Abstract
Gammaretroviral and lentiviral vectors are promising tools for gene therapy, but they can be oncogenic. The development of safer vectors depends on a quantitative assay for insertional mutagenesis. Here we report a rapid, inexpensive, and reproducible assay which uses a murine cell line to measure the frequency of interleukin-3 (IL-3)-independent mutants. Lentiviral and gammaretroviral vectors cause insertional mutagenesis at similar frequencies; however, they use different mechanisms. Human immunodeficiency virus (HIV)-based vectors generate mutants by insertion only into the growth hormone receptor (Ghr) locus. The HIV enhancer/promoter is active in the absence of the HIV Tat protein in this locus, and an HIV/Ghr spliced transcript expresses GHR and cells respond to GH. Deletion of the enhancer/promoter in a self-inactivating HIV-based vector prevents this mechanism of insertional mutagenesis. In contrast, gammaretroviral vectors insert into other loci, including IL-3 and genes identified as common insertion sites in the Retroviral Tagged Cancer Gene Database (RTCGD).
Gammaretroviral and lentiviral vectors are promising tools for the correction of inherited genetic diseases. Because such vectors integrate into cellular DNA, transduction of patients’ bone marrow stem cells has been used to modify progeny cells of the hematopoietic system, leading to correction of inherited immunodeficiencies (1, 6, 23). However, vector integration may also cause tumors by upregulating cellular oncogenes adjacent to the integration site. For example, patients with common gamma-chain deficiency treated with a gammaretroviral vector have developed leukemia following vector insertion in oncogenic loci (13). It remains unclear whether the common gamma-chain itself or the rapid proliferation caused by its restoration predisposes these particular patients to insertional oncogenesis (25, 37). Vector integration can also cause clonal expansion (16); in patients with chronic granulomatous disease, the expansion of vector-modified clones has improved the rate of gene correction (23). However, this is probably not a desirable outcome, as hyperproliferation may select for premalignant or otherwise dysfunctional clones.
The successful clinical studies have all used retroviral vectors based on murine leukemia virus (MLV); the vectors retain the enhancer within the gammaretroviral long terminal repeat (LTR). The most commonly described mechanism of cell transformation by MLV is insertion of the provirus adjacent to cellular protooncogenes, resulting in disregulation of their expression and tumor formation. The activity of the enhancer within the U3 region of the viral LTR controls the rate and type of tumor formation (9, 31, 34, 35). Insertional oncogenesis by wild-type (WT) lentiviruses is not a common mechanism of pathogenesis; a potentially oncogenic insertion of human immunodeficiency virus (HIV) has been reported in rare HIV-associated T-cell lymphomas (28). However, most lentiviral vectors lack viral genes that cause other disease pathologies and also carry an envelope that extends cell tropism, so insertional oncogenesis cannot be ruled out.
Clearly the development of safer vectors requires a reproducible assay that quantitates insertional mutagenesis. Chemical mutagenesis results in DNA damage which can lead to loss of gene function; this can be scored by the selection of cells for the loss of an activity, for example, that of the enzyme thymidine kinase (7). An insertional mutagenesis assay needs to measure gain of gene function; a general assay should screen as many loci as possible, and a disease-specific model could be used to screen cooperating loci. Retroviral-vector insertional oncogenesis has been quantitated in mice either by using vectors expressing an oncogene (11, 18) or mutant, tumor-prone mice (22, 29). However, mouse oncogenesis experiments are expensive and time consuming. One cell culture assay has been developed that measures extended culture of mouse bone marrow primary cells; this selects for gammaretroviral-vector insertion in and upregulation of the Evi1 gene (21).
Here we report a cell culture assay for insertional mutagenesis that uses an immortalized mouse cell line. This is based on the observation that retrovirus insertion can render an interleukin-3 (IL-3)-dependent cell line cytokine independent (32). Most genes identified in such an assay are likely to be specific gain-of-function genes (32), as are the common insertion sites (CIS) in the Retroviral Tagged Cancer Gene Database (RTCGD), although it may be possible that haploid gene inactivation could render cells IL-3 independent. While gammaretroviral and lentiviral vectors generate insertional mutants in this assay at similar frequencies, they do so by different mechanisms.
MATERIALS AND METHODS
Cell lines.
The suspension cell lines BAF3 (24) and Bcl15 (8) were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco) with 10% fetal calf serum (FCS), antibiotics, and 2 to 10% WEHI-3B-cell-conditioned medium.
Vectors.
The HIV type 1 (HIV-1)-derived lentiviral vectors pHV and its self-inactivating (SIN) counterpart pHRSIN-CSGW (10) are described in reference 14; SINLV SFFV IL2RG (SIN lentivirus spleen focus-forming virus IL-2 receptor, gamma chain) is described in reference 39. The gammaretroviral vectors used were pCNCG (30) and pMFG.S eGFP (expressing enhanced green fluorescent protein) (26) (the latter kindly provided by J. S. Lee, Harvard Gene Therapy Initiative). Vesicular stomatitis virus G glycoprotein-pseudotyped vector stocks were produced by transient transfection of 293T cells. The lentiviral transfer vector plasmid were cotransfected with the HIV-1-packaging plasmid pCMVΔR8.91 and vesicular stomatitis virus G glycoprotein plasmid pMDG. The gammaretroviral transfer vector plasmid was cotransfected with MLV packaging plasmid pCMVintron and pMDG. Viral supernatants were concentrated 100-fold by ultracentrifugation. The titers of vector preparations were determined on Bcl15 cells by quantitative PCR (qPCR) for integrated GFP or HIV-1 gag leader sequence (SINLV SFFV IL2RG only) copies. For each sample, TaqMan quantification of 18S rRNA copies was performed in parallel to control for input. (All primer and probe sequences can be found in Table S1 in the supplemental material). The lentiviral- and gammaretroviral-vector titers were in the range of 4 × 107 to 12 × 107 and 1 × 106 to 10 × 106 Bcl15 infectious units/ml, respectively.
Transduction and selection of IL-3-independent cells.
On day 1, Bcl15 target cells were transduced for 4 h at a multiplicity of infection (MOI) of 4 to 12 (experiments LV2 to LV8 [using lentiviral vectors]) or 0.1 to 1.1 (experiments RV1 to RV3 [using gamma retroviral vectors]). Bcl15 cells were transduced at a cell density of 106 cells per ml in T75 flasks; the vector preparation made up 10% of the total volume at the time of transduction. After 4 h, the culture volume was expanded 12- to 15-fold, and Bcl15 target cells were grown in complete medium plus IL-3 for the next 72 h. On day 4 of the IL-3 selection protocol, cells were washed and plated in 24-well plates at 5 × 105 cells/well in 2 ml medium without IL-3. On day 11, the cells were “rescued” by the readdition of IL-3 and maintained in IL-3; many wells reached confluence after 2 to 3 days. On days 14 and 15, the cells in each well were washed free of IL-3 once more and grown in 10 ml complete medium without IL-3 in T25 tissue culture flasks. IL-3-independent clones appeared 7 to 10 days later in a small number of flasks. To select for Ghr insertional mutants, cells were grown from day 4 onwards in 24-well plates in medium containing FCS and 1 μg/ml recombinant bovine growth hormone (bGH) (kind gift of Monsanto). The number of wells in which IL-3-independent clones grew out was scored after 4 to 6 weeks. For both gammaretroviral and lentiviral vectors, transduction at lower MOIs did not give IL-3-independent mutants.
Selection of IL-3-dependent single-cell clones by limiting dilution.
IL-3-dependent single-cell clones were selected from cell populations transduced with gammaretroviral vectors. Cells were plated at 0.3 cells per well in 96-well plates in complete medium containing IL-3. The IL-3 dependence of the clones was confirmed by their inability to grow in the absence of IL-3. The presence of the vector in individual clones was verified by integration site analysis and measuring GFP expression by using flow cytometry.
Statistical analysis.
