Abstract
Tumor necrosis factor (TNF) and members of the interferon (IFN) family have been shown to independently inhibit the replication of a variety of viruses. In addition, previous reports have shown that treatment with various combinations of these antiviral cytokines induces a synergistic antiviral state that can be significantly more potent than addition of any of these cytokines alone. The mechanism of this cytokine synergy and its effects on global gene expression, however, are not well characterized. Here, we use DNA microarray analysis to demonstrate that treatment of uninfected primary human fibroblasts with TNF plus IFN-β induces a distinct synergistic state characterized by significant perturbations of several hundred genes which are coinduced by the individual cytokines alone, as well as the induction of more than 850 novel host cell genes. This synergy is mediated directly by the two ligands, not by intermediate secreted factors, and is necessary and sufficient to completely block the productive replication and spread of myxoma virus in human fibroblasts. In contrast, the replication of two other poxviruses, vaccinia virus and tanapox virus, are only partially inhibited in these cells by the synergistic antiviral state, whereas the spread of both of these viruses to neighboring cells was efficiently blocked. Taken together, our data indicate that the combination of TNF and IFN-β induces a novel synergistic antiviral state that is highly distinct from that induced by either cytokine alone.
Poxviruses are large enveloped DNA viruses whose replication cycle occurs exclusively in the cytoplasm (27). Although many poxviruses are highly species specific, some members, such as monkeypox virus, tanapox virus (TPV), and cowpox virus, are able to readily leap species barriers and initiate productive zoonotic infections in humans (19, 23, 28, 30). Myxoma virus (MV), an example of more restricted host species range, is a member of the Leporipoxvirus genus, which is uniquely restricted to rabbits (10, 16, 47). This host species restriction is so profound that even injection of live MV into any other vertebrate species, including humans, fails to produce a productive infection. The molecular mechanisms behind this host restriction remain largely unknown; however, there is evidence that some of the host barriers to MV permissiveness outside the rabbit are regulated by antiviral cytokines such as interferon (IFN) and tumor necrosis factor (TNF) (50, 51). The issue of restriction of MV replication in primary human cells is of particular interest because MV replicates productively in a wide variety of human cancer cells and is currently being developed as a novel oncolytic virotherapeutic to treat human cancer (46). Consequently, a better understanding of the mechanisms by which MV is restricted in primary human cells and tissues is critical before MV enters human trials.
Previously, our lab has shown that primary mouse fibroblasts are nonpermissive for MV replication because they induce and secrete type I IFN in response to MV infection (51). In addition, mice which lack STAT1 and are therefore unable to manifest the type I and II antiviral IFN responses (40, 48) become fully susceptible to lethal MV infection in vivo (51). In contrast, primary human fibroblasts are completely permissive to MV, and even the addition of high levels of exogenous IFN-β is unable to completely block viral replication. Human fibroblasts become nonpermissive for MV only after the addition of both IFN-β and TNF (50).
It is well documented that replication of many viruses can be synergistically inhibited by the addition of either type I and II IFN together (7, 32, 38, 39, 53) or TNF plus type II IFN (6, 9, 21, 22, 52). The mechanisms by which this antiviral synergy takes place, however, have not been fully elucidated (3). Recently, DNA microarray analysis was used to demonstrate that antiviral synergy between type I and II IFNs could be mediated via two mechanisms (31). The first mechanism, known as “synergy through independent action,” occurs when two cytokines upregulate distinct sets of host genes whose combined functions synergistically enhance a single antiviral pathway. The second mechanism, known as “synergy through cooperative action,” occurs when addition of a single cytokine upregulates a host antiviral gene, but the addition of the combination of two cytokines upregulates that same gene to a greater extent than can be attributed to an additive effect (31). In contrast, very little has been published concerning possible antiviral synergy between TNF and type I IFNs (25, 35). These data, however, suggest that synergy between TNF and type I IFNs is distinct from other antiviral TNF and/or IFN synergies (25).
Our present study uses DNA microarray analysis to elucidate global changes in gene expression of primary human fibroblasts treated with TNF, IFN-β, or the combination of TNF plus IFN-β. Our results demonstrate that treatment with TNF plus IFN-β induces antiviral synergy against poxviruses through both “independent action” and “cooperative action,” as well as a third type of synergy we propose to call “cooperative induction.” This synergistic antiviral state induced by TNF plus IFN-β results in a dramatic early block in MV replication and is able to completely restrict MV replication in primary human fibroblasts. Other poxviruses which are permissive in humans, such as vaccinia virus (VV) and TPV, are also affected by this synergistic antiviral state, although to various degrees compared to MV.
MATERIALS AND METHODS
Cell culture, reagents, and infections.
Primary human GM02504 fibroblasts were obtained from the Coriell Institute for Medical Research and cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 1× penicillin-streptomycin, and 2 mM l-glutamine. Recombinant TNF and IFN-β were obtained from Biosource and PBL Biomedical Laboratories, respectively.
Infections were done by removing existing media and replacing them with a minimal amount of complete DMEM containing the indicated virus. The virus was allowed to adsorb to the cells for 1 h, at which point media containing virus were removed and replaced with fresh media. Except where noted, cytokines were added after the 1-h viral adsorption.
Construction of MV-GFP/TrFP.
Construction of a recombinant MV expressing both green fluorescent protein (GFP) driven by a synthetic early/late (sE/L) poxvirus promoter and tomato red fluorescent protein (TrFP) driven by poxvirus P11 late promoter was performed using Gateway technology (Invitrogen). The sequence encoding GFP was PCR amplified using gene-specific primers with the 5′ primer containing a partial sE/L promoter sequence. The rest of the sE/L primer sequence, as well as the attB3 attachment site sequences, was added in a subsequent PCR step. The resultant sE/L-GFP fragment was then PCR ligated to another PCR fragment representing the MV M136 gene, which was amplified using a downstream primer containing the attB2 attachment site sequences. The resultant attB3-sE/L-GFP-M136-attB2 PCR fragment was recombined into the pDONR221-P3P2 plasmid (Invitrogen) using the BP clonase enzyme mix (Invitrogen). The sequence encoding the TrFP was PCR amplified using gene specific primers with the 5′ primer containing partial P11 primer sequences and the downstream primer containing the attB3r attachment site sequences. The remaining P11 promoter sequence, as well as the attB4r attachment site sequence, was added in a second PCR step using another 5′ primer containing these sequences, along with the original downstream primer. The resultant attB4r-P11-TdFP-attB3r PCR fragment was recombined into the pDONR221-P4rP3r plasmid (Invitrogen) by using the BP clonase enzyme mix. The MV sequences corresponding to M135 and the partial sequence of M134 were PCR amplified using gene specific primers that contained the attB1 and attB4 attachment site sequences. The resultant attB1-134-135-attB4 PCR fragment was recombined into the pDONR221-P1P4 plasmid (Invitrogen) using the BP clonase enzyme mix. After sequence confirmation, all three plasmids, along with the pcDNA3.2/capTEV-CT/V5-DEST_verA destination plasmid (Invitrogen), were then subjected to an LR recombination reaction using LR clonase II (Invitrogen) to generate the final plasmid construct used to construct the double-labeled virus (GFP/TrFP pcDNA3.2).
Subconfluent monolayers of BGMK cells were infected with MV at a multiplicity of infection (MOI) of 0.1. GFP/TrFP pcDNA3.2 DNA was then transfected by using Lipofectamine reagent (Invitrogen) according to the manufacturing's instructions. At 18 h posttransfection, cells were collected, and virus was released after three rounds of freezing and thawing. Fresh BGMK monolayers were then infected with cell lysates and overlaid with 10% DMEM containing 1% low-melting-point agarose. Three days after infection positive recombinants expressing both GFP and TrFP were selected and subjected to four subsequent rounds of plaque purification.
Immunofluorescence and flow cytometry.
A total of 2.5 × 104 GM02504 cells were plated in each well of a 96-well plate. The following day, cells were infected with MV-GFP (15) at an MOI of 0.1. At 24, 48, and 72 h after infection the sizes and shapes of GFP+ foci were observed by using a Leica DMI 6000B microscope. Cells were then harvested using trypsin and fixed in 2% paraformaldehyde, and the percentage of GFP+ cells was quantitated by using flow cytometry on a BD FACSCalibur.
Virus titers.
A total of 2 × 105 GM02504 cells were plated in each well of a 12-well dish. The following day, cells were infected with MV-GFP at an MOI of 0.1. At each indicated time point, cells were harvested using trypsin, resuspended in 500 μl of DMEM, and frozen. After collection at all time points, the cells were lysed via repeated freeze-thawing, and the titer of each virus was determined by using the limiting dilution method (18).
RNA purification.
For purification of total RNA for microarray analysis and real-time PCR, 106 GM02504 cells were plated in each well of a six-well dish. The following day the cells were mock treated or treated with TNF (20 ng/ml), IFN-β (500 U/ml), or the combination of TNF plus IFN-β. At the given time points, cells were lysed directly in RLT buffer (Qiagen), and total RNA was extracted by using an RNeasy kit (Qiagen).
Microarray analysis.
The experiment was carried out with four independent replicates of each condition which consisted of mock, TNF, IFN-β, and TNF plus IFN-β treatments. From each replicate of each treatment group the total RNA was isolated 24 h after cytokine treatment. Starting with 1 μg of total RNA, labeled cRNA was prepared and used to interrogate Affymetrix U133 + two GeneChips using standard protocols recommended by Affymetrix. Arrays were hybridized, washed, and stained using Affymetrix fluidics protocol FS450_004, and the GeneChips were scanned with an Affymetrix G7 scanner.
