Abstract
Currently there is limited information about the quality of immune responses elicited by candidate human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env)-based immunogens in primates. Here we describe a comprehensive analysis of neutralizing antibody and T-cell responses obtained in cynomolgus macaques by three selected immunization regimens. We used the previously described YU2-based gp140 protein trimers administered in an adjuvant, preceded by two distinct priming strategies: either alphavirus replicon particles expressing matched gp140 trimers or gp120 core proteins stabilized in the CD4-bound conformation. The rationale for priming with replicon particles was to evaluate the impact of the expression platform on trimer immunogenicity. The stable core proteins were chosen in an attempt to expand selectively lymphocytes recognizing common determinants between the core and trimers to broaden the immune response. The results presented here demonstrate that the platform by which Env trimers were delivered in the priming (either protein or replicon vector) had little impact on the overall immune response. In contrast, priming with stable core proteins followed by a trimer boost strikingly focused the T-cell response on the core sequences of HIV-1 Env. The specificity of the T-cell response was distinctly different from that of the responses obtained in animals immunized with trimers alone and was shown to be mediated by CD4+ T cells. However, this regimen showed limited or no improvement in the neutralizing antibody responses, suggesting that further immunogen design efforts are required to successfully focus the B-cell response on conserved neutralizing determinants of HIV-1 Env.
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) consists of a trimer of noncovalently associated gp120/gp41 heterodimers, which form the functional viral spike and mediate entry into CD4 and CCR5 receptor-positive host target cells. The exterior envelope glycoprotein, gp120, and the transmembrane glycoprotein, gp41, are the sole virally encoded targets for neutralizing antibodies on the surface of the virus and likely represent a critical immunogenic component for an effective prophylactic vaccine against HIV-1 (7, 23, 33). The HIV-1 Envs are also potential targets for cell-mediated immune responses, and as such, their inclusion in future HIV-1 vaccine candidates may contribute to the induction of both protective antibody and cellular immune responses (25). In attempts to elicit antibodies that recognize the functional Env spike, soluble trimeric molecules containing full-length gp120 covalently linked to the gp41 ectodomain have been designed (5, 24, 40, 46, 53, 54). An incremental advance in neutralizing antibody elicitation using soluble trimeric Env spike mimetics compared to the use of monomeric gp120 was observed (28, 55), but further improvements in trimer immunogen design are still needed both to mimic better the functional viral spike and to elicit broadly neutralizing antibodies (reviewed in reference 7 and 37).
The immunogenicity of cleavage-defective Env trimers derived from the primary R5 isolate YU2, possessing heterologous trimerization motifs derived either from T4 bacteriophage (foldon) or from the transcription factor GCN4, were examined in several small animals studies (6, 15, 28, 55). However, to date these trimeric Env immunogens were not analyzed for their ability to elicit neutralizing antibodies and Env-specific T-cell responses in nonhuman primates. Other oligomeric Env proteins, such as the SF162 gp140 proteins with or without a deletion of the second major variable region (ΔV2), were evaluated with nonhuman primates (1, 12, 51, 52). For example, a recent study demonstrated that gp140SF162ΔV2 administered in the MF59 adjuvant mediated protection against mucosal challenge with the SHIV-162P4 virus (2), implicating Env-directed immune responses in mediating protection against this homologous virus challenge.
The capacity of different Env immunogens to stimulate humoral and cellular responses was also evaluated using genetic means of expression, such as plasmid DNA or recombinant viral vectors, followed by immunization of purified Env protein in an adjuvant to boost antibody responses (15, 34, 45, 51). While such heterologous immunization regimens may enhance Env-directed cellular immune responses, little is known about the quality of neutralizing antibody responses induced by viral vector priming followed by a protein boost or about the relative responses elicited by regimens consisting of purified Env protein in an adjuvant using homologous or heterologous protein priming/boosting. One potential concern when Env is expressed in vivo from DNA or viral vectors is that the actual dose and the antigenic integrity of the immunogen are not easily assessed. For example, incorrectly folded but immunogenic Env protein released from dying cells in vivo may adversely affect the quality of the elicited antibody response. Since many candidate vaccines which are currently moving into clinical trials rely on in vivo genetic expression (9, 10, 18, 38, 39, 48, 49), analysis of the quality of antibody responses elicited by genetic platforms is warranted and is an aim of our present study.
Previously we performed a head-to-head study using rabbits to characterize neutralizing antibody responses stimulated by sequential administrations of YU2 Env protein trimers emulsified in the Ribi adjuvant compared to two immunizations of single-round infectious recombinant Semliki Forest virus (rSFV) particles expressing YU2 trimers followed by a boost with YU2 trimeric proteins in an adjuvant (15). SFV is an alphavirus, genetically related to Sindbis virus and Venezuelan equine encephalitis virus (VEE), two other viruses for which single-round replicon systems have been developed (8, 41). SFV has been used extensively in preclinical immunogenicity experiments (3, 4, 14, 15, 16, 19, 21, 29, 30, 32, 47), human vaccine candidates based on VEE are already in clinical trials (11), and currently the chimeric VEErep/SINenv vector is one of the most frequently used alphavirus-based vector systems for preclinical HIV-1 vaccine studies (35, 51). In our previous study, we demonstrated that rSFV infection of BHK-21 cells in vitro resulted in the secretion of homogeneous and stable gp140 trimers into the culture medium (15). Here, to evaluate recombinant alphavirus priming in greater detail, we performed a comprehensive study with nonhuman primates to examine both antibody and cellular responses induced by gp140 trimers with and without rSFV priming. We inoculated cynomolgus macaques with Env trimers administered as purified protein in the AS01B adjuvant system from GlaxoSmithKline Biologicals (GSK) or expressed in vivo from rSFV particles. We used cleavage-defective (−) gp140 trimers possessing the heterologous foldon (F) trimerization motif (gp140-F) (54), hereafter referred to as gp140-F trimers. As an additional arm of the study, we used a modified gp120 core protein in AS01B as a prime, followed by boosts with Env trimers, both formulated in AS01B. We sought to test the concept of “immunofocusing” by first inoculating with the stable gp120 core, followed by the gp140-F proteins in sequence to focus responses on the shared and conserved core elements of the two forms of Env. The modified gp120 core proteins, hereafter referred to as “stable core,” contain pocket-filling mutations and were stabilized by disulfide-linked cysteine pairs spanning the inner and outer domains of gp120, as previously described (56). These modifications were designed to reduce the flexibility of gp120 and to improve presentation of conserved but discontinuous cross-domain antigenic surfaces of Env, such as the CD4-binding site. We hypothesized that priming with a stable core protein and boosting with gp140-F trimers would favor the expansion of Env-specific B cells recognizing common determinants between the two immunogens and that this might translate into an increased breadth of neutralization. We also wished to determine if priming with stable cores and boosting with trimers would alter the Env-specific T-cell response compared to immunization with gp140-F trimers alone, since this may represent a strategy to focus the cellular response on conserved sequences common to the two immunogens. Our results show that the properties of the Env antigen used to prime the response are more important for shaping the overall Env-specific immune response than the platform by which Env is delivered. Priming with the stable core protein and boosting with trimers strikingly focused the T-cell response on core sequences of Env even as measured after the trimer boost. However, this regimen did not improve the neutralizing antibody response, suggesting that further immunogen design efforts are required to successfully focus the B-cell response on conserved structural determinants on the functional viral spike to stimulate antibodies possessing an increased breadth of neutralization.
MATERIALS AND METHODS
Cells, recombinant viruses, and antibodies.
BHK-21 cells (ATCC) were cultured in Glasgow minimal essential medium (MEM) (Invitrogen, Carlsbad, California), supplemented with 5% fetal calf serum, 10 mM HEPES, 10% tryptose phosphate, 2 mM l-glutamine, penicillin, and streptomycin (Sigma, St. Louis, MO). Sequences encoding YU2gp140(−/FT), flanked by XmaI/XhoI, were amplified by PCR from YU2gp140FT/pCDNA3.1(−), respectively (54). The sequences were inserted into pSFV10 (22) to generate rSFV-gp140-F trimer particles. Single-round infectious recombinant pSFV-gp140-F particles (rSFV-gp140-F) were produced by cotransfecting BHK-21 cells with in vitro-transcribed vector and split-helper mRNA (3) according to the standard protocol, and titration of viral stocks was performed using BHK-21 cells as described previously (22). The monoclonal antibodies (MAbs) used in this study were 17b, F105, and 447-52D, generously obtained from James Robinson, Marshal Posner, and Susan Zolla-Pazner, respectively, and D7324, an antibody directed against the C5 region of gp120, purchased from Aalto Bio Reagents Ltd. (Dublin, Ireland).
Expression and purification of Env immunogens.