Statistical analysis of the mutagenesis frequency data (Table 1 and see Fig. 3b) was performed on the number of wells with IL-3-independent clones or the number of IL-3-independent mutants in the mock- and vector-transduced groups. For unpaired data analysis, the numbers of such wells or mutants from individual experiments using similar vectors were pooled, i.e., the numbers of observations in the mock, WT LTR, and SIN LTR groups as shown in Table 1 were 10, 6, and 5, respectively; the data for mock and RV experiment groups in Fig. 3b include the results from four observations each. Differences in the numbers of such wells or mutants for the three groups (mock, WT LTR, and SIN LTR) for which results are shown in Table 1 and the two groups (mock and RV experiment) for which results are shown in Fig. 3b were compared by using parametric (t test) and nonparametric (Mann-Whitney U test) tests (Table 2 and legend to Fig. 3).
TABLE 1.
Lentiviral mutagenesis frequenciesa
| Type of vector, expt | Selection methodc | No. of target cells | Mock transduction mutants
|
No. of integrants | Vector-transduced mutants
|
||||
|---|---|---|---|---|---|---|---|---|---|
| No. of wells or flasks with clonesd | Mutant cell frequencye | No. of wells or flasks with clonesd | No. of mutantsf | Mutant cell frequencye | Integrant frequencyg | ||||
| WT HIV LTR | |||||||||
| LV1b | IL-3 | 4.0 × 108 | 0 | <2.5 × 10−9 | 1.0 × 109 | 8 | 1 | 2.5 × 10−9 | 1.0 × 10−9 |
| LV2 | IL-3 | 1.0 × 107 | 0 | <1.0 × 10−7 | 1.2 × 108 | 2 | 2 | 2.0 × 10−7 | 1.7 × 10−8 |
| LV3 | IL-3 | 5.0 × 107 | 2 | 4.0 × 10−8 | 1.9 × 108 | 13 | 13 | 2.6 × 10−7 | 6.8 × 10−8 |
| LV3 | bGH | 5.0 × 107 | 0 | <2.0 × 10−8 | 1.9 × 108 | 23 | 18 | 3.6 × 10−7 | 9.5 × 10−8 |
| LV6 | bGH | 3.6 × 107 | 4 | 1.1 × 10−7 | 1.9 × 108 | 13 | 9 | 2.5 × 10−7 | 4.7 × 10−8 |
| LV7 | bGH | 3.6 × 107 | 0 | <2.8 × 10−8 | 2.9 × 108 | 14 | 7 | 1.9 × 10−7 | 2.4 × 10−8 |
| SIN HIV LTR | |||||||||
| LV4 | IL-3 | 5.0 × 107 | 1 | 2.0 × 10−8 | 3.1 × 108 | 7 | 4 | 8.0 × 10−8 | 1.3 × 10−8 |
| LV5 | IL-3 | 5.0 × 107 | 0 | <2.0 × 10−8 | 5.4 × 108 | 0 | 0 | <2.0 × 10−8 | <1.9 × 10−9 |
| LV4 | bGH | 5.0 × 107 | 3 | 6.0 × 10−8 | 3.1 × 108 | 6 | 1 | 2.0 × 10−8 | 3.3 × 10−9 |
| LV5 | bGH | 5.0 × 107 | 0 | <2.0 × 10−8 | 5.4 × 108 | 0 | 0 | <2.0 × 10−8 | <1.9 × 10−9 |
| LV8 | bGH | 3.6 × 107 | ND | ND | 2.1 × 108 | 0 | 0 | 2.7 × 10−8 | <4.8 × 10−9 |
The WT HIV-1 LTR lentiviral vector HV was used in experiments LV1, -2, -3, -6, and -7; the SIN lentiviral vector HR SIN CSGW (expressing GFP) was used in experiments LV4 and -5; and SINLV SFFV IL2RG (expressing IL2RG) was used in experiment LV8.
Pooled data from six independent mutagenesis experiments in BAF3 cells are shown for experiment LV1.
IL-3-independent mutants were either selected according to the IL-3 selection protocol (IL-3) or selected in serum supplemented with 1 μg/ml bGH (bGH).
The number of flasks (IL-3 selection) or wells (bGH selection) in which IL-3-independent clones grew out was scored after 7 to 10 days or 4 to 6 weeks, respectively. ND, not done.
The mutant cell frequency was calculated by dividing the number of independent mutants by the total number of target cells in each experiment. ND, not determined.
Replicate (“sibling”) clones were eliminated following Southern blot and/or vector integration site analysis, and the number of independent mutants obtained in each experiment is shown.
The integrant frequency was calculated by dividing the number of independent mutants by the total number of integrants in each experiment. In experiment LV1, the number of integrants was calculated by multiplying the total number of target cells by the MOI. The MOI was determined by GFP fluorescence-activated cell sorting at 72 h posttransduction. In all other experiments, the average number of vector copies per cell was determined by qPCR at 72 h posttransduction. This number was multiplied by the number of target cells to obtain the number of integrants.
FIG. 3.
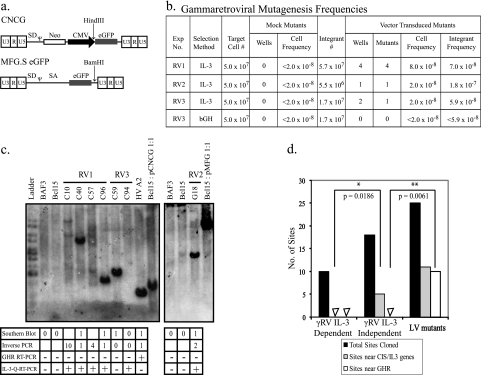
Characterization of gammaretroviral-vector insertional mutants. (a) The gammaretroviral vectors CNCG and MFG.S eGFP are shown. The locations of the unique HindIII and BamHI restriction sites used in Southern blotting of CNCG and MFG.S eGFP insertional mutants, respectively, are also shown. (b) Gammaretroviral-mutagenesis frequencies. The CNCG vector was used in experiments RV1 and RV3; MFG.S eGFP was used in RV2. IL-3-independent mutants were selected either according to the IL-3 selection protocol (IL-3; RV1 to RV3) or in serum supplemented with 1 μg/ml bGH (bGH; RV3 only). The number of flasks (IL-3 selection) or wells (bGH selection) in which IL-3-independent clones grew out was scored after 7 to 10 days and 4 to 6 weeks, respectively (fourth and seventh columns). Observations from these four experiments using gammaretroviral vectors were pooled. Statistically significant differences between the numbers of wells (P = 0.0286) and mutants (P = 0.0286) in the mock- and vector-transduced groups were seen with a Mann-Whitney U test. The cell frequency was calculated by dividing the number of independent mutants by the total number of target cells in each experiment. The integrant frequency was calculated by dividing the number of independent mutants by the total number of integrants in each experiment. (c) Southern blots of gammaretroviral insertional mutants from experiments RV1 to RV3. Genomic DNA was digested with HindIII (RV1 and RV3) or BamHI (RV2). Blots were probed with the cDNA for enhanced GFP (eGFP). The tables below the blots show estimated vector copy numbers and the number of integration sites in each mutant that were cloned by inverse PCR. In addition, RT-PCR for the Ghr transcript and qRT-PCR for the IL-3 transcript were performed as described in the legend for Fig. 1d. Mutant C94 was eliminated from the analysis as no integrated vector copies were demonstrated by qPCR. (d) Comparison of genes targeted by vector integration in gammaretroviral-vector-transduced IL-3-independent mutants (γRV IL-3 independent); gammaretroviral-vector-transduced IL-3-dependent clones, cloned by limiting dilution (γRV IL-3 dependent); and WT LTR lentiviral-vector-transduced IL-3-independent mutants (LV mutants). Sites near CIS genes or the IL-3 gene are significantly more frequent targets of gammaretroviral-vector integration in RV experiment mutants than in RV experiment clones (two-tailed P value, 0.0186; Mann-Whitney U test). The Ghr gene is preferentially targeted by WT LTR lentiviral-vector integration but not gammaretroviral-vector integration in IL-3-independent mutants (two-tailed P value, 0.0061; Mann-Whitney U test).
TABLE 2.
