Abstract
One attractive strategy for the development of a human immunodeficiency virus (HIV) vaccine is the use of viral vectors with a proven safety profile and an absence of preexisting immunity in humans, such as Newcastle disease virus (NDV). Several NDV vaccine vectors have been generated, and their immunogenicities have been investigated with different animal models. However, a systematic study to evaluate the optimal insertion site of the foreign antigens into NDV that results in enhanced immune responses specific to the antigen has not yet been conducted. In this article, we describe the ability of NDV expressing HIV Gag to generate a Gag-specific immune response in mice. We also have determined the optimal insertion site into the NDV genome by generating recombinant NDV-HIVGag viruses in which HIV gag was located at different transcriptional positions throughout the NDV viral genome. All recombinant viruses were viable, grew to similar titers in embryonated chicken eggs, and expressed Gag in a stable manner. Our in vivo experiments revealed that higher HIV Gag protein expression positively correlates with an enhanced CD8+ T-cell-mediated immune response and protective immunity against challenge with vaccinia virus expressing HIV Gag. We also inserted a codon-optimized version of HIV gag in the described best location, between the P and M genes. Virus expressing the codon-optimized version of HIV gag induced a higher expression of the protein and an enhanced immune response against HIV Gag in mice. These results indicate that strategies directed toward increasing antigen expression by NDV result in enhanced immunogenicity and vaccine efficacy.
The human immunodeficiency virus (HIV) pandemic affects approximately 40 million people around the world. The most affected region is sub-Saharan Africa, where two-thirds of the world's infected population live (73). Treatment with antiviral drugs can control the progression of the disease, but the limited access to antiviral drugs (especially in developing countries) and the possible appearance of resistance to these drugs make the development of a vaccine the most realistic hope of bringing the pandemic under control. In order to develop an HIV vaccine, a wide variety of approaches have been pursued. Traditional methods, such as the use of inactivated or attenuated viruses as vaccines, have not been successful and/or present safety concerns (4). Alternative vaccine strategies include DNA vaccines, the use of viral components, recombinant viral or nonviral vectors, and various combinations of these approaches. Several viral vectors to be used in vaccines are currently being developed and tested in preclinical and clinical trials (reviewed in references 6 and 17).
Today, it is mostly accepted that an ideal vaccine against HIV should induce both long-lasting neutralizing antibodies and CD8+ T-cell responses (50). Despite important advances in the study and generation of strong neutralizing antibodies, the development of a vaccine that provides sterilizing immunity still remains elusive. In addition to neutralizing antibodies, a potent specific CD8+ T cell is likely required for elimination of HIV-infected cells, which could limit HIV replication, thereby reducing the infection (27). Several observations confirm the importance of the CD8+ T-cell response. There is a correlation between the control of the viremia in primary HIV infection and the appearance of an HIV-specific cytotoxic-T-lymphocyte response; later in infection, the viral load is inversely related to the level of HIV-specific cytotoxic-T-lymphocyte response (10, 40, 56). During primary and chronic infections, HIV escapes the pressure mediated by HIV-specific CD8+ T cells through mutations within or near epitopes (11, 31, 64). Finally, long-term nonprogressors have been reported to have a highly functional HIV-specific CD8+ T-cell response in comparison to progressors, suggesting a strong relation between the HIV-specific CD8+ T-cell response and the progression of the disease (8).
The Gag, Pol, and Nef proteins are among the most immunogenic proteins during viral infection, (1, 7, 20, 23, 49, 55). Gag-specific CD8+ T-cell responses might play a special role in controlling viral replication (14, 20, 23, 30, 33, 38, 40, 66). In addition, Gag is highly conserved among sequence isolates, making it one of the most attractive candidate antigens in the development of an HIV vaccine.
Newcastle disease virus (NDV) is a member of the Avulavirus genus in the Paramyxoviridae family. NDV is pathogenic in several avian species. Strains of NDV are classified in three pathotypes depending on the severity of disease: lentogenic (avirulent), mesogenic (intermediate), and velogenic (highly virulent) (2). The NDV genome consists of a single-stranded negative-sense nonsegmented RNA that contains six transcriptional units, NP, P, M, F, HN, and L, which encode eight proteins in the order NP-P/V/I-M-F-HN-L. NDV transcription and replication follow the traditional model for single-stranded negative-sense RNA viruses. The viral polymerase transcribes the NDV genome starting at the 3′ end by a stop-and-restart mechanism at each transcriptional unit. This sequential transcription creates a gradient of decreasing abundance of the mRNAs from the NP gene to the L gene (13, 42).
Several characteristics make NDV a promising candidate as a delivery vector for human vaccines. Clinical trials for cancer treatment have demonstrated NDV to be well tolerated with few adverse side effects, since only mild flu-like symptoms and mild conjunctivitis have been reported (3, 21, 54). Since NDV is an avian virus, there is no preexisting immunity in humans which would limit the effectiveness of vaccine vectors (3). A reverse-genetics system for this virus was developed, and it can be used to generate recombinant viruses expressing foreign proteins in a stable manner (12, 16, 28, 35, 48, 51, 52, 59, 63, 67, 74). NDV grows to high titers in embryonated chicken eggs and generates a strong humoral and cellular immune response in murine and nonhuman primate models, thus making this vector an attractive vehicle for the expression of foreign antigens (12, 16, 35, 51, 52, 59). In this study, we first generated recombinant NDVs expressing HIV Gag (NDV-HIVGag) from various positions in the genome, and we determined the optimal location for the gene insert that would result in a maximum Gag-specific immune response. We then characterized the cellular immune responses and the protective efficacy of the HIV Gag-specific immune response elicited by NDV-HIVGag viruses in mice using a challenge model with vaccinia virus expressing Gag (Vac-HIVGag). We also generated a recombinant NDV expressing a synthetic codon-optimized version of HIV gag (NDV-HIVGag-opt) (39, 70). Insertion of HIVGag-opt at the previously determined optimal position revealed a large increase in the levels of expression of HIV Gag compared to the virus containing nonmodified HIV Gag in vitro. These higher expression levels of Gag resulted in an enhanced anti-Gag immune responses in vivo, a greater protective efficacy after challenge with Vac-HIVGag, and a larger Gag-specific cellular response compared with results for nonoptimized NDV-HIVGag.