The gp140-F trimers were produced by transient transfection of adherent 293F cells or Freestyle 293F suspension cells (Invitrogen). Adherent cells were transfected in Dulbecco's modified Eagle medium-10% heat-inactivated fetal bovine serum-0.1 mM MEM nonessential amino acid solution (Invitrogen) media, using LipofectAMINE 2000 (Invitrogen) as per the manufacturer's instructions. One day posttransfection, serum-containing Dulbecco's modified Eagle medium was replaced with serum-free 293 SFM II medium (Invitrogen). Beginning 2 days after transfection, cell culture supernatants were collected daily and fresh 293 SFM II medium added to the cell culture flasks until the sixth day posttransfection. Suspension cells were transfected in Gibco Freestyle293 expression medium by using the 293Fectin transfection reagent in Opti-MEM according to the manufacturer's instructions (Invitrogen). The supernatant was collected 4 days after transfection. Following collection, all supernatants were centrifuged at 3,500 × g, filtered through a 0.22-μm filter, and supplemented with Complete, EDTA-free protease inhibitor cocktail (Roche) and penicillin-streptomycin (Invitrogen) for storage at 4°C until further purification. The highly glycosylated and His6-tag-containing YU2gp140-F trimers were purified from sterile-filtered, serum-free medium in a three-step process. First, the protein was captured via the N-linked glycans with lentil-lectin affinity chromatography (GE Healthcare, Uppsala, Sweden). After extensive washing in phosphate-buffered saline (PBS), the protein was eluted with PBS-1 M α-β-mannopyranoside-0.5 M NaCl-10 mM imidazole, subsequently captured in the second step via the His tag by nickel chelation chromatography (GE Healthcare), and then washed and eluted with a 300 mM imidazole-containing PBS buffer. Finally, the YU2gp140-F trimers were separated from lower-molecular-weight proteins by gel filtration chromatography using a Superdex 200 26/60 prep-grade column and the ÄKTA Fast protein liquid chromatography system (GE Healthcare). The stable core proteins used in the current study are a slightly modified version of the cysteine-stabilized Ds12F123 protein described previously (56). Following analysis of the core+V3 structure (20), the Ds12F123 core protein was redesigned to enhance protein folding by the addition of 13 residues at the base of the V3 loop, with the original GAG tripeptide substitution of the V3 region replaced with 16 residues, RPNNGGSGSGGNMRQA (Ds12F123V3S; B. Dey et al., unpublished data). The Ds12F123V3S stable core protein was produced by transient transfection of adherent 293F cells in serum-free medium as described above. Sterile-filtered supernatant containing the stable core proteins was applied to an immunoglobulin G (IgG) 17b affinity column. After extensive washing with PBS, core protein was eluted from the column with 100 mM glycine-Tris HCl-150 mM NaCl, pH 2.8, immediately neutralized with Tris base, pH 8.5, and then dialyzed against PBS-0.5 NaCl, pH 7.4. All proteins were spin concentrated with Amicon Ultra 30,000-molecular-weight-cutoff centrifugal filter devices (Millipore, Bedford, MA) to a concentration between 1 and 3 mg/ml.
Biochemical analysis of Env immunogens.
Purified proteins were analyzed for conformational integrity and oligomeric status prior to inoculation. Conformational integrity was confirmed by immunoprecipitating purified proteins with two conformation-sensitive MAbs: 17b (coreceptor site directed) and F105 (CD4bs directed), as well as with 447-52D V3-directed antibody. Briefly, 5 μg of protein was coincubated for 1 h at room temperature (RT) with 15 μg antibody and 30 μl Protein A beads (GE Healthcare) in 500 μl PBS. After extensive washing three times with PBS-0.5 M NaCl followed by one wash with PBS, beads with bound antibody-antigen complexes were heated to 100°C for 5 min in 1× NuPAGE lithium dodecyl sulfate sample buffer (Invitrogen), supplemented with 1× NuPAGE sample reducing agent (Invitrogen), and subjected to gel electrophoresis. The proteins were resolved on a NuPAGE 4 to 12% bis-Tris gel. The oligomeric status of the purified proteins was analyzed by resolving 10 μg protein on a NuPAGE 4 to 12% bis-Tris gel under “blue native” conditions (40). Briefly, samples diluted in 2× sample buffer (100 mM Tris HCl, 100 mM morpholinepropanesulfonic acid, 40% glycerol, 0.1% Serva-G, pH 7.7) were analyzed on a NuPAGE 4 to 12% bis-Tris gel (Invitrogen) at 4°C at 50 V for 18 h. The running buffer contained 50 mM Tris-HCl, 50 mM morpholinepropanesulfonic acid, pH 7.7, and to the cathode, 10 mg Serva-G (Serva Electrophoresis GmbH, Heidelberg, Germany) per 500 ml running buffer was added. Thyroglobulin and ferritin (GE Healthcare) were used as molecular weight markers.
Animals and inoculations.
Sixteen female cynomolgus macaques (Macaca fascicularis) of Chinese origin, 5 to 6 years old, were housed in the Astrid Fagraeus laboratory at the Swedish Institute for Infectious Disease Control. Housing and care procedures were in compliance with the provisions and general guidelines of the Swedish Animal Welfare Agency. All procedures were approved by the Local Ethical Committee on Animal Experiments. Animals were housed in pairs in 4-m3 cages, enriched to allow expression of physiological and behavioral needs. They were habituated to the housing conditions for more than 6 weeks before the start of the experiment and subjected to positive-reinforcement training to reduce the stress associated with experimental procedures. All immunizations and blood sampling were performed under sedation with ketamine (10 mg/kg of body weight) given intramuscularly (Ketaminol, 100 mg/ml; Intervet, Sweden). Macaques were weighed and examined for swelling of lymph nodes and spleen at each immunization or sampling occasion. Before entering the study, all animals were confirmed to be negative for simian immunodeficiency virus, simian T-cell lymphotropic virus, and simian retrovirus type D.
The 16 cynomolgus macaques (3 groups of 5 animals and 1 naive control animal) were inoculated five times with the immunogens described above (Table 1). Immunizations were performed at weeks 0, 3, 8, 12, and 18 by intramuscular injection, except for rSFV-gp140-F, which was given subcutaneously. All protein immunizations were administered in combination with the AS01B adjuvant system from GSK. Protein doses were 200 μg per animal for the first inoculation and 100 μg for the following injections. The SFV-gp140-F dose used in the first two immunizations in group 3 was 5 × 108 IU per injection. Vaccines were given in a total volume of 1 ml, divided equally between the left and right hind legs. Blood samples were taken before and 2 weeks after each immunization. As a negative control, one animal was immunized five times with the AS01B adjuvant system alone using the same immunization interval as described above.
TABLE 1.
Immunizationsa
| Group (nb) | Animal identity(ies) | Content of immunization no. (wk):
|
||||
|---|---|---|---|---|---|---|
| 1 (0) | 2 (3) | 3 (8) | 4 (12) | 5 (18) | ||
| 1 (5) | E71, E72, E75, E76, E87 | gp140-F | gp140-F | gp140-F | gp140-F | gp140-F |
| 2 (5) | E89, E90, E91, E95, E96 | SFV-gp140-F | SFV-gp140-F | gp140-F | gp140-F | gp140-F |
| 3 (5) | E73, E74, E77, E78, E88 | Stable core | Stable core | gp140-F | gp140-F | gp140-F |
| 4 (1) | E93 | |||||
All protein immunizations (gp140-F and stable core; groups 1 to 3) and all control immunizations (group 4) were given intramuscularly with the AS01B adjuvant system. SFV-gp140-F immunizations (group 2) were given subcutaneously. Animals were bled 2 weeks after each immunization. For details, see Materials and Methods.
n, no. of animals.
ELISA.