Unpaired data analysis of lentiviral mutagenesis frequency results in Table 1
| Statistical test |
P valuesa for nos. of wells with IL-3-independent clones and nos. of IL-3-independent mutants for:
|
|||||
|---|---|---|---|---|---|---|
| All mock (n = 10) vs WT LTR (n = 6)
|
All mock (n = 10) vs SIN LTR (n = 5)
|
WT LTR (n = 6) vs SIN LTR (n = 5)
|
||||
| Wells | Mutants | Wells | Mutants | Wells | Mutants | |
| Independent t test | 0.0002*** | 0.0036** | 0.2340 | 1 | 0.0220* | 0.0379* |
| Mann-Whitney U test | 0.0017** | 0.0075** | 0.6787 | 1 | 0.0173* | 0.0303* |
*, P < 0.05; **, P < 0.01; ***, P < 0.001.
In each experiment, a single population of Bcl15 target cells was subjected to either mock or vector transduction, which might affect the level of spontaneous transformation to IL-3 independence. Hence, we also carried out paired data analysis. Differences in the numbers of wells with IL-3-independent clones/IL-3-independent mutants obtained in each experiment following mock or vector transduction were analyzed by using parametric (paired t tests) and nonparametric (Wilcoxon signed-rank test) tests (Table 3). Two-tailed P values are shown; these were calculated by using Analyze-it and Microsoft Excel software.
TABLE 3.
Paired data analysis of lentiviral mutagenesis frequency results in Table 1
| Statistical test |
P valuesa for nos. of wells with IL-3-independent clones and nos. of IL-3-independent mutants for:
|
|||
|---|---|---|---|---|
| Mock vs WT LTR (n = 6)
|
Mock vs SIN LTR (n = 4)
|
|||
| Wells | Mutants | Wells | Mutants | |
| Paired t test | 0.0115* | 0.0366* | 0.2152 | 0.8240 |
| Wilcoxon signed-rank test | 0.0313* | 0.0313* | 0.5000 | 1 |
Two-tailed P values are shown; statistically significant differences are shown by asterisks.
Southern blot analysis.
Genomic DNA was digested with BamHI for the HV, CSGW, and MFG.S eGFP vectors and HindIII for the CNCG vector. DNA fragments were blotted by neutral transfer onto a nylon membrane (Hybond-XL; Amersham Bioscience). The blot was hybridized overnight at 65°C with a 720-bp α32P-labeled GFP cDNA probe diluted in Rapid-Hyb buffer (GE Healthcare) supplemented with 10 μg/ml salmon testis DNA (Sigma). After being washed, the blot was exposed to a phosphor screen and visualized by using a Storm PhosphorImager (GE Healthcare). The image was analyzed by using ImageQuant software (GE Healthcare). Some blots were also exposed to Kodak BioMax film.
Insertion site identification.
For inverse PCR (5), genomic DNA was digested with NlaIII (HV vector) or MspI (HV/CNCG/MFG vectors) and then ligated. Nested PCR on circularized DNA was performed by using divergently oriented primers complementary to the HIV-1 or MLV LTR. The CSGW vector integration sites were cloned by using ligation-mediated PCR (LM-PCR) (38); DNA was digested overnight with NlaIII, MspI, or TacI restriction enzyme. A second digest was carried out for 2 h with either EcoRI (for NlaIII or MspI initial digest) or SacI (for TacI initial digest). The inverse/LM-PCR products were size separated on an agarose gel, cloned into pGEM-T Easy vector (Promega), and sequenced (Lark Technologies). Vector-genome junction sequences were blasted against the mouse genome released in August 2007 (www.ensembl.org/Mus_musculus). Some integration sites were confirmed by site-specific PCR. In experiments LV3, -6, and -7, multiplex PCR was used to identify additional HV vector integration sites in the Ghr locus. Seven forward primers hybridizing every 2 kb in the 15-kb region immediately upstream of Ghr exon 2 and an HV vector-specific reverse primer were used.
RNA analysis.
For reverse transcriptase PCR (RT-PCR), RNA was extracted by using an RNeasy kit (Qiagen). RNA was reverse transcribed by using a Protoscript first strand cDNA synthesis kit (New England Biolabs). Either a gene-specific reverse primer or random primers were used to prime the RT reaction. An RT-PCR for the presence of Ghr transcript was then performed. For qRT-PCR, cDNA was generated from RNA by using a QuantiTect reverse transcription kit (Qiagen) and IL-3 transcript was quantified by TaqMan probe qRT-PCR. An IL-3 cDNA in the pGEM-T Easy vector was used for the quantification of IL-3 copies. For each sample, TaqMan quantification of 18S rRNA copies was performed in parallel. A 600-bp fragment of the 18S rRNA gene cloned into pCR-Blunt II-TOPO was used for the quantification of 18S rRNA copies. The ratio of IL-3 copies to 18S copies was calculated from duplicate samples.
For 5′ rapid amplification of cDNA ends-PCR (5′ RACE-PCR), total RNA was extracted from 1 × 107 cells by using RNA-Zol B (Biogenics). 5′ RACE was carried out by using a 5′ RACE system (Invitrogen), using a Ghr exon 4 primer for the RT reaction. The products were separated on a 1% Tris-acetate-EDTA agarose gel, cloned into TOPO TA cloning vector (Invitrogen), and sequenced. For Northern blotting, 20 μg of RNA was loaded onto a morpholinepropanesulfonic acid (MOPS; Sigma) formaldehyde (BDH) RNA gel and blotted overnight by basic transfer on to a Hybond N+ membrane (Amersham Biosciences). The blot was hybridized overnight at 68°C with an α32P-labeled Ghr exon 11-3′ untranslated region probe diluted in SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), milk powder mix (Oxoid), and 25% dextran sulfate supplemented with 10 μg/ml salmon testis DNA (Sigma). After being washed, the blot was exposed either to a storage phosphor screen (Kodak) or to a Kodak BioMax film (Kodak).
Protein analysis.
For Western blotting, 1 × 107 cells were lysed with phosphate-buffered saline (PBS) containing 1% IGEPAL (Sigma) and 1× protease inhibitor cocktail (Roche) and then separated on a 10% polyacrylamide gel, blotted onto Hybond ECL nitrocellulose membrane (Amersham Biotech), and probed with biotinylated anti-mouse GHR antibody (BAF1360; R&D systems) and then streptavidin horseradish peroxidase complex (Amersham Biosciences) in Tris-buffered saline with 5% milk powder (Oxoid). Detection was carried out by using enhanced chemiluminescence Western blot detection reagents (Amersham Biosciences).
For GHR flow cytometry, cells were incubated with 5 μg/ml biotinylated goat anti-mouse GHR antibody (BAF1360; R&D systems) which was diluted in blocking buffer (1× PBS, 2% FCS, 0.01% Na azide). Biotinylated normal goat immunoglobulin G (BAF108; R&D systems) was used as a control. Cells were then incubated with allophycocyanin-conjugated streptavidin (17-4317; eBioscience). The samples were analyzed by flow cytometry using a BD LSR flow cytometer and CellQuest software. For STAT5 staining, cells were washed in serum-free medium and then cultured in serum-free medium plus 0.5% bovine serum albumin (BSA) for 6 h. Cells were then stimulated for 12 min with 400 ng/ml human GH (kind gift of Pharmacia-Upjohn), PBS-0.5% BSA as a negative control, or 100 ng/ml recombinant murine IL-3 (403-ML/CF; R&D systems) as a positive control. Following stimulation, cells were incubated for 3 min at room temperature with 100 μl reagent A fixation medium (Fix and Perm from Caltag or GAS 004 from Invitrogen) and then with 500 μl chilled flow buffer (PBS-0.5% BSA-0.1% Na azide) containing 1 mM pervanadate and 20 mM sodium fluoride and fixed in ice-cold methanol. Cells were then incubated with phycoerythrin-conjugated mouse anti-phospho-Stat5 antibody (612567; BD Phosflow) or phycoerythrin-conjugated mouse immunoglobulin G1κ (555749; BD Pharmingen). Cells were analyzed by flow cytometry.
Proliferation assay.
