Abstract
Kaposi's sarcoma-associated herpesvirus-encoded microRNA (miRNA) MiR-K12-11 was recently shown to be a functional ortholog of miR-155, a miRNA that plays a major role in lymphoid malignancies and the modulation of immune responses. Here we show that miR-M4, encoded by the highly oncogenic Marek's disease virus of chickens, shares common targets with miR-155 and thus is also a functional ortholog of miR-155, the first one identified in an alphaherpesvirus. The observation that two distinct oncogenic herpesviruses associated with distinct types of lymphomas in different species encode functional miR-155 orthologs suggested the importance of this miRNA in regulatory pathways and the biology of lymphomagenesis.
MicroRNAs (miRNAs) constitute a large family of small noncoding RNAs functioning as major regulators of gene expression in several eukaryotes (1, 22) as well as in a number of viruses, particularly in the members of the family Herpesviridae (10, 17, 20). Among the numerous miRNAs expressed in hematopoietic cells, miR-155 was shown to have the most wide-ranging effects on the biology of lymphocytes (7, 29, 30). An association of miR-155 with various types of malignancies has also been demonstrated in several studies (8, 9, 15, 21, 26-28). Although the precise molecular mechanisms by which miR-155 modulates lymphocyte transformation are not clear, it is suggested to be a combinatorial repression of a broad range of genes such as the PU.1, BACH-1, and CEBPβ genes (18, 22).
Compared to the metazoan miRNAs, which are often highly conserved between species, virus-encoded miRNAs generally do not share sequence homologies with other virus- or host-encoded miRNAs (6, 17, 34). However, partial sharing of sequences, particularly in the target interaction region, can result in the conservation of miRNA functions between virus- and host-encoded miRNAs. Recent studies have demonstrated that Kaposi's sarcoma herpesvirus (KSHV)-encoded KSHV-miR-K12-11 can modulate the some of the target genes that are repressed by miR-155, thereby acting as a functional ortholog of miR-155 (11, 16, 24). As part of a study to look at the functional conservation of virus- and host-encoded miRNAs, we examined the miRNAs encoded by the oncogenic Marek's disease virus (MDV) (3-5, 34, 35) for any sequence homologies with miRNAs listed in miRBase (http://microrna.sanger.ac.uk/). One of the MDV type 1 (MDV-1)-encoded miRNAs, MDV-miR-M4, shared perfect seed sequence with gga-miR-155 and with KSHV-miR-K12-11, demonstrating its potential as a functional ortholog of miR-155. We examined whether MDV-1-miR-M4 and gga-miR-155 shared a common set of target genes by use of a recently developed miRNA target prediction algorithm, MirTarget2 (32, 33). Several of the predicted targets of MDV-1-miR-M4 (see Table S1 in the supplemental material) were common to those already identified as gga-miR-155 targets (http://mirdb.org/cgi-bin/search.cgi). Among the predicted targets, PU.1 (SPI-1), C/EBPβ, and HIVEP2 (Schnurri-2) have been validated experimentally as targets of miR-155 and the KSHV-miR-K12-11 ortholog (11, 16, 24, 30, 36). Nearly all of the predicted targets showed high sequence homology to the complementary target miRNA response element (MRE), with the sequences showing conservation between chicken and human genes (see Fig. S1 in the supplemental material), demonstrating the potential of MDV-1-miR-M4 to regulate at least some of the gga-miR-155 target genes.
In order to validate the predicted targets experimentally, we generated expression vectors for both miRNAs (Fig. 1). Sequences of all the oligonucleotides used are shown in Table S2 in the supplemental material. In the gga-miR-155 expression vector, the EF1α promoter drives a partial BIC sequence from exon 2, with sequences 50 bp upstream and ∼300 bp downstream of the miR-155 precursor (Fig. 1E and F). An identical vector driving the expression of MDV-1-miR-M4 from the EF1α promoter was also constructed with sequences ∼100 bp upstream and ∼500 bp downstream of the precursor (Fig. 1C). We also generated an expression vector of the whole miRNA cluster (miR-M12, miR-M5, miR-M3, miR-M2, and miR-M4) driven by the cytomegalovirus (CMV) promoter in the pcDNA3.1/myc-His vector (Fig. 1B). For the construction of a miRNA-negative expression vector, we synthesized a 1,445-nucleotide NgoMIV-EcoRV fragment (CodonDevices) corresponding to the position of 134780 to 136225 in the RB-1B strain (accession number EF523390) of the MDV sequence (25) in which all the miRNAs were mutated to prevent the formation of a miRNA hairpin, at the same time retaining the potential R-LORF8 open reading frame in the antisense direction from that region (Fig. 1D). The mutant region was amplified by PCR using MDV-miR cluster For and Rev primers and cloned into the pcDNA3.1/myc-His vector. The expression of miRNAs from these constructs was confirmed by Northern blotting or quantitative real-time PCR analysis (not shown).