HIV-1 gp120-specific serum IgG was measured by enzyme-linked immunosorbent assay (ELISA). Briefly, Nunc Maxisorp microtiter plates were coated with insect-cell-produced YU2 gp120 protein at 1 μg/ml in 50 mM PBS overnight (ON) at +4°C. After blocking in PBS containing 2% milk, serum samples were added to and incubated for 2 h at RT. gp120-specific IgG was detected by adding secondary biotinylated anti-monkey IgG antibody (Jackson ImmunoReserach, Suffolk, United Kingdom) and streptavidin conjugated to horseradish peroxidase (Mabtech, Stockholm, Sweden) followed by O-phenylenediamine (Sigma, Schnelldorf, Germany). Between each incubation step, the plates were washed six times with PBS supplemented with 0.05% Tween 20. Substrate reactions were terminated with 2.5 M H2SO4, and the optical density (OD) was read at 492 and 650 nm. The OD-at-50-nm (OD50) titers for each sample were calculated by interpolating from the mean OD50 value calculated from controls using the formula [(ODmax − ODmin)/2] + ODmin. When antibody responses against denatured epitopes were investigated, gp120 was denatured by boiling for 2 min in 1% sodium dodecyl sulfate and 50 mM dithiothreitol (Sigma) and then diluted and analyzed as described above. As controls in this assay, we used two monoclonal antibodies, F105, which recognizes a conformation-sensitive epitope in gp120, and an anti-C5 antibody, D7324, which recognizes a linear epitope in gp120, to confirm that the boiling resulted in a loss of conformational but not linear epitopes. The assay with binding antibodies against gp140-F or the stable core protein measured in serum samples after the second and fourth immunizations were performed as described above with the following modifications: specific IgG was detected with horseradish peroxidase-conjugated secondary anti-human IgG (Fc region) (Jackson Laboratories) using 3,3′,5,5′-tetramethylbenzidine (Bio-Rad) as a substrate. The reaction was stopped by adding 1 M H2SO4, and the OD was read at 450 nm.
Virus neutralization assays.
Sera from immunized animals were tested for virus neutralization capacity against a panel of diverse HIV-1 isolates. Neutralization assays were performed using a single-round-infection HIV-1 Env pseudovirus assay and TZM-bl target cells as described previously (27, 45). Env pseudoviruses were prepared by cotransfecting 293T cells with an Env expression plasmid containing a full gp160 env gene and an env-deficient HIV-1 backbone vector (pSG3ΔEnv). To determine the serum dilution that resulted in a 50% reduction in relative luminescence units, serial dilutions of sera were performed and the neutralization dose-response curves were fitted by nonlinear regression using a four-parameter-hill slope equation programmed into the JMP statistical software program (JMP 5.1; SAS Institute Inc., Cary, NC). The results are reported as the serum neutralization ID50, which is the reciprocal of the serum dilution producing 50% virus neutralization. Peptide competition neutralization assays were done in the same assay format as the neutralization assay, except that the control or test peptide was added to serum 30 min prior to the addition of virus. The V3 peptide sequences used in this study were synthesized by SynPep (Dublin, CA) and were based on the YU2 sequence (TRPNNNTRKSINIGPGRALYTTG). A scrambled V3 peptide (IGPGRATRPNNNFYTTGTRKSIH) was used as a negative control.
Diverse HIV-1 virus isolates, including viruses from clades A, B, and C, were used in the neutralization assays. Clade B viruses included a panel of Env pseudoviruses that were recently characterized and recommended for use in assessing neutralization by HIV-1 immune sera (26). Several investigators also provided replication-competent viruses or functional Env plasmids for pseudoviruses. Dana Gabuzda (Dana Farber Cancer Institute) provided the Env plasmids for YU2 and murine leukemia virus. Env plasmids for SF162 and JRFL were provided by Leonidas Stamatatos (Seattle Biomedical Research Institute) and James Binley (Torrey Pines Institute), respectively. The clade A DJ263.8 sequence was cloned from the original peripheral blood mononuclear cell (PBMC)-derived virus provided by Francine McCutchan and Vicky Polonis (U.S. Military HIV Research Program), and the clade C MW965 Env plasmid was obtained from the AIDS Research and Reagent Repository. The BaL.01 Env (45) and SS1196.1 Env (26) plasmids were recently described by our laboratory.
Peptides and cells for enzyme-linked immunospot (ELISPOT) assay.
Peptide pools, comprised of 15-mer peptides overlapping by 10 residues, were purchased (New England Peptide LLC, Gardner, MA). Two different peptide pools based on the YU2 gp140 sequence were used: “gp140-F core” covered the conserved regions of gp120, corresponding to the stable core immunogen, and “gp140-F noncore” covered the remaining parts of YU2 gp140. A peptide pool based on the HXBc2 stable core immunogen sequence was also used. PBMC were isolated from EDTA blood by Ficoll-Paque PLUS (GE Healthcare Biosciences AB, Uppsala, Sweden) separation, frozen in R20 medium with 10% dimethylsulfoxide and stored at −150°C or in liquid nitrogen.
For the ELISPOT assays, MSIPN4550 plates (Millipore, Bedford, MA) were prewetted with 40 μl 35% ethanol, washed six times with distilled H2O, and coated with the anti-gamma interferon (IFN-γ) MAb GZ-4 (Mabtech, Nacka, Sweden) at 10 μg/ml in PBS ON at 4°C. The plates were then blocked with 150 μl R10 medium for 1 h at 37°C. The blocking solution was removed without washing, and monkey PBMC, thawed and rested ON in R10 medium at 37°C with 5% CO2, were added at 2 × 105 cells/well in duplicates with medium alone, phytohemagglutinin (5 μg/ml), or one of three overlapping peptide pools at 2.5 μg/ml. The cell viability was always >90%. After 20 h of incubation in 5% CO2 at 37°C, biotinylated anti-IFN-γ MAb 7-B6-1 (Mabtech) was added at 1 μg/ml and the plates were incubated for 2 h at RT followed by 1 h of incubation at RT with streptavidin-alkaline phosphatase (Mabtech). Then 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate (Sigma-Aldrich, St Louis, MO) was added, and spots were allowed to develop for 30 min before the enzymatic reaction was stopped. Volumes were 100 μl/well, and the plates were washed six times with PBS between all steps except where indicated. The spots were counted using an ImmunoSpot series 4 analyzer (CTL, Aalen, Germany) and expressed as numbers of spot-forming cells per 106 PBMC. For experiments with CD8+ T-cell-depleted PBMC, CD8+ cells were removed by magnetic cell sorting using a nonhuman primate-specific CD8 depletion kit (Miltenyi Biotec Inc., Auburn, CA). The purity of the CD8+-depleted fraction was determined by flow cytometry after staining with anti-CD3-APC-Cy7 (clone SP34-2), anti-CD4-AmCyan (clone L200), and anti-CD8-Pacific Blue (clone RPA-T8) (all from BD Biosciences, San Jose, CA).
RESULTS
Biochemical characterization of Env immunogens and study design.
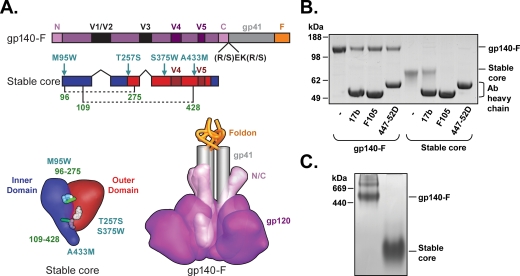
A schematic of the two Env proteins used in this study is shown in Fig. 1A. To characterize the preparations of the gp140-F trimers and stable Ds12F123V3S core proteins used for immunizations in this study, we immunoprecipitated the two protein immunogens with three MAbs: 17b, which binds a determinant overlapping the gp120 coreceptor binding site; F105, which binds to a surface overlapping the CD4 binding site; and 447-52D, which recognizes the V3 region of many isolates possessing a GPGR motif at the tip of their V3 region (including YU2 and HXBc2). Free proteins and protein-antibody complexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by Coomassie blue staining. As expected, gp140-F trimers were recognized by all three antibodies, while the stable core protein was precipitated with 17b but not with 447-52D or F105, consistent with the protein lacking essential 447-52D binding elements in the V3 loop (17) and with previously published data on gp120 proteins harboring Phe 43 cavity-filling mutations (50, 56) (Fig. 1B). Analysis by Blue Native gel electrophoresis confirmed that the mobility of the majority of the gp140-F protein was consistent with that of a trimer while the mobility of the stable core protein was consistent with the size of a loop-deleted gp120 monomer (Fig. 1C). Expression of gp140 trimers from the rSFV particles was analyzed as described previously (15).
FIG. 1.
Design and biochemical characterization of HIV-1 envelope glycoproteins. (A) Linear (sequence) and tertiary (structure) schematic representation of antigens used for immunization. Trimers of gp140-F were based on YU2 and include the full-length gp120 sequence with N and C termini (N and C), variable regions (V1/V2/V3/V4 and V5), and the gp41 ectodomain (gray). They are cleavage defective (the REKR sequence at the cleavage site was changed to SEKS to maintain a covalent gp120-gp41 association) and contain a heterologous foldon trimerization motif (F) (yellow). Stable cores were based on HXBc2 and lack N and C termini, the V1/V2/V3 regions, and gp41. They were modified by structure-based design to contain pocket-filling mutations (M95W, T257S, A433M, and S375W, which eliminates F105 recognition) (13, 56) and extra cysteine bonds between amino acids 96 and 275 and amino acids 109 and 428, as indicated by dotted lines in the linear schematic and green bars in the tertiary cartoon; these disulfides stabilize the CD4-bound conformation of gp120 (56). The inner domain of the HXBc2 core is colored blue and the outer domain red. Numbering of amino acids is based on the HXBc2 numbering convention. (B) Purified proteins were analyzed for their antigenic profiles by immunoprecipitation with the three monoclonal antibodies: 17b (coreceptor site directed), F105 (CD4bs directed), and 447-52D (V3 directed). The eluted proteins were resolved on a NuPAGE 4-to-12% bis-Tris gel (left panel). (C) The oligomeric status of the proteins was analyzed by blue native gel electrophoresis. Thyroglobulin and ferritin (GE Healthcare) were used as molecular mass markers.