Cells were cultured in serum-free medium for 6 h and then plated in 96-well plates in medium with FCS or bGH (see Fig. 2f). On day 2, cells were pulsed with 1 μCi [3H]thymidine per well (ICN Biomedical, High Wycombe, United Kingdom). On day 3, DNA from cells was harvested on a filtermat using a Tomtec harvester model 96. Cell proliferation was measured by liquid scintillation counting (Microbeta Systems).
FIG. 2.
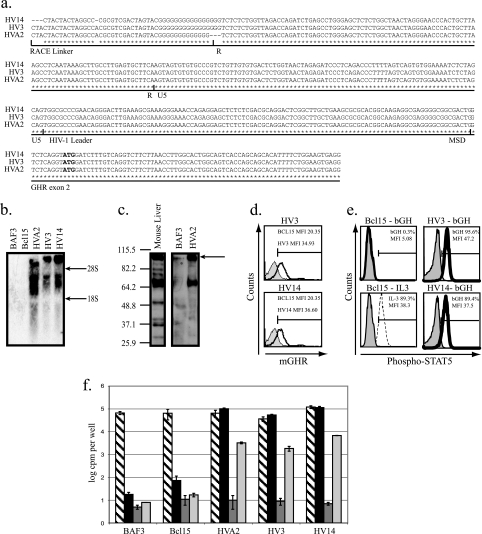
Structure of the Ghr transcript, GHR protein expression, and results of functional studies. (a) 5′ RACE was performed on mutants HV A2, HV3, and HV14. A 590-bp product was cloned and sequenced. An alignment of the first 414 bp of the 5′RACE product is shown here. The RACE linker cassette is 46 bp; the Ghr transcript starts at the HIV-1 R region. It contains 289 bp of HV vector sequence from before it splices from the HIV-1 major splice donor to the splice acceptor of Ghr exon 2. The ATG start codon for Ghr is shown in bold. (b) In mutants HV A2, HV3, and HV14, a 5-kb transcript was detected by Northern blotting using a probe that detects the intracellular domain of Ghr. Also shown are the sizes of mouse 28S (4.7 kb) and 18S (1.7 kb) rRNA. (c) A 103-kDa GHR protein was detected in mutant HV A2 and mouse liver, but not in the parental BAF3 cell. A smaller, ∼70kDa protein, the secreted GH binding protein, is also seen. Immunoblot analysis of total cell lysate was performed using an antibody that recognizes the mouse GHR extracellular domain, common to GHR and GH binding protein. Sizes in kDa are shown on the left. (d) Mutants HV3 and HV14 (black peak) express GHR on the cell surface in FACS, unlike the parental Bcl15 cell (solid gray peak), demonstrated by surface staining using an antibody that recognizes the mouse GHR (mGHR) extracellular domain. (e) Parental cells and mutants were stimulated with PBS-0.2% BSA (solid gray peak) or 400 ng/ml human GH (black peak) for FACS. Intracellular staining for phosphorylated STAT5 was performed. Stimulation with 100 ng/ml IL-3 (dashed black peak) also results in STAT5 phosphorylation and acted as a positive control. Selection method (bGH or IL-3) is indicated. MFI, mean fluorescence intensity. (f) Parental cells and mutants were cultured for 48 h in DMEM, 10% FCS, 10% WEHI-3B-cell-conditioned medium (hatched bars); DMEM, 10% FCS (solid black bars); serum-free medium (solid dark-gray bars); or serum-free medium supplemented with 1 μg/ml bGH (solid light-gray bars). Cell proliferation was measured by [3H]thymidine uptake. Error bars indicate standard errors of the means calculated from the results of two independent experiments.
RESULTS
Our aim was to develop a cell line assay for insertional mutagenesis. This was based on the observation that retrovirus insertion can be used to identify genes involved in growth factor signaling by selection of growth factor-independent mutants from growth factor-dependent hematopoietic cell lines (32). This technique has also been performed using gammaretroviral-vector infection of the murine IL-3-dependent cell line BAF3 (24) to identify bcl-X as a molecule that inhibits apoptosis without stimulating proliferation (33). In our first experiments, we transduced BAF3 cells with the lentiviral vector HV (Fig. 1a) in the presence of IL-3 and then removed IL-3 from the culture medium after 4 days and plated cells at 5 × 105 cells/well in 24-well plates in medium with FCS. None of the wells contained expanding clones of cells by visual inspection after 7 days, so we then readded IL-3 and maintained it in the wells until cells grew in some. We then reselected these growing clones in the absence of IL-3 and isolated a single mutant from a large number of transduced cells (Table 1, experiment LV1, and Fig. 1b, mutant HV A2). This selection protocol, designated IL-3 selection, was used in further experiments where indicated.
FIG. 1.
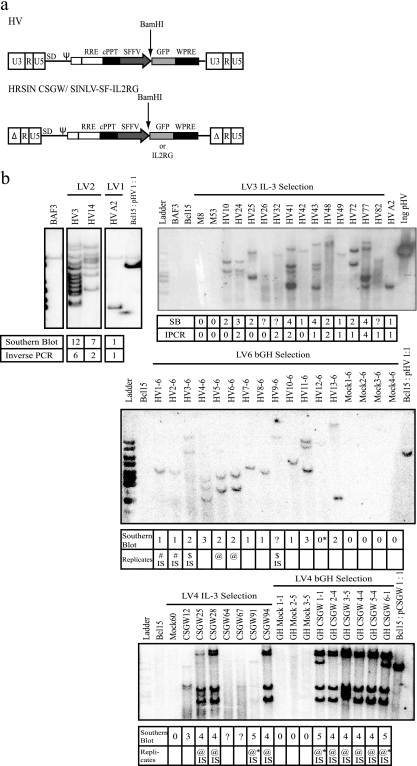
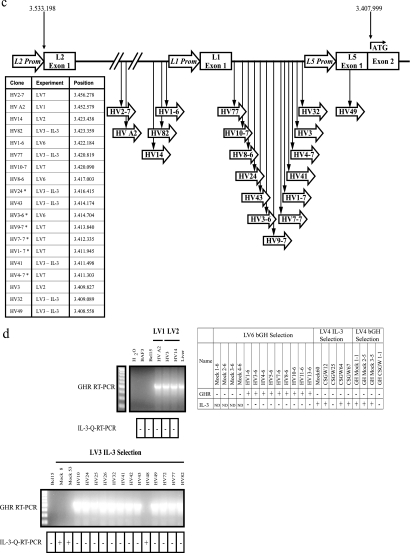
Characterization of lentiviral-vector insertional mutants. (a) HIV-1 derived lentiviral vectors used in insertional mutagenesis experiments shown as proviral genomes. The HV vector has WT HIV-1 LTRs and an internal SFFV LTR promoter driving GFP expression. The SIN lentiviral vectors HRSIN-CSGW and SINLV SFFV IL2RG (SINLV-SF-IL2RG) lack 400 bp of the U3 region of the HIV-1 LTRs; an internal SFFV promoter drives GFP or IL2RG expression. The location of the unique BamHI restriction site used in Southern blotting is also shown. (b) Southern blots (SB) of lentiviral insertional mutants from experiments LV1, LV2, LV3 (IL-3 selection), LV6, and LV4. Genomic DNA was digested with BamHI, and blots were probed with GFP. Phosphorscreen images are shown of all blots except that of LV3 (IL-3 selection), which was exposed to Kodak BioMax film. The first row of the table below each blot shows estimated vector copy numbers from the Southern blot. In experiments LV1, LV2, and LV3 (IL-3 selection), mutants were independent, as defined by different (or additional) bands on Southern blots, combined with integration site cloning. The number of integration sites cloned by inverse PCR from these mutants is shown in the table below the blots. Replicate clones were obtained in experiments LV4 and LV6; the final number of mutants shown in Table 1 does not include replicate clones. The replicates are identified by common symbols. IPCR, number of insertion site loci identified in the mutant by integration site PCR; IS, integration sites shared by these clones were verified by site-specific PCR; ?, vector copy number was not determined; *, mutant HV12-6 from LV6 was eliminated from further analysis: though it shows a faint band on Southern blot, no integrated vector copies were detected by qPCR. Ladder, 1-kbp DNA marker (Fermentas). (c) Locations of insertions into the Ghr allele on mouse chromosome 15. The 19 HV vector insertions that we mapped occurred in the same transcriptional orientation as Ghr and were in the 125-kb region upstream of exon 2, the first coding exon of Ghr. *, Ghr insertion was recovered by multiplex PCR spanning the region between L1 exon 1 and L5 promoter; all other Ghr insertions were obtained by inverse PCR. (d) RT-PCR for the Ghr transcript. A forward primer in exon 4 and a reverse primer in exon 8a were used to amplify a 552-bp section of the Ghr transcript. Representative gels are shown for mutants from experiments LV1, LV2, and LV3 (IL-3 selection). qRT-PCR to determine IL-3 transcript levels was also performed. A positive result (+) indicates >103 IL-3 copies per 109 18S rRNA copies; a negative result (−) indicates <101 IL-3 copies per 109 18S rRNA copies. The table summarizes the expression of these two transcripts by the LV6 and LV4 experiment mutants. ND, not done. Ladder, 100-bp DNA marker (Fermentas).