Microarray normalizations and modeling.
Microarray data were normalized, and a model-based expression matrix was derived using the perfect-match-only algorithms of dChip (20). For unsupervised analysis, probe sets whose hybridization signal intensity varied across the data set with a coefficient of variation of >0.5 were identified and visualized by average linkage hierarchal clustering using the clustering algorithms implemented in dChip. For supervised analysis, probe sets whose hybridization signal intensities showed significant (P < 0.001) variation between the various treatment and control groups were identified by f-test using the class comparison tools implemented in BRB ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools.html). Leave-one-out cross-validation was used to assess the ability of significant probe sets to distinguish between the treatment classes. Each array was left out in turn, and a classifier composed of probe sets significant at P < 0.001 with the remaining 15 arrays was derived. Then, using only the significant probe sets with a nearest-neighbour prediction model, the class identity of the array left out was predicted. In all 16 cross-validation studies, the correct class label was predicted, thus establishing the validity of the probe sets significant at P < 0.001 to distinguish between the treatment groups. Ability of probe sets that were significantly different between the treatment groups was established by using leave-one-out cross-validation studies based on the class prediction tools implemented in BRB ArrayTools. Each array was left out in turn, a classifier was derived composed of probe sets significant at P < 0.001 with the remaining 15 arrays, and then, using only the significant probe sets with nearest-neighbor prediction, models of the class identity of the array left out were established.
Real-time PCR.
A total of 2 μg of total RNA was used to create cDNA. Initially, genomic DNA was removed from total RNA using the DNA-free kit (Ambion) according to the manufacturer's recommendations. After removal of the genomic DNA, 1 μl of the deoxynucleoside triphosphates (100 mM) and 1 μl of random hexamer primers (50 μg/ml) were added, and the mixture was incubated for 5 min at 65°C. After this incubation, the tube was allowed to cool to room temperature, and 6 μl of 5× reaction buffer, 3 μl of dithiothreitol (0.1 M), 1 μl of RNasin (Promega), and 1 μl of Superscript III reverse transcriptase (Invitrogen) were added. The resulting mixture was incubated for 1 h at 42°C and then for 15 min at 72°C. The final reaction was diluted 1:10 with sterile H2O and used for Sybr green-based real-time PCR. Then, 4 μl of diluted cDNA was added to 21 μl of PCR mix containing 0.5 U of Taq polymerase (NEB), 1× Thermo Pol buffer, 0.1× Sybr green dye (Molecular Probes), 0.5× Rox reference dye (Invitrogen), a 160 μM concentration of deoxynucleoside triphosphates (Invitrogen), 4 mM MgCl (Invitrogen), 4 ng of forward primer, and 4 ng of reverse primer. The resulting 25-μl reaction was run on an ABI 7300 real-time PCR machine under the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Primers used in real-time PCR analysis are listed in Table 1.
TABLE 1.
Primers for real-time PCR
| Gene | Primer
|
|
|---|---|---|
| Directiona | Sequence (5′-3′) | |
| CXCL10 | For | AGGAACCTCCAGTCTCAGCA |
| Rev | ATTTTGCTCCCCTCTGGTTT | |
| GAPDH | For | AGGTTGTCTCCTGCGACTTCA |
| Rev | CCAGGAAATGAGCTTGACAAAGTT | |
| TLR3 | For | TGGTTGGGCCACCTAGAAGTA |
| Rev | TCTCCATTCCTGGCCTGTG | |
| RSAD2 | For | CCTGCTTGGTGCCTGAATCT |
| Rev | GCGCATATATTCATCCAGAATAAGG | |
| BST2 | For | CCAGAAGGGCTTTCAGGATGT |
| Rev | AAGCCATTAGGGCCATCACA | |
| TNFSF13B | For | CGCGGGACTGAAAATCTTTG |
| Rev | CACGCTTATTTCTGCTGTTCTGA | |
| IF127 | For | TGCCTCGGGCAGCCT |
| Rev | TTGGTCAATCCGGAGAGTCC | |
| ISG20 | For | AGATCCTGCAGCTCCTGAAA |
| Rev | TGTTCTGGATGCTCTTGTGC | |
| CCL2 | For | GAAGAATCACCAGCAGCAAGTGT |
| Rev | TGGAATCCTGAACCCACTTCTG | |
| BIRC3 | For | GGGAAGAGGAGAGAGAAAGAGCAA |
| Rev | GAATTACACAAGTCAAATGTTGAAAAAGT | |
| IL-8 | For | GAATGGGTTTGCTAGAATGTGATA |
| Rev | CAGACTAGGGTTGCCAGATTTAAC | |
| LIF | For | TGGTTCTGCACTGGAAACATG |
| Rev | TGTAATAGAGAATAAAGAGGGCATTGG | |
| SWAN | For | TGTCAACCATCCTCGGTGTCTA |
| Rev | GCCTAAGGACTTTCAGGTAATCAGAGT | |
| IFNAR2 | For | GAGCACAGTGATGAGCAAGC |
| Rev | TCAAGACTTTGGGGAGGCTA | |
Rev, reverse; For, forward.
RESULTS
DNA microarray analysis of primary human fibroblasts treated with TNF, IFN-β, or TNF plus IFN-β.
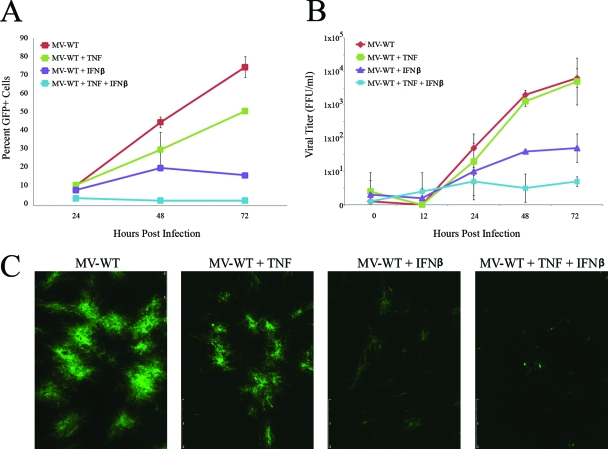
Previously, our lab has shown that CCD-922sk primary human fibroblasts are fully permissive for MV replication. The addition of either TNF or IFN-β to these cells leads to a partial restriction of MV, while the addition of TNF plus IFN-β leads to a complete restriction (50). For the experiments reported here, we utilized primary human GM02504 fibroblasts and observed that the levels of MV inhibition after treatment with TNF or IFN-β varied from experiment to experiment. TNF exhibited only a minor ability to inhibit MV progeny formation (Fig. 1B) but appears to inhibit the cell-to-cell spread of MV to a larger extent (Fig. 1A). This difference is likely caused by the depiction progeny virus on a logarithmic scale and cell-to-cell spread on a linear scale. IFN-β inhibited both MV progeny formation (Fig. 1B) and cell-to-cell spread (Fig. 1A). However, consistent with our previous findings, complete restriction of MV was only observed after the addition of TNF plus IFN-β together. This complete restriction was observed in terms of spread of MV-GFP after infection at a low MOI (Fig. 1A), progeny virus replication (Fig. 1B), and formation of classic MV foci in monolayer cultures (Fig. 1C). To determine the mechanisms responsible for viral inhibition following these three cytokine treatments, we used DNA microarray analysis of GM02504 cells mock treated or treated with TNF (20 ng/ml), IFN-β (500 U/ml), or the combination of TNF (20 ng/ml) plus IFN-β (500 U/ml). Four replicates of each condition were generated to confirm the reproducibility of each cytokine treatment. At 24 h after cytokine treatment, the total cellular RNA was harvested from each sample and used to create cRNAs for DNA microarray analysis. Each sample was then analyzed on an Affymetrix U133+2.0 complete human genome array, which surveys 56,000 human transcripts. The complete data set is available in the Tables S1 to S3 in the supplemental material.
FIG. 1.
TNF and IFN-β synergistically inhibit MV in primary human fibroblasts. Primary human GM02504 fibroblasts were infected with MV-GFP at an MOI of 0.1. After 1 h of viral adsorption, cells were treated with TNF, IFN-β, or the combination of TNF plus IFN-β. To determine the effect of these treatments on viral spread, cells were harvested via trypsin treatment at 24, 48, and 72 h postinfection. The percentage of GFP+ live cells in each sample was then determined by using flow cytometry. (A) Graph depicting a representative experiment run in triplicate. (B) Graph depicting the average of three independent experiments. (C) Prior to each analysis of each experiment, focus sizes were observed by florescence microscopy. To determine the effect of each treatment on virus titer, cells were harvested at the given time points via trypsinization. Cells were then lysed via repeated freeze-thawing, and virus titers were determined as outlined in Materials and Methods.