The design for the present immunogenicity study involving 16 female cynomolgus macaques is outlined in Table 1. Five monkeys (E71, E72, E75, E76, and E87) were inoculated five times with purified gp140-F trimers combined with the GSK AS01B adjuvant system (group 1). Five monkeys (E89, E90, E91, E95, and E96) were inoculated twice with 5 × 108 IU rSFV particles expressing gp140-F trimers, followed by three immunizations with purified gp140-F trimers/adjuvant (group 2). A final five monkeys (E73, E74, E77, E78, and E88) were immunized twice with the stable core proteins followed by three immunizations with gp140-F trimers/adjuvant (group 3). As a negative control for the neutralization assay, one animal (E93) was inoculated with the AS01B adjuvant alone. The interval between inoculations was 1 month, and the animals were bled 2 weeks after each immunization for isolation of PBMC and collection of sera.
Analysis of sera from Env-immunized macaques.
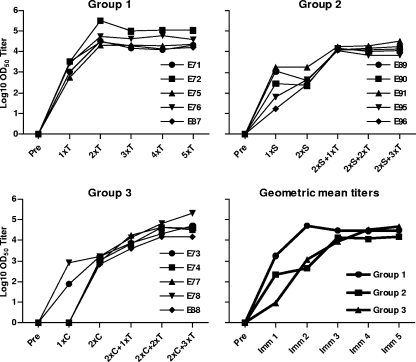
Levels of HIV-1 Env-binding antibodies were measured by standard ELISA using YU2 gp120-coated wells, and OD50 titers were calculated as described in Materials and Methods. All animals in group 1 mounted high antibody responses after a single Env immunization, consistent with the potent immunogenicity provided by the combination of Env and the AS01B adjuvant system (Fig. 2, top left panel). The Env-specific binding response was boosted by the second immunization, after which the antibody response plateaued. In group 2, Env-specific antibody responses were also detectable after the first rSFV-gp140-F immunization, but these responses were considerably lower than group 1 titers (Fig. 2, top right panel). A second rSFV-gp140-F immunization did not boost binding antibody levels, possibly due to the impact of antivector immune responses as previously reported (31, 47). However, antibody responses were readily boosted by one subsequent immunization with gp140-F trimer protein and reached levels similar to those in group 1. The stable core protein used for priming in group 3 elicited lower levels of gp120-binding antibodies than did the trimers (Fig. 2, bottom left panel), despite the same amount of protein being administered and use of the same adjuvant. However, the responses increased after the first boost with gp140-F trimer proteins, and after the second boost with the protein trimers, the responses reached a plateau similar to that observed in the other groups. The geometric mean ELISA titers for the three groups are shown in the lower right panel of the figure. There was no difference in the preimmune reactivity to Env among the animals in the three groups (see Fig S1A in the supplemental material), and an intra-assay control run on all ELISA plates confirmed that all assays were equally sensitive (see Fig S1B in the supplemental material). Full titration curves of all individual animals are presented in Fig S1C in the supplemental material.
FIG. 2.
Env binding antibodies in serum from immunized animals. The titers of Env-binding antibodies were measured in sera 2 weeks after each immunization using a standard ELISA with gp120-coated wells. The OD50 titer for each sample was calculated by interpolating from the mean OD50 value calculated from controls: [(ODmax − ODmin)/2] + ODmin. The results for individual animals, as well as group means, are shown. Preimmune Env-directed reactivity in sera diluted 100-fold was also determined and was shown not to differ between the groups (see Fig. S1A in the supplemental material). “T” indicates gp140-F trimer protein immunization, “S” indicates SFV-gp140-F immunization, and “C” indicates stable core protein immunization.
Neutralizing antibody responses in Env-immunized macaques.
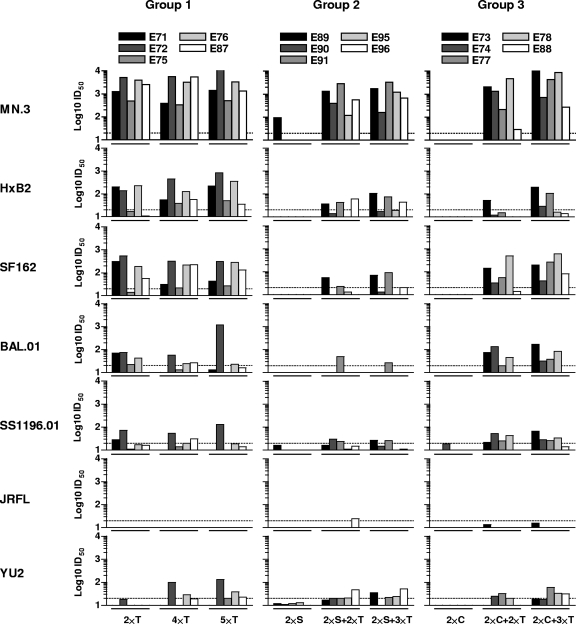
We next measured neutralizing activity in sera collected after the second, fourth, and fifth immunizations. We used a panel of recombinant viruses pseudotyped with the “tier 1” Env glycoproteins MN, HXBc2, SF162, BaL, SS1196 (clade B), and two non-clade B viruses, MW965 (clade C) and DJ263 (clade A) and the “tier 2” Env glycoproteins YU2, JRFL, TRO.11, and 6535.3. The data are presented as 50% neutralization titers (50% inhibitory dilution [ID50]) with the serum ID50 neutralizing titer of 20 highlighted as a dotted line (Fig. 3). The most potent neutralizing activity in the post-2 sera was displayed by animals immunized with the gp140-F trimer protein administered in the AS01B adjuvant (group 1), where relatively high neutralization titers against HXBc2, MN, SF162 (Fig. 3), and the clade C virus MW965 (see Fig S2 in the supplemental material) were elicited in all animals. Weaker or variable neutralizing activity was observed against BaL, YU2, and SS1196, and no neutralization was observed against the tier 2-type viruses, JRFL, 6535.3, and TRO.11 (Fig. 3 and data not shown; see also Fig S2 in the supplemental material).
FIG. 3.
Neutralizing antibody responses against clade B viruses. Serum samples from each animal taken 2 weeks after the second, fourth, and fifth immunizations were analyzed for neutralizing activity using the TZM-bl/Luc neutralization assay and the following Env glycoproteins: MN, HXBc2, SF162, BaL.01, SS1196.01, JRFL, and YU2 (clade B). Serial serum dilutions from individual monkeys were tested, and the data are presented as 50% neutralization titers (ID50). Neutralizing ID50 titers above 20 are considered positive, as indicated by the dotted lines. “T” refers to gp140-F trimer protein immunization, “S” refers to SFV-gp140-F immunization, and “C” refers to stable core protein immunization.
In animals primed with rSFV-expressing gp140-F trimers and boosted with the gp140-F trimer protein in an adjuvant, there was no consistent neutralizing activity after the first two immunizations. After two booster immunizations with the gp140-F protein, there was neutralizing activity against MN, HXBc2, and SF162 in most animals, but the activity against all viruses except MN was generally lower than in group 1. Overall, rSFV priming did not confer an advantage to the neutralizing response elicited by the gp140-F trimeric protein alone. Similarly, there was no detectable neutralizing activity against any of the viruses after two inoculations of the stable core protein in an adjuvant (group 3, post-2). After two booster immunizations with the gp140-F protein in an adjuvant (group 3, post-4), consistent neutralizing activity was observed only against MN and SF162. There was a tendency toward more consistent neutralization of BaL and SS1196 in the post-5 serum from group 3 than in group 1, suggesting a possible advantage with the stable core-prime, trimer-boost regimen. High neutralization titers against the clade C virus, MW965, was observed in the post-4 and post-5 sera from all three groups, while no or very weak neutralization was detected against the tier 2-type viruses, JRFL, 6535, and TRO.11, for most sera (Fig. 3 and data not shown; see also Fig S2 in the supplemental material). Serum from the naive control animal (group 4) tested negative for neutralization of all HIV-1 viruses in the assay, and all samples from groups 1 to 4 tested negative against a murine leukemia virus pseudotype control, confirming that the assay was specific for HIV-1 Env-directed neutralization (data not shown).
