Abstract
The 5′ untranslated region (5′UTR) of the dengue virus (DENV) genome contains two defined elements essential for viral replication. At the 5′ end, a large stem-loop (SLA) structure functions as the promoter for viral polymerase activity. Next to the SLA, there is a short stem-loop that contains a cyclization sequence known as the 5′ upstream AUG region (5′UAR). Here, we analyzed the secondary structure of the SLA in solution and the structural requirements of this element for viral replication. Using infectious DENV clones, viral replicons, and in vitro polymerase assays, we defined two helical regions, a side stem-loop, a top loop, and a U bulge within SLA as crucial elements for viral replication. The determinants for SLA-polymerase recognition were found to be common in different DENV serotypes. In addition, structural elements within the SLA required for DENV RNA replication were also conserved among different mosquito- and tick-borne flavivirus genomes, suggesting possible common strategies for polymerase-promoter recognition in flaviviruses. Furthermore, a conserved oligo(U) track present downstream of the SLA was found to modulate RNA synthesis in transfected cells. In vitro polymerase assays indicated that a sequence of at least 10 residues following the SLA, upstream of the 5′UAR, was necessary for efficient RNA synthesis using the viral 3′UTR as template.
Dengue fever is the most prevalent mosquito-borne viral disease in humans. Any of the four dengue virus (DENV) serotypes (DENV1 to DENV4) can produce clinical illness ranging from dengue fever, a nonspecific flu-like syndrome, to dengue hemorrhagic fever, a severe and sometimes fatal disease (14). The World Health Organization continues to report outbreaks of severe forms of the disease in the Americas and Asia. It is estimated that more than 50 million DENV infections occur annually. Despite the urgent need to control this virus, a licensed vaccine against DENV is not yet available. Incorporation of attenuating mutations into DENV clones has been shown to be a valuable tool for generating live vaccine candidates (32). In this regard, manipulation of the viral 5′ untranslated region (5′UTR) and the 3′UTR has been shown to be a feasible strategy. For instance, Whitehead and collaborators have produced a recombinant DENV that harbors a 30-nucleotide deletion at the 3′UTR. DENV1 Δ30 and DENV4 Δ30 are promising candidates that are being tested in clinical trials (6, 33). In addition, mutations within the 5′UTR have also been explored to generate attenuated DENVs (8, 27). Because a dengue vaccine must be able to protect against all four circulating virus serotypes, dissecting conserved cis-acting elements of the viral genome will aid to define mutations that can alter virulence in the four serotypes.
DENVs are members of the genus Flavivirus in the Flaviviridae family, together with other important human pathogens, such as yellow fever virus, West Nile virus (WNV), and Japanese encephalitis virus (13). The viral genome is a single-stranded RNA molecule with positive polarity, about 11 kb in length, which encodes a long polyprotein that is co- and posttranslationally processed by host and viral proteases, yielding three structural proteins (C, prM, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The coding sequence is flanked by 5′ and 3′UTRs, which contain cis-acting RNA elements that control viral translation, RNA synthesis, and encapsidation (22).
The viral RNA has a type I cap at the 5′ end. The 5′UTR is about 100 nucleotides long and shows high sequence conservation among different DENV serotypes. It contains two RNA domains with distinct functions during viral RNA synthesis. The first domain of ∼70 nucleotides is predicted to fold into a large stem-loop (SLA). A similar structure is present at the 5′UTRs of other members of the Flavivirus genus (7, 12, 21, 28). The DENV SLA has been proposed to act as the promoter for the viral RNA-dependent RNA polymerase (NS5). Direct binding of NS5 to SLA was shown to be necessary for viral RNA synthesis in vitro and viral replication in transfected cells (11, 38). The second domain of the DENV 5′UTR is predicted to form a short stem-loop (SLB). This element contains a 16-nucleotide-long sequence, known as the 5′ upstream AUG region (5′UAR), which is complementary to a region present at the 3′ end of the viral genome (3′UAR) (2, 3). Specific nucleotides at the 3′ end of the viral genome also play a crucial role in viral RNA synthesis. The approximately 450-nucleotide-long DENV 3′UTR lacks a poly(A) tail but ends in a very conserved 3′ stem-loop (3′SL) structure. A detailed functional analysis of the 3′SL revealed its absolute requirement for viral replication (for a review, see references 24 and 26). Specific bulges within the 3′SL as well as nucleotides at the loop and the 3′-terminal sequence are required for RNA replication (16, 29, 30, 37). Moreover, the bottom part of the 3′SL contains the 3′UAR sequence, which is complementary to the 5′UAR region (2, 3). In addition, upstream of the 3′SL, there is another essential cyclization element, known as the 3′ cyclization sequence (3′CS). This region of 11 nucleotides is complementary to a sequence present in the coding sequence of the C protein (1, 10, 15, 17, 23, 25). Hybridization of 5′-3′CS and 5′-3′UAR has been shown to be necessary for genome cyclization and RNA synthesis (1-3, 11, 17, 35, 36, 38, 39).
Here, we analyzed the secondary structure of SLA in solution and defined RNA elements within this structure that are necessary for DENV replication. Using an infectious DENV2 clone, we identified two helical regions, a side stem-loop (SSL), a top loop (TL), and a U bulge as critical structural elements within the SLA for viral replication. In addition, we determined that in vitro SLA-polymerase recognition and viral replication in transfected cells require elements of SLA that can be exchangeable between different DENV serotypes. We believe that dissecting conserved cis-acting RNA elements involved in viral replication will help researchers understand the DENV biology and assist in a rational design of live attenuated vaccines.
MATERIALS AND METHODS
Construction of recombinant DENVs.
The desired mutations were introduced in a DENV type 2 cDNA clone (18) (GenBank accession number U87411) by replacing the SacI-SphI fragment of the wild-type (WT) plasmid with the respective fragment derived from an overlapping PCR. The forward and reverse oligonucleotides used to introduce the specific mutations are given in Table 1. The overlapping PCR was performed with the common outside oligonucleotides AVG 194 (GGAATTCGAGCTCCGCGGACGCGTAAATTTAATACGAC) and AVG 239 (TCTGTGATGGAACTCTGTGG). However, in the case of Mut B3, the outside forward oligonucleotide was AVG 989 (GAATTCGAGCTCACGCGTAAATTTAATACGACTCACTATAAGTTGTTAGCCTACGTGGAC). To generate the mutated replicons, we used the same strategy replacing the WT sequence in the subgenomic DENV2 replicon pDVRep previously described (1) with the fragment derived from the overlapping PCR. In this case, the outside common oligonucleotides used were AVG 194 and AVG 331 (CATTCCAAAACCGTGATGGAATGG).
TABLE 1.
