Abstract
Newcastle disease virus (NDV) entry into host cells is mediated by the hemagglutinin-neuraminidase (HN) and fusion (F) glycoproteins. We previously showed that production of free thiols in F protein is required for membrane fusion directed by F protein (S. Jain et al., J. Virol. 81:2328-2339, 2007). In the present study we evaluated the oxidation state of F protein in virions and virus-like particles and its relationship to activation of F protein by HN protein, F protein conformational intermediates, and virus-cell fusion. F protein, in particles, does not have free thiols, but free thiols were produced upon binding of particles to target cells. Free thiols were produced at 16°C in F protein in virions bound to the target cells. They also appeared in different fusion defective mutant F proteins. Free thiols were produced in the presence of mutant HN proteins that are defective in F protein activation but are attachment competent. These results suggest that free thiols appear prior to any of the proposed major conformational changes in F protein which accompany fusion activation. These results also indicate that HN protein binding to its receptor likely facilitates the interaction between F protein and host cell isomerases, leading to reduction of disulfide bonds in F protein. Taken together, these results show that free thiols are produced in F protein at a very early stage during the onset of fusion and that the production of free thiols is required for fusion in addition to activation by HN protein.
Newcastle disease virus (NDV), an avian paramyxovirus, enters the host cell by fusion of the viral membrane to the plasma membrane. Two virion-associated glycoproteins, the hemagglutinin-neuraminidase (HN) and fusion (F) proteins, are responsible for virion attachment to the target cell receptor and fusion of viral and host cell membranes, respectively. F protein, a trimer, is synthesized as a precursor, F0, which is cleaved into two disulfide linked subunits F1 and F2 (as reviewed in references 8, 21, and 32). The new amino terminus, generated by cleavage of the precursor, is the fusion peptide (FP). The fusion protein also contains two important heptad repeat (HR) domains (reviewed in reference 7). One HR domain (HR1 or HRA) is located just carboxyl terminal to the fusion peptide, and another (HR2 or HRB) is located adjacent to transmembrane (TM) domain. HR1 and HR2 peptides have strong affinity and form a very stable six helical bundle (6HB) (4). Based on studies showing inhibition of cell-cell fusion by each of these peptides, it is thought that HR1 and HR2 domains do not form the coil-coil, 6HB prior to fusion activation and are complexed only in the postfusion form (22, 41, 55).
Subsequent studies of structures of F protein from different paramyxoviruses showed that F protein may exist in two different forms. One form, exemplified by the structures of parainfluenza virus 3 F protein (53) and NDV F protein (6), is proposed to be in postfusion conformation because the structures contain the two HR domains complexed in the 6HB form. Another structure was derived from a soluble form of PIV5 F protein (54), which was stably trimerized by fusing the carboxyl terminus of the HR2 domain to the yeast GCN4 sequence, preventing 6HB formation between HR2 and HR1 domains. This structure was proposed to be the prefusion form of F protein. Changes in F protein conformation were also explored by defining the effects of HR1 and HR2 peptides (41) and mutations in HR1 and HR2 domains (16, 24, 30, 36, 42, 44, 51) on cell-cell fusion at different temperatures. Based on these studies, it was proposed that paramyxovirus F protein undergoes a series of major conformational changes leading to final 6HB formation (41).
The trigger for this major refolding of F protein is thought to be binding of the HN protein to its receptor. Whether other factors, besides interaction of F protein with HN protein, play a role in activation of F protein has not been explored. Nor is it clear how the F protein accomplishes the major refolding proposed to occur concomitant with membrane fusion. One potential mechanism to facilitate this refolding is disulfide bond isomerization or disruption, as suggested by studies of retrovirus envelope proteins. It has been shown that one or multiple disulfide bonds in human immunodeficiency virus (HIV) Env are reduced, at the time of membrane fusion, facilitating refolding of Env (5, 10, 12, 25, 43). The appearance of free cysteine residues in the HIV Env protein is mediated by host protein disulfide isomerase (PDI) or related thiol isomerases that are present on cell surfaces (11, 25). In some other retroviruses, such as murine leukemia virus, the thiol/disulfide isomerization is thought to be mediated by an isomerase motif, Cys-X-X-Cys (CXXC), in the viral Env glycoprotein, the activity of which is triggered by receptor binding (37, 49, 50). Recently, entry of other viruses, for example, Sindbis virus (1) and avian leukosis virus A (45), has been shown to be dependent on appearance of free thiols in viral fusion proteins. It has also been shown that the conserved cysteine residues of the hepatitis B virus envelope protein in hepatitis delta virus are required for virus entry and that entry is inhibited by membrane-impermeable inhibitors of thiol/disulfide isomerases (2).
Thiol/disulfide isomerases, including PDI, are a family of 19 structurally related proteins with a thioredoxin-like domain (as reviewed in reference 3). Most of these isomerases have a CXXC motif that catalyzes formation, reduction, and rearrangement of the disulfide bonds in proteins (3, 9, 34, 52). These isomerases are primarily involved in the folding of proteins in the endoplasmic reticulum, catalyzing formation of disulfide bonds, and most of these proteins have endoplasmic reticulum retention signals (3). However, in recent years, isomerases from the PDI family have also been shown to be present on cell surfaces, both in functional and in biochemical assays (reviewed in reference 11). The mechanisms involved in the expression and retention of these proteins at cell surfaces are unknown, but it has been speculated that they may be bound to resident host cell surface proteins (3, 11, 14, 48). Cell surface disulfide isomerases are proposed to be involved in processes such as cell adhesion, nitric oxide signaling, and reduction of disulfide bonds in cell entry proteins of viruses (reviewed in references 11, 19, and 48).
We have reported previously that free thiols are detected in cell surface-expressed NDV F protein and that the presence of free thiols is required for virus entry into cells, as well as for cell-cell fusion in cells expressing NDV glycoproteins (18). We also reported that the fusion of membranes in cells expressing NDV glycoproteins is inhibited by free thiol blockers at a very early step, before hemifusion (18). How the appearance of free thiols correlates with F protein activation or with conformational changes during membrane fusion is unknown.
In this report we show that free thiols are not present in F protein on the surface of virions or virus-like particles (VLPs) but are produced after binding of virus or VLPs to the target cells. We also correlated the appearance of free thiols in F protein in virus and VLPs to the conformational changes proposed to occur in F protein and to the activation of F protein by HN protein. Our results suggest that free thiols are produced in F protein on surfaces of virus or VLPs only after attachment to cells but before any major proposed conformational changes in F protein and that the appearance of free thiols is independent of activation of F protein by HN protein.
MATERIALS AND METHODS
Cells, virus, and antibodies.
COS-7 cells, obtained from American Type Culture Collection, were grown in Dulbecco modified Eagle medium (DMEM) (Gibco) supplemented with nonessential amino acids, vitamins, penicillin, and streptomycin, and 10% fetal calf serum (FCS). East Lansing Line (ELL-0) avian fibroblasts, obtained from American Type Culture Collection, were maintained in DMEM supplemented with penicillin-streptomycin and 10% FCS. Guinea pig red blood cells (RBCs) were obtained from Bio-Link, Inc.