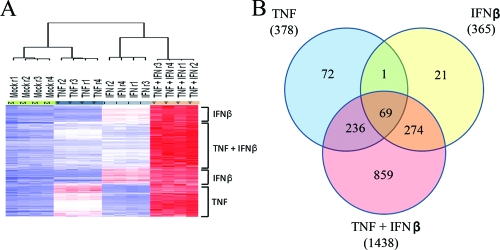
After the DNA microarray analysis was completed, apparent gene expression profiles among the treatment groups were analyzed by using both unsupervised and supervised methods of analyses. Supervised analysis between the treatment groups identified 14,663 probe sets that were significant at the P < 0.001 significance threshold, in terms of significant variation from the group mean in at least one of the four cytokine treatment sets. A heat map displaying the probe sets from this analysis that exhibited a level of induction >2-fold compared to the mock-treated group revealed that these genes fell into three main categories: (i) genes that were induced by TNF or by TNF plus IFN-β, (ii) genes that were induced by IFN-β or by TNF plus IFN-β, and (iii) genes that were induced only in the presence of TNF plus IFN-β (Fig. 2A). Inspection of the dendrogram obtained after hierarchical clustering of these probe sets showed that all four biological replicates of each sample clustered tightly together, but each treatment was highly distinct from the others. The major node of separation placed mock- and TNF-treated samples on one side of the dendrogram and IFN-β- and TNF+IFN-β-treated samples on the other side of the dendrogram. In the unsupervised analysis, probe sets that varied the most across the data set were identified by using a variation filter. This analysis identified 2,222 probe sets with a coefficient of variation of >0.5 (data not shown).
FIG. 2.
Treatment with TNF plus IFN-β causes upregulation of a large set of human genes not induced by TNF or IFN-β alone. An f-test was preformed to identify probe sets perturbed significantly (P < 0.001) among the four treatment groups. (A) The heat map displays the probe sets from this analysis, which also displayed a level of induction of >2-fold compared to mock treatment groups. The general grouping of these genes into sets that are induced by TNF, IFN-β, or TNF plus IFN-β is shown on the right of the heat map. Inspection of the dendrogram obtained after hierarchical clustering showed that for all replicates of each treatment class clustered tightly together. (B) Venn diagram analysis to illustrate the number of genes that overlapped between multiple treatment classes. Note that the Venn diagram displays genes that were significantly perturbed from the mock-treated sample (P < 0.001).
Synergy of TNF and IFN-β by “independent action.”
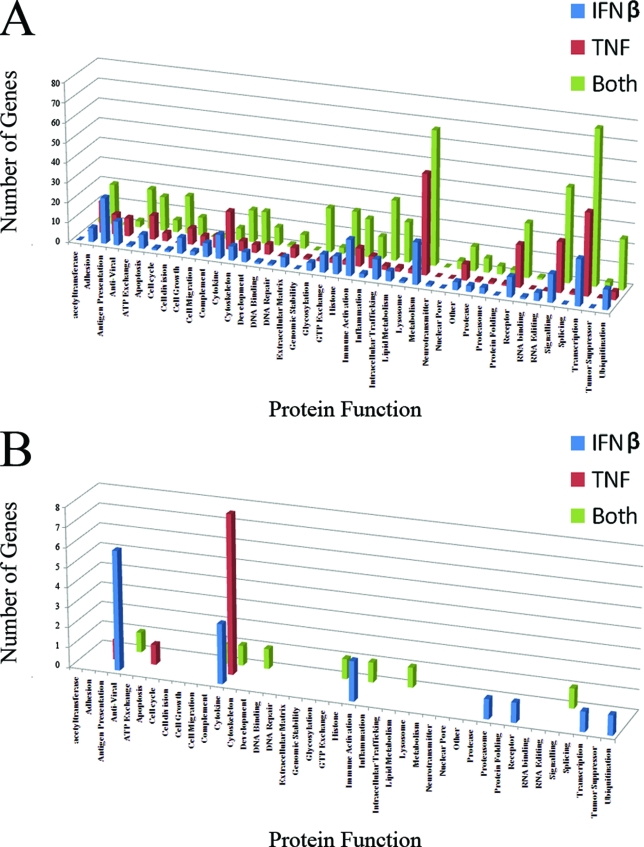
Previous reports have hypothesized that the antiviral synergy observed after the addition of multiple cytokines can occur through two distinct mechanisms: “synergy by independent action” and “synergy by cooperative action” (31). To determine whether synergy through independent action was playing a role in the synergistic inhibition of MV by TNF plus IFN-β, we grouped genes which were upregulated by each cytokine treatment into general functional categories based on their annotation in the Swiss-Prot/TrEMBL database (http://ca.expasy.org/). Consistent with previously published papers (5, 11, 42, 54), our results indicate that treatment of primary fibroblasts with TNF or IFN-β alone leads to upregulation of ∼350 distinct host genes, which were significant at P < 0.001 and displayed a level of induction of >2-fold (Fig. 2). The subsets of genes upregulated by TNF or IFN-β, however, are remarkably distinct and largely exclusive from one another (Fig. 2B). In fact, only 70 of 743 (∼9.4%) genes upregulated by TNF or IFN-β are upregulated by both cytokines, suggesting that the antiviral states induced in primary human cells by each treatment are very different from one another. Interestingly, while most genes upregulated by TNF or IFN-β are not upregulated by the other, both cytokines activate many genes within the same functional categories (Fig. 3A). For example, TNF upregulates 12 genes involved in regulation or execution of apoptosis, while IFN-β upregulates seven genes involved in this cellular pathway; none of these apoptotic genes, however, is upregulated by both TNF and IFN-β. In addition, TNF upregulates 21 cellular receptors, whereas IFN-β upregulates only 10. Again, however, none of these genes are commonly upregulated by both TNF and IFN-β.
FIG. 3.
Functional categories of upregulated human genes. Each human gene that was scored as upregulated in the DNA microarray analysis was grouped into a functional category based on its annotation in the Swiss-Prot database. Graphs depict the total number of genes in each category (A), as well as the number of genes in each category which were significant at P < 0.001 and were upregulated by >50-fold (B).
In contrast, the human genes that are most highly upregulated by TNF, IFN-β, or TNF plus IFN-β (i.e., P < 0.001 and fold change of >50) tend to fall into largely unique functional categories (Fig. 3B and Table 2). Treatment with TNF upregulates 11 human genes by >50-fold; of these, 8 (∼72%) encode secreted cytokines or chemokines. In contrast, treatment with IFN-β highly induced (by >50-fold) 24 genes; but only 2 of these genes, CXCL11 and ANGPTL1, encode secreted proteins. The remaining 22 IFN-inducible genes encode proteins that fall into a wide range of functional categories, including antigen presentation, known antiviral effectors, immune system activators, surface receptors, transcription regulators, and protein degradation pathway members, as well as nine proteins with currently unknown function. These data strongly suggest that synergy between TNF and IFN-β likely can occur via “independent action” and that this type of synergy most likely occurs between series of genes which, individually, display low fold inductions in response to TNF or IFN-β alone.
TABLE 2.
Classes of highly upregulated genes
| Gene class and gene | Cytokine (fold increase)
|
Function | ||
|---|---|---|---|---|
| TNF+IFNβ | TNF | IFN-β | ||
| Distinct genes | ||||
| CCL5 | 140 | 0 | 0 | Cytokine |
| CD38 | 112 | 0 | 0 | Signaling |
| DHX58 | 101 | 0 | 0 | Antiviral |
| APOL1 | 82 | 0 | 0 | Lipid metabolism |
| VCAM1 | 66 | 0 | 0 | Immune activation |
| GBP4 | 66 | 0 | 0 | GTP exchange |
| CR1 | 61 | 0 | 0 | Complement |
| DMRTAI | 52 | 0 | 0 | Development |
| Partially distinct genes (TNF) | ||||
| IL-8 | 1,575 | 1,411 | 0 | Cytokine |
| IL-32 | 500 | 212 | 0 | Cytokine |
| CCL2 | 360 | 167 | 0 | Cytokine |
| BIRC3 | 248 | 128 | 0 | Cytokine |
| CXCL2 | 234 | 181 | 0 | Cytokine |
| C8orf4 | 215 | 97 | 0 | Apoptosis |
| 116 | 191 | 118 | 0 | Cytokine |
| CXCL1 | 143 | 112 | 0 | Cytokine |
| CXCLS | 72 | 211 | 0 | Unknown |
| CXCL6 | 9 | 59 | 0 | Cytokine |
| Partially distinct genes (IFN) | ||||
| CXCL11 | 2,004 | 0 | 69 | Antiviral |
| RSAD2 | 1,065 | 0 | 679 | Immune activation |
| BST2 | 867 | 0 | 410 | Unknown |
| TNFSF13B | 620 | 0 | 120 | Receptor |
| 15G20 | 330 | 0 | 117 | Cytokine |
| IF127 | 280 | 0 | 139 | Antiviral |
| NPBWR1 | 205 | 0 | 121 | Unknown |
| ANGPTLI | 168 | 0 | 69 | Immune activation |
| IF144L | 163 | 0 | 107 | Unknown |
| TLR3 | 152 | 0 | 79 | Cytokine |
| BATF2 | 131 | 0 | 54 | Unknown |
| LOC727996 | 124 | 0 | 64 | Transcription |
| IL18BP | 94 | 0 | 51 | Cytokine |
| Shared genes | ||||
| CXCL10 | 954 | 6 | 4 | Cytakine |
| OAS1 | 606 | 6 | 397 | Antiviral |
| IFIT2 | 394 | 4 | 186 | Unknown |
| MX2 | 337 | 6 | 248 | Unknown |
| IL-411 | 293 | 51 | 25 | Antigen presentation |
| TNFAIP6 | 274 | 17 | 3 | Adhesion |
| OAS2 | 185 | 6 | 137 | Antiviral |
| IFIH1 | 185 | 9 | 59 | Antiviral |
| EPSTI1 | 150 | 7 | 67 | Unknown |
| LOC129607 | 139 | 4 | 95 | Unknown |
| IFIT1 | 137 | 3 | 106 | Unknown |
| GCH1 | 136 | 13 | 4 | Metabolism |
| HERC6 | 132 | 5 | 84 | Ubiquitination |
| ICAM1 | 130 | 41 | 4 | Immune activation |
| C15orf48 | 119 | 23 | 7 | Unknown |
| CT55 | 118 | 15 | 21 | Antigen presentation |
| CFB | 115 | 3 | 14 | Complement |
| APOL3 | 104 | 4 | 38 | Intracellular trafficking |
| PSMB9 | 102 | 6 | 76 | Proteasome |
| GBP5 | 87 | 7 | 9 | GTP exchange |
| IFIT3 | 60 | 3 | 40 | Unknown |
| MX1 | 56 | 5 | 52 | Antiviral |
| DDX58 | 54 | 2 | 24 | Antiviral |
| OAS3 | 51 | 4 | 36 | Antiviral |
| CCL7 | 51 | 15 | 5 | Cytokine |
We clustered the significantly induced genes (P < 0.001) that were induced by >50-fold into three categories based on the ability of each gene to distinguish an antiviral state (Table 2). “Distinct” genes were upregulated only in the presence of TNF plus IFN-β. Since these genes are only upregulated in the presence of both cytokines, they can be used to identify conditions in which novel cytokine synergy occurs. Of particular interest in this category are DHX58 and GBP4. DHX58 (LGP2) is an adaptor molecule that negatively regulates signaling through RIG-I (36) and thus influences cellular monitoring of viral infection. GBP4 is a largely unstudied member of the guanylate binding protein family. Other members of this family, however, have been shown to have antiviral properties (2, 4), suggesting that GBP4 could play a role in mediating the unique antiviral state induced by TNF plus IFN-β. In contrast to “distinct” genes, “partially distinct” genes could only distinguish between two of the three antiviral states: (i) TNF alone and TNF plus IFN-β or (ii) IFN-β alone and TNF plus IFN-β. These genes are able to differentiate between the TNF and IFN-β antiviral responses and are generally well characterized. “Shared” genes are upregulated in all three antiviral states. This set of genes includes the most known antiviral genes (such as OAS1, -2, and -3, DDX58, and MX1); however, it also includes the most uncharacterized genes. Several of these uncharacterized genes are likely to have antiviral potential. For example, the murine homologue of MX2 has been shown to inhibit both rhabdovirus and vesicular stomatitis virus (24, 55). Since so many genes in the “shared” category have been shown to inhibit viral replication, currently uncharacterized genes in this category such as IFIT1 and IFIT2, EPSTI1, and LOC129607 represent likely candidates for novel antivirals.