Oligonucleotides used for overlapping PCRs
| Mutation | Orientationa | Sequence |
|---|---|---|
| Mut S2.1 | F | ACAGATTCTTTGAGGGAGCTAAGC |
| R | CTCCCTCAAAGAATCTGTCTTTGTCGGTCCACCACGACTAACAACTTATAGTG | |
| Mut S2.2 | F | GACAGATTCTTTGAGGGAGCTAAGCTCAACCATGTTCTAACAGTTTTTTAATTAG |
| R | CTCCCTCAAAGAATCTGTCTTTGTCGGTCCACCACGACTAACAACTTATAGTG | |
| Mut S3.1 | F | CGTTGAGCTTAGCTCCCTCATTCAATCTGTGAATGTC |
| R | GTTAGTCTACGTGGACCGACATTCACAGATTGAATG | |
| Mut S3.2 | F | CGACAAAGACAGATTGAATGAGGGAGCTAAGCT |
| R | AGCTTAGCTCCCTCATTCAATCTGTCTTTGTCG | |
| Mut S3.3 | F | CGTTGAGCTTAGCTCCCTCATTCAATCTGTGAATGTC |
| R | GTTAGTCTACGTGGACCGACATTCACAGATTGAATG | |
| Mut S3.4 | F | TACGTGGACCGACAATCTCAGATTCTTTGAGGG |
| R | CCCTCAAAGAATCTGAGATTGTCGGTCCACGTA | |
| Mut S3.5 | F | GACAATCTCAGATAGATTGAGGGAGCTAAGCTC |
| R | CTCAATCTATCTGAGATTGTCGGTCCACGTA | |
| Mut ΔS3 | F | CTACGTGGACCGACGACAGATTCGAGGGAGCTAAGC |
| R | GCTTAGCTCCCTCGAATCTGTCGTCGGTCCACGTAG | |
| Mut C | F | ACAGATTCTTTGAGGGAGCTAAGC |
| R | CTCCCTCAAAGAATCTGTCTTTGTCGGATGACGTAGACTAACAACTTATAG | |
| Mut B1 | F | GCTCAACGTAGAACTAACAGTTTTTTAATTAGAG |
| R | CTGTTAGTTCTACGTTGAGCTTAGCTCC | |
| Mut B2 | F | GGGAGCTAAGCTCAACGTAGACTAACAGTTTTTTAATTAGAGAG |
| R | CTCTCTAATTAAAAAACTGTTAGTCTACGTTGAGCTTAGCTCCC | |
| Mut B3 | F | TAAGCTCAACGTAGTGCTAACAGTTTTTTAA |
| R | TTAAAAAACTGTTAGCACTACGTTGAGCTTA | |
| Mut TL | F | CAAAGAAGACTTCTTTGAGGGAGCTAAGCTC |
| R | CTCAAAGAAGTCTTCTTTGTCGGTCCACGTAGAC | |
| Mut ΔSSL | F | GACAGATTCTTTGAGGGAACGTAGTTCTAACAG |
| R | CTGTTAGAACTACGTTCCCTCAAAGAATCTGTC | |
| Mut SSL-1 | F | GACAGATTCTTTGAGGGCGGCTAAGCCGCAACGTAGTTCTAACAGTT |
| R | AACTGTTAGAACTACGTTGCGGCTTAGCCGCCCTCAAAGAATCTGTC | |
| Mut SSL-2 | F | GACAGATTCTTTGAGGGCTAAGCAACGTAGTTCTAACAGTT |
| R | AACTGTTAGAACTACGTTGCTTAGCCCTCAAAGAATCTGTC | |
| Mut SSL-3 | F | GACAGATTCTTTGAGGCTCGTAAGCTCAACGTAGT |
| R | ACTACGTTGAGCTTACGAGCCTCAAAGAATCTGTC | |
| Mut SSL-4 | F | CTTTGAGGGAGCTAACGAGAACGTAGTTCTAACAG |
| R | CTGTTAGAACTACGTTCTCGTTAGCTCCCTCAAAG | |
| Mut SSL-3/4 | F | GACAGATTCTTTGAGGCTCGTAACGAGAACGTAGTTCTAACAG |
| R | CTGTTAGAACTACGTTCTCGTTACGAGCCTCAAAGAATCTGTC | |
| Mut SSL-5 | F | CTTTGAGGGAGCATTGCTCAACGTAGTTCTAAC |
| R | CTACGTTGAGCATTGCTCCCTCAAAGAATCTG | |
| Mut Δ6U | F | ACGTAGTTCTAACAGAATTAGAGAGCAGAT |
| R | ATCTGCTCTCTAATTCTGTTAGAACTACGT | |
| Mut 6U/6A | F | CGTAGTTCTAACAGAAAAAAAATTAGAGAGCAGA |
| R | TCTGCTCTCTAATTTTTTTTCTGTTAGAACTACG | |
| Mut DENV1/DENV2 | F | GAACAGTTTCGAATCGGAAGCTTGCTTAACGTAGTTCTAACAG |
| R | CTTCCGATTCGAAACTGTTCTTGTCGGTCCACGTAGACTAAC |
F, forward; R; reverse.
RNA transcription and transfections.
To obtain infectious and replicon DENV RNAs, in vitro transcription by T7 RNA polymerase in the presence of an m7GpppA cap analog was used as previously described (3). RNA transcripts (3 μg) were transfected with Lipofectamine 2000 (Invitrogen) into BHK-21 cells. As a control, 100 ng of Renilla luciferase RNA was cotransfected, and the luciferase activities for both were quantified using a dual luciferase assay kit, following the manufacturer's instructions (Promega).
For enzymatic probing and RdRp activity assays, RNAs corresponding to the first 160 nucleotides of the viral genome carrying WT or mutated sequences were obtained by in vitro transcription with T7 RNA polymerase (90 min, 37°C). The templates were obtained by PCR with oligonucleotides AVG 1 (TCGTTAATACGACTCACTATAGGAGTTGTTAGTC) and AVG 130 (GTTTCTCTCGCGTTTCAGCATATTG). The RNAs were treated with RNase-free DNase I to remove the templates and purified using an RNeasy minikit (Qiagen, Inc.) to eliminate free nucleotides. The products were quantified spectrophotometrically, and the integrity of the RNAs was verified by electrophoresis on agarose gels.
IF.
Transfected cells with WT or mutated full-length DENV RNA were used for an immunofluorescence (IF) assay. BHK-21 cells were grown in 35-mm-diameter tissue culture dishes containing a 1-cm2 coverslip inside. The coverslips were removed and directly used for IF analysis. The transfected cells were trypsinized on day 3, and two-thirds of the total cells were reseeded to a 35-mm-diameter tissue culture dish containing a coverslip. This procedure was repeated every 3 days until a cytopathic effect was observed. At each time point, a 1:200 dilution of murine hyperimmune ascitic fluid against DENV2 in phosphate-buffered saline-0.2% gelatin was used to detect viral antigens. Cells were fixed in paraformaldehyde. Alexa Fluor 488 rabbit anti-mouse immunoglobulin G and Alexa Fluor 488 goat anti-rabbit immunoglobulin G conjugates (Molecular Probes) were used as detector antibodies at a 1:500 dilution. Photomicrographs (×200 magnification) were acquired with an Olympus BX60 microscope coupled to a CoolSnap-Pro digital camera (Media Cybernetics) and analyzed with Image-Pro Plus software.
Viral RNA extraction and sequencing.
Viral RNA was extracted with TRIzol (Invitrogen) from a 300-μl aliquot of the medium from transfected cells. The RNA was reverse transcribed by Superscript II reverse transcriptase (Invitrogen) for 1 h at 42°C using oligonucleotide AVG 239 (see above). In order to sequence the complete 5′UTR of the viral genome, a reaction using 5′ rapid amplification of cDNA ends was performed as follows. The cDNA was purified by precipitation with ethanol-NH4Cl and resuspended in 11 μl of Milli-Q water. A poly(A)-tailing reaction was performed by adding 2 mM dATP, 10 U of terminal deoxynucleotidyl transferase (New England Biolabs) to give a final volume of 20 μl and allowing the reaction to proceed for 1 h at 37°C. The reaction was stopped by heating for 3 min at 95°C, and 5 μl was used as a template for amplification by a PCR using oligonucleotides AVG 423 (TTTTTTTTTTTTTTTAGTTG) and AVG 239. In each case, reverse transcription-PCR (RT-PCR) products were sequenced using an ABI 377 automated DNA sequencer and BigDye Terminator chemistry (Applied Biosystems).
RNase digestions and chemical modification.
In vitro-transcribed RNAs were heated for 5 min to 85°C and slow cooled to room temperature. The RNAs were then treated with different concentrations of ribonucleases to determine the proper concentration to use. The following concentrations were employed: 0.001 U of RNase V1 (Ambion), 0.01 U of RNase A (Ambion), and 0.4 U of RNase PhyM (Pierce) (in a total volume of 10 μl). The reactions were performed at 25°C for 15 min in buffer containing 10 mM Tris-HCl, pH 7.0, 100 mM KCl, and 10 mM MgCl2 and were stopped by addition of 20 μl of RNase inactivation/precipitation buffer (Ambion). Reaction products were purified by incubation at −20°C for 15 min, followed by centrifugation at 15,700 × g for 5 min. Pellets were washed in 70% ethanol and resuspended in 5 μl Milli-Q water. Control reactions were carried out in parallel under the same conditions, without addition of RNases.
The ribose 2′ positions of exposed RNA nucleotides were acylated with the addition of N-methylisatoic acid (NMIA) as described by Wilkinson et al. (34). In vitro-transcribed 5′ DENV RNA was modified with 13 mM NMIA dissolved in dimethyl sulfoxide. The reactions were performed at 37°C for 45 min in buffer containing 100 mM HEPES (pH 8.0), 100 mM NaCl, and 6 mM MgCl2. Control reactions were carried out in the presence of dimethyl sulfoxide without NMIA. The modified RNAs were precipitated with ethanol to eliminate buffers that could be detrimental to primer extension.
Primer extension.
The digested or modified RNA products were used for primer extension. As primer, we used a 5′ end fluorescently labeled oligonucleotide (5′-cy5-TCATCAGAGATCTGCTCTCTAATTAAAAA 3′) (IDT). Reactions were performed with 20 U ArrayScript reverse transcriptase (Ambion) at 42°C for 50 min with a 10-μl mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 mM dithiothreitol, and 500 μM of each of the four deoxynucleoside triphosphates. cDNA products were ethanol precipitated, resuspended in 90% formamide, and heat denatured for 3 min at 95°C immediately prior to electrophoresis. Dideoxy sequencing reactions were carried out on unmodified RNAs and run in parallel with the primer extension products on 10% polyacrylamide-7 M urea sequencing gels. The gels were then analyzed directly using a Storm 840 imager (Molecular Dynamics).
RdRp in vitro assay.