NDV, strain Australia-Victoria (AV), was propagated in embryonated chicken eggs by standard protocols (26) in BCL-3 containment.
Anti-HR2 antibody was raised against peptide with sequence from the NDV F protein HR2 domain (29). Anti-NDV antibody was raised in rabbits against UV-inactivated stocks of NDV (strain AV), by standard protocols as previously described (28). Mouse monoclonal Anti-PDI antibody was purchased from Abnova Corp. Anti-AS, an antibody specific for the HN protein, was raised against a sequence in the NDV HN protein as previously described (29).
Plasmids.
NDV HN, F, M, and NP cDNAs, subcloned in pCAGGS vector as described previously (29, 35), were used for transfection. F protein gene with mutations in the HR1 domain (N147K) and the HR2 domain (L488,495K) regions have been described previously (40, 44). HN protein gene mutants (HN I133L and HN V81A/L110A) have been described previously (13, 47).
Transfections.
Transfection of cells was accomplished by using Lipofectamine (Invitrogen) as recommended by the manufacturer. For each transfection, a mixture of DNA (0.5 μg/35 mm plate) and 7 μl of Lipofectamine in OptiMEM (Gibco/Invitrogen) was incubated at room temperature and added to cells previously washed with OptiMEM. The cells were incubated for 5 h, and OptiMEM was replaced with 2 ml of supplemented DMEM.
Production and purification of VLPs.
VLPs were generated from avian cells and purified as described previously (35). Briefly, cells were transfected with cDNAs (4 μg of each/100-mm plate) encoding NDV HN, F, M, and NP proteins. Supernatant from the cells was collected at 24, 48, and 72 h and was clarified by centrifugation at 5,000 rpm for 5 min at 4°C. The clarified supernatant was overlaid on top of a step gradient consisting of 1 ml of 20% and 0.5 ml of 65% sucrose solutions in TNE buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA), and centrifuged at 24,000 rpm for 12 h at 4°C using an SW50.1 rotor (Beckman). The interface (containing concentrated particles) was collected in a 1-ml volume using a polystaltic pump, mixed with 1 ml of 80% sucrose, and overlaid on top of a 1-ml 80% sucrose cushion. Additional layers of sucrose (1 ml of 50% and 0.5 ml of 10% sucrose) were layered on top of the particle containing interface. The gradient was centrifuged at 38,000 rpm for 16 h at 4°C. The top 2 ml of the gradient containing particles was collected by using a polystaltic pump, mixed with 3 ml of TNE buffer, and pelleted by centrifuging at 38,000 rpm for 6 h at 4°C. The pellet was dissolved in 100 μl of TNE and used as VLP stock.
MPB labeling of virions and VLPs.
Equal quantities of NDV (AV strain) virions or VLPs were divided into aliquots into three separate tubes. Set 1 was treated with 5 mM dithiothreitol (DTT; Sigma-Aldrich, Inc.) for 1 h at 37°C, while the other two sets received no treatment. To remove DTT, virions or VLPs were purified by centrifugation on a step gradient consisting of 1-ml 20% and 0.5-ml 65% sucrose solutions in TNE buffer at 24,000 rpm for 8 h at 4°C. The interface containing purified particles was collected and set 1 and 2 particles were incubated with MPB [3-(N-maleimidylpropionyl)biocytin; Molecular Probes] at room temperature for 30 min. Set 3 particles were negative controls. To remove unbound MPD, virions or VLPs were purified by centrifugation through layer of 20% sucrose solution in TNE buffer at 24,000 rpm for 8 h at 4°C. The pellet was dissolved in 100 μl of RSB lysis buffer (0.01 M Tris-HCl [pH 7.4], 0.01 M NaCl, 1.5 mM MgCl2) containing 1% Triton X-100, 0.5% sodium deoxycholate, 2.5 mg of N-ethylmaleimide per ml, and 0.2 mg of DNase per ml, and the proteins were precipitated with neutravidin-agarose that had been washed sequentially with phosphate-buffered saline (PBS) containing 0.5% Tween 20 and 5 mg of bovine serum albumin/ml and with PBS containing 0.5% Tween 20 and 1 mg of BSA/ml and that contained 0.3% sodium dodecyl sulfate (SDS). Precipitates were washed three times with PBS containing 0.5% Tween 20 and 0.4% SDS and resolved by SDS-polyacrylamide gel electrophoresis (PAGE).
Labeling of virions and VLPs with MPB in the presence of RBCs.
Guinea pig RBCs were washed in PBS containing 0.1 mM CaCl2 and 1 mM MgCl2 (PBS-CM) and resuspended in PBS-CM to give a 0.4% solution by volume. RBCs were counted, and nearly 106 cells were incubated with NDV (AV strain) (multiplicity of infection of ∼100) or VLPs in the presence or absence of MPB (0.5 mM) for 1 h. Unbound virions or VLPs along with free MPB were removed by washing RBCs three times with PBS-CM. Washed RBC pellet was lysed with RSB lysis buffer and analyzed for labeling by MPB as described above.
PAGE and Western blot analysis.
Proteins in extracts of cells, virions, or VLPs lysed with RSB buffer or immunoprecipitates, diluted in gel sample buffer (125 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol) with 0.7 M β-mercaptoethanol, were resolved on 10% polyacrylamide gels. After electrophoresis, gels were equilibrated in transfer buffer (25 mM Tris [pH 8.2], 192 mM glycine, 15% methanol) and transferred to Immobilon-P (Millipore Corp.) membranes. The membranes were blocked overnight at 4°C in PBS containing 0.5% Tween 20 and 10% nonfat milk, washed with PBS-Tween 20, and incubated for 1 h at room temperature with primary antibody diluted to 1:1,000 in PBS-Tween 20. Membranes were then washed, incubated for 1 h at room temperature with secondary antibody (goat anti-rabbit immunoglobulin G [IgG] or anti-mouse IgG coupled to horseradish peroxidase) (Amersham Biosciences) diluted to 1:40,000 in PBS-Tween 20 and then washed extensively in PBS-Tween 20. Bound antibody was detected by using the ECL Western blotting detection reagent system (Amersham Biosciences). Quantification of the signal was accomplished by using a Fluor-S imager (Bio-Rad).
Plaque assay.
NDV virus (AV strain) was incubated with MPB and purified by centrifugation through sucrose gradients as described above. Purified virus was serially diluted in Ca2+-rich medium and then added to confluent avian cells. As a control, unlabeled virus was similarly purified, serially diluted, and added to the cells with or without MPB (0.5 mM). After adsorption for 45 min at 37°C, unbound virus was removed and agar diluted to 1% in DMEM and supplemented with nonessential amino acids, vitamins, penicillin-streptomycin, sodium bicarbonate, and 10% FCS was then placed over the monolayers. After 48 h of incubation, the plaques were counted.
RESULTS
Detection of free thiols in F protein in virions upon cell binding.