MATERIALS AND METHODS
Cells, viruses, and animals.
Chicken embryo fibroblast cells were prepared from specific-pathogen-free 10-day-old embryonated eggs (Charles River SPAFAS, North Franklin, CT). Chicken embryo fibroblasts were maintained in minimal essential medium containing 10% fetal bovine serum (FBS). A549, HEp-2, Vero, CV-1, and P815 cells were maintained in Dulbecco modified Eagle medium with 10% FBS. The recombinant NDV-GFP virus (bearing the green fluorescent protein [GFP] gene) was generated previously (58). Wild-type New York City Board of Health vaccinia virus (Vac/wt) was kindly provided by Gómez Yafal at Therion Biologicals, and recombinant vaccinia virus expressing HIV Gag (Vac-HIVGag) (vP1287) was kindly provided by Matthias J. Schnell. Both vaccinia viruses were grown and titrated in CV-1 cells. Six- to eight-week-old female BALB/c mice were used in the animal experiments. All animal care and procedures were in accordance with the animal experimentation guidelines of the National Institutes of Health (NIH).
Construction and growth of NDV-HIVGag viruses.
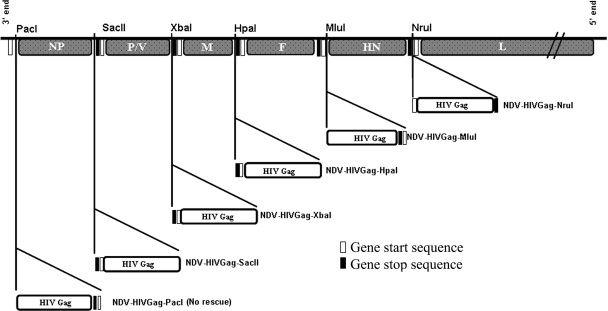
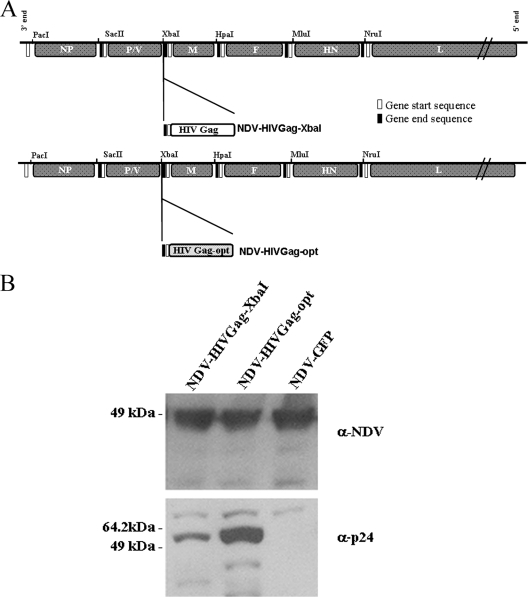
Full-length HIV gag (1,503 nucleotides) was obtained by PCR using as a template a plasmid containing HIV gag, NL4-3 strain, kindly provided by Matthias J. Schnell. A potential gene end sequence for NDV polymerase (5′-333TAAGAAAAAA342-3′) was present in the original gag cDNA. This possible gene end sequence was eliminated by site-directed mutagenesis with the following primers: SDM (+), 5′-328CAAAGCAAGAAGAAGGCACAGC349-3′, and SDM (−), 5′-349GCTGTGCCTTCTTCTTGCTTTTG328-3′; nucleotide modifications are shown in bold. HIV gag, codon optimized (HIVGag-opt) (1,503 nucleotides), was obtained by PCR using as a template a plasmid containing the full length of a clade B mammalian codon-optimized synthetic construct, kindly provided by Yaoxing Huang (39, 70). Both HIV Gag and HIVGag-opt have an amino acid identity greater than 98%; only 11 amino acids are different between the two HIV Gag proteins. These changes are located outside of the Gag-specific H-2Kd peptide AMQMLKETI. The plasmid used to generate the recombinant NDVs contains a full-length NDV infectious cDNA derived from the genome of the NDV strain Hitchner B1 (51). In order to facilitate the cloning of HIV gag in different positions in the NDV genome, three different enzyme restriction sites, PacI (inserted in front of NP), HpaI (between M and F), and MluI (between F and HN), in addition to the already-inserted XbaI site between the P/V and M genes and NruI site between the HN and L genes, were added by site-directed mutagenesis. Site-directed mutagenesis was facilitated by subcloning small portions of the NDV cDNA in the pSL1180 (Amersham Pharmacia Biotech) plasmid (51). Site-directed mutagenesis was performed using the following primers: SMD-PacI (+), 5′-94CAAACTCGAGGATTAATTAAGCCAACATGTCTTCC131, and SMD-PacI (−), 5′-131GGAAGACATGTTGGCTTAATTAATCCTCGAGTTTG94; SMD-HpaI (+), 5′-4454 GGATTATTTACAGTTAACTTACCTG4477, and SMD-HpaI (−), 5′-4477CAGGTAAGTTAACTTAAATAATC4454; SMD-MluI (+), 5′-6374CCTCCGTTCTACGCGTTCACCGAC6397, and SMD-MluI (−) 5′-GTCGGTGAACGCGTAGAACGGAGG. The enzyme restriction site is shown in italics, and nucleotide modifications are shown in bold. The HIV gag cDNA was cloned into NDV transcriptional units containing NDV-specific gene start and gene end sequences. An optimal Kozak translation sequence was also included just upstream of ATG. In order to follow the “rule of six,” necessary for NDV replication, a few nucleotides were added after the stop codon of HIV gag. The gag transcriptional units were inserted in front of NP (pNDV-HIVGag-PacI), between the NP and P/V genes (pNDV-HIVGag-SacII), the P/V and M genes (pNDV-HIVGag-XbaI), the M and F genes (pNDV-HIVGag-HpaI), the F and HN genes (pNDV-HIVGag-MluI), and the HN and L genes (pNDV-HIVGag-NruI). The obtained plasmids were used to rescue the recombinant viruses NDV-HIVGag-SacII, NDV-HIVGag-XbaI, NDV-HIVGag-HpaI, NDV-HIVGag-MluI, and NDV-HIVGag-NruI by using previously described methods (51, 58) (see Fig. 1). The transcriptional unit containing the HIV gag codon-optimized cDNA was constructed as aforementioned and then inserted between the P/V and M genes (pNDV-HIVGag-opt). This plasmid was used to rescue the recombinant virus NDV-HIVGag-opt.