Synergy of TNF and IFN-β by “cooperative action.”
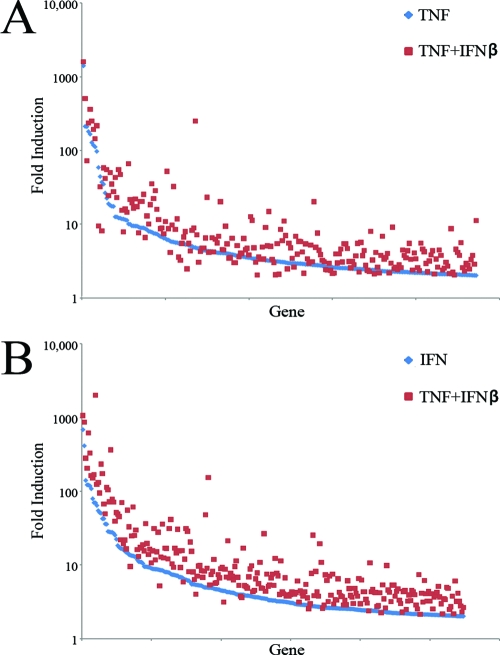
Simultaneous treatment with TNF plus IFN-β leads to the upregulation of 1,438 human genes that were significant at P < 0.001 and displayed a level of induction of >2-fold, including the majority of genes upregulated by either TNF (306 of 378 [∼81%]) or IFN-β alone (344 of 365 [∼94%]). To determine whether simultaneous treatment with TNF plus IFN-β leads to “synergy by cooperative action,” we compared the fold induction of all genes that were upregulated by a single cytokine and the combination of both cytokines (Fig. 4 and Table 2). In the set of 344 human genes upregulated by IFN-β or TNF plus IFN-β, 332 genes displayed a higher fold induction after treatment with TNF plus IFN-β, whereas only 12 genes displayed a lower fold induction after treatment with both cytokines. Treatment with TNF plus IFN-β upregulated this gene set by an average of ∼36-fold, while treatment with IFN-β upregulated this set by only 13-fold (P = 0.008), suggesting that this increased modulation of expression after treatment with TNF plus IFN-β was not due to random fluctuation (Fig. 4B). A similar result was observed for the set of 306 genes upregulated by the addition of TNF alone or TNF plus IFN-β. In this gene set, 263 genes displayed higher levels of induction after treatment with TNF plus IFN-β, while 43 genes displayed lower levels of induction. The average fold induction of genes in this set after the addition of TNF alone was ∼16-fold compared to 24-fold after the addition of TNF plus IFN-β (Fig. 4A). Although this difference failed to reach statistical significance (P = 0.19), these data, combined with the data from the IFN-β induced gene set, argue for an overall trend in which treatment with TNF plus IFN-β results in generally higher fold inductions than treatment with either cytokine alone. Interestingly, although representing only a minority of genes uniquely regulated by IFN plus TNF, ca. 8% of human genes are either upregulated less or actually downregulated in the synergistic antiviral state compared to either cytokine alone. For example, CXCL5 and CXCL6 are upregulated 210- and 78-fold, respectively, by TNF alone but only 70- and 10-fold by TNF plus IFN-β (see Tables S1 to S3 in the supplemental material).
FIG. 4.
Treatment with TNF plus IFN-β results in “synergy by cooperative action.” (A) Graph depicting the 236 human genes upregulated by TNF alone or TNF plus IFN-β; (B) graph depicting the 274 human genes upregulated by either IFN-β alone or TNF plus IFN-β. Each gene is shown twice on each graph. The fold change induced by the single cytokine alone is shown in blue, while the fold change induced by the combination of both cytokines is shown in red.
Addition of TNF plus IFN-β frequently alters the kinetics of human gene upregulation.
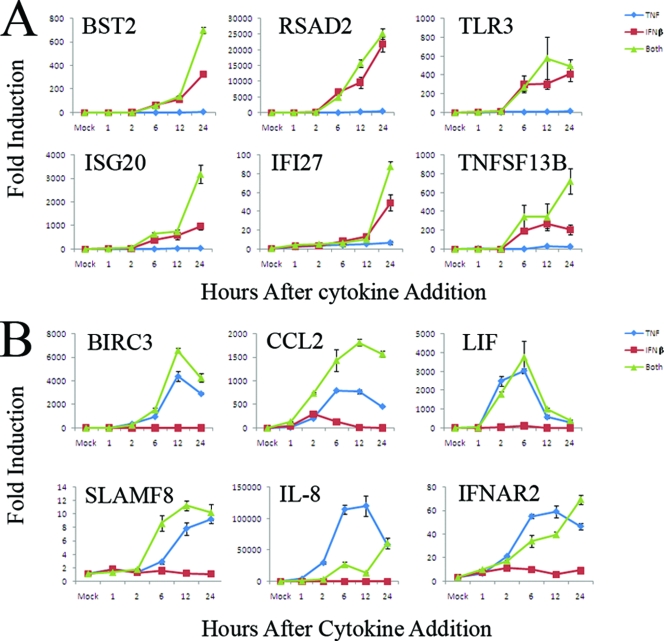
To determine the mechanism by which addition of TNF plus IFN-β results in “synergy by cooperative action,” we analyzed the expression kinetics for a representative sampling of human genes by real-time PCR. GM02504 cells were mock treated or treated with TNF, IFN-β, or TNF plus IFN-β. At the indicated times after cytokine treatment, cells were harvested, and RNA was extracted and used to create cDNA for real-time PCR analysis as detailed in Materials and Methods. The expression kinetics of 12 human genes that are strongly induced by TNF plus IFN-β were analyzed: 6 that are predominantly induced by IFN-β and 6 that are predominantly induced by TNF.
All six genes predominantly induced by IFN-β showed a similar pattern of kinetic expression. In samples treated with IFN-β alone, these genes were initially induced between 2 and 6 h after the addition of IFN-β and then continued to increase until the final 24-h time point (Fig. 5A). In samples treated with TNF plus IFN-β, all six analyzed genes were induced with identical kinetics; however, the level of induction was significantly higher for five of the six genes compared to samples treated with IFN-β alone. Analysis of the six selected genes that were predominantly induced by TNF resulted in far more varied results (Fig. 5B). For example, interleukin-8 (IL-8), SLAMF8, BIRC3, and CCL2 were all induced between 2 and 6 h after the addition of TNF. Though IL-8 expression continued to increase until 24 h, however, the expression of SLAMF8, BIRC3, and CCL2 peaked at 6 to 12 h and began to decrease by 24 h. In contrast, LIF was initially induced between 1 and 2 h after the addition of TNF, peaked at 6 h, and had returned to near baseline levels by 24 h. Interestingly, expression of IL-8 and IFNAR2, which peak at 6 to 12 h after the addition of TNF, remained low during these times after the addition of TNF plus IFN-β and instead showed a major increase in expression between 12 and 24 h (Fig. 5B). Thus, addition of TNF plus IFN-β not only enhances individual gene induction but also can alter the kinetics of that induction.