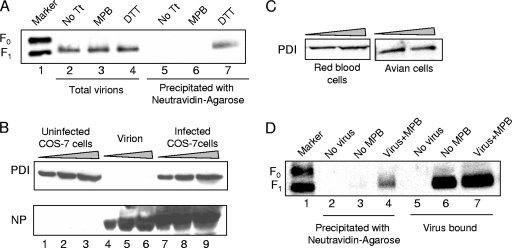
To determine whether free thiols are present in F protein in virions, we utilized MPB, which is a membrane-impermeable, thiol-reactive, biotin-containing compound that links to free thiols and biotinylates proteins with free thiols (18). We have previously reported that NDV F protein, expressed on the surfaces of cells transfected with cDNA encoding the F protein, is labeled with this reagent, indicating the presence of free thiols in the molecule (18). Virions were incubated with MPB and then repurified to remove unbound MPB and lysed. Proteins in lysed virions were analyzed for MPB labeling by precipitating with neutravidin-agarose. As shown in Fig. 1A (lane 6), F protein in virions was not labeled by MPB, whereas in virions treated with DTT, a reducing agent (lane 7), prior to incubation with MPB, F protein was labeled with MPB. This result indicated that F protein in virions does not have free thiols.
FIG. 1.
Detection of free thiols in virions after cell binding. (A) Virions were incubated with MPB (lanes 3 and 6), analyzed for MPB labeling of F protein by precipitating with neutravidin-agarose (lanes 5 to 7), and detected by Western blotting with anti-HR2 antibody. Untreated virions (No Tt, lanes 2 and 5) and virions treated with DTT prior to incubation with MPB (lanes 4 and 7) were used as controls. Total F protein in extracts from virions (lanes 2 to 4) was resolved as a control for the amounts of virions. (B) Increasing amounts of extracts from virions (lanes 4 to 6) and COS-7 cells either uninfected (lanes 1 to 3) or infected with NDV (lanes 7 to 9) were resolved by SDS-PAGE and detected for PDI (top panel) and NP protein (bottom panel). (C) Increasing amounts of extracts from avian cells and guinea pig RBCs were resolved by SDS-PAGE and detected for PDI. (D) Virions were incubated with RBCs and MPB at 4°C for 1 h, followed by 22°C for 15 min, shown in lanes 4 and 7. RBCs incubated with MPB alone (lanes 2 and 5) or virus alone (lanes 3 and 6) were used as controls. Unbound virions and MPB were removed, and F protein in bound virions was analyzed for MPB labeling by precipitation with neutravidin-agarose (lanes 2 to 4), followed by detection with anti-HR2 antibody. Extracts from RBCs (lanes 5 to 7) were resolved as a control for the amount of virus bound to RBCs.
We have previously shown that detection of free thiols in surface-expressed F protein was suppressed by membrane impermeable inhibitors of isomerases belonging to PDI family (18). One possibility for the absence of free thiols in F protein in virions is that PDI-like isomerases are not packaged into the virions. To determine whether PDI is present in virions, we analyzed virion-associated proteins by Western analysis using anti-PDI antibody. As shown in Fig. 1B (lanes 4 to 6), PDI was not detected in virion extracts, while PDI was detected in extracts from infected (lanes 7 to 9) or uninfected (lanes 1 to 3) COS-7 cells. Similarly, we also detected PDI in extracts from avian cells and guinea RBCs (Fig. 1C).
We have previously reported that isomerase inhibitors block virus infection (18). Thus, failure to detect free thiols in virion-associated F protein suggested that free thiols may be produced in virion-associated F protein only after binding to target cells. To test this possibility, we incubated virions with guinea pig RBCs in the presence of MPB. RBCs were washed to remove unbound virus, as well as soluble MPB. F protein in bound virions present in the RBCs extract was then analyzed for MPB labeling. As shown in Fig. 1D (lane 4), virion F protein was labeled by MPB in the presence of RBCs. This result suggested that free thiols are produced in F protein only after binding to target cells.
Effect of MPB labeling of virions on infectivity.
We have previously shown that MPB linkage to F protein inhibits cell-cell fusion (18). If free thiols in virion F protein are produced only after the addition of target cells, then incubation of virions with MPB prior to adding target cells should not have any effect on their infectivity, while incubation of virus with MPB during virus attachment to cells should inhibit infection. To test this proposal, we incubated virions with MPB and then repurified virions to remove unbound MPB. Infectivity of repurified virions was compared to infectivity of virions not previously treated with MPB and to infectivity of virus added to target cells in the presence of MPB. A plaque assay was conducted in the presence of MPB, yielding the following results in PFU/ml for the indicated virus treatment groups: no treatment, 1.45 ± 0.2 × 109; MPB before infection, 1.17 ± 0.14 × 109; and MPB during infection, 1.07 ± 0.83 × 108. Virions in the “MPB before infection” group were treated with MPB and purified before being added to the cells. For the “MPB during infection” group, MPB was added to the cells at the time of infection, and unbound virions as well as MPB were removed before the agar overlay was added. These results show that infectivity of virions pretreated with MPB was not significantly different from that of unlabeled virions consistent with absence of MPB labeling. In contrast, the infectivity of virions added to target cells in the presence of MPB was ∼10-fold lower than that of unlabeled virions, a result consistent with the generation of free thiols upon virus binding.
Detection of free thiols in F protein in VLPs after cell binding.
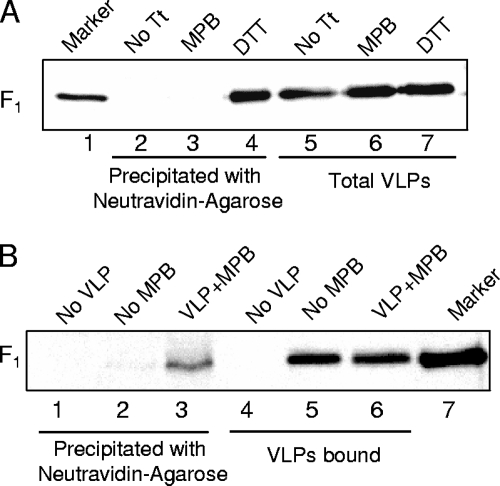
We have previously reported that avian cells expressing the NDV NP (nucleocapsid protein), M (membrane), HN, and F proteins release VLPs that resemble virus both biochemically and morphologically (35). Because we wanted to explore MPB labeling of different F protein mutants and F protein in the presence of HN protein mutants in particles, we determined whether free thiols are present in F protein in VLPs. NDV VLPs were incubated with MPB, and VLPs were then repurified to remove unbound MPB. Proteins in VLP extracts were analyzed for MPB binding by precipitating with neutravidin-agarose. The results in Fig. 2A (lane 3) show that, similar to F protein in virions, F protein in VLPs was not labeled by MPB, whereas F protein in VLPs treated with DTT (lane 4) prior to incubation was labeled with MPB.
FIG. 2.