Mapping specificities of anti-Env serum antibody responses.
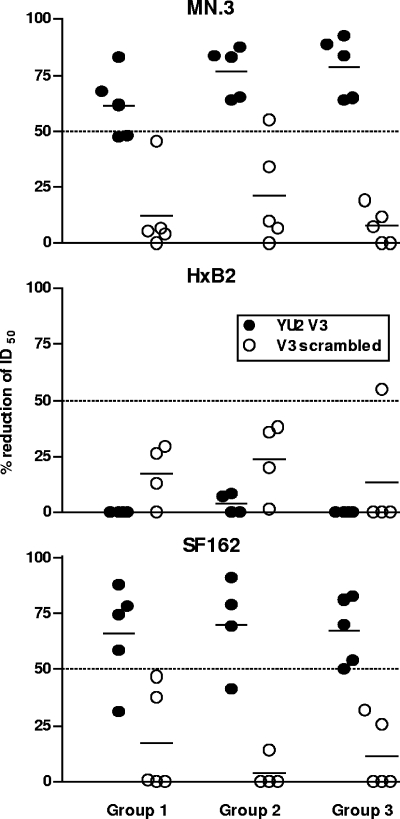
To better understand what antibody specificities mediated the neutralizing responses against MN, SF162, HXBc2, and MW965, we performed peptide inhibition assays using the post-5 serum from each hyperimmune animal. The results from these experiments are shown as percent reduction of the ID50 neutralization values, where only values greater than a 50% reduction in neutralizing activity were considered reliable. Values in excess of a 50% reduction in ID50 neutralization are shown in Fig. 4. By using a YU2-derived V3 peptide and a scrambled control V3-derived peptide, we found that V3-specific antibodies were responsible for a significant fraction (up to 90% in some sera) of the neutralizing activity against SF162 and MN, and this was similar in all three groups (Fig. 4, top and lower panel). It was previously shown that SF162 is a V3-sensitive virus (12), and our results are consistent with this observation. In contrast, there was no detectable reduction of serum neutralizing activity against HXBc2 (Fig. 4, middle panel) in the V3 peptide competition assay and only sporadic reduction against MW965 (data not shown), perhaps due to V3 mismatch. Additional attempts to map the neutralizing antibody response against HXBc2 were made using a peptide that spans the linear epitope of the gp41-broadly neutralizing antibody 2F5 to probe for antibodies directed against the membrane-proximal external region of gp41, which contains the 2F5 epitope. No inhibition of neutralization was observed in any of the 2F5-related mapping experiments (data not shown). It is possible that antibodies recognizing the CD4-induced coreceptor binding site of gp120 are responsible for the neutralizing activity observed against HXBc2, since such antibodies were detected in the sera from these animals (16a).
FIG. 4.
V3 peptide competition mapping of neutralizing activity. V3 peptide mapping was performed on individual serum samples against MN, HXBc2, and SF162. The V3 peptide sequence used to map neutralizing activity targeting the V3 loop was based on the YU2 sequence (TRPNNNTRKSINIGPGRALYTTG) (filled circles). A scrambled V3 peptide (IGPGRATRPNNNFYTTGTRKSIH) was used as a negative control (open circles). The results from these experiments are shown as percent inhibition of the ID50 neutralization values where only values over 50% are considered a reliable signal. Values in excess of a 50% reduction of ID50 neutralization are indicated by the dotted line.
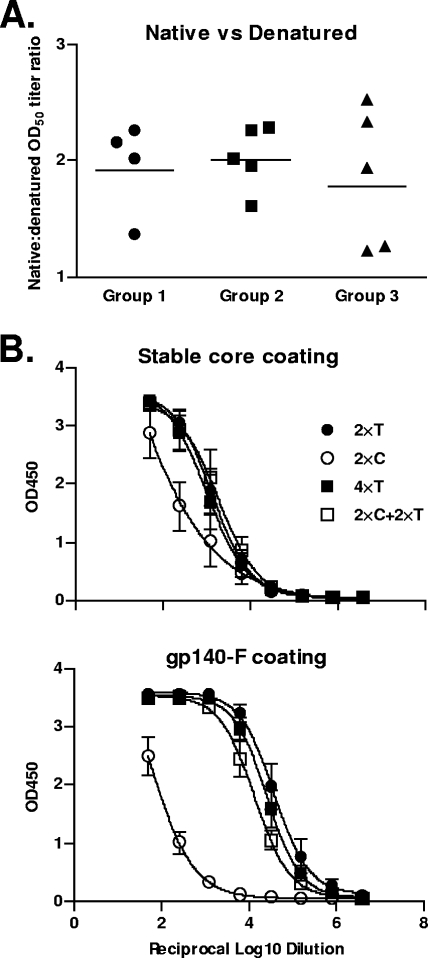
As mentioned above, the overall neutralizing activities were qualitatively quite similar in three groups, but the magnitude of the responses was somewhat lower in group 2 than in group 1 even at time points when the gp120 ELISA binding titers were comparable. This prompted us to ask if the proportion of antibodies recognizing denatured gp120 was greater in group 2 than in the other groups. It is possible, for example, that immature, misfolded Env released from dying infected cells contributed to the elicitation of antibody responses in rSFV-primed animals. To address this, we analyzed all post-5 sera in an ELISA using denatured gp120 antigen for coating side-by-side with the conventional ELISA using native gp120 for coating (Fig. 5A). As controls, we used two monoclonal antibodies: F105, which recognizes a conformation-sensitive epitope in gp120, and D7324, which recognizes a linear epitope in the C-terminal region of gp120. F105 was used to confirm that the gp120 used for coating was denatured, and D7324 confirmed that similar levels of protein were used in the two assays (see Fig S3 in the supplemental material). Using this assay, there was no marked difference between the sera in their relative ability to recognize misfolded gp120 compared to the native protein.
FIG. 5.
ELISA using different Env proteins for coating. (A) Serum samples from 2 weeks after the fifth immunization were tested in an ELISA using either native or denatured gp120 for coating. The native versus denatured OD50 titer ratio for each individual animal is shown. (B) Binding antibodies against the stable core protein (upper panel) or gp140-F (lower panel) used for coating in the ELISA plates were measured in serum samples from individual animals 2 weeks after the second and fourth immunizations and plotted as group means. “T” refers to gp140-F trimer protein immunization, “S” refers to SFV-gp140-F immunization, and “C” refers to stable core protein immunization.
We next asked why the regimen used in group 3 (stable core priming, gp140-F trimer boosting) did not yield sera possessing a greater increase in the breadth of neutralization. The rationale behind this regimen was that common epitopes between the two proteins would allow elicitation (by the priming) and selective expansion (by the boosting) of the B-cell response recognizing conserved surfaces, which might translate into increased neutralization breadth. Since this was not observed, we performed an ELISA using either the stable core protein or gp140-F trimers for coating, and we analyzed the sera obtained from the animals in group 1 and group 3 after the second and fourth immunizations. The results are presented as absorbance against log reciprocal serum dilution for group means (Fig. 5B). When the stable core was used for coating, there was only a modest boosting effect by the gp140-F trimers in group 3, while in contrast, when the trimers were used for coating, there was a substantial boost effect by the trimers in group 3. These data suggest that perhaps the cross-reactivity between the stable core protein and gp140-F is limited and that the increase in titers after gp140-F trimer immunizations likely represents new reactivities against trimer-specific determinants rather than boosting of memory B cells initially stimulated by the stable core prime. Nevertheless, trimers were able to modestly boost the stable core-directed antibody responses, suggesting that the strategy of immunofocusing may be beneficial with an improved immunogen design (37).
Env-specific T-cell responses in immunized macaques.
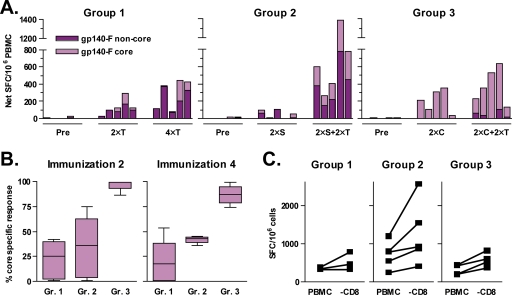
An additional goal of this study was to analyze Env-specific T-cell responses and particularly to determine whether priming with the stable core protein conferred an advantage in focusing the T-cell responses on more conserved regions of Env. Env-specific T-cell responses were measured using an IFN-γ ELISPOT assay with cells isolated before immunization or after the second or fourth immunization. Total PBMC were stimulated with two pools of 15-mer peptides spanning the gp140-F protein sequence. The “gp140-F core” peptide pool covered the conserved regions of gp120, corresponding to the stable core immunogen, while the “gp140-F noncore” peptide pool covered the remaining parts of gp140. Results are shown as total spot-forming cells per 1 million PBMC (Fig. 6A). High Env-specific IFN-γ responses were obtained in all three groups, and a clear increase in the magnitude of the responses was measured between the second and fourth immunizations. These data demonstrate the ability of gp140-F trimers administered in the AS01B adjuvant system to boost T-cell responses elicited either by prior immunizations with protein or by rSFV-gp140-F particles.