FIG. 1.
Schematic diagram of miRNA expression and luciferase reporter vectors. (A) Genomic organization of MDV miRNA cluster 1, with miRNA names indicated above the hairpin loops. The position of the antisense R-LORF8 transcript is indicated. (B) Expression vector construct of MDV miRNA cluster 1 in the pcDNA3.1-V5/His vector under the control of the CMV promoter. (C) MDV miR-M4 cloned into the pEF6-TOPO expression vector driven from the EF1α promoter. (D) Expression vector of mutant MDV cluster 1, showing mutated miRNAs but retaining the RLORF-8 open reading frame. (E) Diagram representing the chicken BIC transcript showing the miR-155 loop structure. (F) miR-155 cloned into the pEF6-TOPO expression vector downstream of the EF1α promoter. (G) Luciferase reporter vector containing three tandem MRE sequences downstream of the simian virus 40 promoter-driven Renilla luciferase cassette from the psiCHECK-2 vector. The firefly (Firfly) luciferase cassette driven independently by the TK promoter was used as the internal control. SV40, simian virus 40.
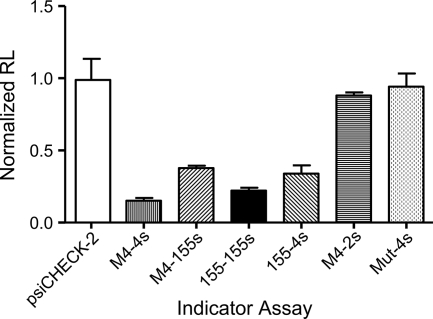
We next examined the biological activity of a specific miRNA by use of an indicator reporter assay by placing perfect complementary target sequence of the miRNA into the 3′ untranscribed region (3′UTR) of a reporter gene so that it can function as a small interfering RNA target. We constructed sensor vectors for both miR-M4 and miR-155 by introducing antisense complementary binding sites within the 3′UTR of the Renilla luciferase reporter in the psiCHECK-2 vector (Fig. 1G). Luciferase reporter assays were carried out with DF-1 cells (12) in 96-well plates (105 cells/well) transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. Firefly and Renilla luciferase activities were measured consecutively with the dual luciferase reporter assay system (Promega) by use of a Lucy 1 luminometer (Anthos Labtec). In all cases, a constitutively expressed firefly luciferase activity in the psiCHECK-2 vector served as a normalization control for transfection efficiency. Cotransfection of each miRNA expression vector with the respective miR-M4 and miR-155 sensor vectors (10:1 ratio) resulted in 75 to 85% knockdown of Renilla luciferase activity for pEF6-miR-M4:pmiR-M4 sensor (M4-4s) or pEF6-miR-155:pmiR-155 sensor (155-155s) combinations (Fig. 2). The pEF6-miR-M4:pmiR-155 sensor (M4-155s) and pEF6-miR-155:pmiR-M4 sensor (155-4s) combinations showed reciprocal knockdown efficiencies of approximately 60 to 65%. Notably, the negative controls, pEF6miR-M4:pmiR-M2 sensor (M4-2s) and mutant miRNA:pmiR-M4 sensor (Mut-4s), showed no repression effect (Fig. 2). These assays demonstrated that miR-M4 and miR-155 can inhibit the expression of their own and each other's sensor vectors.
FIG. 2.
miRNA indicator assay. The sensor plasmid with three-repeat reverse complementary sequence corresponding to miR-M4 (4s) or miR-155 (155s) or control miR-M2 (2s) was inserted into the end of Renilla luciferase of the psiCHECK-2 vector, as indicated in Fig. 1G. The normalized Renilla luciferase levels (RL) of the different miRNA-sensor pairs relative to that seen for the empty vector are shown.