The standard RdRp in vitro assay was performed as previously described (11). Briefly, the reaction was carried out with a total volume of 25 μl in buffer containing 50 mM HEPES, pH 8.0, 10 mM KCl, 5 mM MgCl2, 2 mM MnCl2, 10 mM dithiothreitol, 4 U RNase inhibitor, 500 μM (each) ATP, CTP, and UTP, 10 μM [α-32P]GTP, 0.5 μg template RNA, and 0.15 μg of recombinant purified NS5. The reaction mixture was incubated at 30°C for 30 min and the reaction stopped by adding a denaturing solution to give final concentrations of 7% (wt/vol) trichloroacetic acid (TCA) and 50 mM H3PO4 at 0°C. The TCA-precipitated RNAs were then collected by vacuum filtration using a V-24 apparatus, with the mixture carefully added onto the center of a Millipore filter (type HAWP, 0.45-μm pore size). The filters were washed eight times with 5 ml each of cold 7% (wt/vol) TCA-50 mM H3PO4 and dried, and the radioactivity was measured. For polyacrylamide gel electrophoresis analysis of the RNA products and the trans initiation assay, the standard mixture was the same as the one described above except that the reaction was ended by phenol extraction followed by ethanol precipitation. The 5′RNA templates 5′RNAΔ3, 5′RNAΔ6, 5′RNAΔ10, 5′RNA+6, and 5′RNA+10 were obtained by overlapping PCR and in vitro transcription. The sequences of the mutated RNAs are listed in Table 2.
TABLE 2.
Sequences of mutated RNAs
| 5′RNA template | Sequencea |
|---|---|
| WT | UUUUUUAAUU |
| 5′RNAΔ3 | UUUAAUU |
| 5′RNAΔ6 | AAUU |
| 5′RNAΔ10 | - |
| 5′RNA+6 | UUUUUUUUUUUUAAUU |
| 5′RNA+10 | UUUUUUUUUUUUUUUUAAUU |
Following position 70.
The RNA products of the polymerase assay were resuspended in Tris-borate-EDTA containing formamide (80%) and heated for 5 min at 65°C. The samples were then analyzed by electrophoresis on a 5% denaturing polyacrylamide gel-6 M urea and visualized by autoradiography.
RNA binding assays.
RNA-RNA interactions were analyzed by electrophoretic mobility shift assays. Uniformly 32P-labeled 3′SL probe was obtained by in vitro transcription using T7 RNA polymerase and purified on 5% polyacrylamide gels and 6 M urea. The binding reaction mixtures contained 5 mM HEPES, pH 7.9, 100 mM KCl, 5 mM MgCl2, 3.8% glycerol, 2.5 μg tRNA, uniformly 32P-labeled 3′SL (0.1 nM, 0.013 μCi), and 0, 2, 4, 8, 16, 40, 80, and 160 nM of 5′RNA WT and Mut Δ6U in a final volume of 25 μl. RNA samples were heat denatured at 85°C for 5 min and slow cooled to room temperature. RNA-RNA complexes were analyzed by electrophoresis through native 5% polyacrylamide gels supplemented with 5% glycerol. Gels were prerun for 30 min at 4°C at 150 V, and then 20 μl of the sample was loaded and electrophoresis was allowed to proceed for 4 h at constant voltage. Gels were dried and visualized by autoradiography.
RESULTS
Secondary structure of the 5′ end of the DENV genome.
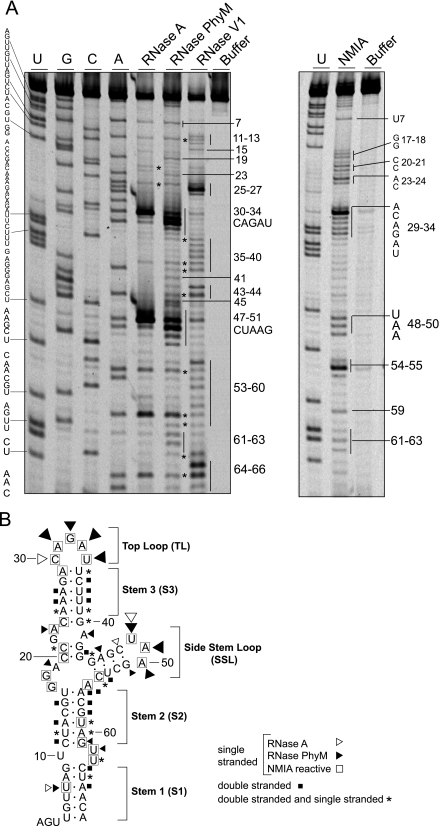
The 70-nucleotide-long structure present at the 5′ end of the DENV genome plays a crucial role in viral RNA synthesis (9, 11). This cis-acting RNA element was predicted to form a large stem-loop with a side loop (SL) in a Y-shaped structure (12). Here, we examined the secondary structure of this region by RNase probing analysis. The RNA corresponding to the first 160 nucleotides of the viral genome was digested with double-stranded specific RNase V1 or single-stranded specific RNase A or PhyM or treated with the chemical reagent NMIA. NMIA reacts with the 2′ position of the ribose of any flexible nucleotide, forming 2′-O adducts. Nucleotides forming base pairings or tertiary interactions are unreactive (34). The sites cleaved by RNases and the sites of 2′-O adducts were identified as stops to primer extension by reverse transcriptase, using fluorescent probes. A representative sequencing gel is shown in Fig. 1A. A summary of the probing results are mapped onto the predicted structure (Fig. 1B). Two regions highly sensitive to single-stranded specific RNases were observed at nucleotides 30 to 34 and 48 to 50, which correspond to the TL and SL, respectively, supported by the prediction analysis. These nucleotides also reacted with NMIA: in the TL, nucleotides 29 to 34 were exposed, in which C30 was the most reactive, and in the SL, nucleotides 48 to 50 were sensitive to the reagent (Fig. 1A). In addition, nucleotides in the three predicted helical regions named stem 1 (S1), stem 2 (S2), and stem 3 (S3) (Fig. 1B) were sensitive to RNase V1; however, certain nucleotides within these regions were observed to be sensitive to both single- and double-stranded specific RNases (Fig. 1). Single-stranded regions were also detected in predicted bulges and mismatches (Fig. 1B).
FIG. 1.
Enzymatic and chemical probing of the 5′-terminal stem-loop of the DENV genome. (A) An RNA corresponding to the first 160 nucleotides of the viral genome was subjected to RNase A, RNase PhyM, RNase V1, or NMIA treatment, and the cleaved or modified RNAs were analyzed by primer extension. The products of primer extension were analyzed with a sequencing gel along with a control sample without treatment and a sequencing ladder. The nucleotide sequence is indicated on the left, and specific cleavages are indicated on the right by numbers. (B) Optimal MFOLD-predicted RNA secondary structure model of SLA that includes a summary of detected enzymatic and chemical modifications. Nucleotides labeled with asterisks were observed to be sensitive to both single- and double-stranded specific RNases.
Although the sensitivity to RNases and NMIA of certain nucleotides suggested differences from the predicted SLA, the overall secondary structure obtained in solution is consistent with the predicted Y-shaped structure at the 5′ end of the DENV genome.
Helical region S2 but not S3 within SLA is essential for DENV2 replication.
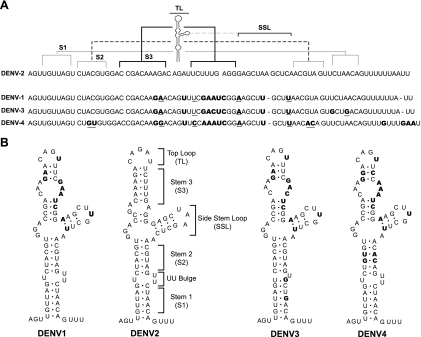
To gain further insight into the structural elements of SLA required for viral replication, sequence analysis of the first 70 nucleotides of the four DENV serotypes was carried out. Alignment of about 200 available sequences was performed. In Fig. 2A, a summary comparing the consensus sequences of DENV1, DENV2, DENV3, and DENV4 is shown. The bottom part of SLA, including S1 and S2, displays the highest conservation. In contrast, the SSL and the S3 region show both sequence and predicted structure variation, particularly when DENV2 is compared with the other three serotypes (Fig. 2B).
FIG. 2.
Comparison of the nucleotide sequences and secondary structures of the SLA structures of different DENV serotypes. (A) Alignment of the consensus sequences of the first 70 nucleotides of DENV1, DENV2, DENV3, and DENV4. The regions corresponding to the predicted secondary structures S1, S2, S3, TL, and SSL of DENV2 are indicated on the top. Nucleotide variation between DENV2 and the other three serotypes is indicated in bold. Underlined nucleotides indicate covariations. (B) Comparative RNA secondary structure analysis predicted for the SLA structures of the four DENV serotypes. The nucleotide differences between DENV2 and the other three serotypes are indicated in bold.