To improve the frequency of mutagenesis, we reasoned that cells already overexpressing a bcl-2 transgene, the BAF3 derivative Bcl15 (8), might be more robust during selection, allowing us to rescue more mutants. This proved to be the case, and we isolated a further 15 mutants from two HV vector-transduced populations at a cell frequency higher than the background frequency of spontaneous IL-3-independent mutants (Table 1, experiments LV2 and LV3 [IL-3 selection], and Fig. 1b, mutants HV3, -10, -14, -24, -25, -32, -41, -42, -43, -48, -49, -72, -77, and -82). We estimated the number of HV integrants in these mutants by Southern blot analysis (Fig. 1b) and cloned HV integration sites from many of them by inverse PCR. Table 4 shows that 10 of these mutants contained an HV insertion in the GH receptor (Ghr) locus. Mutants HV A2 and HV49 apparently contain a single HV insertion in Ghr, suggesting that this event is capable of making both BAF3 and Bcl15 cells IL-3 independent. Ghr is also likely to participate in primary cell transformation by retroviruses as it is defined as a CIS, appearing six times in the RTCGD (2).
TABLE 4.
Lentiviral vector integration sites in IL-3-independent mutants (experiments LV1 to LV4)a
| Expt, clone | Copy no. in SBb | No. of ISc | In gened | Genee | Entrez Gene ID | Chromosome | Distance to TSSf | Orientationg | Predicted gene functionh | No. of RTCGD hit(s)i |
|---|---|---|---|---|---|---|---|---|---|---|
| LV1 | ||||||||||
| HVA2 | 1 | 1 | Y | Ghr | 14600 | 15 | + | Growth hormone receptor | 6 CIS | |
| LV2 | ||||||||||
| HV3 | 12 | 6 | Y | Ghr | 14600 | 15 | + | Growth hormone receptor | 6 CIS | |
| Y | Dock2 | 94176 | 11 | 323 kb, intron 27-28 | + | Cytoskeletal organization | ||||
| Y | Stag1 | 20842 | 9 | 149 kb, intron 7-8 | + | Cell cycle, cell division | 2 CIS | |||
| Y | Hcfc2 | 67933 | 10 | 5,867 bp, intron 3-4 | − | |||||
| Y | Plekha5 | 109135 | 6 | 103 kb, intron 5-6 | − | |||||
| Y | 2610020 H08 Rik | 434234 | 7 | 48 kb, intron 1-13 | + | |||||
| HV14 | 7 | 2 | Y | Ghr | 14600 | 15 | + | Growth hormone receptor | 6 CIS | |
| Y | Usp45 | 77593 | 4 | 34 kb, intron 9-10 | + | Ubiquitin cycle | ||||
| LV3 | ||||||||||
| HV24 | 3 | 2 | Y | Ghr | 14600 | 15 | + | Growth hormone receptor | 6 CIS | |
| N | 14 | |||||||||
| HV32 | 2 | Y | Ghr | 14600 | 15 | + | Growth hormone receptor | 6 CIS | ||
| N | 7 | |||||||||
| HV41 | 4 | 2 | Y | Ghr | 14600 | 15 | + | Growth hormone receptor | 6 CIS | |
| Y | Suclg2 | 20917 | 6 | 26 kb, intron 1-2 | + | Metabolic process, GDP forming activity, tricarboxylic acid cycle | 1 | |||
| HV43 | 4 | 1 | Y | Ghr | 14600 | 15 | + | Growth hormone receptor | 6 CIS | |
| HV48 | 2 | Y | Senp1 | 223870 | 15 | 7 kb, intron 2-3 | − | Multicellular organismal development, protein desumoylation | ||
| Y | Plekha5 | 109135 | 6 | 3 kb, intron 1-2 | + | |||||
| HV49 | 1 | 1 | Y | Ghr | 14600 | 15 | + | Growth hormone receptor | 6 CIS | |
| HV72 | 2 | 1 | Y | Ccdc111 | 408022 | 8 | 10 kb, intron 4-5 | − | DNA replication | |
| HV77 | 4 | 4 | Y | Ghr | 14600 | 15 | Growth hormone receptor | 6 CIS | ||
| Y | Gmps | 229363 | 3 | 1,217 bp, intron 1-2 | + | Purine nucleotide biosynthetic process, glutamine metabolic process | ||||
| N | 16 | |||||||||
| Y | Hif1a | 15251 | 12 | 15 kb, intron 8-9 | + | Angiogenesis, cartilage development | 1 | |||
| HV82 | 1 | Y | Ghr | 14600 | 15 | + | Growth hormone receptor | 6 CIS | ||
| LV4 | ||||||||||
| GH CSGW 1-1 | 5 | 4 | Y | Zfp407 | 240476 | 18 | 23 kb, intron 1-2 | + | ||
| Y | Cnot4 | 53621 | 6 | 32 kb, intron 1-2 | − | CCR4 NOT transcription complex, subunit 4 | ||||
| Y | Gmeb2 | 229004 | 2 | 26 kb, intron 4-5 | − | Glucocorticoid modulatory element binding protein 2 | ||||
| Y | Wipi2 | 74781 | 5 | 12 kb, intron 2-3 | + | WD repeat domain, phosphoinositide interacting 2 | ||||
| GH CSGW 2-4 | 4 | 3 | Y | Cnot4 | 53621 | 6 | 32 kb, intron 1-2 | − | CCR4 NOT transcription complex, subunit 4 | |
| Y | Gmeb2 | 229004 | 2 | 26 kb, intron 4-5 | − | Glucocorticoid modulatory element binding protein 2 | ||||
| Y | Wipi2 | 74781 | 5 | 12 kb, intron 2-3 | + | WD repeat domain, phosphoinositide interacting 2 |
Integration sites were cloned by inverse PCR or LM-PCR from mutants obtained in experiments LV1, LV2, LV3 (IL-3).
SB, Southern blotting. Indicates estimated vector copy number determined by Southern blotting.
IS, insertion site(s). Data show the number of insertion site loci identified in the mutant by integration site PCR.
Y, yes; N, no.
If an insertion is not in a gene, the nearest gene(s) within 30 kb up- or downstream of the insertion site is listed. The Ghr insertion sites in mutants HV24 and HV77 were cloned by multiplex PCR; all other Ghr insertions sites were obtained by inverse PCR.
TSS, transcription start site. For insertion sites, the distance to the transcription start site of the nearest gene and the intron into which the vector is inserted, where applicable (between numbered exons), are shown, except in the case of Ghr insertions.
Indicates whether the vector insertion occurred in the same (+) or in the opposite (−) orientation as the gene's transcript.
Predicted gene function is shown where known.
The number of appearances in the RTCGD of each insertion site locus is shown where applicable, and loci defined as CIS (http://RTCGD.ncifcrf.gov) are indicated.