FIG. 1.
Scheme of the different recombinant NDVs expressing HIV Gag protein. Transcriptional units containing 1.5 kb HIV gag were inserted in different positions in the NDV genome. In every new gag transcriptional unit, nucleotides were added in the untranslated region to follow the rule of six. The representation of the different genes is not to scale.
Growth kinetics of NDV in embryonated chicken eggs and Vero cells.
Ten-day-old embryonated chicken eggs were inoculated with the different NDV viruses at 100 PFU per egg. Viral titers present in the allantoic fluids at different time points were determined by indirect immunofluorescence in Vero cells infected with different dilutions of the allantoic fluids, using an anti-NDV rabbit serum followed by incubation with fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G (IgG) (39, 40). Vero cells were inoculated with the different viruses at an MOI of 1. Viral titers in the media at 13, 24, 36, and 42 h postinfection were established by indirect immunofluorescence in Vero cells as we described above.
Stability and expression of HIV Gag by NDV.
The stability of Gag expression by the different NDV-HIVGag viruses was evaluated by using five serial passages in eggs. Ten-day-old embryonated eggs were inoculated with allantoic fluid containing the different viruses at a 1/100 dilution at each passage. Allantoic fluids obtained after the last passage were tested for the ability to express HIV Gag by using indirect immunofluorescence in Vero cells infected at a low MOI. The presence of NDV antigens was determined using an anti-HN mouse monoclonal antibody (7B1) (18, 59), and the expression of HIV Gag was detected by using the anti-p24 human monoclonal antibody 71-31 (29), obtained from Susan Zolla-Pazner through the AIDS Research and Reference Reagent Program. As secondary antibodies, Texas Red-conjugated anti-mouse Ig and fluorescein isothiocyanate-conjugated anti-human IgG were used (Dako, Carpinteria, CA). Expression of the HIV Gag protein was also determined by Western blotting. Vero cells were infected at an MOI of 1 with each recombinant NDV virus. Cells were harvested and lysed in radioimmunoprecipitation assay buffer for 30 min in the presence of protease inhibitors at different time points following infection. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. Membranes were incubated with an anti-NDV rabbit serum, followed by incubation with an anti-rabbit IgG peroxidase-labeled antibody (Amersham, Piscataway, NJ) and anti-p24 human monoclonal antibody 71-31 or anti-p24 mouse monoclonal antibody 24-4 (19, 71), obtained from Susan Zolla-Pazner and Michael H. Malim, respectively, through the AIDS Research and Reference Reagent Program, followed by an anti-human IgG peroxidase-labeled antibody (Dako, Carpinteria, CA) or anti-mouse IgG peroxidase-labeled antibody (Amersham, Piscataway, NJ), respectively.
Immunizations in mice.
Groups of six mice each were intranasally immunized with 5 × 105 PFU of recombinant viruses. The mice were boosted with 106 PFU of the recombinant viruses, also intranasally, 3 weeks after the first immunization.
Challenge infections with Vac-HIVGag.
Three weeks after the second immunization, half of the mice in each group were challenged with 106 PFU of HIV Gag-expressing recombinant vaccinia virus (Vac-HIVGag) intranasally. The remaining animals of the groups were challenged intranasally with the same dose of wild-type vaccinia virus (Vac/wt). Five days after challenge, mice were sacrificed and their lungs were extracted and homogenized for vaccinia virus titration in CV-1 cells. Two days after infection, CV-1 cells were stained with 0.1% crystal violet solution to count the number of vaccinia virus plaques.
Preparation of CD8+ T cells.
Lymphocyte populations were isolated from spleens of immunized mice collected at different time points after the initial infection and boost with NDV or after challenge with vaccinia. Pooled spleens from two mice were dissociated into single-cell suspensions by grinding. Red blood cells in the suspension were lysed using a NH4Cl-Tris solution. Cells were washed, and the total number of splenocytes was counted. In some of the experiments, CD8α (Ly-2) magnetic microbeads (Miltenyi Biotec, Auburn, CA) were used to isolate the CD8+ T cells from the splenocytes, following the manufacturer's instructions.
ELISPOT assay for the detection of IFN-γ-producing cells.