Free thiols appear in VLPs after cell binding. (A) VLPs were incubated with MPB (lanes 3 and 6) and analyzed for MPB labeling of F protein by precipitation with neutravidin-agarose (lanes 2 to 4), followed by Western analysis with anti-HR2 antibody. Untreated VLPs (No Tt, lanes 2 and 5) and VLPs treated with DTT prior to incubation with MPB (lanes 4 and 7) were used as controls. Total F protein in extracts from VLPs (lanes 5 to 7) was resolved as a control for the amounts of VLPs. (B) VLPs were incubated with RBCs and MPB at 4°C for 1 h, followed by 22°C for 15 min (lanes 3 and 6). RBCs incubated with MPB alone (lanes 1 and 4) or VLPs without MPB (lanes 2 and 5) were used as controls. Unbound VLPs and MPB were removed, and F protein in bound VLPs was analyzed for MPB labeling by precipitation with neutravidin-agarose (lanes 1 to 3). Extracts from RBCs (lanes 4 to 6) were resolved as a control for the amount of VLPs bound to RBCs.
To determine whether free thiols are produced in F protein in VLPs after the addition of target cells, VLPs were incubated with RBCs in the presence of MPB. RBCs were washed to remove unbound VLPs, as well as soluble MPB. F protein in bound VLPs present in RBC extracts was then analyzed for MPB labeling. As shown in Fig. 2B (lane 3), F protein in VLPs was labeled by MPB in the presence of RBCs. These results suggested that free thiols, similar to virions, are produced in F protein in VLPs only after binding to target cells.
Production of free thiols in F protein in different temperature-arrested conformations.
It has been previously documented that conformational changes in paramyxovirus F protein upon activation can be arrested at low temperatures (41). The first intermediate, susceptible to HR1 peptide binding, was proposed to exist at 15°C, while another intermediate form, susceptible to both HR1 and HR2 peptides, was shown to exist only at 37°C.
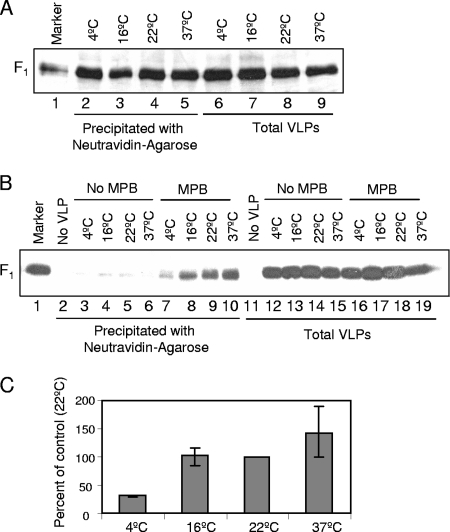
To determine the earliest form of F protein with detectable free thiols, we characterized MPB labeling of temperature arrested intermediates of F protein in VLPs and in virions. For these studies, we first determined the efficiency of MPB labeling at different temperatures by incubating DTT-treated VLPs with MPB at 4, 16, 22, and 37°C. As shown in Fig. 3A (lanes 2 to 5), labeling of F protein in VLPs by MPB was comparable at all of the temperatures.
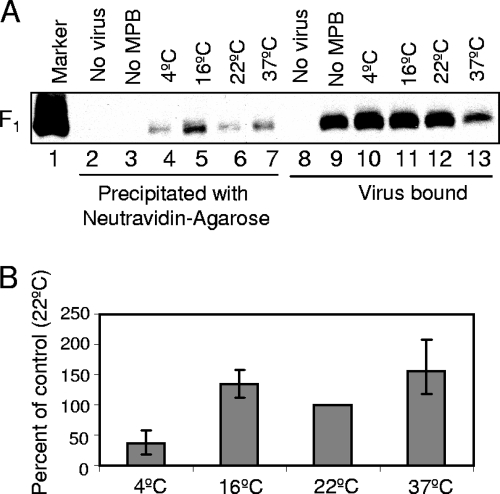
FIG. 3.
MPB labeling of temperature arrested intermediates of F protein in VLPs. (A) VLPs were treated with DTT and labeled with MPB at 4°C (lanes 2 and 6), 16°C (lanes 3 and 7), 22°C (lanes 4 and 8), or 37°C (lanes 5 and 9). F protein in VLPs was analyzed for labeling with MPB by precipitation with neutravidin-agarose (lanes 2 to 5), and extracts from VLPs (lanes 6 to 9) were resolved as a control for amount of VLPs. (B) VLPs were incubated with RBCs with (lanes 7 to 10 and lanes 16 to 19) or without MPB (lanes 3 to 6 and lanes 12 to 15) at the indicated temperatures. RBCs incubated with MPB in the absence of VLPs (lanes 2 and 11) were used as a control. Unbound VLPs and MPB were removed, and F protein in bound VLPs was analyzed for MPB labeling by precipitation with neutravidin-agarose (lanes 2 to 10), followed by Western analysis with anti-HR2 antibody. Extracts from RBCs (lanes 11 to 19) were resolved as control for the amount of VLPs bound to RBCs. (C) Quantification of MPB labeling of temperature arrested intermediates of F protein in VLPs. F protein bands in Western blots (B) were quantified by using a densitometer, and the values for MPB labeling (lanes 2 to 10) were normalized for binding (lanes 11 to 19) at respective temperatures and expressed as a percentage of labeling at 22°C. Error bars indicate the range obtained in three independent experiments. Quantification was accomplished using blots exposed in linear range of film.
Next, we assessed MPB labeling of F proteins in VLPs when incubated with RBCs at 4, 16, 22, and 37°C. RBCs were washed to remove unbound VLPs, as well as MPB. F protein in bound VLPs present in RBC extracts was then analyzed for MPB labeling. A representative result is shown in Fig. 3B, and the quantification of three similar experiments is shown in Fig. 3C. As shown in Fig. 3B (lanes 7 to 10), F protein in VLPs was labeled by MPB at all of the incubation temperatures. Labeling of F protein in VLPs incubated at 4°C was significantly decreased, while labeling at other temperatures was comparable when normalized to VLP binding to RBCs (Fig. 3C).
Similar results were seen after binding of virions to RBCs at different temperatures. As shown in Fig. 4A (lanes 4 to 7), F protein in virions was labeled by MPB at all of the incubation temperatures. Labeling of F protein in RBC-bound virions incubated at 4°C was significantly decreased, while labeling at other temperatures was comparable when normalized to virus binding (Fig. 4B). These results suggest that free thiols are produced in F protein before the major proposed conformational changes in F protein.
FIG. 4.
MPB labeling of temperature-arrested intermediates of F protein in virions. (A) Virions were incubated with RBCs and MPB (lanes 4 to 7 and lanes 10 to 13) at the indicated temperatures. RBCs incubated with MPB in the absence of virions (lanes 2 and 8) and virions without MPB at 22°C (lanes 3 and 9) were used as controls. Unbound virions and MPB were removed, and F protein in bound virions was analyzed for MPB labeling by precipitation with neutravidin-agarose (lanes 2 to 7). Extracts from RBCs (lanes 8 to 13) were resolved as a control for the amount of virions bound to RBCs. (B) Quantification of MPB labeling of temperature-arrested intermediates of F protein in virions. F protein bands in Western blot (A) were quantified by using a densitometer, and the values for MPB labeling (lanes 2 to 7) were normalized for binding (lanes 8 to 13) at respective temperatures and expressed as a percentage of labeling at 22°C. Error bars indicate the range obtained in three independent experiments. Quantification was accomplished by using blots exposed in a linear range of film.