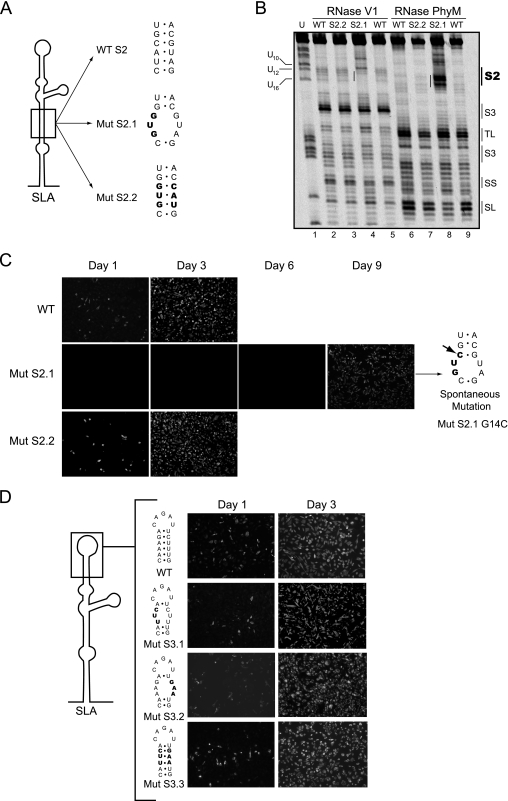
Previously, it was shown that mutations at the bottom of SLA (corresponding to S1) impaired viral replication (11). A possible requirement of helical region S2 for viral replication is supported by 2-nucleotide covariation in DENV4 (Fig. 2). Thus, we examined the importance of S2 in DENV replication by introducing mutations disrupting or reconstituting this helical region in the context of the infectious cDNA of DENV2 16681 (18). Two mutants were designed. In one, S2 was disrupted by a 3-nucleotide substitution (Fig. 3A, Mut S2.1), and in the other, 3 nucleotides on each side of the stem were changed, maintaining the stem complementarity (Fig. 3A, Mut S2.2). This mutant included six substitutions, three of which were the same as the ones incorporated in Mut S2.1. Before introducing these mutations into the infectious clone, the predicted disruption of S2 in Mut S2.1 and the reconstitution of the base pairings in Mut S2.2 were analyzed using RNase probing analysis. The RNase V1 and PhyM digestion patterns of the mutated RNAs confirmed the predicted changes within S2 (Fig. 3B), while the overall secondary structure of SLA (S3, TL, side stem [SS], and SL) was similar to that of the WT RNA.
FIG. 3.
Role of helical regions S2 and S3 in viral replication. (A) Schematic representation of specific mutations within S2 of SLA. The sequences of the WT and the mutants Mut S2.1 and Mut S2.2 are shown, and the nucleotide changes are indicated in bold. (B) Enzymatic probing of WT and Mut S2.1 and Mut S2.2 mutant RNAs. The regions corresponding to the predicted secondary structures S2, S3, TL, SL, and SS are indicated on the right. (C) Expression of DENV proteins in BHK cells transfected with WT and mutated viral RNAs was monitored by IF at 1, 3, 6, and 9 days posttransfection. The spontaneous mutation of a recovered virus obtained after transfection of the Mut S2.1 RNA is indicated with an arrow. (D) Viral replication tolerates variations within the helical S3 region. (Left) Schematic representation of SLA showing specific nucleotides modified in Mut S3.1, Mut S3.2, and Mut S3.3 disrupting or reconstituting the helical region. (Right) Expression of DENV proteins in BHK cells transfected with WT or mutated RNAs. Viral replication was monitored by IF at 1 and 3 days after transfection.
Replication of Mut S2.1 and Mut S2.2 was investigated by IF of transfected BHK cells with full-length viral RNAs. The mutation Mut S2.1 caused a dramatic delay in viral replication compared with the level for parental RNA (Fig. 3C). Transfection of Mut S2.1 RNA resulted in negative IF until day 6. After day 7, positive cells became evident, and on day 9, viral replication spread throughout the monolayer, suggesting that viral revertants may have arisen. In contrast, transfection of Mut S2.2 showed viral replication comparable to the level for parental RNA (Fig. 3C). These results indicate that base pairings within S2 and not the substituted nucleotide in Mut S2.1 were required for SLA function. To determine whether spontaneous mutations emerged after transfection of Mut S2.1, the viral RNA was extracted from the medium and analyzed by RT-PCR and DNA sequencing using 5′ rapid amplification of cDNA ends. A G14C mutation was observed in three independent transfections, which restored a base pair within S2 (Fig. 3C, Mut S2.1 G14C). The revertant virus Mut S2.1 G14C replicated to high titers (∼107 PFU/ml) but displayed a small plaque phenotype compared with that of the parental virus (data not shown). These results suggest an important role for helical region S2 in viral replication.
To investigate the importance of helical region S3, substitutions on each side of the stem disrupting three predicted base pairings and substitutions on both sides reconstituting the stem were also introduced (Fig. 3D, Mut S3.1, Mut S3.2, and Mut S3.3). Interestingly, the mutants with disrupted S3 replicated similarly to the parental virus (Fig. 3D). Viral stocks were obtained from these mutants, and the plaque phenotype as well as the growth curves was indistinguishable from that of the parental virus (data not shown). The results indicate that while base pairings in S1 and S2 are required structural elements for SLA function in vivo, helical region S3 tolerates large variations.
Common promoter activities and structures of SLAs from different DENV serotypes.
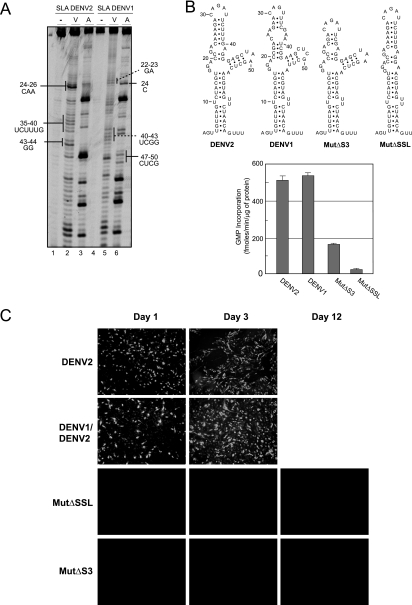
Analysis of the sequences and predicted structures of SLAs from the four DENV serotypes showed variations within the SSL and the S3 region. The sizes of the stems and the sequences of the loops of the SSL and helical region S3 differed in different serotypes. For instance, serotypes 1, 3, and 4 bear three mismatches in the central region of S3, while this region is base paired in DENV2 (Fig. 2B). RNase probing analysis comparing the secondary structures in a solution of the SLAs from DENV2 and DENV1 confirmed these predicted differences (Fig. 4A, compare lanes 2 and 3 with lanes 5 and 6). On the basis of this observation, we asked whether the SLA structure contains serotype-specific determinants for polymerase recognition and RNA synthesis. To address this question, we analyzed the ability of the recombinant NS5 protein from DENV2 to use the SLA from DENV1 as a promoter for in vitro RNA synthesis. The polymerase activity was evaluated as previously described, using filter binding assays (11). It was observed that the DENV2 polymerase was able to recognize and copy an RNA template carrying the SLA from DENV1 as efficiently as the DENV2 RNA (Fig. 4B). This result suggests that the elements involved in promoter recognition are common in the two serotypes.
FIG. 4.
Functional in vivo and in vitro compatibility of SLAs from different DENV serotypes. (A) Enzymatic probing of the first 70 nucleotides of DENV1 and DENV2. The RNAs were subjected to RNase V1 (V) or RNase A (A) digestion, and the cleaved RNAs were analyzed by primer extension. Differences within the S3 and SSL regions of SLAs from DENV1 and DENV2 are indicated. (B) In vitro activity of viral RdRp for WT and mutated RNA templates with 160 nucleotides. (Top) Sequences of SLAs of the RNAs used as templates for RNA synthesis. (Bottom) Viral polymerase activity, expressed in fmol [α-32P]GMP incorporated into acid-insoluble RNA per minute and per μg of protein, for each RNA template. The reaction was carried out as described in Materials and Methods. Error bars indicate the standard deviations of results from three experiments. (C) Expression of DENV proteins in BHK cells transfected with RNA of WT DENV2, a chimeric DENV carrying the SLA from DENV1 (DENV1/DENV2), and mutants with deletions of the SSL or S3 (Mut ΔSSL or Mut ΔS3, respectively) was determined. Viral replication was monitored by IF for up to 12 days posttransfection.
We further investigated the role of S3 and the SSL (which are the most variable regions between phenotypes) in polymerase recognition and activity. RNA templates carrying a deletion of 3 bp in the central region of S3 or a deletion of the SSL were tested. Both mutations greatly decreased SLA promoter activity. The RNA synthesis with the mutants was between 4- and 10-fold lower than that observed with the WT SLA (Fig. 4B), suggesting that although the S3 and SSL elements tolerate variations, the presence of these structures is important for SLA function in vitro.