Having shown that miR-M4 and miR-155 are functional in the chicken cells through indicator assays, we asked whether these miRNAs can downregulate any of the predicted targets. For the functional identification of miRNA target genes, we used reporter constructs containing three copies of the MRE from the 3′UTR of the predicted target gene downstream of the Renilla luciferase in the psiCHECK-2 vector (Fig. 1G). The use of such reporter constructs with multiple MREs has proven to be a valid functional method for the identification of several miRNA targets (13, 14, 19, 23, 24, 31). Reporter assays using high ratios (20:1) of miRNA expression to reporter plasmids showed very good silencing of the predicted targets for both gga-miR-155 and MDV-1-miR-M4, respectively, when expressed either individually or as part of the MDV-1-miR cluster 1 (Fig. 3). Restoration of the Renilla luciferase levels in the mutant reporter constructs with MRE (seed mutant) or miRNA (miR mutant) mutations confirmed the specificity of the suppression of luciferase by the two miRNAs. Thus, the reporter assays demonstrated that targets such as PU.1, CEBPβ, HIVEP2, BCL2L13, and PDCD6 can be negatively regulated by both gga-miR-155 and MDV-1-miR-M4.
FIG. 3.
Luciferase reporter assay of predicted targets of miR-M4 and miR-155. Three tandem repeats of MRE sequences or their seed mutants (mut) were cloned in psiCHECK-2 at the end of Renilla luciferase gene, as shown in Fig. 1G. DF-1 cells were cotransfected with the reporter constructs with plasmids expressing miR-M4, MDV miR cluster 1, gga-miR-155, or MDV miR cluster mutants. Firefly and Renilla luciferase activities were measured consecutively with the dual luciferase reporter system (Promega). The Renilla luciferase levels (RL) were normalized using the firefly luciferase readings. The average Renilla luciferase level from the empty psiCHECK-2 vector was set at 1. Error bars are derived from four replicates.
In order to obtain more evidence that MDV-miR-M4 and miR-155 are true functional orthologs, we carried out further analysis on one of the predicted targets, PU.1. First, we repeated the reporter assay using a 110-bp fragment (see Table S1 in the supplemental material) of the chicken PU.1 3′UTR transcript (accession no. NM_205023) that contained the predicted MRE. A second potential MRE in the 468-nucleotide-long 3′UTR with only a partial seed match at positions 2 to 7 that failed to match the MirTarget2 prediction criteria was not included in the assay. A mutant reporter construct (PU.1-mu2) in which two nucleotide positions of the MRE were mutated was included for validating the specificity of the assay. Reporter assays with DF-1 cells transfected with the miRNA expression constructs showed that miR-M4 expressed alone or as part of the miR cluster, as well as miR-155, induced nearly 50% silencing of luciferase (Fig. 4a), confirming the findings obtained from the PU.1 multi-MRE reporter construct shown in Fig. 3. The absence of silencing of the mutant PU.1-mu2 reporter further validated the specificity of the assay. Next, we measured miR-M4- and miR-155-mediated silencing by directly measuring the levels of PU.1 expression in the chicken myelomonocytic cell line HD11 (2), stably expressing these miRNAs. Western blot analysis of PU.1 levels using rabbit anti-SPIB polyclonal antibody (Aviva Systems Biology) showed that miR-M4 expressed alone or as part of the cluster and miR-155 had a negative effect on PU.1 levels compared to what was seen for control HD11 cells or mutant constructs (Fig. 4b and c).
FIG. 4.
Repression of endogenous PU.1 levels by miR-M4 and miR-155. (a) Repression of luciferase reporter constructs of the MRE-containing region of the 3′UTR of the PU.1 transcript or seed mutant (PU.1-mu2) cloned in psiCHECK-2 at the end of the Renilla luciferase gene. The histogram shows the relative Renilla luciferase activity in DF-1 cells transfected with empty vector or miRNA expression constructs as indicated. (b) Western blot analysis of PU.1 protein expression in stably selected HD11 cells. After transfection with expression vectors of miR-M4, miR-155, viral miR cluster 1, or a miR mutant, HD11 cells were selected with appropriate antibiotics. The untransfected HD11 cells were included as controls. (Top) A Western blot assay was carried out with polyclonal anti-SPIB antibodies; (bottom); for the loading control, the same blot was reprobed with antitubulin (α-Tubulin) antibody. (c) Relative signal intensities of the PU.1 Western blot band were quantified using ImageQuant and normalized against the corresponding signal from the antitubulin band. The signal from untransfected control HD11 cells was set as 1.