Production of free thiols in HR1 and HR2 mutant F proteins.
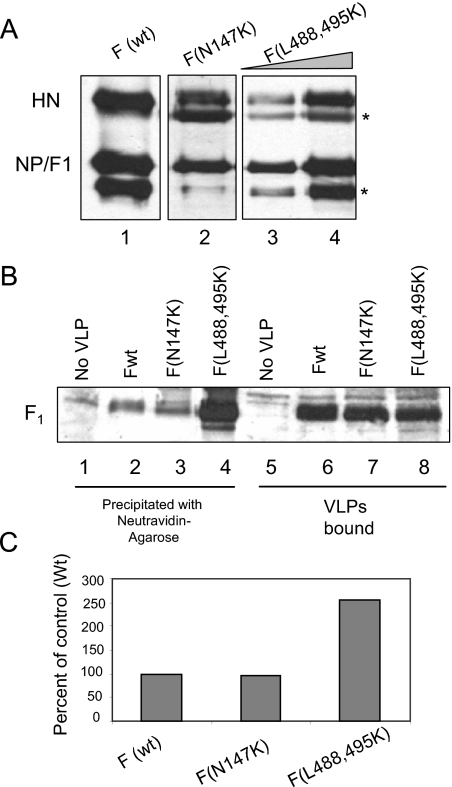
HR2 and HR1 mutant F proteins are thought to be defective in attaining the prehairpin (diagramed in Fig. 7C) intermediate conformation (27, 36, 42, 44, 51). To determine whether these mutant proteins can form free thiols, we generated VLPs containing wild-type F protein or HR1 (N147K) or HR2 (L488,495K) mutant F proteins. Proteins in VLPs containing wild-type F protein or F protein with mutations in HR1 (N147K) or HR2 (L488,495K) domain are shown in Fig. 5A. The amounts of VLPs were equalized by normalizing to the NP/F1 protein band. Equivalent amounts of VLPs were labeled by MPB in the presence of RBCs and analyzed. That the amounts of VLPs used were comparable is confirmed by comparable amounts of F protein in bound VLPs containing wild-type F protein (Fig. 5B, lane 6) or HR1 or HR2 mutant F proteins (lanes 7 or 8). As shown in Fig. 5B, HR1 mutant F protein (lane 3) was labeled by MPB as well as wild-type F protein (lane 2). The HR2 mutant F protein (lane 4) was also labeled with MPB. Interestingly, this mutant F protein was labeled with MPB at significantly higher levels compared to the wild-type F protein. That both mutant proteins had free cysteine residues suggested that free thiols are produced before any major conformational change in F protein. These results also suggest that F protein with an HR2 mutation may be more accessible to isomerases than wild-type or HR1 mutant proteins.
FIG. 7.
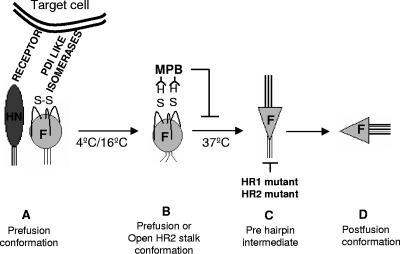
Model for the role of free thiols in virion-associated F protein. (A) F protein in prefusion conformation. HN protein binds to the receptor and facilitates interaction between F protein and target cell isomerases. This binding leads to cleavage of disulfide bond in F protein (B) in prefusion or open stalk conformation, which has been shown to be present at lower temperatures. (C) F protein in prehairpin conformation, which has been shown to require 37°C. (D) F protein in postfusion conformation. HR1 and HR2 mutants are proposed to be defective in attaining form C and are arrested in form B, an open stalk conformation.
FIG. 5.
MPB labeling of F mutants in VLPs. (A) VLPs were generated using Fwt (lane 1) or HR1 mutant, F (N147K) (lane 2) or HR2 mutant, F (L488,495K) (lanes 3 and 4). The amounts of VLPs generated in each case were estimated by detecting particle associated proteins by a Western blot using a mix of anti-NDV antibody, anti-F antibody, and anti-HN (anti-AS) antibody. The protein band below the NP protein (*) is a degradation product of NP variably seen in different preparations of virus and VLPs (unpublished observation). The protein band below the HN protein (*) is a nonspecific protein band variably detected by anti-AS antibody, an anti-HN antibody. (B) Equivalent amounts of VLPs (normalized to NP protein shown in panel A) with Fwt (lanes 2 and 6) or HR1 mutant F (N147K) (lanes 3 and 7) or HR2 mutant F (L488,495K) (lanes 4 and 8) were incubated with RBCs and MPB at 4°C for 1 h, followed by 22°C for 15 min. RBCs incubated with MPB in the absence of VLPs (lanes 1 and 5) were used as a control. Unbound VLPs and MPB were removed, and F protein in bound VLPs was analyzed for MPB labeling by precipitation with neutravidin-agarose (lanes 1 to 4), followed by Western analysis with anti-HR2 antibody. Extracts from RBCs (lanes 5 to 8) were resolved as a control for the amount of VLPs bound to RBCs. (C) Quantification of MPB labeling in panel B. The results are representative of two similar experiments. Quantification was accomplished using blots exposed in a linear range of film. As shown in Fig. 1 to 4 and in other experiments, there was no nonspecific precipitation of wild-type or mutant F protein with neutravidin-agarose in the absence of MPB binding.
Role of HN protein in production of free thiols in F protein.
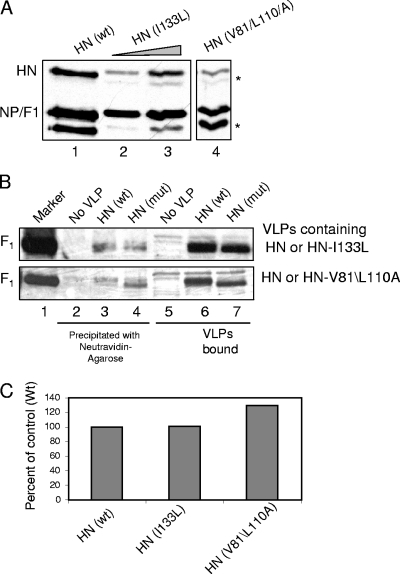
We have previously reported that, in transfected cells, free thiols were detected in F protein when it was expressed alone, without HN protein (18). This result suggested that production of free thiols in F protein is independent of HN protein in transfected cells. To determine the role of HN protein in production of free thiols in F protein in VLPs, we analyzed VLPs containing mutant HN protein (HN I133L and HN V81A/L110A) for labeling of F protein by MPB. These mutant HN proteins have alterations in the stalk domain and have been shown to be defective in fusion promotion but are comparable to wild-type HN protein in attachment activity (13, 46). Figure 6A shows the proteins incorporated in VLPs containing wild-type or mutant HN proteins. The amounts of VLPs were equalized by normalizing to the NP/F1 protein band. Equivalent amounts of VLPs were labeled by MPB in the presence of RBCs and analyzed. Figure 6B, lanes 6 and 7, show that the amounts of VLPs used were comparable since the amounts of F protein in bound VLPs were similar in VLPs containing wild-type or mutant HN protein. The F protein in VLPs containing mutant HN protein migrates slightly faster on the gel than F protein in VLPs with wild-type HN protein (Fig. 6B, lanes 6 and 7). This difference may be due to the treatment of cells with neuraminidase during production of VLPs containing mutant HN proteins since these mutants are defective in neuraminidase activity (13, 46). Lower neuraminidase activity inhibits release of VLPs from cells (unpublished observation). As shown in Fig. 6B (lane 4), F protein in VLPs containing these mutant HN proteins was labeled by MPB at levels similar to VLPs containing wild-type HN protein. This result suggested that the production of free thiols in F protein in VLPs is independent of HN protein activation.