The observation that the DENV2 NS5 was able to recognize the DENV1 SLA as a promoter for in vitro RNA synthesis prompted us to investigate SLA-NS5 compatibility in vivo in the context of the infectious viral RNA. The cDNA of DENV2 was used to swap the SLA from the corresponding region of DENV1. After RNA transfection, replication of the chimeric virus DENV1/DENV2 was as efficient as that of parental DENV2, indicating that the foreign SLA was fully competent to promote viral RNA replication (Fig. 4C). In addition, the deletions of the SSL or S3, which compromised promoter activity in vitro, also abolished viral replication in transfected cells (Fig. 4C).
In summary, the data indicate that the important structural determinants for promoter activity are conserved within SLAs of different DENV serotypes despite the sequence/structure differences within S3 and the SSL.
The essential structural elements of DENV SLA are found in other flavivirus genomes.
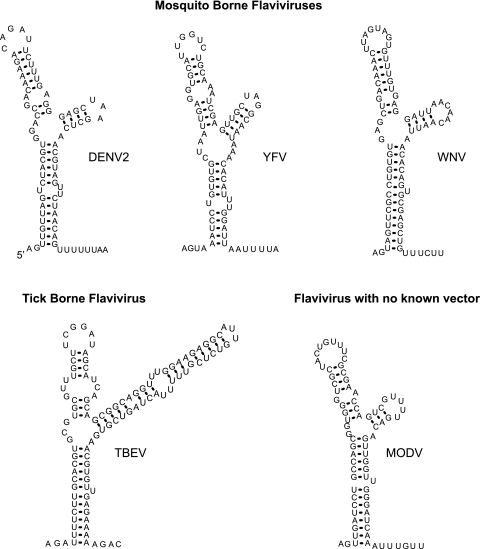
We found that the requirement of SLA for polymerase activity is conserved between different DENV serotypes. Interestingly, an RNA structure similar to SLA can be found in all flavivirus genomes (7, 12, 21, 28), suggesting a common role for this element in different members of the genus. A comparison of the predicted stem-loop structures present at the 5′ ends of mosquito-borne flaviviruses (DENV, yellow fever virus, and WNV), tick-borne encephalitis virus, and a flavivirus with no known vector (Modoc virus) shows similar structural elements (Fig. 5). These structures contain two helical regions resembling S1 and S2 of DENV, which are always separated by a U bulge or a U-U mismatch, and an S3 region interrupted by mismatches in different locations. In addition, all genomes bear a TL as well as an SSL with stems of different lengths (Fig. 5). In general, the 5′SL is followed by a short track of U residues, except in the tick-borne encephalitis virus genome. To study whether the conserved elements within the SLA were required for viral replication, we designed new mutations altering each of these common structures.
FIG. 5.
Predicted 5′-end stem-loop structures of flaviviruses. The optimal MFOLD-predicted RNA secondary-structure models for mosquito-borne and tick-borne flaviviruses and a flavivirus with no known vector are shown. Results are shown for WNV M12294, yellow fever virus (YFV) NC-002031, tick-borne encephalitis virus (TBEV) U27495, and Modoc virus (MODV) AJ242984.
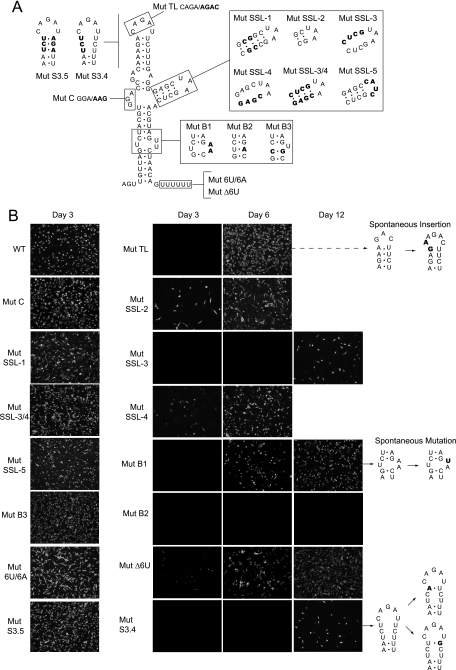
First, to address the importance of the bulges, the UU62-63 sequence that separates S1 and S2 was replaced by AA, and the GGA17-19 sequence was replaced by AAG. Viral replication was analyzed after RNA transfection by IF assays. The RNA with the mutation in the GGA bulge replicated very efficiently (Fig. 6B, Mut C), and high viral titers were obtained (∼5 × 107 PFU/ml). Sequencing analysis revealed that the recovered viruses retained the introduced mutation. In contrast, the replication of the RNA with the mutation in the UU bulge (Fig. 6B, Mut B1) was delayed. The IF for DENV antigens was evident by day 6, and by day 12, about 40% of the monolayer showed a positive signal. From three independent transfections, viruses were recovered in two cases, suggesting spontaneous mutations. To analyze whether revertant viruses had emerged, viral RNA was purified from the medium and used for RT-PCR and sequencing analysis. The same single spontaneous mutation appeared in both transfections, in which an A at position 62 reverted to the parental U nucleotide (Fig. 6B). The sequence of SLA found in the revertant virus was reintroduced into the parental clone. Transfection of this RNA confirmed the phenotype observed with the revertant virus (data not shown). This result suggests that at least one U in the bulge (present in all flavivirus genomes) is required for viral replication. Because some flaviviruses contain only a single U bulge, we analyzed whether a UU mismatch or a U bulge was required for DENV replication. To this end, new mutants were constructed. A mutant in which the UU bulge (positions 62 and 63) was replaced by a single A residue, resulting in a UA base pair (Fig. 6A, Mut B2), and a mutant in which the UU mismatch (positions 10 and 63) was replaced by a CG base pair, resulting in a single U bulge (Fig. 6A, Mut B3), were designed. Transfection of these RNAs indicated that Mut B2 did not replicate for up to 12 days after transfection, while the Mut B3 replicated efficiently. Together, these results confirm that a single U bulge is essential and sufficient for DENV replication.
FIG. 6.
Functions of structural elements of SLA common in flavivirus genomes. (A) Schematic representation of specific mutations within SLA. The modified nucleotides are indicated for each case: in Mut TL, the sequence of the loop CAGA was replaced by AGAC; in Mut B1, the sequence of the UU bulge was replaced by AA; in Mut B2, the UU bulge was replaced by a single A residue; in Mut B3, a UU mismatch was replaced by a CG base pair; in Mut C, the sequence of the GGA bulge was replaced by AAG; in Mut SSL-1, the stem of the SSL was stabilized by 5 GC base pairs; in Mut SSL-2, the stem of the SSL includes a 2-base-pair deletion; in Mut SSL-3, the 4 nucleotides GAGC on one side of the stem of the SSL were replaced by CUCG to disrupt the stem; in Mut SSL-4, the 4 nucleotides CUCG on the other side of the SSL were replaced by GAGC to disrupt the stem; in Mut SSL-3/4, the nucleotides modified in Mut SSL-3 and Mut SSL-4 were introduced together to reconstitute the stem; in Mut SSL-5, the sequence of the SL UAA was replaced by CAUC; in Mut S3.4, the upper half of S3 was disrupted; in Mut S3.5, the predicted base pairs in S3 were reconstituted; in Mut Δ6U, the 6 U residues downstream of SLA were deleted; and in Mut 6U/6A, the oligo(U) track was replaced by an oligo(A) sequence. (B) Expression of DENV proteins in BHK cells transfected with WT and SLA-mutated RNAs described for panel A. Viral replication was monitored by an IF assay as a function of time after RNA transfection using specific anti-DENV antibodies. Viruses recovered in the supernatants from transfected cells were used to purify the RNA for sequencing analysis. In the right panel, the spontaneous mutations are shown in bold.
We have shown that deletion of the SSL abolishes viral replication (Fig. 4C). To further study the importance of this element, we generated a set of six mutants altering the stem stability and the sequence of the loop. Two mutants were designed to increase or decrease the length of the stem (Mut SSL-1 and Mut SSL-2, respectively). Other mutants with substitutions on each side of the stem were designed to disrupt predicted base pairings (Mut SSL-3 and Mut SSL-4). In addition, these substitutions were introduced together to reconstitute the stem (Mut SSL3/4). Replication of Mut SSL-1 was similar to that of the WT virus; however, replication of Mut SSL-2 was delayed (Fig. 6B). Disruption of the stem in Mut SSL-3 and Mut SSL-4 decreased viral replication. However, while Mut SSL-4 showed IF-positive cells at 3 days after transfection, Mut SSL-3 displayed a sublethal phenotype (Fig. 6B). Importantly, the reconstitution of the stem in Mut SSL-3/4 (carrying eight substitutions) replicated similarly to the control virus. To determine whether the sequence of the loop of the SSL was important, the UAA sequence was replaced by the unrelated sequence CAUC (Mut SSL-5). Transfection of this RNA resulted in viruses that replicated as efficiently as the parental virus (Fig. 6B). These results indicate that although the stem of the SSL tolerates variations of sequence and length, the presence of this element is necessary for SLA function in vivo, which is in agreement with the in vitro requirement observed for the viral RNA polymerase (Fig. 4B).