FIG. 6.
Vaccine-induced T-cell responses. HIV-1 Env-directed T-cell responses were measured by IFN-γ ELISPOT after restimulation of PBMC with overlapping peptides. Two different peptide pools based on the YU2 gp140 sequence were used: “gp140-F core” covered the conserved regions of gp120, corresponding to the stable core immunogen, and “gp140-F non-core” covered the remaining parts of YU2 gp140. (A) gp140-F core (light purple) and noncore (dark purple) responses before immunization (Pre) and 2 weeks after the second (2×T) and fourth (4×T) immunizations are shown. “T” refers to gp140-F trimer protein immunization, “S” refers to SFV-gp140-F immunization, and “C” refers to stable core protein immunization. (B) The percent core-specific response of the total gp140-F response after the second and fourth immunizations is shown. (C) PBMC collected 2 weeks after immunization 4 were depleted of CD8+ T cells by magnetic cell sorting. Total PBMC (PBMC) and CD8+ T-cell-depleted PBMC (−CD8) were restimulated with the gp140-F core and gp140-F noncore peptide pools. Shown are the cumulative gp140-F core and noncore responses. The viability in both total PBMC and CD8+ T-cell-depleted PBMC was >93%. The CD8+ T-cell-depleted PBMC contained <0.2% CD8+ T cells, as measured by flow cytometry.
The most striking outcome from the T-cell analysis was the clear difference in the response to the different peptide pools observed between the groups (Fig. 6A). The animals in group 1 and group 2 had a dominant IFN-γ response to the noncore peptide pool and a relatively weak response to the core peptide pool. In contrast, IFN-γ responses to the core peptide pool dominated in group 3 even after two boosts with gp140-F trimers with no or very low responses against the noncore pools. These data demonstrate very clearly that heterologous prime-boost regimens can be used to focus the T-cell response on conserved epitopes of HIV-1 Env. When the frequencies of core-specific responses of total Env-specific T-cell responses were analyzed, the animals in group 1 and group 2 showed similar patterns despite the two different platforms by which the trimers were administered (Fig. 6B). In contrast, the animals in group 3 were strikingly different, with 80 to 100% of the IFN-γ T-cell responses directed against the core (Fig. 6B). Similar results were obtained when the HXBc2 core peptide pool was used for stimulation (data not shown), indicating that the measured differences in the distribution of the core-specific responses were not biased by the use of YU2 peptides.
To determine the relative contributions of CD4+ versus CD8+ T cells for the overall Env-specific IFN-γ T-cell responses, the CD8+ T-cell fraction was depleted using magnetic bead separation and the IFN-γ ELISPOT assay was repeated using the total PBMC fraction and the fraction depleted of CD8+ T cells (Fig. 6C). This resulted in an unaltered or increased IFN-γ response for all animals, suggesting that CD4+ T cells were responsible for the IFN-γ response. The increased response can be explained by a greater relative number of CD4+ T cells in the PBMC population after CD8+ T-cell depletion, indicating that the AS01B adjuvant system potently induced Env-specific CD4+ T-cell responses.
DISCUSSION
In this study, we characterized neutralizing antibody and T-cell responses elicited by soluble YU2-based gp140-F trimers administered in the AS01B adjuvant system (GSK) in nonhuman primates. We also examined the consequences of priming with a heterologous immunogen, either an alphavirus replicon vector expressing matched gp140-F trimers or a monomeric gp120 core protein stabilized in the CD4-bound conformation, the latter administered in the AS01B adjuvant system. The stable core protein was selected in an attempt to focus the immune response on conserved determinants of Env that are common between the stable core and the gp140-F proteins. High titers of antibody were elicited with primates immunized with soluble gp140-F trimers in adjuvant, with OD50 titers of approximately 104, which corresponds to endpoint titers of about 106, at the peak of the response. Protein trimers in the AS01B adjuvant system also efficiently boosted antibody responses primed by rSFV-expressed gp140-F trimers, demonstrating that this adjuvant also is highly efficacious following a viral vector prime. The stable core protein appeared less immunogenic than the trimers, as measured by lower antibody responses after the first and second immunizations. We initially hypothesized that this might be explained by fewer CD4 T-cell helper epitopes in the stable cores than in the trimer proteins, but our subsequent T-cell analysis showed that the T-cell response against the stable core protein was in fact higher than that observed against the trimer, as discussed below.
Attempts to focus the immune response on shared neutralizing determinants between the stable core and the gp140-F trimers did not markedly improve the neutralizing antibody response. One possible explanation for this is that broadly neutralizing determinants, such as the CD4 binding site, are not similar enough between the two proteins. For example, both proteins bind CD4, but the stable cores are, by design, deficient in their ability to bind many of the nonneutralizing CD4-binding site antibodies. Also, this version of the stable core was reported to display a slightly decreased affinity for the broadly neutralizing CD4 binding site antibody, b12, as well as a somewhat increased affinity for CD4 itself (56). These changes in ligand affinities for the CD4-binding site might indicate that at least a portion of this surface is different between the core and the trimer. Poor cross-reactivity between the stable cores and gp140-F was also observed in the ELISA analysis shown in Fig. 5B, which demonstrates that gp140-F immunizations poorly boosted the stable core-specific antibody responses. In contrast, responses against the trimer were markedly increased by gp140-F immunizations (Fig. 5B), suggesting stimulation of new antibody reactivities, such as those directed against the immunogenic variable regions of Env, rather than boosting of responses previously elicited by the stable cores. Although not studied here, longer intervals that lead to a waning of the serum antibody levels between each immunization may also favorably affect the distribution of the memory B-cell compartment, with an impact on the expansion of B-cell clones that recognize common determinants between the proteins.
Binding antibody titers against gp120 after a single rSFV-gp140-F immunization were surprisingly high, reaching OD50 titers of between 102 and 103, and this response was enhanced by subsequent inoculations of gp140-F trimers in AS10B. Overall, responses in the three groups were qualitatively similar after the gp140-F trimer boosts. Significant neutralization titers were detected against HXBc2, MN, SF162 (all clade B), and the clade C virus MW965. Our results with the gp140-F trimers are consistent with the results obtained in other recent studies with nonhuman primates using the TZM-bl-cell single-cycle infectivity assay (42). ID50 titers against SF162 were around 100 to 1,000, which is the same range as the responses associated with protection against vaginal SHIV-SF162P4 challenge (2). We found that a considerable fraction of the neutralizing activity against SF162 and MN was mediated by antibodies against the V3 region of Env, as determined by peptide competition studies, consistent with cross-reactivity between the YU2 and the V3 region of these viruses. Since the majority of V3-directed antibodies tend to be strain specific rather than broadly neutralizing and most primary isolates are selected to occlude their V3 loops, a dominant antibody response against the V3 region is not desired for a broadly protective HIV-1 vaccine. Modifications designed to dampen the V3-directed response elicited by the gp140-F trimers are therefore warranted, such as the hyperglycosylation strategies described by Selvarajah et al. (43, 44).
We observed a striking difference in the specificities of the Env-specific T-cell responses when the three groups were compared side-by-side using pools of peptides spanning core versus noncore regions of the gp140-F trimers. While the dominant response in groups 1 and 2 was directed against epitopes in the noncore peptide pool, the response in group 3, even after the gp140-F boost, was almost exclusively (around 90%) directed against epitopes contained in the core peptide pool. These data highlight the potential of priming with Env core proteins to focus the cellular response on conserved regions of Env, presumably by increasing the frequencies of primed T cells against the conserved core determinants. Strategies to focus on the immune response in such a manner may add value to current attempts to induce protective immune responses to HIV-1. Finally, we also asked if there was a difference between the groups in terms of how much of the response was mediated by CD4+ T cells compared to CD8+ T cells. Detectable Env-directed IFN-γ T-cell responses appeared to be mediated by CD4+ T-cell responses in all three groups.
In conclusion, the inability of the core prime/trimer boost approach to broaden the neutralizing antibody responses may suggest that the stable core protein shares too few common immunogenic B-cell epitopes with gp140-F or that one or both of these immunogens are insufficient mimics of the functional viral spike complex. However, a marked shift in the specificity of the T-cell response was observed with this regimen. The utility of Env as a T-cell immunogen was recently called into question (36). Perhaps creative and rational attempts to focus both the B- and T-cell responses on conserved determinants of Env, such as those presented here, are worthy of consideration in the design of vaccine regimens capable of stimulating protective immune responses against HIV-1.