FIG. 5.
The combination of TNF plus IFN-β alters the kinetics of human gene induction. Primary human GM02504 fibroblasts were treated with TNF, IFN-β, or TNF plus IFN-β. At the listed time points, RNA was extracted from each sample and used to synthesize cDNA. The expression of each gene at each time point was assayed with gene-specific primers using Sybr green-based real-time PCR as detailed in Materials and Methods. The graphs in panels A and B show the average of two independent experiments each done in triplicate.
Note that the levels of induction obtained using real-time PCR are significantly higher for many genes than those obtained from the microarray analysis. Many of these genes are not expressed in the mock-treated samples to any detectable level; thus, the denominator when calculating fold induction for these genes is close to zero and varies depending on which technique is used to measure it. This leads to very high fold inductions of these genes when values are measured using real-time PCR and accounts for the difference in the fold induction levels between the two techniques.
Synergy of TNF and IFN-β by “cooperative induction.”
In addition to the 650 human genes that display “synergy by cooperative action,” treatment with TNF plus IFN-β also upregulates a novel set of 859 genes that are not upregulated after the addition of either cytokine alone (Fig. 3). This form of synergy is technically a subset of “synergy by cooperative action.” However, we believe it to be distinct enough from the previously mentioned synergies, in which the combination of cytokines enhances induction of a gene that is already induced by a single cytokine, to warrant a unique name. Therefore, we propose to term this form of synergy “synergy by cooperative induction”.
Similar to treatment with TNF or IFN-β alone, treatment with both cytokines upregulates human genes across a wide variety of functional categories. Several of these categories show significant overlap with those upregulated by TNF or IFN-β alone. For example, treatment with TNF plus IFN-β upregulates all 19 genes involved in apoptosis upregulated by TNF or IFN-β alone. Treatment with both cytokines, however, also upregulates an additional 17 genes involved in apoptosis that were not upregulated by either cytokine alone. Likewise, treatment with TNF and IFN-β upregulates 24 of the 31 cellular receptors upregulated by TNF or IFN-β alone, as well as an additional 27 receptors.
Treatment with TNF plus IFN-β, however, also activates genes in functional categories only minimally activated by either cytokine alone; for example, treatment with TNF plus IFN-β upregulates 22 genes whose products are involved in protein glycosylation. In contrast, TNF upregulates only one glycosylation factor, while IFN-β upregulates four. Similarly, TNF plus IFN-β upregulate 20 genes that function in lipid metabolism, while TNF upregulates only 2 such genes and IFN-β upregulates only 5. These data collectively support the hypothesis that the intracellular antiviral states induced by TNF, IFN-β, or TNF plus IFN-β are highly distinct from one another.
To determine whether “cooperative induction” played a role in the synergistic antiviral state caused by TNF and IFN-β, we used small interfering RNA to inhibit induction of the eight “distinct” genes identified in our microarray analysis (Table 2). Although successful knockdown of these genes did not allow MV to grow in the presence of TNF and IFN-β, this does not necessarily mean that they do not play a role in the synergistic antiviral state. More experiments will be needed to fully elucidate whether each of these genes plays a role in inhibiting MV replication.
TNF mediates synergy with IFN-β directly and not via secreted intermediates.
Previously, it has been demonstrated that synergistic antiviral states can be induced by a variety of cytokines other than TNF and IFN-β. For example, IL-1β, in combination with IFN-γ, has been shown to synergistically inhibit replication of both herpes simplex virus type 1 (HSV-1) (1, 13) and measles virus (29). Importantly, our DNA microarray results indicated that IL-1β was upregulated 35-fold in primary human fibroblasts in response to TNF (see Table S1 in the supplemental material). This raises the possibility that the antiviral effects of TNF and the synergy we observed after the addition of TNF plus IFN-β might be mediated through a third induced cytokine, such as IL-1β.
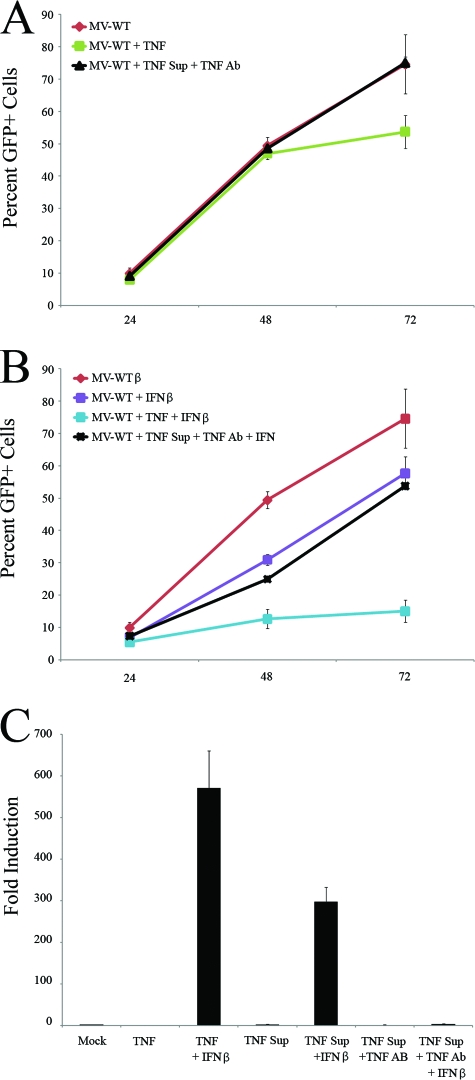
To determine whether the anti-MV effects of TNF were mediated directly or through a secreted intermediate factor, we assessed whether supernatants from TNF-treated human fibroblasts contained any factor that inhibited MV replication. GM02504 cells were treated with 1 ng of TNF/ml for 24 h, after which the resulting supernatant was removed and TNF was depleted using an anti-TNF neutralizing antibody (Biosource). The ability of this depleted supernatant to inhibit MV replication was then determined by adding this TNF-depleted supernatant to fresh GM02504 cells infected with MV-GFP at an MOI of 0.1. The spread of MV-GFP infection in the fresh GM02504 cell culture was then measured by using flow cytometry. Although fresh cells treated directly with TNF displayed reduced the numbers of GFP+ cells at 72 h, cells treated with the TNF-depleted supernatant were indistinguishable from mock-treated cells (Fig. 6A). The observation that treatment with TNF only significantly reduced MV infection at 72 h was consistently observed in five independent experiments. We hypothesize that this is a result of using lower levels of TNF used in these experiments. These data suggest that the anti-MV effect of TNF is not mediated through any non-TNF secreted factor.
FIG. 6.
Synergy between TNF and IFN-β is not mediated by a secreted intermediate factor. To generate TNF-treated supernatants, GM02504 primary human fibroblasts were treated with 1 ng of TNF/ml for 24 h. TNF was then removed from the resulting supernatant by using a depleting anti-huTNF antibody. To determine whether this TNF-depleted supernatant had any anti-MV properties, fresh GM02504 cells were infected with MV-GFP at an MOI of 0.1 and then either mock treated, treated with 1 ng of TNF/ml, or treated with the TNF supernatant. (A) At 24, 48, and 72 h postinfection, cells were harvested, and the ability of MV-GFP to spread cell to cell was assayed by flow cytometry. To determine whether secreted factor(s) in the TNF-treated supernatant was able to synergize with IFN-β, fresh GM02504 cells were infected with MV at an MOI of 0.1 and then either mock treated or treated with IFN-β, IFN-β + 1 ng of TNF/ml, or IFN-β + TNF supernatant. (B) At 24, 48, and 72 h, the ability of MV-GFP to spread through these cultures was assayed by using flow cytometry. To further analyze whether the TNF supernatants could induce synergy with IFN-β, the ability of these supernatants to induce “synergy by cooperative induction” was tested by using real-time PCR. Fresh GM02504 cells were treated as indicated. At 24 h after treatment, RNA was harvested from cells and used to make cDNA. Levels of gene expression were assayed with gene specific primers using Sybr green-based real-time PCR as detailed in Materials and Methods. (C) Graph showing the induction of CXCL10.
To determine whether IFN-β synergized directly with TNF, we measured the ability of TNF-treated supernatants to synergistically inhibit cell-to-cell spread of MV and to induce “synergy by cooperative induction” when combined with IFN-β. TNF-treated supernatants were generated as described above and then depleted of TNF. Fresh GM02504 cells were then infected with MV-GFP at an MOI of 0.1 and treated with IFN-β, IFN-β plus TNF, or IFN-β plus TNF supernatant. The cell-to-cell spread of MV-GFP infection in the GM02504 cell culture was then measured by using flow cytometry. While MV-GFP was effectively unable to spread through fresh cells treated with TNF plus IFN-β, cells treated with IFN-β plus TNF supernatant displayed only a partial block (Fig. 6B). This block was similar to that observed after treatment with IFN-β alone, suggesting that the TNF supernatant did not contain any factor that could synergize with IFN-β.
To determine whether TNF supernatants could induce “synergy by cooperative induction” when combined with IFN-β, GM02504 cells were treated with IFN-β or IFN-β plus TNF-induced (but then TNF-depleted) supernatant. At 24 h after cytokine treatment, RNA was recovered and used to synthesize cDNA for real-time PCR analysis. While IFN-β, in combination with either TNF or TNF supernatants from which TNF had not been depleted, was fully able to induce the representative marker genes for “synergy by cooperative induction,” these genes were not induced after the addition of IFN-β and TNF-depleted supernatants (Fig. 6C and data not shown). These data suggest that both TNF's antiviral effect and its ability to induce “synergy by cooperative induction” with IFN-β are mediated directly by TNF and not through a third-party secreted cytokine.