The enzyme-linked immunospot (ELISPOT) assay was performed according to a previously described protocol (52, 53). Briefly, 96-well mixed cellulose ester membrane plates (Millipore Corp., Bedford, MA) were coated with a phosphate-buffered saline (PBS) solution containing 7 μg/ml of the anti-mouse gamma interferon (IFN-γ) monoclonal antibody R4-6A2 (BD Pharmingen, San Diego, CA). After overnight incubation at 4°C, wells were washed four times with 150 μl of RPMI without serum (Invitrogen). After the final wash, wells were incubated for 3 h at 37°C in 100 μl of RPMI supplemented 10% serum. One million CD8+ T cells or 1 million splenocytes were placed in triplicate in the coated wells and were cocultured with P815 (105 cells per well) pulsed with 5 μg of the Gag-specific H-2Kd peptide AMQMLKETI. Nonpulsed P815 cells were used as negative controls. Following a 36-h incubation at 37°C and 5% CO2, the plates were washed with a solution of Tween 20 (0.05%) in PBS, and then 100 μl of 2-μg/ml biotinylated rat anti-mouse IFN-γ monoclonal antibody XMG1.2 (BD Pharmingen, San Diego, CA) was added to each well. Plates were then incubated overnight at 4°C, washed with Tween 20 (0.05%) in PBS, and then incubated with peroxidase-labeled streptavidin (1:800 in PBS-Tween 20; BD Pharmingen, San Diego, CA) for 1 h at room temperature. Wells were washed, and the substrate was added at a concentration of 1 μg/ml (Sigma Fast 3,3′-diaminobenzidine tablets; Sigma, St. Louis, Mo). The reaction was stopped with double-distilled H20 when the spots were visible, and an ELISPOT reader (Cellular Technology Ltd., Shaker Heights, OH) was used for counting.
In vivo cytotoxicity assay.
Six- to eight-week-old female BALB/c mice were intranasally immunized once with NDV-GFP, NDV-HIVGag-XbaI, or NDV-HIVGag-opt at two different doses, 5 × 105 PFU and 5 × 106 PFU. Splenocytes were isolated from naive BALB/c mice, and single-cell suspensions were prepared. Naive splenocytes were exogenously pulsed with the HIV-1 H-2Kd peptide (10 μg/ml) and washed to remove excess peptides. To distinguish between exogenously peptide-pulsed splenocytes (targets) and their unpulsed counterparts (controls), two concentrations of carboxyfluorescein diacetate succinmidyl ester (CFSE) were used for their labeling. Whereas targets were labeled at 0.2 μM CFSE, the unpulsed controls received 2 μM CFSE, thus making both populations easily discerned by flow cytometric analysis using a FACS FC500 Benchtop cytometer, (Becton Dickinson, Franklin Lakes, NJ). Labeled cells were washed with an equal volume of fetal calf serum, followed by two washes in complete medium (RPMI containing 10% FBS). The two populations were mixed equally, and 2.5 × 106 mixed cells were injected intravenously into the immunized mice 8 days after priming. Spleens from recipient mice were harvested 18 h later, and survival of the pulsed and unpulsed populations was assessed by flow cytometry.
Intracellular IFN-γ staining.
Six- to eight-week-old female BALB/c mice were immunized twice with NDV-GFP, NDV-HIVGag-SacII, NDV-HIVGag-XbaI, or NDV-HIVGag-opt, and 3 weeks after the second immunization, mice were challenged with Vac-HIVGag. Five days postchallenge, splenocytes were extracted and incubated with the previously used HIV Gag CD8-specific peptide for 20 h at 37°C. Four hours before the end of the incubation, brefeldin A (Sigma, St. Louis, MO) was added at 5 μg/ml. Cells were harvested and incubated for 30 min on ice with anti-mouse CD8 conjugated to Alexa 647 (eBioscience, San Diego, CA). Splenocytes were fixed and permeabilized using a Cytofix/Citoperm kit (BD Pharmingen, Franklin Lakes, NJ). Cells were stained with anti-mouse IFN-γ conjugated to phycoerythrin (BD Pharmingen, Franklin Lakes, NJ) for 15 min at room temperature. Finally, samples were analyzed by flow cytometry.
RESULTS
Generation of NDV-HIVGag viruses expressing HIV Gag within the various intergenic regions.
Following previously described reverse-genetics approaches for the rescue of NDV (51, 58), five different NDVs expressing HIV Gag were generated in the backbone of the avirulent strain Hitchner B1: NDV-HIVGag-SacII, NDV-HIVGag-XbaI, NDV-HIVGag-HpaI, NDV-HIVGag-MluI, and NDV-HIVGag-NruI (Fig. 1). Rescued virus clones were isolated by limiting dilution in eggs. Viral stocks grown in embryonated eggs were titrated, and the regions containing the inserted gag gene were amplified by reverse transcription-PCR and sequenced to confirm the presence of the correct 1.5-kb insert (data not shown). Despite several attempts, we were not able to rescue a virus expressing HIV Gag in front of the NP gene of NDV.
Replication and stability of NDV-HIVGag viruses in embryonated chicken eggs.
In order to investigate the consequences that the insertion of HIV gag could have for the growth of the recombinant viruses, 100 PFU of each NDV-HIVGag virus was inoculated into 10-day-old eggs. As a control, eggs were inoculated with NDV-GFP, bearing the GFP gene at the XbaI site between the P/V and M transcriptional units. Eggs were incubated at 37°C during 96 h, and allantoic fluid samples were collected every 24 h. The viruses in the samples were titrated in Vero cells by indirect immunofluorescence. Both the kinetics and the magnitude of the replication of the six viruses were similar, which indicated that the growth of the recombinant viruses in eggs was not affected by the insertion of HIV gag in any specific position in comparison to GFP (Fig. 2). The fact that the embryonated eggs were alive after 96 h postinoculation confirmed the attenuation of these viruses (24).
FIG. 2.