At least two oncogenic herpesviruses are known to exploit miR-155 regulatory pathways. Epstein-Barr virus upregulates cellular miR-155 expression in latently infected B cells (37), and KSHV encodes a functional miR-155 ortholog (11, 24). Here we demonstrate that MDV also utilizes miR-155 pathways by encoding the functional ortholog miR-M4. Demonstration of a functional ortholog of miR-155 encoded by two distantly related herpesviruses associated with two distinct types of tumors in different host species strongly indicated the significance of the miR-155 pathway in lymphomagenesis. Interestingly, the expression of endogenous miR-155 was significantly reduced in cells transformed by both these viruses (reference 24 and our unpublished data). The reasons for the downregulation of endogenous miR-155 in these transformed cells are not known. However, a demonstration of the activation of miR-155 expression through conserved AP-1 elements would suggest a role for autoregulatory mechanisms (37). Although the precise roles and molecular pathways of miR-155 in neoplastic transformation are not fully known, its repressive function on transcriptional factors such as PU.1 can have wide-ranging effects on the cellular milieu and the global gene expression profiles seen for lymphocytes. Similarly, the repression of some of the other target genes, such as the CEBPβ, HIVEP2, BCL2L13, and PDCD6 genes, are also likely to contribute to the induction of hematopoietic cell malignancy.
Supplementary Material
Acknowledgments
We thank Malik Yousef for the prediction of targets using NBmiRTAR, Mick Watson and Dennis Prickett for organizing the chicken 3′UTR dataset, and Mick Gill for assistance in digital imaging and graphics.
This work was funded by BBSRC, United Kingdom.
Footnotes
Published ahead of print on 22 October 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Baek, D., J. Villen, C. Shin, F. D. Camargo, S. P. Gygi, and D. P. Bartel. 2008. The impact of microRNAs on protein output. Nature 45564-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beug, H., A. von Kirchbach, G. Doderlein, J. F. Conscience, and T. Graf. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18375-390. [DOI] [PubMed] [Google Scholar]
- 3.Burnside, J., E. Bernberg, A. Anderson, C. Lu, B. C. Meyers, P. J. Green, N. Jain, G. Isaacs, and R. W. Morgan. 2006. Marek's disease virus encodes MicroRNAs that map to meq and the latency-associated transcript. J. Virol. 808778-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnside, J., and R. W. Morgan. 2007. Genomics and Marek's disease virus. Cytogenet. Genome Res. 117376-387. [DOI] [PubMed] [Google Scholar]
- 5.Burnside, J., M. Ouyang, A. Anderson, E. Bernberg, C. Lu, B. C. Meyers, P. J. Green, M. Markis, G. Isaacs, E. Huang, and R. W. Morgan. 2008. Deep sequencing of chicken microRNAs. BMC Genomics 9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, X., A. Schafer, S. Lu, J. P. Bilello, R. C. Desrosiers, R. Edwards, N. Raab-Traub, and B. R. Cullen. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathogens 2e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calame, K. 2007. MicroRNA-155 function in B cells. Immunity 27825-827. [DOI] [PubMed] [Google Scholar]
- 8.Costinean, S., N. Zanesi, Y. Pekarsky, E. Tili, S. Volinia, N. Heerema, and C. M. Croce. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA 1037024-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eis, P. S., W. Tam, L. Sun, A. Chadburn, Z. Li, M. F. Gomez, E. Lund, and J. E. Dahlberg. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 1023627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottwein, E., and B. R. Cullen. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3375-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottwein, E., N. Mukherjee, C. Sachse, C. Frenzel, W. H. Majoros, J. T. Chi, R. Braich, M. Manoharan, J. Soutschek, U. Ohler, and B. R. Cullen. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 4501096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248295-304. [DOI] [PubMed] [Google Scholar]
- 13.Ko, M. H., S. Kim, W. Hwang do, H. Y. Ko, Y. H. Kim, and D. S. Lee. 2008. Bioimaging of the unbalanced expression of microRNA9 and microRNA9* during the neuronal differentiation of P19 cells. FEBS J. 2752605-2616. [DOI] [PubMed] [Google Scholar]