TNF plus IFN-β induces an early block against MV replication in human fibroblasts.
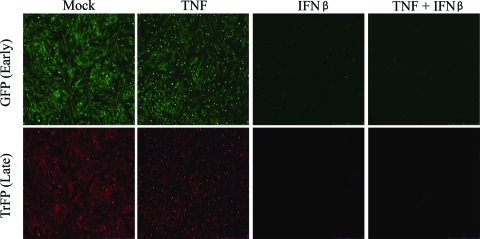
To determine what MV replication stage was blocked by TNF plus IFN-β in human fibroblasts, we utilized a recombinant MV that expresses GFP under a sE/L promoter and TrFP under the P11 late promoter. GM02504 cells were mock treated or pretreated with TNF, IFN-β, or TNF plus IFN-β for 24 h. Treated cells were infected with MV-GFP/TrFP at an MOI of 1, and the infection was allowed to progress for 24 h in the presence of TNF, IFN-β, or TNF plus IFN-β. Expression of GFP and TrFP was then analyzed by using a Leica DMI 6000B microscope. Although both GFP and TrFP are easily observed in mock- and TNF-treated samples, samples treated with either IFN-β or the combination of TNF plus IFN-β show strikingly reduced levels of both GFP and TrFP (Fig. 7). These data are consistent with a block in MV replication either at or before early viral gene expression.
FIG. 7.
The combination of TNF plus IFN-β blocks MV replication prior to, or at the level of, early gene expression. GM02504 primary human fibroblasts were treated with TNF, IFN-β, or TNF plus IFN-β. At 24 h after the addition of these cytokines, cells were infected with a recombinant MV that expresses both GFP, under an sE/L promoter, and TrFP, under a viral P11 late promoter. At 24 h after infection, the levels of both GFP and TrFP were measured via florescence microscopy.
The antiviral state induced by TNF plus IFN-β variably affects different poxviruses.
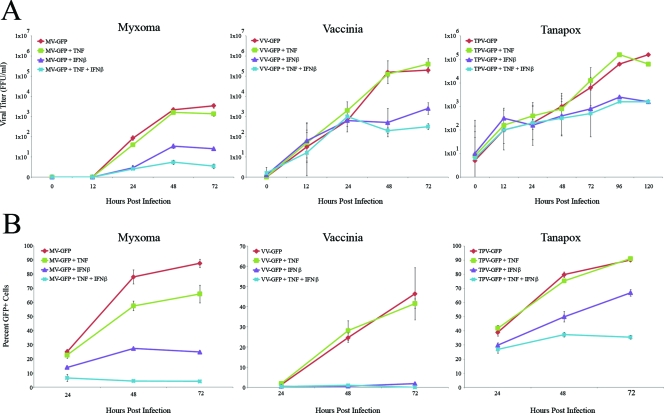
Since the restriction of MV replication in primary human fibroblasts is so pronounced after the addition of TNF plus IFN-β, we were interested to determine whether this cytokine combination also comparably inhibited other poxviruses capable of infecting human cells. To address this question, we infected GM02504 cells with GFP-expressing versions of either MV, the orthopoxvirus VV or the yatapoxvirus TPV at an MOI of 0.1. Immediately after viral adsorption, infected cells were mock treated or treated with TNF, IFN-β, or TNF plus IFN-β. The effect of cytokine treatment on the replication and spread of each GFP-tagged virus was then measured by using limiting dilution titer assay to assess progeny virus production and flow cytometry to assess cell-to-cell spread.
To analyze the effects of TNF, IFN-β, or TNF plus IFN-β on the productive replication of various poxviruses, cells were treated as described above and then harvested at 12, 24, 48, 72, 96, or 120 h after infection. After the samples had been collected, the titer of each virus was determined on permissive cells as described in Materials and Methods. All three viruses exhibited either no or very modest reduction in progeny virus production after the addition of TNF. Similarly, all three viruses exhibited larger but still incomplete inhibition of viral replication after treatment with IFN-β. Although treatment with TNF plus IFN-β completely inhibited replication of MV, both VV and TPV were still able to replicate to an appreciable degree even in the presence of both cytokines (Fig. 8A, middle and right panels).
FIG. 8.
The combination of TNF plus IFN-β uniquely affects multiple poxviruses. Primary human GM02504 fibroblasts were infected with MV-GFP, VV-GFP, or TPV-GFP at an MOI of 0.1. After 1 h of viral adsorption, cells were treated with TNF, IFN-β, or the combination of TNF plus IFN-β. To determine the effect of each treatment on the progeny virus titer, cells were harvested at the given time points via trypsinization. Cells were then lysed via repeated freeze-thawing, and virus titers were determined as outlined in Materials and Methods. (A) Graph depicting the average of three independent experiments. To determine the effect of these treatments on viral spread, cells were harvested via trypsinization at 24, 48, and 72 h postinfection. The percentage of GFP+ live cells in each sample was then determined by using flow cytometry. (B) Graph depicting a representative experiment run in triplicate.
To measure the effect of each cytokine treatments on the ability of the viruses to spread from cell to cell, we used flow cytometry of human fibroblasts infected with a low MOIs of MV-GFP, VV-GFP, or TPV-GFP. Cells were infected as described above and then harvested at 24, 48, and 72 h. The percentage of cells that were infected at each time point was determined by measuring the number of GFP+ cells in each sample by flow cytometry. For samples treated with TNF, IFN-β, or TNF plus IFN-β and infected with MV-GFP, this assay revealed a pattern similar to that observed with progeny viral titration. The appearance that TNF has a much larger effect on MV cell-to-cell spread assay is likely caused by this assay being depicted on a linear scale compared to the logarithmic scale used for the progeny titer assay. Treatment with TNF or IFN-β alone partially reduced viral spread, while treatment with TNF plus IFN-β completely eliminated expansion of GFP+ cells (Fig. 8B, left panel). The spread of VV-GFP was unaffected by TNF alone but was largely abrogated in the presence of IFN-β or the combination of TNF plus IFN-β (Fig. 8B, middle panel). The spread of TPV was also relatively unaffected by TNF alone. Interestingly, however, although IFN-β only partially reduced TPV spread, the combination of TNF plus IFN-β inhibited spread to a greater degree (Fig. 8B, right). This is in contrast to the TPV progeny titer assay in which IFN-β alone and the combination of TNF plus IFN-β had a similar inhibitory effect. The differences between each virus observed at 24 h are likely due to slight variations in the virus titer for each virus.
DISCUSSION
Our data demonstrate that complete restriction of MV in GM02504 primary human fibroblasts requires a novel synergistic antiviral state induced by addition of TNF plus IFN-β. Similar results were obtained using three other primary fibroblast lines, suggesting that this observation is not specific to GM02504 cells (50; unpublished observations). These data highlight the species distinction that complete restriction of MV in primary mouse fibroblasts requires only type I IFNs, whereas in primary human cells it requires TNF plus IFN-β (50, 51). Similar to the case with MV-infected mouse cells, our data also indicate that BHK hamster cells, which are largely permissive for MV replication, become completely nonpermissive after treatment with IFN-β alone (data not shown). Thus, complete restriction of MV by IFN-β appears to be generally applicable to rodent cells but not to primary human cells. We hypothesize that the divergence of these results between host species is due either to activation of unique antiviral signaling pathways in rodent versus human cells after addition of the two cytokines (34, 41) or murine versus human species-specific differences for some of the virus-encoded anti-cytokine immunomodulators (43).
Similarly, it has been previously demonstrated that human cancer cells frequently respond differently to IFN than human primary cells (33). In fact, compromised IFN signaling has been a widely recognized consequence of oncogenic transformation of human cells (8, 45). These distinctions in the cellular IFN response can have dramatic effects on the outcome of viral infections (17). To determine whether this was true for MV infection, we tested a variety of human cancer lines for their responsiveness to TNF, IFN-β, or the combination of both cytokines. Consistent with previous literature, most human cancer cell lines display a compromised response to the addition of TNF or IFN-β. Interestingly, while several human cancer cell lines were identified that clearly responded to both TNF and IFN-β, no cell line we have tested to date has displayed a level of antiviral synergy comparable to primary human fibroblasts after the addition of TNF plus IFN-β (data not shown). This defect could result from a signaling pathway required for TNF and IFN-β synergy also playing a key role in tumorigenesis. Alternatively, one side effect of fully functional TNF and IFN-β synergy could be the inhibition of tumorigenesis. In either case, this would likely lead to the inability of many human cancer lines to synergistically respond to TNF plus IFN-β.
Regardless of the mechanism behind many human cancer cells inability to synergize in response to TNF plus IFN-β, this inability has strong implications for the use of MV as a therapeutic oncolytic virus. One of the criteria required for a good oncolytic virus is that its replication be restricted to the target tumor cells. This frequently requires genetic alteration of the oncolytic virus candidate such that its general replicative robustness is impaired even in permissive cells. In contrast, the strict block against MV caused by TNF plus IFN-β inhibits wild-type MV from successfully replicating in primary human tissues, while having very little effect on the ability of the virus to replicate and spread in human tumor cells.