Growth kinetics of NDVs in embryonated chicken eggs. Ten-day-old embryonated eggs were inoculated with 100 PFU of the indicated NDV viruses. Eggs were incubated for 96 h, and allantoic fluids were harvested at different time points (24, 48, 72, and 96 h postinfection). NDV titers in the allantoic fluids were determined by measuring indirect inmunofluorescence in Vero cells.
The stability of expression of HIV Gag by the recombinant viruses was tested after five serial passages in eggs. Allantoic fluids obtained from the fifth passage were quantified and were used to infect Vero cells at a low MOI. The correlation between the presence of the virus and the expression of HIV Gag was determined by indirect immunofluorescence using specific antibodies. HIV Gag expression was stable in all NDV viruses after five passages in eggs (Fig. 3).
FIG. 3.
Recombinant NDV viruses stably express HIV Gag. NDV-HIVGag viruses were passaged five times in 10-day-old embryonated eggs. Allantoic fluids obtained after the fifth passage were used to infect Vero cells, and expression of Gag in NDV-infected cells was visualized by indirect immunofluorescence. Anti-HN monoclonal antibody detected the presence of NDV-infected cells (B to F; mock, A). An anti-p24 monoclonal antibody (71-31) detected the expression of HIV Gag (panels H to L; mock, G).
Levels of expression of HIV Gag at different intergenic positions in NDV.
To determine the level of expression of the Gag protein driven by the five NDV-HIVGag viruses in vitro, Vero cells were infected at an MOI of 1 with the recombinant viruses. NDV-GFP was used as a control. Sixteen hours postinfection, cells were harvested and lysed and Western blot analyses were performed (Fig. 4A). The presence of the NP protein of NDV was detected in all the lysates at similar levels. We also found the Gag protein in every lysate with the exception of the negative control (NDV-GFP). It would be expected that infected cells would have different levels of expression of Gag, according to the theoretical expression gradient of NDV; i.e., we expect to observe higher levels of expression of Gag as the insertion is closer to the 3′ end. This gradient of expression was the case when Gag was inserted between the P and M genes and positions located after the M gene (in order of greatest to least expression, NDV-HIVGag-XbaI, NDV-HIVGag-HpaI, NDV-HIVGag-MluI, and NDV-HIVGag-NruI). Unexpectedly, the level of Gag expression was considerably lower when the gene was located between the NP and P genes (NDV-HIVGag-SacII). To assess the kinetics of the production of HIV Gag driven by the NDV-HIVGag viruses, Vero cells were infected at an MOI of 1 with NDV-HIVGag-SacII, NDV-HIVGag-XbaI, NDV-HIVGag-HpaI, or NDV-HIVGag-MluI. NDV-GFP was used as a control virus. Cells were harvested and lysed at 12, 24, 36, or 42 h after infection, and Western blot analysis was performed (Fig. 4B). We observed an increase in the expression of the NP and HIV Gag proteins at different time points. In the case of NP, the amounts of protein were approximately the same at every time point; however, we again observed a gradient in the expression of HIV Gag (in order of greatest to least expression, NDV-HIVGag-XbaI, NDV-HIVGag-HpaI, and NDV-HIVGag-MluI). As we observed in the previous experiment, the expression of HIV Gag obtained by infection with NDV-HIVGag-SacII was lower than expected, especially at early time points. Furthermore, at 12, 24, 36, and 42 h, medium from Vero-infected cells with the viruses was harvested and virus titers were measured by immunofluorescence in Vero cells. These viruses displayed similar growth kinetics (Fig. 4C). In summary, the location of gag between the P and M genes (NDV-HIVGag-XbaI) induced the highest expression of this foreign protein in NDV-infected cells.
FIG. 4.
Expression levels of HIV Gag, driven by the different recombinant NDVs. (A) Lysates from Vero cells infected with the NDV-HIVGag and NDV-GFP viruses were subjected to SDS-PAGE and were transferred to nitrocellulose membranes. Membranes were immunoblotted using an anti-NDV rabbit serum (α-NDV) and an anti-p24 (α-p24) human monoclonal antibody (71-31), followed by incubation with an anti-rabbit IgG peroxidase-labeled antibody and an anti-human IgG peroxidase-labeled antibody, respectively. (B) Lysates from Vero cells infected with the NDV-HIVGag and NDV-GFP viruses at 12, 24, 36, or 42 h postinfection were separated by SDS-PAGE, and Western blotting was performed using anti-NDV rabbit serum and anti-p24 mouse monoclonal antibody (24-4), followed by incubation with an anti-rabbit IgG peroxidase-labeled antibody and an anti-mouse IgG peroxidase-labeled antibody, respectively. (C) Vero cells were infected at an MOI of 1 with the indicated NDV viruses. Media from the infected cells were harvested at the indicated time points, and virus titers were measured by indirect immunofluorescence in Vero cells.
Protection against challenge infection with Vac-HIVGag in mice immunized with NDV-HIVGag.