FIG. 6.
(A) MPB labeling of F protein in VLPs with HN mutants. (A) VLPs were generated using HNwt (lane 1) or HN (I133L) (lanes 2 and 3) or HN (V81A/L110A) (lane 5) separately. The amounts of VLPs generated in each case were estimated by detecting proteins associated to VLPs by SDS-PAGE and analyzing them by Western blotting using a mix of anti-NDV antibody, anti-F antibody, and anti-HN antibody. The protein band below NP protein (*) is a degradation product of NP variably seen in different preparations of virus and VLPs (unpublished observation). The protein band below HN protein (*) is a nonspecific protein band variably detected by anti-AS antibody. (B) Equivalent amounts of VLPs (normalized to NP protein shown in A) with HNwt (lanes 3 and 6) or HN mutant HN I133L (top panel) or HN V81A/L110A (bottom panel) (lanes 4 and 7) were incubated with RBCs and MPB at 4°C for 1 h, followed by 22°C for 15 min. RBCs incubated with MPB in the absence of VLPs (lanes 1 and 5) were used as a control. Unbound VLPs and MPB were removed, and F protein in bound VLPs was analyzed for MPB labeling by precipitation with neutravidin-agarose (lanes 2 to 4), followed by Western analysis with anti-HR2 antibody. Extracts from RBCs (lanes 5 to 7) were resolved as a control for the amount of VLPs bound to RBCs. (C) Quantification of MPB labeling in panel B. The results are representative of two similar experiments. Quantification was accomplished using blots exposed in a linear range of film. As shown in Fig. 1 to 4 and in other experiments, there was no nonspecific precipitation of F protein with neutravidin-agarose in the absence of MPB binding.
DISCUSSION
Paramyxovirus F proteins are thought to undergo major conformational changes during membrane fusion. These changes are required for the fusion process and are triggered by HN protein binding to the host cell receptors. How F protein accomplishes this refolding is not clear. One possible mechanism proposed to facilitate refolding of the F protein is reduction of disulfide bonds in F protein by host cell thiol/disulfide isomerases (18). We have previously reported that free thiols are present in cell surface-expressed F protein and that these free thiols are required for fusion mediated by F protein (18). In the present study we found that free thiols cannot be detected in purified virions or VLPs. However, free thiols could be detected upon binding of the particles to cell surfaces. We further defined the relationship of the appearance of these reduced forms of particle-associated F protein to F protein activation by HN protein and the conformational changes in F protein upon activation.
The position and numbers of cysteine residues in paramyxovirus F proteins are highly conserved (31). In Sendai virus F protein, it has been shown that all of the cysteines in virion associated F protein are involved in disulfide bonds (17). Although the linkages between the extracellular cysteine residues in the NDV F protein have not been directly demonstrated, it is likely that the disulfide bonding is similar to that of Sendai virus F protein. Indeed, our results showing lack of MPB labeling of F protein in purified virions suggest that there are no free thiols in virion-associated F protein and all cysteines are involved in disulfide bonds (Fig. 1). Consistent with this observation is the absence of PDI isomerase in virus membranes. It has been reported that cell surface PDI is not associated with lipid raft domains (25). We have previously reported that NDV is assembled in lipid raft domains (20); thus, the failure to package PDI into virions is not surprising.
Free thiols were, however, detected in virions and VLPs bound to RBCs and incubation with MPB could inhibit plaque formation only if it was present during particle binding to cell surfaces (Fig. 1 and 2 and Table 1). These results suggested that particles must bind to cell membranes to be exposed to the cell surface host cell isomerases and only then are F protein disulfide bonds reduced. Although thiol isomerases have not been reported on RBC surfaces, surface expression of these enzymes has been documented in numerous cell types (reviewed in reference 11), including COS-7 cell surfaces (18a).
According to current models proposed for paramyxovirus membrane fusion, F protein undergoes major refolding upon activation (41, 54). The first proposed conformational change in F protein is the disassociation of HR2 domains in the trimer. Subsequently, the HR1 domain unfolds and then refolds into an extended trimeric helix resulting in the insertion of the fusion peptide, which is at the tip of the helix, into target cell membranes. F protein in this state is referred to as prehairpin intermediate. After insertion of fusion peptide into the target membrane, the next proposed major conformational change involves refolding of F protein into a stable trimer of hairpins formed by complexing of the HR1 and HR2 domains into a 6HB. The refolding of F protein into this postfusion form leads to the close approach and fusion of the target and effector membranes.
Conformational intermediates in F protein refolding have been defined previously by evaluating inhibition of cell-cell fusion after HR1 or HR2 peptide binding at temperatures lower than 37°C, reasoning that, at lower temperatures, the energy available for F protein refolding is suboptimal (41). Based on their results, Russell et al. (41) have proposed that F protein at 4 or 15°C is arrested in a prefusion or open stalk state as shown in Fig. 7. Our results (Fig. 3 and 4) that F protein in virions or VLPs is labeled by MPB at 16°C, at levels comparable to those at 22 or 37°C, suggest that free thiols are produced in F protein before any major conformational changes in F protein take place. The decrease in MPB labeling seen at 4°C compared to 16°C could be due to lower activity of host cell isomerases at this temperature.
We also observed a decrease in binding of virions to RBCs at 37°C in the presence of MPB (Fig. 4). One possibility for this decrease in binding is that neuraminidase activity of HN protein at this temperature leads to disassociation of bound virions. Since MPB inhibits fusion (18), the F protein with bound MPB may not insert its fusion peptide into the target membranes. In this case, the F protein may not anchor the virus to target cells facilitating release of the particles from the cells by the HN protein neuraminidase (38). A failure of F protein to anchor to target cells in the presence of MPB is consistent with the proposal that free thiols are produced before the prehairpin intermediate of F protein is formed. It is this form of F protein, with the FP domain extended toward the target membrane, which is thought to insert into target membranes.