Figure 1c shows that all the HV vector integrants were upstream of the first coding exon of Ghr (exon 2) and in the same orientation as the Ghr transcript. The mouse L2 promoter is normally predominantly active in the liver, and the L1 and L5 promoters are more active in the placenta (12, 19). Fig. 1d shows that the 10 mutants from experiments LV2 and LV3 with HV inserted in the Ghr locus all expressed Ghr RNA, detected by RT-PCR. In contrast, the parental cells, the two spontaneous mutants, and HV48, which did not contain a Ghr insertion, did not express Ghr transcripts. However, we did detect IL-3 mRNA by qRT-PCR in the two spontaneous mutants and in HV48 (Fig. 1d). This suggests that the upregulation of IL-3 gene expression is a common cause of “background” spontaneous mutants in this assay. It is possible that HV48 is a spontaneous mutant; however, it also contains one insertion site in common with HV3, in the Plekha5 gene, which may contribute to the IL-3-independent phenotype.
To determine the mechanism of Ghr gene regulation by HV insertion, we cloned the 5′ end of the major Ghr transcripts in the HV A2, HV3, and HV14 cells by using 5′ RACE-PCR. Each mutant yielded a single predominant PCR product; the sequences are shown in Fig. 2a. The transcripts start at the 5′ end of the R region of the HIV LTR and contain vector sequences to the point of the HIV major splice donor; they then splice to the Ghr splice acceptor at the start of exon 2. This structure suggests that the HIV enhancer and promoter, within the HIV U3 region, are active in the HV vectors integrated at this locus and that this drives expression from the HIV transcription start site at the start of the R region. In agreement with this, we did not detect transcription of Ghr exon 1 or the HIV U3 region by RT-PCR (data not shown). Also supporting this hypothesis, these three clones contain a Ghr transcript of approximately 5 kb, detected by Northern blotting (Fig. 2b), consistent in size with a fused vector Ghr transcript. We can detect two proteins of sizes consistent with GHR and the smaller, secreted GH binding protein, generated by alternative splicing of the 3′ end of the Ghr transcript, by immunoblotting (15) (Fig. 2c), and we can detect surface GHR expression by fluorescence-activated cell sorting (FACS) (Fig. 2d). IL-3 stimulated rapid STAT5 phosphorylation in the parental cells, and GH stimulated STAT5 phosphorylation to a similar extent in the GHR-expressing HV mutants (Fig. 2e).
Hematopoietic cells engineered to express GHR proliferate in GH. In BAF3 cells, this effect is due to a direct stimulation by GH, not to induction of insulin-like growth factor secretion (3). Figure 2f shows that the GHR-expressing HV mutants proliferated in medium with FCS (which contains some bGH), explaining their growth in the absence of IL-3. They also proliferated in serum-free medium supplemented with recombinant bGH, though FCS was more potent, suggesting that it contains additional growth factors (Fig. 2f).
As the predominant HV mechanism of mutagenesis involved GHR expression, we also selected three HV-transduced populations of cells in medium supplemented with both FCS and additional recombinant bGH. In this case, after cells were plated in 24-well plates, wells contained expanding clones without IL-3 readdition (Table 1, LV3, LV6, and LV7, selection in GH). Southern blot analysis demonstrates that these included a total of 34 independent insertional mutants (Fig. 1b; see Fig. S1 in the supplemental material), and the results in Fig. 1d (also see Fig. S1 in the supplemental material) show that they all expressed Ghr mRNA. Analysis of the number of mutants induced by the HV vector compared to the number of mock mutants shows a significant difference considering either the number of wells containing mutants or the number of independent mutants (Tables 2 and 3).
This rapid selection of cells in GH allowed us to screen variants of the HV vector to determine which components are necessary for insertional mutagenesis in the Ghr locus. We initially tested the vector HRSIN-CSGW or a derivative expressing IL2RG (Fig. 1a), both of which contain a deletion in the HIV enhancer/promoter, as we thought that the HIV enhancer/promoter was important for the expression of the Ghr transcripts. Table 1 shows that these vectors did not generate mutants compared to the background of spontaneous mutants when cells were selected either under IL-3 selection (experiments LV4 and LV5) or in the presence of GH (experiments LV4, LV5, and LV8). Analysis of the five CSGW vector-transduced mutants that we isolated shows that four (CSGW12, -25, -64, and -67) were potentially different (Fig. 1b). GH CSGW2-4 is identical to CSGW25, and CSGW1-1 is a subclone (Table 4). The CSGW mutants did not express Ghr mRNA (Fig. 1d); however, the spontaneous mutants and three of the CSGW mutants (CSGW12, -64, and -67) expressed IL-3 mRNA. It remains possible that the CSGW vector causes a low level of insertional mutagenesis, and we are continuing to investigate these mutants.
Finally, we measured insertional mutagenesis by gammaretroviral vectors in the Bcl15 cell assay. We used two gammaretroviral vectors (Fig. 3a), one being MFG.S, the backbone used in the common gamma-chain clinical trials (6). We isolated seven mutants after IL-3 selection at integrant frequencies similar to those observed with the HV vector (Fig. 3b). Southern blot analysis (Fig. 3c) demonstrates that these were independent clones, though C94 was apparently a spontaneous mutant having no vector insert (Fig. 3c) and yielding no integration sites by inverse PCR (data not shown). The integration sites in these gammaretroviral mutants were distinct from those in the HV mutants. Mutant G18 had an insertion just downstream of the IL-3 gene, and three of the other clones that we have analyzed, C40, C57, and C96, had insertions at CIS listed in the RTCGD (Table 5); note that Osbpl3 is adjacent to the HoxA locus, which is Evi7, with 25 hits in the RTCGD. The gammaretroviral insertions in Sema4b, Plekhg2, and IL-3 were within or downstream of the coding sequences, suggesting an enhancer insertion effect. Analysis of the 11 gammaretroviral-vector insertion sites from eight control cell clones that remained IL-3 dependent detected some genes listed in the RTCGD but none defined as CIS (Fig. 3d). No gammaretroviral-vector insertions were detected in GHR (Fig. 3d); we also selected some gammaretroviral-vector-transduced Bcl15 cells in GH and did not isolate mutants (Fig. 3b). This suggests that the insertions in CIS may contribute to the IL-3-independent phenotype (Table 5). None of the gammaretroviral-vector mutants expressed Ghr mRNA, although all expressed IL-3 mRNA. This suggests either that some or all of the integrations have an effect on IL-3 gene expression or that spontaneously upregulated IL-3 expression cooperates with retroviral-vector insertion to generate robust IL-3-independent cells. An interesting precedent which could be explained by either of these mechanisms was described for the WEHI-3B myeloid leukemia cell line, which has intracisternal A particle insertions in both the HoxB locus and the IL-3 locus and secretes IL-3 (4).
TABLE 5.