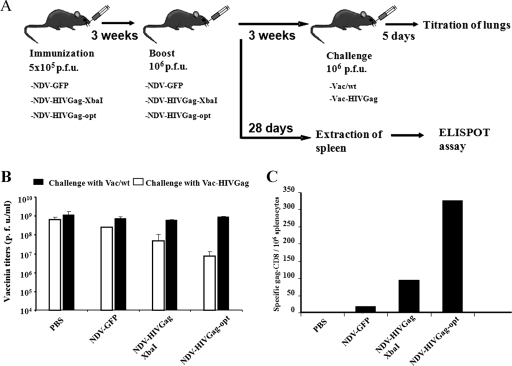
Next, we evaluated the ability of NDV-HIVGag viruses to induce a protective immune response in mice. In order to ascertain the NDV that induces the best protective immune response, each NDV-HIVGag virus was used to immunize BALB/c mice intranasally (5 × 105 PFU). Three weeks after the immunization, animals were boosted with a second dose of each virus (106 PFU). Mice did not show a loss of body weight or overt symptoms of disease after either the first or second immunization, indicating that these viruses were not virulent in mice. Three weeks after the second immunization, intranasal challenge infections with Vac-HIVGag or Vac/wt were performed (Fig. 5A). Animals were sacrificed at day five postchallenge, and the titers of vaccinia virus in lungs were examined. As shown in Fig. 5B, animals challenged with Vac/wt raised titers close to 109 PFU/ml of vaccinia virus in lungs, irrespective of their immunization schedule. In contrast, most animals vaccinated with the recombinant viruses expressing HIV Gag showed a reduced titer of Vac-HIVGag compared to that of unvaccinated controls. Vac-HIVGag virus titers were more than 10-fold lower in mice immunized with NDV-HIVGag-XbaI than in controls. Additionally, we observed a gradient in the ability to inhibit vaccinia replication: in order of greatest to least inhibition, NDV-HIVGag-XbaI, NDV-HIVGag-HpaI, NDV-HIVGag-MluI, and NDV-HIVGag-NruI. Finally, we did not observe a clear decrease in Vac-HIVGag virus titers in mice vaccinated with NDV-HIVGag-SacII. These results demonstrate that intranasal administration of NDV-HIVGag viruses induces a Gag-specific antiviral response in mice. Furthermore, we showed that the optimal insertion of HIV gag to induce the strongest immune response is between the P and M genes (NDV-HIVGag-XbaI). We also observed a strong correlation between the levels of the HIV Gag protein produced by the viruses in vitro (Fig. 4) and their ability to inhibit Vac-HIVGag replication in mice.
FIG. 5.
Challenge with Vac-HIVGag in mice immunized with NDV-HIVGag viruses. (A) Groups of nine mice were immunized intranasally with 5 × 105 PFU of each NDV-HIVGag virus. Control mice were inoculated with 5 × 105 PFU of NDV-GFP or with PBS. Three weeks after, animals were boosted with 106 PFU of the same virus. Three weeks after the boost, three mice per group were challenged with 106 PFU of Vac/wt or Vac-HIVGag. The rest of the mice were left for 51 days, after which they were sacrificed and their spleens were removed for quantification of Gag-specific CD8 T cells. (B) Five days after the infection with vaccinia viruses, mice were sacrificed, lungs were homogenized in 1 ml of PBS, and vaccinia titers in CV-1 cells were determined. (C) Fifty-one days after the second immunization, CD8+ T cells were selected and incubated with HIV Gag-specific peptide-pulsed cells in an ELISPOT assay to determine the number of HIV Gag-specific IFN-γ-secreting cells.
Induction of cellular immune responses against HIV Gag after immunization with NDV-HIVGag.
To quantify the cellular immune response generated in mice vaccinated with the NDV-HIVGag viruses, ELISPOT assays were performed. Groups of BALB/c mice were immunized intranasally as in the previous vaccinia virus challenge study (Fig. 5A). Fifty-one days after the second immunization, mice were sacrificed and spleens were extracted. The number of Gag-specific CD8+ IFN-γ-producing cells in the spleen was quantified by ELISPOT assay (Fig. 5C). We detected CD8+ cells specific for HIV Gag in the spleens of four of the NDV-HIVGag-immunized mice. Control groups (PBS or NDV-GFP) yielded negative results, as expected. NDV-HIVGag-SacII-immunized mice also yielded negative results. Our results show the following gradient in the generation of Gag-specific CD8+ T cells: in order of greatest to least number of cells, NDV-HIVGag-XbaI, NDV-HIVGag-HpaI, NDV-HIVGag-MluI, and NDV-HIVGag-NruI. Thus, the highest number of Gag-specific CD8+ IFN-γ-producing cells occurred in the mice vaccinated with NDV-HIVGag-XbaI, confirming that the insertion between the P and M genes of the NDV genome is the optimal location for the antigen in order to obtain the greatest cellular immune response.
Levels of expression of a recombinant virus expressing a synthetic codon-optimized version of HIV gag.
Following the previously described reverse-genetics protocol, we generated a recombinant NDV expressing a synthetic codon-optimized version of HIV gag (NDV-HIVGag-opt). The transcriptional unit containing HIVGag-opt was introduced between the P and M genes, a position portrayed to induce the strongest immune response (Fig. 6A). To study the level of expression of the Gag protein driven by NDV-HIVGag-opt virus in vitro, Vero cells were infected with the same doses of NDV-HIVGag-opt and NDV-HIVGag-XbaI; NDV-GFP was used as a control. After 16 h, the cells were collected and lysed and proteins were analyzed by Western blotting. We detected comparable levels of the NP protein in every lysate. As expected, we observed signal corresponding to the HIV Gag protein only in lysates from cells infected with NDV-HIVGag-opt and NDV-HIVGag-XbaI; however, the levels of expression of Gag were different between these two viruses. Cells infected with NDV-HIVGag-opt showed considerably higher levels of expression of HIV Gag than cells infected with NDV-HIVGag-XbaI (Fig. 6B). This increase in the expression of HIV Gag did not affect the replication of the virus, since all the viruses grew at same level in eggs (data not shown).
FIG. 6.