- 14.Kong, Y. W., I. G. Cannell, C. H. de Moor, K. Hill, P. G. Garside, T. L. Hamilton, H. A. Meijer, H. C. Dobbyn, M. Stoneley, K. A. Spriggs, A. E. Willis, and M. Bushell. 2008. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc. Natl. Acad. Sci. USA 1058866-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrie, C. H., S. Soneji, T. Marafioti, C. D. Cooper, S. Palazzo, J. C. Paterson, H. Cattan, T. Enver, R. Mager, J. Boultwood, J. S. Wainscoat, and C. S. Hatton. 2007. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int. J. Cancer 1211156-1161. [DOI] [PubMed] [Google Scholar]
- 16.McClure, L. V., and C. S. Sullivan. 2008. Kaposi's sarcoma herpes virus taps into a host microRNA regulatory network. Cell Host Microbe 31-3. [DOI] [PubMed] [Google Scholar]
- 17.Nair, V., and M. Zavolan. 2006. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 14169-175. [DOI] [PubMed] [Google Scholar]
- 18.O'Connell, R. M., D. S. Rao, A. A. Chaudhuri, M. P. Boldin, K. D. Taganov, J. Nicoll, R. L. Paquette, and D. Baltimore. 2008. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamura, K., W. J. Chung, J. G. Ruby, H. Guo, D. P. Bartel, and E. C. Lai. 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453803-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2269-276. [DOI] [PubMed] [Google Scholar]
- 21.Rai, D., S. Karanti, I. Jung, P. L. Dahia, and R. C. Aguiar. 2008. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet. Cytogenet. 1818-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selbach, M., B. Schwanhausser, N. Thierfelder, Z. Fang, R. Khanin, and N. Rajewsky. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 45558-63. [DOI] [PubMed] [Google Scholar]
- 23.Shah, C., and K. Forstemann. 2008. Monitoring miRNA-mediated silencing in Drosophila melanogaster S2-cells. Biochim. Biophys. Acta. doi: 10.1016/j.bbagrm.2008.06-008. [DOI] [PubMed]
- 24.Skalsky, R. L., M. A. Samols, K. B. Plaisance, I. W. Boss, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 8112836-12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spatz, S. J., Y. Zhao, L. Petherbridge, L. P. Smith, S. J. Baigent, and V. Nair. 2007. Comparative sequence analysis of a highly oncogenic but horizontal spread-defective clone of Marek's disease virus. Virus Genes 35753-766. [DOI] [PubMed] [Google Scholar]
- 26.Tam, W., D. Ben-Yehuda, and W. S. Hayward. 1997. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol. Cell. Biol. 171490-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam, W., and J. E. Dahlberg. 2006. miR-155/BIC as an oncogenic microRNA. Genes Chromosomes Cancer 45211-212. [DOI] [PubMed] [Google Scholar]
- 28.Tam, W., S. H. Hughes, W. S. Hayward, and P. Besmer. 2002. Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding RNA, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis. J. Virol. 764275-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner, M., and E. Vigorito. 2008. Regulation of B- and T-cell differentiation by a single microRNA. Biochem. Soc. Trans. 36531-533. [DOI] [PubMed] [Google Scholar]
- 30.Vigorito, E., K. L. Perks, C. Abreu-Goodger, S. Bunting, Z. Xiang, S. Kohlhaas, P. P. Das, E. A. Miska, A. Rodriguez, A. Bradley, K. G. Smith, C. Rada, A. J. Enright, K. M. Toellner, I. C. Maclennan, and M. Turner. 2007. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27847-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakiyama, M., K. Takimoto, O. Ohara, and S. Yokoyama. 2007. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 211857-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, X. 2008. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA 141012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, X., and I. M. El Naqa. 2008. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics 24325-332. [DOI] [PubMed] [Google Scholar]
- 34.Yao, Y., Y. Zhao, H. Xu, L. P. Smith, C. H. Lawrie, A. Sewer, M. Zavolan, and V. Nair. 2007. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 817164-7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao, Y., Y. Zhao, H. Xu, L. P. Smith, C. H. Lawrie, M. Watson, and V. Nair. 2008. MicroRNA profile of Marek's disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J. Virol. 824007-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin, Q., J. McBride, C. Fewell, M. Lacey, X. Wang, Z. Lin, J. Cameron, and E. K. Flemington. 2008. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 825295-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin, Q., X. Wang, J. McBride, C. Fewell, and E. Flemington. 2008. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J. Biol. Chem. 2832654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.