Gammaretroviral vector integration sites in IL-3-independent mutants (experiments RV1 and RV2)a
| Expt, clone | No. of ISb | In genec | Gened | Entrez Gene ID | Chromosome | Distance to TSSe | Orientationf | Predicted gene functiong | No. of RTCGD hit(s)h |
|---|---|---|---|---|---|---|---|---|---|
| RV1 | |||||||||
| C10 | 10 | Y | Slc36a3 | 215332 | 11 | 3,539 bp, intron 3-4 | + | Proton amino acid transporter | 1 |
| Y | Mamdc2 | 71738 | 19 | 96 kb, intron 8-9 | − | ||||
| N | 16 | ||||||||
| N | Srm | 20810 | 4 | 1,003 bp upstream | − | Spermidine synthase | |||
| Y | Psma6 | 26443 | 12 | 685 bp, intron 1-2 | + | Endopeptidase/hydrolase activity | 1 | ||
| N | 13 | ||||||||
| N | Zdhhc4 | 72881 | 5 | 165 bp upstream | − | Zinc ion binding, acetyltransferase activity | |||
| N | 4 | ||||||||
| N | Mvd | 192156 | 8 | 1,030 bp upstream | − | Diphosphomevalo-nate decarboxylase activity | |||
| N | Dusp6 | 67603 | 10 | 4,740 bp downstream | Dual specificity phosphatase 6 | 4 | |||
| C40 | 1 | N | Osbpl3 | 71720 | 6 | 46 kb upstream | + | Lipid transport, steroid metabolism | 4 CIS |
| C57 | 4 | Y | Myl4 | 17896 | 11 | 802 bp, intron 1-2 | + | Motor activity | 1 |
| N | Cdc27 | 217232 | 1,067 bp upstream | − | Cell division | ||||
| Y | Sema4b | 20352 | 7 | 21 kb, intron 2-3 | + | Cell differentiation, nervous system development | 5 CIS | ||
| N | 9 | ||||||||
| Y | Gata1 | 14460 | X | 4,241 bp, intron 1-2 | + | Transcription factor activity | |||
| C96 | 1 | Y | Rps16 | 20055 | 7 | 779 bp, intron 2-3 | + | Structural constituent of ribosome | |
| N | Zfp36/Plekhg2 | 101497 | 21 kb downstream | − | Guanyl nucleotide exchange factor activity | 9 CIS | |||
| RV2 | |||||||||
| G18 | 2 | N | Il3 | 16187 | 11 | 2,607 bp downstream | + | Cytokine/growth factor activity | |
| N | Fbxo9 | 71538 | 9 | 3,183 bp upstream | + | Ubiquitin-protein ligase activity | |||
| N | Ick | 56542 | 5,744 bp upstream | − | Protein kinase activity, signal transduction | ||||
| Control | |||||||||
| CNCG 2 | 1 | Y | Tpm3 | 59069 | 3 | 8,408 bp, intron 2-3 | + | Regulation of muscle contraction | 1 |
| CNCG 6 | 1 | N | Mll2 | 381022 | 15 | 1,564 bp upstream | − | Mixed-lineage leukemia 2 | |
| Rhebl1 | 69159 | 8,515 bp downstream | − | Ras homolog enriched in brain like 1 | |||||
| Dhh | 13363 | 26 kb downstream | − | 1 | |||||
| CNCG 14 | 1 | N | 8430427 H17Rik | 329540 | 2 | 13.6 kb upstream | − | 2 | |
| CNCG 15 | 2 | N | 18 | ||||||
| Sh3gl1 | 20405 | 17 | 653 bp upstream | − | SH3 domain-containing GRB2-like protein 1 | ||||
| Chaf1a | 27221 | 3,186 bp upstream | + | ||||||
| Mpnd | 68047 | 28 kb upstream | + | ||||||
| CNCG17 | 1 | N | Cnbp2 | 75064 | X | 12.8 kb downstream | − | 1 | |
| CNCG18 | 2 | N | 16 | ||||||
| N | TRAF3 | 22031 | 12 | 720 bp upstream | − | Tumor necrosis factor receptor-associated factor 3 | 2 | ||
| MFG4 | 1 | N | 12 | ||||||
| MFG26 | 1 | N | Dnmt3a | 13435 | 12 | 17 kb upstream | + | DNA methyltransferase | 1 |
Integration sites were cloned by inverse PCR or LM-PCR from mutants obtained in experiments RV1 and RV2. Mutants from experiments RV1 and RV2 were IL-3 independent. Clones from control experiments were IL-3 dependent.
IS, insertion site(s). Data show the number of insertion site loci identified in the mutant by integration site PCR.
Y, yes; N, no.
If an insertion is not in a gene, the nearest gene(s) within 30 kb up- or downstream of the insertion site is listed. The Ghr insertion sites were obtained by inverse PCR.
TSS, transcription start site. For insertion sites, the distance to the transcription start site of the nearest gene and the intron into which the vector is inserted, where applicable (between numbered exons), are shown, except in the case of Ghr insertions.
Indicates whether the vector insertion occurred in the same (+) or in the opposite (−) orientation as the gene's transcript.
Predicted gene function is shown where known.
The number of appearances in the RTCGD of each insertion site locus is shown where applicable, and loci defined as CIS (http://RTCGD.ncifcrf.gov) are indicated.
DISCUSSION
This cell line assay has detected similar rates but different mechanisms of mutagenesis by lentiviral and gammaretroviral vectors. The major mechanism used by the lentiviral vector involves an HIV LTR-initiated transcript that expresses Ghr. A similar insertional mutagenesis mechanism has been reported for retroviruses, using, for example, a cryptic splice donor within the virus to express a Myb protein (27). The activity of the HIV LTR in the Ghr locus in the absence of the HIV Tat protein is surprising, although LTR activity in the absence of Tat is required to reactivate the virus from latency (36). Why this particular locus favors lentiviral-vector mutagenesis in BAF3 cells is unclear, and it is likely that this is a cell-specific phenomenon. We do not know whether it is a preferred integration site, a preferred site for HIV LTR activity, a large intronic region preceding the first coding exon, or a combination of all these factors. However, the fact that this mechanism of mutagenesis can occur demonstrates that the HIV LTR should be inactivated by enhancer/promoter deletion, which we have shown reduces the rate of mutagenesis, to improve safety. Deletion of the enhancer/promoter is readily achieved, whereas deletion of the splice donor site is difficult as it is required to inhibit vector polyadenylation at the 5′ LTR. In a recent AIDS gene therapy clinical trial, a WT HIV-1 LTR lentiviral vector was used to drive antisense RNA against the HIV-1 envelope, so that Tat would activate expression in HIV-infected cells (17). In this type of vector, a splice acceptor is also present, which may prevent lentiviral-vector activation of downstream genes.
In comparison with the lentiviral vector, one mechanism of mutagenesis by MLV retroviral vectors in these cells appears to be that of enhancer insertion into the IL-3 gene or CIS. These results suggest that this cell line is a reasonable surrogate assay for the transformation of primary hematopoietic cells by MLV vectors. Why MLV vectors do not cause insertional mutagenesis by Ghr upregulation, either by MLV enhancer activity on the Ghr promoter or by a mechanism similar to that of the HIV vector, is also unclear. The MLV vectors do contain a splice donor site, and the CNCG vector does not contain a splice acceptor. The two types of vector show different integration site preferences; both favor integration into coding rather than noncoding regions. However, MLV tends to integrate near the transcription start site, and HIV shows a stronger preference for expressed genes (20, 38). These general preferences do not explain why Ghr in mouse cells is not a target for MLV.
This type of assay does not provide a clinically relevant safety measure. An animal disease model, perhaps engineered to accelerate tumor formation, might be more clinically relevant. However, our assay does provide a general method to quantitate insertional mutagenesis. It can be used to test a number of other types of retroviral vectors, and it can also be adapted for human cells by using a human cytokine-dependent cell line.
Supplementary Material
Acknowledgments
This work was funded by the United Kingdom Department of Health, the United Kingdom Health and Safety Executive, and the International Journal of Experimental Pathology.
We thank Mustafa Ceylan for IL-3 qRT-PCR analysis; Abhinav Gupta for mutagenesis experiments; and Giada Mattiuzzo, Conrad Vink and David Escors for help with experiments. We thank Gregg Bogosian, Research Director in Agricultural Biotechnology, Monsanto, for bGH.
M.B. designed and performed most experiments; S.L.S. contributed inverse PCR and Fig. 2a, b, and c; S.K. contributed to Table 5 and to the HV mutagenesis experiments; E.F.G. contributed advice and Fig. 2f; I.C.R. contributed advice; Y.T. conceived and supervised the work; M.K.C. conceived and supervised the work and wrote the paper.