Generation of recombinant NDV containing a mammalian codon-optimized version of HIV gag. (A) A recombinant virus expressing a synthetic codon-optimized version of HIV gag (NDV-HIVGag-opt) inserted between the P/V and M genes was rescued following the same protocol described for earlier experiments. (B) Lysates from Vero cells infected with NDV-GFP, NDV-HIVGag-XbaI, or NDV-HIVGag-opt at an MOI of 1 were separated in an SDS-PAGE gel and transferred to nitrocellulose membranes. Membranes were immunoblotted using an anti-NDV rabbit serum and an anti-p24 mouse monoclonal antibody, 24-4, followed by incubation with an anti-rabbit IgG peroxidase-labeled antibody and an anti-mouse IgG peroxidase-labeled antibody, respectively.
Protection against challenge infection with Vac-HIVGag of mice immunized with NDV-HIVGag-opt.
We studied the ability of NDV-HIVGag-opt virus to induce a protective immune response in mice compared to that of the virus expressing the original HIV Gag. Mice were intranasally immunized twice with NDV-GFP, NDV-HIVGag-XbaI, or NDV-HIVGag-opt as described in Fig. 7A. Animals were sacrificed at day five postchallenge, and the titers of vaccinia virus in lungs were measured. As shown in Fig. 7B, animals challenged with Vac/wt raised similar titers after vaccination with every virus. As we previously showed, animals vaccinated with NDV-HIVGag-XbaI displayed approximately one log reduction in Vac-HIVGag titers, but this reduction was bigger than two logs in the case of mice vaccinated with NDV-HIVGag-opt (Fig. 7B). The reduction correlates with the amounts of the HIV Gag protein observed after infection in Vero cells with NDV-HIVGag-opt and NDV-HIVGag-XbaI (Fig. 6B).
FIG. 7.
Challenge with Vac-HIVGag in mice immunized with NDV-HIVGag-opt virus. (A) Groups of nine mice were immunized intranasally with 5 × 105 PFU of NDV-HIVGag-XbaI or NDV-HIVGag-opt virus. Control mice were inoculated with 5 × 105 PFU of NDV-GFP or PBS. Three weeks afterward, animals were boosted with 106 PFU of the same viruses. Three weeks after the boost, three mice per group were challenged with 106 PFU of Vac/wt or Vac-HIVGag. The rest of the mice were left for 28 days, after which they were sacrificed and their spleens were removed for quantification of Gag-specific CD8 T cells. (B) Five days after the infection with vaccinia viruses, mice were sacrificed and vaccinia titers in lungs were determined in CV-1 cells. (C) Twenty-eight days after the second immunization, splenocytes were isolated and incubated with HIV Gag-specific peptide-pulsed cells. ELISPOT assay was performed to determine the number of HIV Gag-specific IFN-γ-secreting splenocytes.
Induction of cellular immune response and cytotoxic activity in vivo after vaccination with NDV-HIVGag-opt.
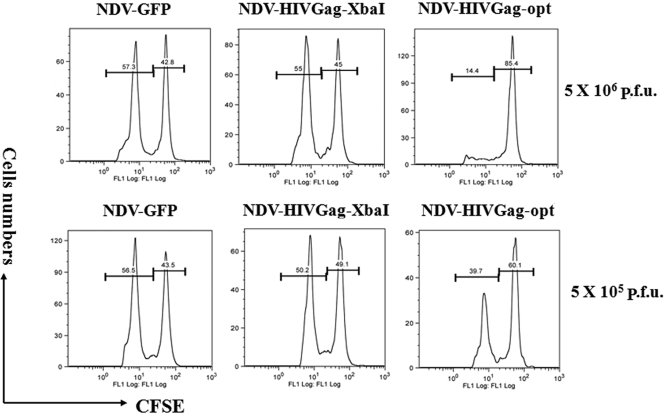
Next, we characterized the cellular immune response generated in mice immunized with NDV-HIVGag-opt, NDV-HIVGag-XbaI, and NDV-GFP as described in the legend to Fig. 7A. Twenty-eight days after the second immunization, animals were euthanized and the number of Gag-specific CD8+ IFN-γ-producing cells in total spleen tissue was quantified by ELISPOT assay (Fig. 7C). No significant levels of Gag-specific CD8+ T cells were detected in animals immunized with either NDV-GFP or PBS. Our results showed that immunization with NDV-HIVGag-opt induced more than three times as many Gag-specific CD8+ cells than immunization with NDV-HIVGag-XbaI. To assess the functional activity of the Gag-specific CD8+ T cells induced after immunization with NDV-HIVGag viruses, a “cytotoxicity assay in vivo” was performed (Fig. 8). Groups of BALB/c mice were immunized once with NDV-GFP, NDV-HIVGag-XbaI, or NDV-HIVGag-opt at two different doses, 5 × 105 PFU and 5 × 106 PFU, using the intranasal route. Eight days later, naive splenocytes were labeled with different concentrations of CFSE and then either pulsed with H-2Kd peptide or left unpulsed. Equivalent amounts of the two populations of splenocytes were injected intravenously into immunized mice. Eighteen hours later, splenocytes of the receiver mice were extracted and the percentage of survival of each cell population was measured by flow cytometry. As expected, the unpulsed splenocytes were detected in all the animals, and no visible cytotoxic activity was observed. Mice inoculated with any of the two doses of NDV-HIVGag-opt showed a remarkable decrease in specific Gag peptide-pulsed splenocytes (Fig. 8). The decline correlates with the amount of NDV-HIVGag-opt used to immunize the animals. In contrast, we did not observe any cytotoxic effect in specific Gag peptide-pulsed splenocytes in mice immunized with NDV-GFP, and surprisingly, no decrease was detected after immunization with NDV-HIVGag-XbaI, indicating that two doses are most likely needed to induce a detectable response in animals immunized with the viruses using the in vivo cytotoxicity assay. Altogether, these data showed that immunization of mice with NDV-HIVGag-opt induced better protection against challenge with Vac-HIVGag than immunization with NDV expressing nonoptimized HIV Gag. This higher level of protection correlated with an increase in specific Gag-specific CD8+ responses generated by the NDV-HIVGag-opt.