Supplementary Material
Acknowledgments
We thank Mats Spångberg, Helene Fredlund, and the personnel at Astrid Fagraeus laboratory at the Swedish Institute for Infectious Disease Control for expert assistance and Eva Hansson Pihlainen, Ulrika Edbäck, and Irene Silhammar for skilled technical assistance. We also thank Chih-chin Huang for contributions to the structure-based additions to V3 in the stable core context and Brenda Hartman, Mike Cichanowski, and Jonathan Stuckey for help with Fig. 1.
This study was supported by a grant from the Swedish International Development Agency (Sida)/Department of Research Cooperation (SAREC) to G.K.H. and R.T. and by the National Institute of Allergy and Infectious Diseases, National Institutes of Health intramural research program, for P.D.K., J.R.M., and R.T.W. Funding was also received from the Bill and Melinda Gates Foundation (P.D.K., J.R.M., and R.T.W.), the International AIDS Vaccine Initiative (P.D.K., R.T.W., and G.K.H.), and the Swedish Research Council (G.K.H.).
Footnotes
Published ahead of print on 12 November 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 755526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, S. W., I. K. Srivastava, E. Kan, F. Zhou, A. Goodsell, A. D. Cristillo, M. G. Ferrai, D. E. Weiss, N. L. Letvin, D. Montefiori, R. Pal, and M. Vajdy. 2008. Protection of macaques against vaginal SHIV challenge by systemic or mucosal and systemic vaccinations with HIV-envelope. AIDS 22339-348. [DOI] [PubMed] [Google Scholar]
- 3.Berglund, P., M. N. Fleeton, C. Smerdou, and P. Liljestrom. 1999. Immunization with recombinant Semliki Forest virus induces protection against influenza challenge in mice. Vaccine 17497-507. [DOI] [PubMed] [Google Scholar]
- 4.Berglund, P., M. Quesada-Rolander, P. Putkonen, G. Biberfeld, R. Thorstensson, and P. Liljestrom. 1997. Outcome of immunization of cynomolgus monkeys with recombinant Semliki Forest virus encoding human immunodeficiency virus type 1 envelope protein and challenge with a high dose of SHIV-4 virus. AIDS Res. Hum. Retrovir. 131487-1495. [DOI] [PubMed] [Google Scholar]
- 5.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bower, J. F., X. Yang, J. Sodroski, and T. M. Ross. 2004. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J. Virol. 784710-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5233-236. [DOI] [PubMed] [Google Scholar]
- 8.Caley, I. J., M. R. Betts, N. L. Davis, R. Swanstrom, J. A. Frelinger, and R. E. Johnston. 1999. Venezuelan equine encephalitis virus vectors expressing HIV-1 proteins: vector design strategies for improved vaccine efficacy. Vaccine 173124-3135. [DOI] [PubMed] [Google Scholar]
- 9.Catanzaro, A. T., M. Roederer, R. A. Koup, R. T. Bailer, M. E. Enama, M. C. Nason, J. E. Martin, S. Rucker, C. A. Andrews, P. L. Gomez, J. R. Mascola, G. J. Nabel, and B. S. Graham. 2007. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine 254085-4092. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Z., Y. Huang, X. Zhao, L. Ba, W. Zhang, and D. D. Ho. 2008. Design, construction, and characterization of a multigenic modified vaccinia Ankara candidate vaccine against human immunodeficiency virus type 1 subtype C/B′. J. Acquir. Immune Defic Syndr. 47412-421. [DOI] [PubMed] [Google Scholar]
- 11.Davis, N. L., A. West, E. Reap, G. MacDonald, M. Collier, S. Dryga, M. Maughan, M. Connell, C. Walker, K. McGrath, C. Cecil, L. H. Ping, J. Frelinger, R. Olmsted, P. Keith, R. Swanstrom, C. Williamson, P. Johnson, D. Montefiori, and R. E. Johnston. 2002. Alphavirus replicon particles as candidate HIV vaccines. IUBMB Life 53209-211. [DOI] [PubMed] [Google Scholar]
- 12.Derby, N. R., Z. Kraft, E. Kan, E. T. Crooks, S. W. Barnett, I. K. Srivastava, J. M. Binley, and L. Stamatatos. 2006. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J. Virol. 808745-8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey, B., M. Pancera, K. Svehla, Y. Shu, S. H. Xiang, J. Vainshtein, Y. Li, J. Sodroski, P. D. Kwong, J. R. Mascola, and R. Wyatt. 2007. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J. Virol. 815579-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleeton, M. N., M. Chen, P. Berglund, G. Rhodes, S. E. Parker, M. Murphy, G. J. Atkins, and P. Liljestrom. 2001. Self-replicative RNA vaccines elicit protection against influenza A virus, respiratory syncytial virus, and a tickborne encephalitis virus. J. Infect. Dis. 1831395-1398. [DOI] [PubMed] [Google Scholar]
- 15.Forsell, M. N., Y. Li, M. Sundback, K. Svehla, P. Liljestrom, J. R. Mascola, R. Wyatt, and G. B. Hedestam. 2005. Biochemical and immunogenic characterization of soluble human immunodeficiency virus type 1 envelope glycoprotein trimers expressed by Semliki Forest virus. J. Virol. 7910902-10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsell, M. N., G. M. McInerney, P. Dosenovic, A. S. Hidmark, C. Eriksson, P. Liljestrom, C. Grundner, and G. B. Karlsson Hedestam. 2007. Increased human immunodeficiency virus type 1 Env expression and antibody induction using an enhanced alphavirus vector. J. Gen. Virol. 882774-2779. [DOI] [PubMed] [Google Scholar]
- 16a.Forsell, M. N. E., B. Dey, A. Mörner, K. Svehla, S. O'dell, C.-M. Högerkorp, G. Voss, R. Thorstensson, G. M. Shaw, J. R. Mascola, G. B. Karlsson Hedestam, and R. T. Wyatt. 2008. B cell recognition of the conserved HIV-I co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 4c1000171. doi: 10.1371/journal.ppat.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J. Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 667538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harari, A., P. A. Bart, W. Stohr, G. Tapia, M. Garcia, E. Medjitna-Rais, S. Burnet, C. Cellerai, O. Erlwein, T. Barber, C. Moog, P. Liljestrom, R. Wagner, H. Wolf, J. P. Kraehenbuhl, M. Esteban, J. Heeney, M. J. Frachette, J. Tartaglia, S. McCormack, A. Babiker, J. Weber, and G. Pantaleo. 2008. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J. Exp. Med. 20563-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hidmark, A. S., G. M. McInerney, E. K. Nordstrom, I. Douagi, K. M. Werner, P. Liljestrom, and G. B. Hedestam. 2005. Early alpha/beta interferon production by myeloid dendritic cells in response to UV-inactivated virus requires viral entry and interferon regulatory factor 3 but not MyD88. J. Virol. 7910376-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 3101025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huckriede, A., L. Bungener, M. Holtrop, J. de Vries, B. L. Waarts, T. Daemen, and J. Wilschut. 2004. Induction of cytotoxic T lymphocyte activity by immunization with recombinant Semliki Forest virus: indications for cross-priming. Vaccine 221104-1113. [DOI] [PubMed] [Google Scholar]
- 22.Karlsson, G. B., and P. Liljestrom. 2003. Live viral vectors: Semliki Forest virus. Methods Mol. Med. 8769-82. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson Hedestam, G. B., R. A. Fouchier, S. Phogat, D. R. Burton, J. Sodroski, and R. T. Wyatt. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6143-155. [DOI] [PubMed] [Google Scholar]
- 24.Kim, M., Z. S. Qiao, D. C. Montefiori, B. F. Haynes, E. L. Reinherz, and H. X. Liao. 2005. Comparison of HIV type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res. Hum. Retrovir. 2158-67. [DOI] [PubMed] [Google Scholar]
- 25.Letvin, N. L., Y. Huang, B. K. Chakrabarti, L. Xu, M. S. Seaman, K. Beaudry, B. Korioth-Schmitz, F. Yu, D. Rohne, K. L. Martin, A. Miura, W. P. Kong, Z. Y. Yang, R. S. Gelman, O. G. Golubeva, D. C. Montefiori, J. R. Mascola, and G. J. Nabel. 2004. Heterologous envelope immunogens contribute to AIDS vaccine protection in rhesus monkeys. J. Virol. 787490-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 7910108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 131032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y., K. Svehla, N. L. Mathy, G. Voss, J. R. Mascola, and R. Wyatt. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 801414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris-Downes, M. M., K. V. Phenix, J. Smyth, B. J. Sheahan, S. Lileqvist, D. A. Mooney, P. Liljestrom, D. Todd, and G. J. Atkins. 2001. Semliki Forest virus-based vaccines: persistence, distribution and pathological analysis in two animal systems. Vaccine 191978-1988. [DOI] [PubMed] [Google Scholar]