Whether the synergistic antiviral state induced in primary human fibroblasts by TNF plus IFN-β is distinct from that induced by other combinations of TNF and/or various IFNs remains to be determined. Recently, Peng et al. reported that the combination of IFN-β and IFN-γ synergistically inhibits the replication of HSV-1 in primary human fibroblasts (31). To our knowledge, this was the first study to use DNA microarray analysis to study the genomic expression profile of cells treated with multiple antiviral cytokines. Interestingly, the genomic expression pattern obtained in their study bears some similarities to our results, even though different cytokines were used. For example, many of the genes that Peng et al. describe as dominantly upregulated by IFN-γ are induced in our study by the combination of TNF plus IFN-β. In contrast, multiple previous studies have shown that the antiviral state induced by the combination of either IFN-β and IFN-γ or TNF and IFN-γ is mediated primarily through upregulation of the tryptophan-degrading enzyme INDO (9, 25). While the addition of TNF plus IFN-β does induce INDO (see Table S3 in the supplemental material), our results indicate that the antiviral state induced by these cytokines is not mediated by tryptophan degradation (our unpublished results). In addition, Peng et al. demonstrated that the synergistic antiviral effect of IFN-β and IFN-γ on HSV is partially mediated through increased cellular apoptosis (31), while our results have not revealed any role for induced apoptosis in the synergistic anti-MV state induced by TNF and IFN-β. These data suggest that, while the antiviral state induced by IFN-β plus IFN-γ might have significant overlap with the state induced by TNF plus IFN-β, the two are not identical.
The signaling pathways and transcription factors responsible for synergy between TNF and various IFNs have not been well defined. Since many of these factors are activated on a posttranscriptional level, our present DNA microarray study is not well suited to elucidating their identity. Using the website Babelomics (http://babelomics.bioinfo.cipf.es/), we addressed whether genes induced only in the presence of TNF plus IFN-β shared common transcription factors. Analysis of the promoters (within 5 kb) of all genes induced >10-fold only in the presence of TNF plus IFN-β, however, did not return any transcription factors that were significantly overrepresented in this set of genes with a P of <0.05 compared to the rest of the genome. In contrast, the transcription factors NF-κB p65 (P = 0.00001), cREL (P = 0.0005), and AR (P = 0.0005) are overrepresented in the promoters of genes induced by TNF alone, whereas the transcription factors ISRE (P = 0.0001), IRF1 (P = 0.0003), and IRF7 (P = 0.0005) are overrepresented in the promoters of genes induced by IFN-β alone compared to the rest of the genome. This analysis suggests that the antiviral state induced by TNF plus IFN-β is not the result of inducing transcription from a single transcription factor. It is interesting, however, that the two transcription factors most highly upregulated after addition of TNF plus IFN-β, ATF3 and ATF5, have similar functions. Both of these proteins repress transcription from promoters containing a consensus cAMP response element (CRE) (5′-GTGACGT[AC][AG]-3′) (12). The CRE is found in a wide variety of both cellular and viral promoters (26, 44, 49). The altered kinetic expression of IL-8 and IFNAR2 (Fig. 5B) suggest that increased expression of ATF3 and ATF5 is not the only mechanism responsible for the synergistic antiviral state, since there is currently no evidence that the promoters of IL-8 or IFNAR2 contain a CRE or respond to either ATF3 or ATF5. These data suggest that other alterations in the cellular transcription environment exist and are likely to play a role in the induction of the synergistic antiviral state.
Our results indicate that the synergy between TNF and IFN-β is mediated directly by TNF acting on cells and not through secretion of another TNF-induced intermediate factor (Fig. 6). These results are important for several reasons. As previously mentioned, other groups have shown that the combination of IL-1β and IFN-γ is able to induce a synergistic antiviral state against several different viruses (1, 13, 29). While IL-1β was induced by TNF in our microarray study, however, our results indicate that it probably does not play a major role in inducing a synergistic anti-MV state (Fig. 6). Several other groups have noted that TNF's synergy with IFN-γ is largely mediated through induction of IFN-β (14, 37). These groups showed that depletion of IFN-β from their systems resulted in a loss of synergy. This led to both groups concluding that the synergy they observed was really between IFN-γ and IFN-β. In view of our results, it is possible that the synergies these groups observed were in fact mediated by TNF and autocrine IFN-β and not through the addition of both type I and type II IFNs.
Combinations of various cytokines have been shown to inhibit a wide variety of viruses, including HSV (32, 38), human cytomegalovirus (39), murine cytomegalovirus (21), varicella-zoster virus (7), pseudorabies virus (53), adenovirus (22), and other viruses. To determine whether the combination of TNF plus IFN-β was able to synergistically inhibit poxviruses capable of infecting human cells, we tested three different GFP-tagged poxviruses—MV, VV, and TPV—from distinct poxviral genera. Each of these viruses was found to respond differently to treatment with the combined cytokines. Replication of MV was completely eliminated only in the presence of TNF plus IFN; VV spread and titer were largely identical to treatment with IFN-β alone, while the spread of TPV was synergistically inhibited by TNF plus IFN-β, but the virus titer following multistep infection was not (Fig. 8). These data suggest that while the combination of TNF plus IFN-β is effective against different poxviruses, the extent of its antiviral effects will vary greatly depending on the virus. Since no virus showed significant differences in either titer or spread at 24 h when cytokine treatment was done simultaneously with infection, the data also suggest that cells must establish the antiviral state induced by TNF plus IFN-β prior to infection to exhibit maximal viral inhibition. This hypothesis is supported by the observation that cells pretreated with TNF plus IFN-β display a strong block in MV replication at an early stage of viral replication (Fig. 7). Preliminary data further indicate that if viral infection is allowed to progress for several hours prior to the addition of TNF plus IFN-β, both VV and TPV are able to effectively overcome the synergy of these cytokines. Interestingly, MV is unable to inhibit the synergy of TNF plus IFN-β, even if these cytokines are added after viral infection (data not shown). This observation could explain why MV is so exquisitely sensitive to the synergistic antiviral state induced by the combination of TNF plus IFN-β in human fibroblasts and provides a more rational basis for understanding why MV is restricted in primary human cells and tissues but permissive in so many human cancer cells.
Supplementary Material
Acknowledgments
G.M. holds an International Scholarship from the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 29 October 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adam, R., D. Russing, O. Adams, A. Ailyati, K. Sik Kim, H. Schroten, and W. Daubener. 2005. Role of human brain microvascular endothelial cells during central nervous system infection: significance of indoleamine 2,3-dioxygenase in antimicrobial defense and immunoregulation. Thromb. Haemost. 94341-346. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, S. L., J. M. Carton, J. Lou, L. Xing, and B. Y. Rubin. 1999. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 2568-14. [DOI] [PubMed] [Google Scholar]
- 3.Bartee, E., M. R. Mohamed, and G. McFadden. 2008. Tumor necrosis factor and interferon: cytokines in harmony. Curr. Opin. Microbiol. 11378-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter, C. C., V. Y. Gorbacheva, and D. J. Vestal. 2005. Inhibition of VSV and EMCV replication by the interferon-induced GTPase, mGBP-2: differential requirement for wild-type GTP binding domain. Arch. Virol. 1501213-1220. [DOI] [PubMed] [Google Scholar]
- 5.Chiu, J. J., P. L. Lee, S. F. Chang, L. J. Chen, C. I. Lee, K. M. Lin, S. Usami, and S. Chien. 2005. Shear stress regulates gene expression in vascular endothelial cells in response to tumor necrosis factor-alpha: a study of the transcription profile with complementary DNA microarray. J. Biomed. Sci. 12481-502. [DOI] [PubMed] [Google Scholar]
- 6.Davignon, J. L., P. Castanie, J. A. Yorke, N. Gautier, D. Clement, and C. Davrinche. 1996. Anti-human cytomegalovirus activity of cytokines produced by CD4+ T-cell clones specifically activated by IE1 peptides in vitro. J. Virol. 702162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desloges, N., M. Rahaus, and M. H. Wolff. 2005. Role of the protein kinase PKR in the inhibition of varicella-zoster virus replication by beta interferon and gamma interferon. J. Gen. Virol. 861-6. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, G. P., C. M. Koebel, and R. D. Schreiber. 2006. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 6836-848. [DOI] [PubMed] [Google Scholar]
- 9.Feduchi, E., M. A. Alonso, and L. Carrasco. 1989. Human gamma interferon and tumor necrosis factor exert a synergistic blockade on the replication of herpes simplex virus. J. Virol. 631354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenner, F., and F. N. Ratcliffe. 1965. Myxomatosis. Cambridge University Press, Cambridge, United Kingdom.