Previous studies have shown a requirement of specific nucleotides at the TL of SLA for DENV replication (11). Point mutations in this region, which maintained the secondary structure of SLA, impaired polymerase activity and viral replication, giving rise to spontaneous mutations that restored the parental phenotype (11). Here, we modified the TL, combining the previously reported mutations into a single RNA (Fig. 6A, Mut TL). Replication of Mut TL RNA was delayed relative to that of the parental virus; however, IF-positive cells for DENV antigens were detected at 4 days posttransfection, and at 6 days, viral replication spread throughout the monolayer (Fig. 6B). RT-PCR and sequencing analysis of the recovered viruses showed a simple solution for this drastic change. A duplication of 2 nucleotides (GA28-29) was observed (Fig. 6B). Infection of fresh cells with a stock of the recovered virus showed delayed replication and a small plaque phenotype compared with that of the parental virus, confirming that the revertant virus was replication impaired. The TL sequence obtained in the revertant virus was reintroduced into the parental clone. Transfection of this RNA showed the same attenuated phenotype as that observed with the revertant virus, confirming that the GA insertion in the TL was responsible for rescuing replication of Mut TL (data not shown). It was intriguing that, while the TL plays an essential role in replication, S3 tolerated disruptions. Therefore, we decided to analyze the replication of new mutants carrying substitutions that open the upper half of S3, including the closing UA base pair, and substitutions on both sides of the stem that would reconstitute the predicted structure (Fig. 6A, Mut S3.4 and Mut S3.5). Replication of Mut S3.4 was greatly delayed. Infected cells were detected by IF only 12 days after transfection. Interestingly, RT-PCR and sequencing analysis of the recovered viruses showed two types of spontaneous mutations, each with a single-nucleotide change at the top of S3 (Fig. 6B). The WT closing AU base pair, which was replaced by a UU mismatch in Mut S3.4, reverted to an AU or mutated to a UG base pair in the recovered viruses. The selection of viruses with a base pair at the top of S3, regardless of the nucleotide sequence in that position, indicates a structural requirement of this element for viral viability. In addition, the efficient replication of Mut S3.5, which bears a reconstituted S3, is in agreement with the observed genotype of the revertant viruses (Fig. 6B).
A U-rich track is present just downstream of the SLA structure in the four DENV serotypes. In order to study the requirement of this element, the six U residues following the SLA in the infectious DENV2 clone were deleted (Mut Δ6U). Replication of Mut Δ6U RNA was greatly delayed. About 30% of IF-positive cells were observed 6 days after transfection. RT-PCR and sequencing analysis of recovered viruses indicated that the oligo(U) track deletion was maintained after 2 weeks in culture. In addition, infection of fresh cells with this mutant virus showed a slow-replication phenotype. Furthermore, to examine whether the nucleotide sequence was important, the oligo(U) was replaced by an oligo(A) track of the same length. Replication of this RNA was very efficient (Fig. 6B, Mut 6U/6A), and the plaque morphology was indistinguishable from that of the parental virus (data not shown). These results indicate that either an oligo(U) or an oligo(A) track allows efficient viral replication, perhaps acting as spacer between the two 5′UTR elements.
Viral translation is not impaired in the SLA mutants.
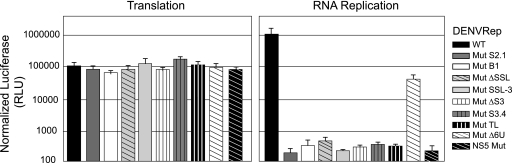
It has previously been reported that mutations within the SLA did not affect translation of the viral RNA but impaired RNA synthesis (11). In this work, we introduced new mutations within the SLA that resulted in drastic viral phenotypes. Because the 5′UTR must be scanned by ribosomes during translation initiation, we analyzed the effects of the mutations that impaired DENV replication on viral translation and RNA synthesis. To this end, mutations in the relevant structural regions within SLA were introduced in a previously reported DENV replicon system. Constructs carrying mutations in S2 (Mut S2.1), in the UU bulge (Mut B1), in the SSL (Mut ΔSSL and Mut SSL-3), in S3 (Mut ΔS3 and Mut S3.4), and in the TL (Mut TL) were designed. In addition, a mutant replicon carrying the deletion of the oligo(U) track downstream of the SLA, which displayed delayed replication, was also analyzed. Replicon RNAs corresponding to the WT and the mutants were cotransfected into BHK cells, with an RNA carrying Renilla used as a control for transfection. A replicon containing a substitution in the catalytic site of the viral polymerase NS5 (Mut NS5) was included as a negative control for RNA amplification. The luciferase determined at 10 h reflected translation, and the measurement at 72 h was used to evaluate RNA amplification (1).
Luciferase activity measured at 10 h posttransfection showed similar levels of translation for all replicons, indicating that the modifications within the 5′UTR did not affect translation of input RNAs (Fig. 7). In contrast, all the mutations within SLA resulted in defects on RNA synthesis. The luciferase levels observed with these replicons at 72 h were similar to that observed in the Mut NS5 control. In addition, the deletion of the oligo(U) track showed about 40-fold-lower RNA replication, in agreement with the attenuated phenotype observed, with the infectious RNA carrying the same mutation (Fig. 6). These results indicate that the impaired replication of the SLA mutants and the Mut Δ6U can be explained by defects in RNA synthesis and not by alteration of the translation process.
FIG. 7.
Mutations within SLA impair RNA synthesis without affecting translation of the input RNA. Shown are translation and RNA replication of WT and mutant DENV replicons in BHK cells. Normalized luciferase levels (in relative luciferase units [RLU]) are shown using a logarithmic scale at 10 h after transfection to estimate translation of input RNA and at 3 days after transfection to evaluate RNA replication. Error bars indicate the standard deviations of results from three independent transfections.
The oligo(U) track between the SLA and the 5′UAR influences the efficiency of RNA synthesis.
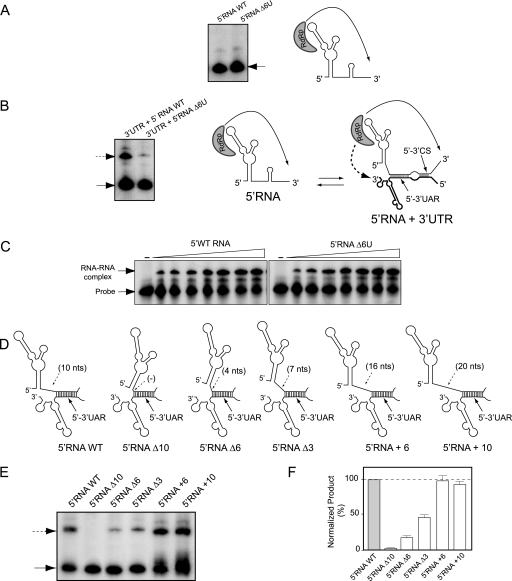
The mutant Δ6U used in this study carries a deletion in a region of the viral 5′UTR that has not previously been examined. Therefore, it was interesting to investigate why this deletion causes low levels of RNA amplification and viral attenuation. We hypothesized that the nucleotides between the SLA and the 5′UAR could modulate polymerase activity by altering RNA structures recognized by the polymerase or by interfering with the long-range 5′-3′UAR interaction. To study these possibilities, we employed different in vitro assays. First, we analyzed whether the mutant Δ6U altered RNA synthesis. To this end, the recombinant DENV NS5 polymerase was incubated with an RNA template corresponding to the first 160 nucleotides of the WT or the mutant Δ6U. The assay was performed as previously described (11), and the products were analyzed with denaturing polyacrylamide gels. Both RNAs, those of the WT and the mutant Δ6U, were efficient templates for RNA synthesis (Fig. 8A), suggesting that the deletion did not affect promoter recognition or polymerase activity. In addition, similar levels of GMP incorporation were observed for the two templates in filter binding assays (data not shown).
FIG. 8.
