cis inhibition of antigen processing by the latency-associated nuclear antigen I of Kaposi sarcoma herpes virus. Mol. Immunol. 221352-1360. [DOI] [PubMed] [Google Scholar]