- 11.Geiss, G. K., V. S. Carter, Y. He, B. K. Kwieciszewski, T. Holzman, M. J. Korth, C. A. Lazaro, N. Fausto, R. E. Bumgarner, and M. G. Katze. 2003. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J. Virol. 776367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hai, T., and M. G. Hartman. 2001. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 2731-11. [DOI] [PubMed] [Google Scholar]
- 13.Heseler, K., K. Spekker, S. K. Schmidt, C. R. Mackenzie, and W. Daubener. 2008. Antimicrobial and immunoregulatory effects mediated by human lung cells: role of IFN-gamma-induced tryptophan degradation. FEMS Immunol. Med. Microbiol. 52273-281. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, T. K., T. A. Kaspar, and D. H. Coppenhaver. 1988. Synergy of antiviral actions of TNF and IFN-gamma: evidence for a major role of TNF-induced IFN-beta. Antivir. Res. 101-9. [DOI] [PubMed] [Google Scholar]
- 15.Johnston, J. B., J. W. Barrett, W. Chang, C. S. Chung, W. Zeng, J. Masters, M. Mann, F. Wang, J. Cao, and G. McFadden. 2003. Role of the serine-threonine kinase PAK-1 in myxoma virus replication. J. Virol. 775877-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerr, P., and G. McFadden. 2002. Immune responses to myxoma virus. Viral Immunol. 15229-246. [DOI] [PubMed] [Google Scholar]
- 17.Krishnamurthy, S., T. Takimoto, R. A. Scroggs, and A. Portner. 2006. Differentially regulated interferon response determines the outcome of Newcastle disease virus infection in normal and tumor cell lines. J. Virol. 805145-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaBarre, D. D., and R. J. Lowy. 2001. Improvements in methods for calculating virus titer estimates from TCID50 and plaque assays. J. Virol. Methods 96107-126. [DOI] [PubMed] [Google Scholar]
- 19.Lewis-Jones, S. 2004. Zoonotic poxvirus infections in humans. Curr. Opin. Infect. Dis. 1781-89. [DOI] [PubMed] [Google Scholar]
- 20.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 9831-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucin, P., S. Jonjic, M. Messerle, B. Polic, H. Hengel, and U. H. Koszinowski. 1994. Late phase inhibition of murine cytomegalovirus replication by synergistic action of interferon-gamma and tumour necrosis factor. J. Gen. Virol. 75 (Pt. 1)101-110. [DOI] [PubMed] [Google Scholar]
- 22.Mayer, A., H. Gelderblom, G. Kumel, and C. Jungwirth. 1992. Interferon-gamma-induced assembly block in the replication cycle of adenovirus 2: augmentation by tumour necrosis factor-alpha. Virology 187372-376. [DOI] [PubMed] [Google Scholar]
- 23.McFadden, G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier, E., G. Kunz, O. Haller, and H. Arnheiter. 1990. Activity of rat Mx proteins against a rhabdovirus. J. Virol. 646263-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mestan, J., M. Brockhaus, H. Kirchner, and H. Jacobsen. 1988. Antiviral activity of tumour necrosis factor. Synergism with interferons and induction of oligo-2′,5′-adenylate synthetase. J. Gen. Virol. 69(Pt. 12)3113-3120. [DOI] [PubMed] [Google Scholar]
- 26.Millhouse, S., J. J. Kenny, P. G. Quinn, V. Lee, and B. Wigdahl. 1998. ATF/CREB elements in the herpes simplex virus type 1 latency-associated transcript promoter interact with members of the ATF/CREB and AP-1 transcription factor families. J. Biomed Sci. 5451-464. [DOI] [PubMed] [Google Scholar]
- 27.Moss, B. 2007. Poxviridae: the viruses and their replication, p. 2849-2855. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 28.Nazarian, S. H., J. W. Barrett, M. M. Stanford, J. B. Johnston, K. Essani, and G. McFadden. 2007. Tropism of tanapox virus infection in primary human cells. Virology 36832-40. [DOI] [PubMed] [Google Scholar]
- 29.Obojes, K., O. Andres, K. S. Kim, W. Daubener, and J. Schneider-Schaulies. 2005. Indoleamine 2,3-dioxygenase mediates cell type-specific anti-measles virus activity of gamma interferon. J. Virol. 797768-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker, S., A. Nuara, R. M. Buller, and D. A. Schultz. 2007. Human monkeypox: an emerging zoonotic disease. Future Microbiol. 217-34. [DOI] [PubMed] [Google Scholar]
- 31.Peng, T., J. Zhu, Y. Hwangbo, L. Corey, and R. E. Bumgarner. 2008. Independent and cooperative antiviral actions of beta interferon and gamma interferon against herpes simplex virus replication in primary human fibroblasts. J. Virol. 821934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrera, E., and C. E. Coto. 2006. The synergistic effect of IFN-alpha and IFN-gamma against HSV-2 replication in Vero cells is not interfered by the plant antiviral 1-cinnamoyl-3,11-dihydroxymeliacarpin. Virol. J. 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfizenmaier, K., H. Bartsch, P. Scheurich, B. Seliger, U. Ucer, K. Vehmeyer, and G. A. Nagel. 1985. Differential gamma-interferon response of human colon carcinoma cells: inhibition of proliferation and modulation of immunogenicity as independent effects of gamma-interferon on tumor cell growth. Cancer Res. 453503-3509. [PubMed] [Google Scholar]
- 34.Rangarajan, A., and R. A. Weinberg. 2003. Opinion: comparative biology of mouse versus human cells: modeling human cancer in mice. Nat. Rev. Cancer 3952-959. [DOI] [PubMed] [Google Scholar]
- 35.Reis, L. F., T. Ho Lee, and J. Vilcek. 1989. Tumor necrosis factor acts synergistically with autocrine interferon-beta and increases interferon-beta mRNA levels in human fibroblasts. J. Biol. Chem. 26416351-16354. [PubMed] [Google Scholar]
- 36.Rothenfusser, S., N. Goutagny, G. DiPerna, M. Gong, B. G. Monks, A. Schoenemeyer, M. Yamamoto, S. Akira, and K. A. Fitzgerald. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 1755260-5268. [DOI] [PubMed] [Google Scholar]
- 37.Ruggiero, V., G. Antonelli, G. Conciatori, M. Gentile, J. Van Damme, and F. Dianzani. 1989. The in vitro antiviral activity of tumor necrosis factor (TNF) in WISH cells is mediated by IFN-beta induction. Antivir. Res. 1177-88. [DOI] [PubMed] [Google Scholar]
- 38.Sainz, B., Jr., and W. P. Halford. 2002. Alpha/Beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1. J. Virol. 7611541-11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sainz, B., Jr., H. L. LaMarca, R. F. Garry, and C. A. Morris. 2005. Synergistic inhibition of human cytomegalovirus replication by interferon-alpha/beta and interferon-gamma. Virol. J. 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuringa, J. J., S. van der Schaaf, E. Vellenga, B. J. Eggen, and W. Kruijer. 2002. LIF-induced STAT3 signaling in murine versus human embryonal carcinoma (EC) cells. Exp. Cell Res. 274119-129. [DOI] [PubMed] [Google Scholar]
- 42.Schwamborn, J., A. Lindecke, M. Elvers, V. Horejschi, M. Kerick, M. Rafigh, J. Pfeiffer, M. Prullage, B. Kaltschmidt, and C. Kaltschmidt. 2003. Microarray analysis of tumor necrosis factor alpha induced gene expression in U373 human glioblastoma cells. BMC Genomics 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seet, B. T., J. B. Johnston, C. R. Brunetti, J. W. Barrett, H. Everett, C. Cameron, J. Sypula, S. H. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21377-423. [DOI] [PubMed] [Google Scholar]
- 44.Sjoblom, A., W. Yang, L. Palmqvist, A. Jansson, and L. Rymo. 1998. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J. Virol. 721365-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smyth, M. J., G. P. Dunn, and R. D. Schreiber. 2006. Cancer immunosurveillance and immunoediting: the roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv. Immunol. 901-50. [DOI] [PubMed] [Google Scholar]
- 46.Stanford, M. M., and G. McFadden. 2007. Myxoma virus and oncolytic virotherapy: a new biologic weapon in the war against cancer. Expert Opin. Biol. Ther. 71415-1425. [DOI] [PubMed] [Google Scholar]
- 47.Stanford, M. M., S. J. Werden, and G. McFadden. 2007. Myxoma virus in the European rabbit: interactions between the virus and its susceptible host. Vet. Res. 38299-318. [DOI] [PubMed] [Google Scholar]
- 48.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67227-264. [DOI] [PubMed] [Google Scholar]
- 49.Wang, D., M. X. Guo, H. M. Hu, Z. Z. Zhao, H. L. Qiu, H. J. Shao, C. G. Zhu, L. Xue, Y. B. Shi, and W. X. Li. 2008. Human T-cell leukemia virus type 1 oncoprotein tax represses ZNF268 expression through the CREB/ATF pathway. J. Biol. Chem. 28316299-16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, F., X. Gao, J. Barrett, Q. Shao, E. Bartee, M. Mohamed, M. Rahman, S. Werden, T. Irvine, J. Cao, G. Dekaban, and G. McFadden. 2008. RIG-I mediates the co-induction of tumor necrosis factor and type I interferon elicited by myxoma virus in primary human macrophages. PLoS Pathog. 4e1000099. doi: 10.1371/journal.ppat.1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, F., Y. Ma, J. W. Barrett, X. Gao, J. Loh, E. Barton, H. W. Virgin, and G. McFadden. 2004. Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 51266-1274. [DOI] [PubMed] [Google Scholar]
- 52.Wong, G. H., and D. V. Goeddel. 1986. Tumour necrosis factors alpha and beta inhibit virus replication and synergize with interferons. Nature 323819-822. [DOI] [PubMed] [Google Scholar]
- 53.Yao, Q., P. Qian, Y. Cao, Y. He, Y. Si, Z. Xu, and H. Chen. 2007. Synergistic inhibition of pseudorabies virus replication by porcine alpha/beta interferon and gamma interferon in vitro. Eur. Cytokine Netw. 1871-77. [DOI] [PubMed] [Google Scholar]
- 54.Zimmer, R., and P. Thomas. 2002. Expression profiling and interferon-beta regulation of liver metastases in colorectal cancer cells. Clin. Exp. Metastasis 19541-550. [DOI] [PubMed] [Google Scholar]
- 55.Zurcher, T., J. Pavlovic, and P. Staeheli. 1992. Mouse Mx2 protein inhibits vesicular stomatitis virus but not influenza virus. Virology 187796-800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.