Studies of HR1 and HR2 F protein mutants from different paramyxoviruses have shown that mutations in these domains affect its fusion activity (16, 24, 30, 36, 42, 44, 51). Mutations in HR2 domain were proposed to be arrested in an early intermediate form, before prehairpin formation (42), and mutations in HR1 domain were proposed to be defective in attaining a stable prehairpin conformation (24, 42, 51). Our results (Fig. 5) showed that both HR1 and HR2 mutant F proteins have free thiols and suggest that free thiols are produced in either a prefusion form or a form with open HR2 stalk domain of F protein. That is, the appearance of free thiols occurs in a conformation that forms prior to the point at which the mutant proteins are arrested. The increase in labeling of HR2 mutant F protein suggests that a larger percentage of the total HR2 mutant F protein contains free thiols. The increased amount of HR2 mutant protein in a reduced form could be due to the packaging of a reduced form of this protein into VLPs. However, this explanation is less likely since there are not increased levels of reduced forms of this protein on surfaces of cells transfected with this mutant protein (unpublished observations). Rather, it is possible that the HR2 mutant F protein in bound VLPs may be more accessible to isomerases or that this mutant protein is arrested in an early reduced form while the HR1 mutant proteins may be arrested in a later stage and would include both reduced and disulfide linked forms as does the wild-type protein.
For paramyxovirus F protein activity, binding of HN protein to its receptor has been shown to be essential (33, 39). This requirement for HN protein for fusion is virus specific, suggesting that there is an essential interaction between HN and F proteins (15). The details of this interaction are not clear, and whether other factors are involved in F protein activation is not yet known. We have shown previously that while the production of free thiols in F protein is required for cell-cell and virus-cell fusion, free thiols can be detected in F protein expressed on cell surfaces without HN protein (18). These results suggest that reduction of disulfides and interactions with HN protein are independently required for fusion activation. This conclusion is supported by our finding that MPB linkage to F protein was unaffected by the presence of HN proteins which were attachment competent but defective in fusion activation (Fig. 6). This finding suggests that during virus entry, HN protein binding to its receptor facilitates production of free thiols in F protein by bringing the F protein to proximity of cell surface isomerases and that interaction between HN and F proteins leading to activation of F protein is not required for the production of free thiols in F protein. Upon making contact with cell surfaces or in transfected cell membranes, the prefusion F protein may be in equilibrium between oxidized and reduced forms. If HN protein is present, the F protein with free thiols may proceed to refold, resulting in membrane fusion. Refolding is blocked if the free cysteines are blocked with MPB (Fig. 7), and fusion is inhibited as shown here and in previous results (18).
Comparisons of the positions of the cysteine residues in the proposed pre- and postfusion structures of paramyxovirus F proteins suggest that the disulfide bond linkages in the two forms are identical. That is, there is no evidence for a reshuffling of disulfide bonds upon the change from the prefusion to postfusion forms of the protein. Thus, it may only be the transient disruption of the bonds that facilitates the refolding of the F protein. Disruption of some bonds may decrease the activation energy required or may facilitate the refolding of the molecule. Another possible mechanism may be a necessary transient covalent disulfide bond between the fusion protein and a host cell protein. Actual reshuffling of disulfide bonds has been demonstrated in the murine leukemia virus Env protein. It is not clear whether reshuffling of disulfide bonds occurs in the avian leukosis virus Env, the HIV Env, the alphavirus envelope protein, or the hepatitis delta virus envelope protein.
It has been recently reported that F protein in Sendai virus virions is a mixed population of prefusion and postfusion forms (23). If the NDV F protein in particles exists in both forms, then our results indicate that neither the pre- nor the postfusion virion-associated F protein have free thiols. We have found that overexpression of PDI-like isomerases in cells expressing both HN and F proteins leads to enhanced cell-cell fusion, significantly decreased MPB labeling of F protein (18a), and increased concentrations of the postfusion form of F protein. Thus, the postfusion form of F protein may not be a substrate for isomerases.
Taken together, our results show that free thiols are produced in virion-associated F protein only after binding of virus to cells (diagramed in Fig. 7). These free thiols appear in F protein prior to any major conformational changes in the protein upon fusion activation and are independent of the activation of F protein by HN protein.
Acknowledgments
We thank Homer Pantua for advice and help with VLPs and Jason Laliberte for help with the BSL-3 facility.
This study was supported in part by grant AI 30572 from the National Institutes of Health.
Footnotes
Published ahead of print on 15 October 2008.
REFERENCES
- 1.Abell, B. A., and D. T. Brown. 1993. Sindbis virus membrane fusion is mediated by reduction of glycoprotein disulfide bridges at the cell surface. J. Virol. 675496-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou-Jaoude, G., and C. Sureau. 2007. Entry of hepatitis delta virus requires the conserved cysteine residues of the hepatitis B virus envelope protein antigenic loop and is blocked by inhibitors of thiol-disulfide exchange. J. Virol. 8113057-13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appenzeller-Herzog, C., and L. Ellgaard. 2008. The human PDI family: versatility packed into a single fold. Biochim. Biophys. Acta 1783535-548. [DOI] [PubMed] [Google Scholar]
- 4.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3309-319. [DOI] [PubMed] [Google Scholar]
- 5.Barbouche, R., R. Miquelis, I. M. Jones, and E. Fenouillet. 2003. Protein-disulfide isomerase-mediated reduction of two disulfide bonds of HIV envelope glycoprotein 120 occurs post-CXCR4 binding and is required for fusion. J. Biol. Chem. 2783131-3136. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9255-266. [DOI] [PubMed] [Google Scholar]
- 7.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell. Biol. 4309-319. [DOI] [PubMed] [Google Scholar]
- 8.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 28525-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellgaard, L., and L. W. Ruddock. 2005. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 628-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenouillet, E., R. Barbouche, J. Courageot, and R. Miquelis. 2001. The catalytic activity of protein disulfide isomerase is involved in human immunodeficiency virus envelope-mediated membrane fusion after CD4 cell binding. J. Infect. Dis. 183744-752. [DOI] [PubMed] [Google Scholar]
- 11.Fenouillet, E., R. Barbouche, and I. M. Jones. 2007. Cell entry by enveloped viruses: redox considerations for HIV and SARS-coronaviruses. Antioxid. Redox Signal 91009-1034. [DOI] [PubMed] [Google Scholar]
- 12.Gallina, A., T. M. Hanley, R. Mandel, M. Trahey, C. C. Broder, G. A. Viglianti, and H. J. Ryser. 2002. Inhibitors of protein-disulfide isomerase prevent cleavage of disulfide bonds in receptor-bound glycoprotein 120 and prevent HIV-1 entry. J. Biol. Chem. 27750579-50588. [DOI] [PubMed] [Google Scholar]
- 13.Gravel, K. A., and T. G. Morrison. 2003. Interacting domains of the HN and F proteins of Newcastle disease virus. J. Virol. 7711040-11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogg, P. J. 2003. Disulfide bonds as switches for protein function. Trends Biochem. Sci. 28210-214. [DOI] [PubMed] [Google Scholar]