- 30.Morris-Downes, M. M., B. J. Sheahan, M. N. Fleeton, P. Liljestrom, H. W. Reid, and G. J. Atkins. 2001. A recombinant Semliki Forest virus particle vaccine encoding the prME and NS1 proteins of louping ill virus is effective in a sheep challenge model. Vaccine 193877-3884. [DOI] [PubMed] [Google Scholar]
- 31.Mossman, S. P., F. Bex, P. Berglund, J. Arthos, S. P. O'Neil, D. Riley, D. H. Maul, C. Bruck, P. Momin, A. Burny, P. N. Fultz, J. I. Mullins, P. Liljestrom, and E. A. Hoover. 1996. Protection against lethal simian immunodeficiency virus SIVsmmPBj14 disease by a recombinant Semliki Forest virus gp160 vaccine and by a gp120 subunit vaccine. J. Virol. 701953-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordstrom, E. K., M. N. Forsell, C. Barnfield, E. Bonin, T. Hanke, M. Sundstrom, G. B. Karlsson, and P. Liljestrom. 2005. Enhanced immunogenicity using an alphavirus replicon DNA vaccine against human immunodeficiency virus type 1. J. Gen. Virol. 86349-354. [DOI] [PubMed] [Google Scholar]
- 33.Pantophlet, R., and D. R. Burton. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24739-769. [DOI] [PubMed] [Google Scholar]
- 34.Patterson, L. J., N. Malkevitch, D. Venzon, J. Pinczewski, V. R. Gomez-Roman, L. Wang, V. S. Kalyanaraman, P. D. Markham, F. A. Robey, and M. Robert-Guroff. 2004. Protection against mucosal simian immunodeficiency virus SIVmac251 challenge by using replicating adenovirus-SIV multigene vaccine priming and subunit boosting. J. Virol. 782212-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perri, S., C. E. Greer, K. Thudium, B. Doe, H. Legg, H. Liu, R. E. Romero, Z. Tang, Q. Bin, T. W. Dubensky, Jr., M. Vajdy, G. R. Otten, and J. M. Polo. 2003. An alphavirus replicon particle chimera derived from Venezuelan equine encephalitis and Sindbis viruses is a potent gene-based vaccine delivery vector. J. Virol. 7710394-10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peut, V., and S. J. Kent. 2007. Utility of human immunodeficiency virus type 1 envelope as a T-cell immunogen. J. Virol. 8113125-13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phogat, S., and R. Wyatt. 2007. Rational modifications of HIV-1 envelope glycoproteins for immunogen design. Curr. Pharm. Des. 13213-227. [DOI] [PubMed] [Google Scholar]
- 38.Priddy, F. H., D. Brown, J. Kublin, K. Monahan, D. P. Wright, J. Lalezari, S. Santiago, M. Marmor, M. Lally, R. M. Novak, S. J. Brown, P. Kulkarni, S. A. Dubey, L. S. Kierstead, D. R. Casimiro, R. Mogg, M. J. DiNubile, J. W. Shiver, R. Y. Leavitt, M. N. Robertson, D. V. Mehrotra, and E. Quirk. 2008. Safety and immunogenicity of a replication-incompetent adenovirus type 5 HIV-1 clade B gag/pol/nef vaccine in healthy adults. Clin. Infect. Dis. 461769-1781. [DOI] [PubMed] [Google Scholar]
- 39.Robinson, H. L., S. Sharma, J. Zhao, S. Kannanganat, L. Lai, L. Chennareddi, T. Yu, D. C. Montefiori, R. R. Amara, L. S. Wyatt, and B. Moss. 2007. Immunogenicity in macaques of the clinical product for a clade B DNA/MVA HIV vaccine: elicitation of IFN-gamma, IL-2, and TNF-alpha coproducing CD4 and CD8 T cells. AIDS Res. Hum. Retrovir. 231555-1562. [DOI] [PubMed] [Google Scholar]
- 40.Sanders, R. W., M. Vesanen, N. Schuelke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, P. J. Maddon, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 768875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlesinger, S., and T. W. Dubensky. 1999. Alphavirus vectors for gene expression and vaccines. Curr. Opin. Biotechnol. 10434-439. [DOI] [PubMed] [Google Scholar]
- 42.Seaman, M. S., D. F. Leblanc, L. E. Grandpre, M. T. Bartman, D. C. Montefiori, N. L. Letvin, and J. R. Mascola. 2007. Standardized assessment of NAb responses elicited in rhesus monkeys immunized with single- or multi-clade HIV-1 envelope immunogens. Virology 367175-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selvarajah, S., B. Puffer, R. Pantophlet, M. Law, R. W. Doms, and D. R. Burton. 2005. Comparing antigenicity and immunogenicity of engineered gp120. J. Virol. 7912148-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selvarajah, S., B. A. Puffer, F. H. Lee, P. Zhu, Y. Li, R. Wyatt, K. H. Roux, R. W. Doms, and D. R. Burton. 2008. Focused dampening of antibody response to the immunodominant variable loops by engineered soluble gp140. AIDS Res. Hum. Retrovir. 24301-314. [DOI] [PubMed] [Google Scholar]
- 45.Shu, Y., S. Winfrey, Z. Y. Yang, L. Xu, S. S. Rao, I. Srivastava, S. W. Barnett, G. J. Nabel, and J. R. Mascola. 2007. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine 251398-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srivastava, I. K., L. Stamatatos, E. Kan, M. Vajdy, Y. Lian, S. Hilt, L. Martin, C. Vita, P. Zhu, K. H. Roux, L. Vojtech, C. D. Montefiori, J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2003. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J. Virol. 7711244-11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundback, M., I. Douagi, C. Dayaraj, M. N. Forsell, E. K. Nordstrom, G. M. McInerney, K. Spangberg, L. Tjader, E. Bonin, M. Sundstrom, P. Liljestrom, and G. B. Karlsson Hedestam. 2005. Efficient expansion of HIV-1-specific T cell responses by homologous immunization with recombinant Semliki Forest virus particles. Virology 341190-202. [DOI] [PubMed] [Google Scholar]
- 48.Thongcharoen, P., V. Suriyanon, R. M. Paris, C. Khamboonruang, M. S. de Souza, S. Ratto-Kim, C. Karnasuta, V. R. Polonis, L. Baglyos, R. E. Habib, S. Gurunathan, S. Barnett, A. E. Brown, D. L. Birx, J. G. McNeil, and J. H. Kim. 2007. A phase 1/2 comparative vaccine trial of the safety and immunogenicity of a CRF01_AE (subtype E) candidate vaccine: ALVAC-HIV (vCP1521) prime with oligomeric gp160 (92TH023/LAI-DID) or bivalent gp120 (CM235/SF2) boost. J. Acquir. Immune Defic. Syndr. 4648-55. [DOI] [PubMed] [Google Scholar]
- 49.Wang, S., J. S. Kennedy, K. West, D. C. Montefiori, S. Coley, J. Lawrence, S. Shen, S. Green, A. L. Rothman, F. A. Ennis, J. Arthos, R. Pal, P. Markham, and S. Lu. 2008. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 261098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang, S. H., P. D. Kwong, R. Gupta, C. D. Rizzuto, D. J. Casper, R. Wyatt, L. Wang, W. A. Hendrickson, M. L. Doyle, and J. Sodroski. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 769888-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, R., I. K. Srivastava, C. E. Greer, I. Zarkikh, Z. Kraft, L. Kuller, J. M. Polo, S. W. Barnett, and L. Stamatatos. 2006. Characterization of immune responses elicited in macaques immunized sequentially with chimeric VEE/SIN alphavirus replicon particles expressing SIVGag and/or HIVEnv and with recombinant HIVgp140Env protein. AIDS Res. Hum. Retrovir. 221022-1030. [DOI] [PubMed] [Google Scholar]
- 52.Xu, R., I. K. Srivastava, L. Kuller, I. Zarkikh, Z. Kraft, Z. Fagrouch, N. L. Letvin, J. L. Heeney, S. W. Barnett, and L. Stamatatos. 2006. Immunization with HIV-1 SF162-derived envelope gp140 proteins does not protect macaques from heterologous simian-human immunodeficiency virus SHIV89.6P infection. Virology 349276-289. [DOI] [PubMed] [Google Scholar]
- 53.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 745716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 764634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 751165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.