Footnotes
Published ahead of print on 22 October 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aiuti, A., S. Slavin, M. Aker, F. Ficara, S. Deola, A. Mortellaro, S. Morecki, G. Andolfi, A. Tabucchi, F. Carlucci, E. Marinello, F. Cattaneo, S. Vai, P. Servida, R. Miniero, M. G. Roncarolo, and C. Bordignon. 2002. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science 2962410-2413. [DOI] [PubMed] [Google Scholar]
- 2.Akagi, K., T. Suzuki, R. M. Stephens, N. A. Jenkins, and N. G. Copeland. 2004. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 32D523-D527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baixeras, E., S. Jeay, P. A. Kelly, and M. C. Postel-Vinay. 2001. The proliferative and antiapoptotic actions of growth hormone and insulin-like growth factor-1 are mediated through distinct signaling pathways in the Pro-B Ba/F3 cell line. Endocrinology 1422968-2977. [DOI] [PubMed] [Google Scholar]
- 4.Blatt, C., D. Aberdam, R. Schwartz, and L. Sachs. 1988. DNA rearrangement of a homeobox gene in myeloid leukaemic cells. EMBO J. 74283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carteau, S., C. Hoffmann, and F. Bushman. 1998. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol. 724005-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavazzana-Calvo, M., S. Hacein-Bey, G. de Saint Basile, F. Gross, E. Yvon, P. Nusbaum, F. Selz, C. Hue, S. Certain, J. L. Casanova, P. Bousso, F. L. Deist, and A. Fischer. 2000. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288669-672. [DOI] [PubMed] [Google Scholar]
- 7.Clive, D., W. G. Flamm, M. R. Machesko, and N. J. Bernheim. 1972. A mutational assay system using the thymidine kinase locus in mouse lymphoma cells. Mutat. Res. 1677-87. [DOI] [PubMed] [Google Scholar]
- 8.Collins, M. K., J. Marvel, P. Malde, and A. Lopez-Rivas. 1992. Interleukin 3 protects murine bone marrow cells from apoptosis induced by DNA damaging agents. J. Exp. Med. 1761043-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, B., E. Linney, and H. Fan. 1985. Suppression of leukaemia virus pathogenicity by polyoma virus enhancers. Nature 314550-553. [DOI] [PubMed] [Google Scholar]
- 10.Demaison, C., K. Parsley, G. Brouns, M. Scherr, K. Battmer, C. Kinnon, M. Grez, and A. J. Thrasher. 2002. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 13803-813. [DOI] [PubMed] [Google Scholar]
- 11.Du, Y., S. E. Spence, N. A. Jenkins, and N. G. Copeland. 2005. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood 1062498-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edens, A., and F. Talamantes. 1998. Alternative processing of growth hormone receptor transcripts. Endocr. Rev. 19559-582. [DOI] [PubMed] [Google Scholar]
- 13.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302415-419. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, Y., Y. Takeuchi, F. Martin, F. L. Cosset, K. Mitrophanous, and M. Collins. 2003. Continuous high-titer HIV-1 vector production. Nat. Biotechnol. 21569-572. [DOI] [PubMed] [Google Scholar]
- 15.Kopchick, J. J., and J. M. Andry. 2000. Growth hormone (GH), GH receptor, and signal transduction. Mol. Genet. Metab. 71293-314. [DOI] [PubMed] [Google Scholar]
- 16.Kustikova, O., B. Fehse, U. Modlich, M. Yang, J. Dullmann, K. Kamino, N. von Neuhoff, B. Schlegelberger, Z. Li, and C. Baum. 2005. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science 3081171-1174. [DOI] [PubMed] [Google Scholar]
- 17.Levine, B. L., L. M. Humeau, J. Boyer, R. R. MacGregor, T. Rebello, X. Lu, G. K. Binder, V. Slepushkin, F. Lemiale, J. R. Mascola, F. D. Bushman, B. Dropulic, and C. H. June. 2006. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. USA 10317372-17377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Z., O. S. Kustikova, K. Kamino, T. Neumann, M. Rhein, E. Grassman, B. Fehse, and C. Baum. 2007. Insertional mutagenesis by replication-deficient retroviral vectors encoding the large T oncogene. Ann. N. Y. Acad. Sci. 110695-113. [DOI] [PubMed] [Google Scholar]
- 19.Menon, R. K., A. Shaufl, J. H. Yu, D. A. Stephan, and R. P. Friday. 2001. Identification and characterization of a novel transcript of the murine growth hormone receptor gene exhibiting development- and tissue-specific expression. Mol. Cell. Endocrinol. 172135-146. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modlich, U., J. Bohne, M. Schmidt, C. von Kalle, S. Knoss, A. Schambach, and C. Baum. 2006. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood 1082545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montini, E., D. Cesana, M. Schmidt, F. Sanvito, M. Ponzoni, C. Bartholomae, L. Sergi Sergi, F. Benedicenti, A. Ambrosi, C. Di Serio, C. Doglioni, C. von Kalle, and L. Naldini. 2006. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 24687-696. [DOI] [PubMed] [Google Scholar]
- 23.Ott, M. G., M. Schmidt, K. Schwarzwaelder, S. Stein, U. Siler, U. Koehl, H. Glimm, K. Kuhlcke, A. Schilz, H. Kunkel, S. Naundorf, A. Brinkmann, A. Deichmann, M. Fischer, C. Ball, I. Pilz, C. Dunbar, Y. Du, N. A. Jenkins, N. G. Copeland, U. Luthi, M. Hassan, A. J. Thrasher, D. Hoelzer, C. von Kalle, R. Seger, and M. Grez. 2006. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 12401-409. [DOI] [PubMed] [Google Scholar]
- 24.Palacios, R., and M. Steinmetz. 1985. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell 41727-734. [DOI] [PubMed] [Google Scholar]
- 25.Pike-Overzet, K., D. de Ridder, F. Weerkamp, M. R. Baert, M. M. Verstegen, M. H. Brugman, S. J. Howe, M. J. Reinders, A. J. Thrasher, G. Wagemaker, J. J. van Dongen, and F. J. Staal. 2006. Gene therapy: is IL2RG oncogenic in T-cell development? Nature 443E5. [DOI] [PubMed] [Google Scholar]
- 26.Riviere, I., K. Brose, and R. C. Mulligan. 1995. Effects of retroviral vector design on expression of human adenosine deaminase in murine bone marrow transplant recipients engrafted with genetically modified cells. Proc. Natl. Acad. Sci. USA 926733-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen-Ong, G. L., H. C. Morse III, M. Potter, and J. F. Mushinski. 1986. Two modes of c-myb activation in virus-induced mouse myeloid tumors. Mol. Cell. Biol. 6380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiramizu, B., B. G. Herndier, and M. S. McGrath. 1994. Identification of a common clonal human immunodeficiency virus integration site in human immunodeficiency virus-associated lymphomas. Cancer Res. 542069-2072. [PubMed] [Google Scholar]
- 29.Shou, Y., Z. Ma, T. Lu, and B. P. Sorrentino. 2006. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc. Natl. Acad. Sci. USA 10311730-11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stocking, C., R. Kollek, U. Bergholz, and W. Ostertag. 1985. Long terminal repeat sequences impart hematopoietic transformation properties to the myeloproliferative sarcoma virus. Proc. Natl. Acad. Sci. USA 825746-5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stocking, C., C. Loliger, M. Kawai, S. Suciu, N. Gough, and W. Ostertag. 1988. Identification of genes involved in growth autonomy of hematopoietic cells by analysis of factor-independent mutants. Cell 53869-879. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, J., Y. Leverrier, and J. Marvel. 1998. Bcl-X is the major pleiotropic anti-apoptotic gene activated by retroviral insertion mutagenesis in an IL-3 dependent bone marrow derived cell line. Oncogene 161399-1408. [DOI] [PubMed] [Google Scholar]
- 34.Vogt, M., C. Haggblom, S. Swift, and M. Haas. 1985. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J. Virol. 55184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber, F., and W. Schaffner. 1985. Enhancer activity correlates with the oncogenic potential of avian retroviruses. EMBO J. 4949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams, S. A., H. Kwon, L. F. Chen, and W. C. Greene. 2007. Sustained induction of NF-κB is required for efficient expression of latent human immunodeficiency virus type 1. J. Virol. 816043-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woods, N. B., V. Bottero, M. Schmidt, C. von Kalle, and I. M. Verma. 2006. Gene therapy: therapeutic gene causing lymphoma. Nature 4401123. [DOI] [PubMed] [Google Scholar]
- 38.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 3001749-1751. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, F., S. I. Thornhill, S. J. Howe, M. Ulaganathan, A. Schambach, J. Sinclair, C. Kinnon, H. B. Gaspar, M. Antoniou, and A. J. Thrasher. 2007. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood 1101448-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.