Role of the region between SLA and the 5′UAR in viral RNA synthesis. (A) In vitro RdRp activity of the recombinant NS5 for the first 160 nucleotides of the DENV genome WT or the mutant Δ6U as a template (5′RNA WT or 5′RNA Δ6U, respectively, as indicated above the gel). The radiolabeled RNA products were analyzed with denaturing 5% polyacrylamide gels. The arrow indicates the mobility of the product. On the right, a schematic representation of the initiation process is shown. (B) The efficiency of trans initiation polymerase activity depends on the oligo(U) track. The 5′RNA WT or the 5′RNA Δ6U mutant (160 nucleotides) was incubated with the WT 3′UTR molecule (458 nucleotides) and the viral polymerase. The radiolabeled products were analyzed with denaturing 5% polyacrylamide gels, as previously described (2, 11). The arrows indicate the mobilities of the two products. On the right, a schematic representation of the interaction of the RNA molecules representing the 5′ and 3′ ends of DENV RNA and the viral polymerase initiating RNA synthesis at the 3′ end of the template is shown. The hybridized 5′-3′UAR and 5′-3′CS are also shown. (C) RNA mobility shift assays showing the interaction between the 3′SL WT probe and the 5′RNA WT or 5′RNA Δ6U, as indicated at the top. The mobilities of the 3′SL probe and the RNA-RNA complex are indicated on the left. (D) Schematic representation of the hybridized 5′- and 3′-terminal regions of the viral genome for the WT and the mutants with deletions or insertions in the SLA-5′UAR spacer region: 5′RNAΔ10, 5′RNAΔ6, 5′RNAΔ3, 5′RNA+6, and 5′RNA+10. (E) Efficiency of trans initiation polymerase activity for different 5′RNA molecules. The 5′RNA molecules described for panel D were used for the trans initiation assay. The products were analyzed as described above. (F) The two radiolabeled products obtained in panel E were quantified, and the products obtained by trans initiation are expressed relative to the amount of product obtained by cis initiation for each mutant and referred to as percentages relative to WT levels [(3′RNA value/5′RNA value) × (5′RNA WT value/3′RNA WT value) × 100]. Error bars indicate the standard deviations of results from three independent experiments.
In the in vitro polymerase assay used here, the SLA functions as a promoter but the 5′UAR is not hybridized with the 3′UAR; instead, it is predicted to form the SLB. Therefore, to analyze whether the distance between the SLA and the hybridized 5′-3′UAR is important for RNA synthesis, we used a previously reported polymerase assay that depends on the SLA and the cyclization elements (2). In this assay, two RNA molecules (the first 160 nucleotides of the genome and the viral 3′UTR) are incubated with the viral polymerase. Under these conditions, it was shown that the SLA promotes RNA synthesis in cis and in trans, with the trans initiation being absolutely dependent on 5′-3′UAR hybridization (2). Thus, we analyzed the polymerase activity by incubating the WT 3′UTR RNA with either the WT or the mutant 5′RNA Δ6U. While the polymerase efficiently copied both 5′RNA molecules, the synthesis of the 3′RNA molecule was greatly reduced when the 5′RNA Δ6U was used (Fig. 8B). To determine whether the Δ6U mutation altered 5′RNA-3′RNA interaction, the formation of RNA-RNA complexes was also examined by gel shift assays. The dissociation constants estimated for the complex 5′-3′RNA WT and the 5′Mut Δ6U-3′RNA WT were similar (10 and 12 nM, respectively) (Fig. 8C), indicating that the Δ6U mutation did not affect RNA-RNA interaction.
The results suggested that when the 5′UAR was hybridized, the distance between the SLA and the 5′UAR influences the efficiency of polymerase activity. To further analyze this possibility, we designed a set of RNA molecules carrying different deletions or insertions in this position (Fig. 8D) (for sequences, see Materials and Methods). The WT and mutated 5′RNA molecules were incubated with the 3′UTR and the viral polymerase. The results indicated that the length of the spacer between the SLA and the UAR was critical for RNA synthesis in trans. A direct correlation between the length of the spacer and the level of RNA synthesis was observed (Fig. 8E). Quantification of the level of RNA synthesis of the 3′RNA molecule was normalized by the amount of the 5′RNA synthesized in each case (Fig. 8F). Deletion of the 10-nucleotide spacer (5′RNAΔ10) completely impaired RNA synthesis, while deletions of 6 or 3 nucleotides (5′RNAΔ6 or 5′RNAΔ3) showed levels 22 or 44% relative to the WT levels, respectively. In addition, the insertion of 6 or 10 U residues in this region (5′RNA+6 or 5′RNA+10) showed high levels of RNA synthesis. Together, the results indicate that sequences downstream of SLA modulate RNA synthesis and that a minimum of 10 residues between the SLA and the 5′UAR are necessary for an efficient process.
DISCUSSION
Here, we dissected structural elements of the SLA that are required for DENV replication. Defined helical regions, specific bulges, an SSL structure, and a TL were found to be important elements for DENV RNA synthesis. Interestingly, similar structures are predicted to occur at the 5′ ends of all flavivirus genomes, highlighting the ancestral origin of the promoter for RNA replication and providing evidence for a common mechanism of RNA synthesis.
The predicted Y-shaped structure of SLA was confirmed by RNases and chemical probing. A requirement of an SSL structure during viral replication was demonstrated by deletion of the complete element (Fig. 4C, Mut ΔSSL). However, changing the size of the stem or the sequence of the loop resulted in replication-competent viruses (Fig. 6). In agreement with previous observations (mutant M342 in reference 11), these results indicate that the function of the SLA in viral replication tolerates large variations within the SSL. Interestingly, great variability can be observed in the sequences and structures of the SSLs of different flavivirus 5′SLs (Fig. 5). In addition, an important role for the TL of SLA was also confirmed. Previous studies indicated that the sequence of the TL plays a crucial role in SLA promoter activity (11). Here, we found that mutations altering the sequence of the TL gave rise to spontaneous mutations that rescued viral replication by partially reconstituting the sequence and the structure of this element (Fig. 6B).
Helical regions within S1 and S2, rather than the nucleotide sequence per se, were required for efficient DENV2 replication (Fig. 3) (11). These findings are further supported by covariation in the SLAs of the four DENV serotypes. By using RNAZ software (31), two different types of base pairs were observed at two positions within S2 and at one position within S1 (Fig. 2A). Interestingly, a nucleotide change in S2 was previously reported to be a crucial determinant for attenuation of the DENV2 PDK-53 vaccine candidate (8, 18). A single-nucleotide substitution, C57U, resulted in a small plaque phenotype and attenuation of neurovirulence in mice (8). In addition, a more recent study found a second mutation (G58C), introducing a mismatch in S2 that increased viral replication of the C57U virus (27). Although unexpected, this result is in agreement with the efficient replication observed for a revertant virus obtained after transfection of the Mut S2.1 RNA, in which a spontaneous mutation restored only one base pair of S2, and the virus replicated with two mismatches within this region (Fig. 3). The results indicate that even though the structure of S2 is required for efficient viral replication, certain mismatches are tolerated. A correlation between sequences within the 5′UTR and pathogenesis was previously suggested by sequence comparison between an American and a Southeast Asian DENV2 isolate, which were associated with dengue fever and dengue hemorrhagic fever, respectively. This study showed a consistent nucleotide change at position 77 (A → G), just after the oligo(U) track, and a change at position 69 (A → U), generating a mismatch within S1 of SLA (20). On the basis of this information, it will be important to investigate the correlation between SLA recognition by the viral RdRp, viral replication capacity, and pathogenesis by studying the promoter activities of SLAs obtained from different field isolates.
Interestingly, despite the sequence and structural differences observed between the SLA structures of DENV1 and DENV2, RdRp of DENV2 was able to recognize and efficiently use the SLA of DENV1 both in vitro and in vivo (Fig. 4), suggesting that the determinants for SLA-polymerase recognition are conserved elements. Because the SLA of DENV2 is the most divergent, it is likely that the promoter element for polymerase activity can be exchangeable between all DENV serotypes. DENV 5′UTR contains two defined domains that are crucial for viral RNA synthesis, SLA and SLB. As defined here, the structure of SLA is necessary for viral replication. In contrast, we recently found that the structure of SLB is not essential (2); instead, a sequence within this SLB, known as the 5′UAR cyclization region, has to be complementary to a sequence present within the bottom of the 3′SL. Between the SLA and the SLB, there is an oligo(U) track that was found to play a role in viral replication. Deletion of this element results in viral attenuation (Fig. 6). Because it can be replaced by an oligo(A) sequence without loss of infectivity, it is likely that this region works as a spacer between SLA and the 5′UAR, perhaps providing the correct conformation of the RNA in the initiation complex. In vitro studies showed that the oligo(U) deletion did not alter 5′RNA-3′RNA interaction or promoter activity in cis (Fig. 8A and C). However, a direct correlation between polymerase activity and the length of the spacer between the SLA and the hybridized 5′-3′UAR was observed, indicating that sequences downstream of the promoter element modulate RNA synthesis at the authentic 3′UTR (Fig. 8E and F). This observation provides an explanation for the defect in RNA replication and attenuation of mutant Δ6U (Fig. 6 and 7).
The role of the SLA during DENV RNA synthesis involves binding of the polymerase to this element together with cyclization of the viral genome by long range RNA-RNA interactions (11). It is proposed that the circular conformation of the RNA allows the polymerase to reach the 3′-end initiation site. The presence of common structural elements at the 5′SLs of different flaviviruses and the crucial role of genome cyclization during flavivirus RNA replication (2, 3, 10, 17, 19, 38, 39) suggest fundamentally similar mechanisms of RNA synthesis in the members of this genus. In support of this idea, a recent report demonstrated SLA-viral polymerase compatibility between components of WNV and DENV (38). It is still unclear why flaviviruses evolved to have the promoter element for RNA synthesis at the 5′ end of the genome. Perhaps locating this RNA structure at the opposite end of the initiation site could be a simple way to preserve the integrity of the full-length genome.