- 15.Hu, X. L., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 661528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, Y., H. Komada, S. Kusagawa, M. Tsurudome, H. Matsumura, M. Kawano, H. Ohta, and M. Nishio. 1992. Fusion regulation proteins on the cell surface: isolation and characterization of monoclonal antibodies which enhance giant polykaryocyte formation in Newcastle disease virus-infected cell lines of human origin. J. Virol. 665999-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwata, S., A. C. Schmidt, K. Titani, M. Suzuki, H. Kido, B. Gotoh, M. Hamaguchi, and Y. Nagai. 1994. Assignment of disulfide bridges in the fusion glycoprotein of Sendai virus. J. Virol. 683200-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain, S., L. W. McGinnes, and T. G. Morrison. 2007. Thiol/disulfide exchange is required for membrane fusion directed by the Newcastle disease virus fusion protein. J. Virol. 812328-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Jain, S., L. W. McGinnes, and T. G. Morrison. 2008. Overexpression of thiol/disulfide isomerases enhances membrane fusion directed by the Newcastle disease virus fusion protein. 8212039-12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan, P. A., and J. M. Gibbins. 2006. Extracellular disulfide exchange and the regulation of cellular function. Antioxid. Redox Signal 8312-324. [DOI] [PubMed] [Google Scholar]
- 20.Laliberte, J. P., L. W. McGinnes, M. E. Peeples, and T. G. Morrison. 2006. Integrity of membrane lipid rafts is necessary for the ordered assembly and release of infectious Newcastle disease virus particles. J. Virol. 8010652-10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb, R. A., and T. S. Jardetzky. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17427-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 932186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludwig, K., B. Schade, C. Bottcher, T. Korte, N. Ohlwein, B. Baljinnyam, M. Veit, and A. Herrmann. 2008. Electron cryomicroscopy reveals different F1+F2 protein states in intact parainfluenza virions. J. Virol. 823775-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luque, L. E., and C. J. Russell. 2007. Spring-loaded heptad repeat residues regulate the expression and activation of paramyxovirus fusion protein. J. Virol. 813130-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markovic, I., T. S. Stantchev, K. H. Fields, L. J. Tiffany, M. Tomic, C. D. Weiss, C. C. Broder, K. Strebel, and K. A. Clouse. 2004. Thiol/disulfide exchange is a prerequisite for CXCR4-tropic HIV-1 envelope-mediated T-cell fusion during viral entry. Blood 1031586-1594. [DOI] [PubMed] [Google Scholar]
- 26.McGinnes, L. W., J. Reiter, H. Pantua, and T. G. Morrison. 2006. Newcastle disease virus propagation, quantification, and storage, p. 15F. 2.3-15F. 2.11. In Current protocols in microbiology, vol. 1. John Wiley & Sons, Inc., New York, NY. [DOI] [PubMed] [Google Scholar]
- 27.McGinnes, L., T. Sergel, J. Reitter, and T. Morrison. 2001. Carbohydrate modifications of the NDV fusion protein heptad repeat domains influence maturation and fusion activity. Virology 283332-342. [DOI] [PubMed] [Google Scholar]
- 28.McGinnes, L. W., K. Gravel, and T. G. Morrison. 2002. Newcastle disease virus HN protein alters the conformation of the F protein at cell surfaces. J. Virol. 7612622-12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinnes, L. W., and T. G. Morrison. 2006. Inhibition of receptor binding stabilizes Newcastle disease virus HN and F protein-containing complexes. J. Virol. 802894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGinnes, L. W., T. Sergel, H. Chen, L. Hamo, S. Schwertz, D. Li, and T. G. Morrison. 2001. Mutational analysis of the membrane proximal heptad repeat of the Newcastle disease virus fusion protein. Virology 289343-352. [DOI] [PubMed] [Google Scholar]
- 31.Morrison, T., and A. Portner. 1991. Structure, function, and intracellular processing of the glycoproteins of Paramyxoviridae. Plenum Press, Inc., New York, NY.
- 32.Morrison, T. G. 2003. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta 161473-84. [DOI] [PubMed] [Google Scholar]
- 33.Moscona, A., and R. W. Peluso. 1991. Fusion properties of cells persistently infected with human parainfluenza virus type 3: participation of hemagglutinin-neuraminidase in membrane fusion. J. Virol. 652773-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noiva, R. 1999. Protein disulfide isomerase: the multifunctional redox chaperone of the endoplasmic reticulum. Semin. Cell Dev. Biol. 10481-493. [DOI] [PubMed] [Google Scholar]
- 35.Pantua, H., L. W. McGinnes, J. Leszyk, and T. G. Morrison. 2005. Characterization of an alternate form of Newcastle disease virus fusion protein. J. Virol. 7911660-11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 27017-30. [DOI] [PubMed] [Google Scholar]
- 37.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 718073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porotto, M., M. Murrell, O. Greengard, L. Doctor, and A. Moscona. 2005. Influence of the human parainfluenza virus 3 attachment protein's neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 792383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porotto, M., M. Murrell, O. Greengard, and A. Moscona. 2003. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J. Virol. 773647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reitter, J., T. Sergel, and T. Morrison. 1995. Mutational analysis of the leucine zipper motif in the Newcastle Disease virus fusion protein. J. Virol. 695995-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 204024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell, C. J., K. L. Kantor, T. S. Jardetzky, and R. A. Lamb. 2003. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 163363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryser, H. J., E. M. Levy, R. Mandel, and G. J. DiSciullo. 1994. Inhibition of human immunodeficiency virus infection by agents that interfere with thiol-disulfide interchange upon virus-receptor interaction. Proc. Natl. Acad. Sci. USA 914559-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2001. Mutations in the fusion peptide and adjacent heptad repeat inhibit folding or activity of the Newcastle disease virus fusion protein. J. Virol. 757934-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, J. G., and J. M. Cunningham. 2007. Receptor-induced thiolate couples Env activation to retrovirus fusion and infection. PLoS Pathog. 3e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 716287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stone-Hulslander, J., and T. G. Morrison. 1999. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 733630-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turano, C., S. Coppari, F. Altieri, and A. Ferraro. 2002. Proteins of the PDI family: unpredicted non-ER locations and functions. J. Cell Physiol. 193154-163. [DOI] [PubMed] [Google Scholar]
- 49.Wallin, M., M. Ekstrom, and H. Garoff. 2004. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 2354-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallin, M., R. Loving, M. Ekstrom, K. Li, and H. Garoff. 2005. Kinetic analyses of the surface-transmembrane disulfide bond isomerization-controlled fusion activation pathway in Moloney murine leukemia virus. J. Virol. 7913856-13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West, D. S., M. S. Sheehan, P. K. Segeleon, and R. E. Dutch. 2005. Role of the simian virus 5 fusion protein N-terminal coiled-coil domain in folding and promotion of membrane fusion. J. Virol. 791543-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson, B., and H. F. Gilbert. 2004. Protein disulfide isomerase. Biochim. Biophys. Acta 169935-44. [DOI] [PubMed] [Google Scholar]
- 53.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 1029288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 43938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, J. K., D. Li, M. C. Abramowitz, and T. G. Morrison. 1999. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J. Virol. 735945-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]