A rational DENV vaccine design requires an understanding of the structure-function of the viral RNA elements. On the basis of our finding that the SLA can be exchangeable between different serotypes, and with the knowledge that the 5′UAR is 100% conserved, it is possible to design a unique functional 5′UTR for the four DENV serotypes. This information opens the possibility of designing a modified 5′UTR sequence that decreases the replication of the four serotypes. In this regard, the presence of the oligo(U) track downstream of the SLA in all DENV serotypes, the delayed replication of DENV2 carrying a deletion of this region, and the stability of this mutant virus in culture provide an attractive possibility of generating a universal 5′UTR with mutations or deletions within the oligo(U) track, which can be used to exchange the corresponding regions of different DENVs. A similar strategy, swapping the 3′UTR of DENV3 with that of DENV4 carrying the Δ30 mutation, has been reported (5). Currently, this virus is being evaluated in clinical trials (for a review, see reference 4). Because the 5′UTR is considerably shorter than the 3′UTR and since each of the two RNA domains (SLA and SLB) is well characterized, manipulation of this region could be a strategy for rationally designing new attenuated viruses.
In summary, in the present article we present new information about the structure-function of the SLA of DENV and provide evidence for a general role for this element in the replication of flaviviruses. In addition, we report specific RNA elements of the 5′UTR that can be modified to generate attenuation. We believe that uncovering the molecular details of DENV replication will aid the search for new antiviral strategies.
Acknowledgments
We thank Richard Kinney for DENV cDNA infectious clones, members of A. V. Gamarnik's laboratory for helpful discussions, and Marta Bravo for technical assistance.
This work was supported by grants from HHMI, DENFRAME, PICT-2003, the Bunge & Borne Foundation, and ICGEB-OPS-RELAB. A.V.G. is a member of the Argentinean Council of Investigation (CONICET). M.F.L. was funded by CABBIO and C.V.F. by CONICET.
Footnotes
Published ahead of print on 12 November 2008.
REFERENCES
- 1.Alvarez, D. E., A. L. De Lella Ezcurra, S. Fucito, and A. V. Gamarnik. 2005. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339200-212. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, D. E., C. V. Filomatori, and A. V. Gamarnik. 2008. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology 375223-235. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez, D. E., M. F. Lodeiro, S. J. Luduena, L. I. Pietrasanta, and A. V. Gamarnik. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 796631-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaney, J. E., Jr., A. P. Durbin, B. R. Murphy, and S. S. Whitehead. 2006. Development of a live attenuated dengue virus vaccine using reverse genetics. Viral Immunol. 1910-32. [DOI] [PubMed] [Google Scholar]
- 5.Blaney, J. E., Jr., N. S. Sathe, L. Goddard, C. T. Hanson, T. A. Romero, K. A. Hanley, B. R. Murphy, and S. S. Whitehead. 2008. Dengue virus type 3 vaccine candidates generated by introduction of deletions in the 3′ untranslated region (3′-UTR) or by exchange of the DENV-3 3′-UTR with that of DENV-4. Vaccine 26817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaney, J. E., Jr., N. S. Sathe, C. T. Hanson, C. Y. Firestone, B. R. Murphy, and S. S. Whitehead. 2007. Vaccine candidates for dengue virus type 1 (DEN1) generated by replacement of the structural genes of rDEN4 and rDEN4Delta30 with those of DEN1. Virol. J. 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinton, M. A., and J. H. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162290-299. [DOI] [PubMed] [Google Scholar]
- 8.Butrapet, S., C. Y. Huang, D. J. Pierro, N. Bhamarapravati, D. J. Gubler, and R. M. Kinney. 2000. Attenuation markers of a candidate dengue type 2 vaccine virus, strain 16681 (PDK-53), are defined by mutations in the 5′ noncoding region and nonstructural proteins 1 and 3. J. Virol. 743011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cahour, A., A. Pletnev, M. Vazielle-Falcoz, L. Rosen, and C. J. Lai. 1995. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology 20768-76. [DOI] [PubMed] [Google Scholar]
- 10.Corver, J., E. Lenches, K. Smith, R. A. Robison, T. Sando, E. G. Strauss, and J. H. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 772265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filomatori, C. V., M. F. Lodeiro, D. E. Alvarez, M. M. Samsa, L. Pietrasanta, and A. V. Gamarnik. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 202238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gritsun, T. S., and E. A. Gould. 2007. Origin and evolution of flavivirus 5′UTRs and panhandles: trans-terminal duplications? Virology 3668-15. [DOI] [PubMed] [Google Scholar]
- 13.Gubler, D., G, Kuno, and L. Markoff. 2007. Flaviviruses, vol. 1. Lippincott-Raven, Philadelphia, PA.
- 14.Gubler, D. J. 2006. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found. Symp. 2773-22, 71-73, 251-253. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 19833-41. [DOI] [PubMed] [Google Scholar]
- 16.Khromykh, A. A., N. Kondratieva, J. Y. Sgro, A. Palmenberg, and E. G. Westaway. 2003. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 7710623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 756719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinney, R. M., S. Butrapet, G. J. Chang, K. R. Tsuchiya, J. T. Roehrig, N. Bhamarapravati, and D. J. Gubler. 1997. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230300-308. [DOI] [PubMed] [Google Scholar]
- 19.Kofler, R. M., V. M. Hoenninger, C. Thurner, and C. W. Mandl. 2006. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J. Virol. 804099-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leitmeyer, K. C., D. W. Vaughn, D. M. Watts, R. Salas, I. Villalobos de Chacon, C. Ramos, and R. Rico-Hesse. 1999. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 734738-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leyssen, P., N. Charlier, P. Lemey, F. Billoir, A. M. Vandamme, E. De Clercq, X. de Lamballerie, and J. Neyts. 2002. Complete genome sequence, taxonomic assignment, and comparative analysis of the untranslated regions of the Modoc virus, a flavivirus with no known vector. Virology 293125-140. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach, B., H. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1133. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 23.Lo, M. K., M. Tilgner, K. A. Bernard, and P. Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 7710004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markoff, L. 2003. 5′ and 3′ NCRs in flavivirus RNA, vol. 60. Elsevier Academic Press, New York, NY.
- 25.Men, R., M. Bray, D. Clark, R. M. Chanock, and C. J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 703930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romero, T. A., E. Tumban, J. Jun, W. B. Lott, and K. A. Hanley. 2006. Secondary structure of dengue virus type 4 3′ untranslated region: impact of deletion and substitution mutations. J. Gen. Virol. 873291-3296. [DOI] [PubMed] [Google Scholar]
- 27.Sirigulpanit, W., R. M. Kinney, and V. Leardkamolkarn. 2007. Substitution or deletion mutations between nt 54 and 70 in the 5′ non-coding region of dengue type 2 virus produce variable effects on virus viability. J. Gen. Virol. 881748-1752. [DOI] [PubMed] [Google Scholar]
- 28.Thurner, C., C. Witwer, I. L. Hofacker, and P. F. Stadler. 2004. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 851113-1124. [DOI] [PubMed] [Google Scholar]
- 29.Tilgner, M., T. S. Deas, and P. Y. Shi. 2005. The flavivirus-conserved penta-nucleotide in the 3′ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology 331375-386. [DOI] [PubMed] [Google Scholar]
- 30.Tilgner, M., and P. Y. Shi. 2004. Structure and function of the 3′ terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 788159-8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Washietl, S., I. L. Hofacker, and P. F. Stadler. 2005. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. USA 1022454-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehead, S. S., J. E. Blaney, A. P. Durbin, and B. R. Murphy. 2007. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 5518-528. [DOI] [PubMed] [Google Scholar]
- 33.Whitehead, S. S., B. Falgout, K. A. Hanley, J. E. Blaney, Jr., L. Markoff, and B. R. Murphy. 2003. A live, attenuated dengue virus type 1 vaccine candidate with a 30-nucleotide deletion in the 3′ untranslated region is highly attenuated and immunogenic in monkeys. J. Virol. 771653-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson, K. A., E. J. Merino, and K. M. Weeks. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 11610-1616. [DOI] [PubMed] [Google Scholar]
- 35.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 27615581-15591. [DOI] [PubMed] [Google Scholar]
- 36.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 27433714-33722. [DOI] [PubMed] [Google Scholar]
- 37.Yu, L., and L. Markoff. 2005. The topology of bulges in the long stem of the flavivirus 3′ stem-loop is a major determinant of RNA replication competence. J. Virol. 792309-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, L., M. Nomaguchi, R. Padmanabhan, and L. Markoff. 2008. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology 374170-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, B., H. Dong, D. A. Stein, P. L. Iversen, and P. Y. Shi. 2008. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology 3731-13. [DOI] [PubMed] [Google Scholar]