FIG. 8.
In vivo cytotoxicity assay. Groups of three mice were immunized using 5 × 105 PFU or 5 × 106 PFU of NDV-GFP, NDV-HIVGag-XbaI, or NDV-HIVGag-opt. Eight days later, spleens from naive mice were harvested and were stained either with 0.2 μM or 2 μM CFSE and then cultured with HIV Gag CD8-specific peptides or without peptides, respectively. The two splenocyte populations were mixed equally and injected intravenously into previously immunized mice. Eighteen hours later, spleens from recipient mice were harvested and survival of each population of stained splenocytes was assessed by flow cytometry.
Induction of cellular immune responses against HIV Gag at different time points after immunization and challenge.
To study the kinetics in the generation of cellular immune responses in mice after immunization with the NDV viruses, we measured the Gag-specific CD8+ T cells by ELISPOT assay at different time points after the prime, boost, and challenge. Mice were intranasally immunized twice with NDV-HIVGag-SacII, NDV-HIVGag-XbaI, NDV-HIVGag-opt, and NDV-GFP, used as a control (Fig. 9A). Animals were euthanized 10, 28, 35, 42 and 47 days after the first immunization, CD8+ T cells were isolated from the spleen, and the number of Gag-specific CD8+ IFN-γ-producing cells was quantified by ELISPOT assay (Fig. 9B). We detected CD8+ IFN-γ-producing cells specific for HIV Gag in NDV-HIVGag-XbaI and NDV-HIVGag-opt at every time point. Mice immunized with NDV-HIVGag-SacII yielded low levels of Gag-specific CD8 cells only 10 days after the prime and 7 days after the boost in parallel with the expansion peak phase of CD8+ T cells after every immunization. As expected, no Gag-specific CD8 responses were detected against NDV-GFP. The total values of HIV Gag CD8+ T cells obtained fluctuated during the time points agreeing with the expansion-contraction-memory phases of the activation of the CD8+ T-cell responses (32). The magnitude of the CD8 response in mice immunized with NDV-HIVGag-opt was at least twice as high at each time point (Fig. 9B), confirming once again the advantage of using the virus expressing codon-optimized HIV Gag. Gag-specific CD8+ IFN-γ-producing cells before and after challenge with Vac-HIVGag were also monitored by intracellular staining (Fig. 9C). In both cases, mice immunized with NDV-HIVGag-XbaI or NDV-HIVGag-opt showed a higher percentage of Gag-specific CD8+ T cells than control mice. In concurrence with previous results, we observed a low level of Gag-specific response in mice immunized with NDV-HIVGag-SacII both before and after challenge. Challenge with Vac-HIVGag resulted in an increase between five and six times in the number of Gag-specific CD8+ cells in mice immunized with NDV-HIVGag-XbaI or NDV-HIVGag-opt compared with results for the nonchallenged mice. Both before and after challenge, the number of Gag-specific CD8+ T cells obtained after inoculation of NDV-HIVGag-opt was approximately three to five times higher, respectively, than that with immunization with NDV-HIVGag-XbaI (Fig. 9C). In summary, we found that NDV-HIVGag-opt clearly induced higher levels of Gag-specific cellular response than NDV-HIVGag-XbaI. That supports earlier results on the use of codon-optimized cDNAs to express antigens in vivo in order to generate an optimal immune response (22, 25, 65).
FIG. 9.
Induction of cellular immune responses in mice at different times postvaccination and postchallenge. (A) Mice were immunized intranasally with 5 × 105 PFU of NDV-GFP, NDV-HIVGag-SacII, NDV-HIVGag-XbaI, or NDV-HIVGag-opt virus. Three weeks afterward, animals were boosted with 106 PFU of the same virus. Three weeks after the boost, mice were challenged with 106 PFU of Vac-HIVGag. (B) Spleens were harvested at 10 days after the first immnunization, 7, 14, and 21 days after the boost, and 5 days after challenge. CD8+ T cells were selected and incubated with HIVGag-specific peptide-pulsed cells in an ELISPOT assay to determine the number of HIV Gag-specific IFN-γ-secreting cells. (C) Before and 5 days after challenge, splenocytes were harvested, incubated with the HIV Gag peptide, and subjected to intracellular staining after incubation with anti-mouse Alexa 647-conjugated CD8 and anti-mouse phycoerythrin-conjugated IFN-γ. Samples were analyzed by flow cytometry.
DISCUSSION
Despite more than 20 years of HIV research and considerable effort, the development of an HIV vaccine remains elusive. The lack of robust immunogenicity, safety concerns, and ultimately the lack of protection have been discouraging. Among several strategies, one of the most promising is the use of live viral vectors as vaccines. The potential advantage of a viral vector strategy is to reproduce as closely as possible the efficacy of HIV live-attenuated virus vaccines and at the same time to be safe. NDV has some interesting properties as a delivery viral vaccine vector. The existence of a reverse-genetics system makes possible the insertion of relatively large antigens (approximately 4 kb) (15, 26, 51, 62, 67). NDV can be grown to high titers in embryonated chicken eggs and in cell lines such as Vero cells, both substrates approved for virus vaccine production. In contrast to other viral vectors with large genomes (>100 kbp), such as poxviruses and herpesviruses, NDV encodes fewer proteins that may compete for immunodominant epitopes between the viral vector proteins and the expressed foreign antigens. Moreover, the NDV life cycle occurs in the cytoplasm of infected cells, and in contrast to DNA virus vectors, NDV never integrates into the host cell DNA, eliminating potential oncogenic properties associated with viral vectors with a DNA-integrating phase. Another advantage derived from the cytoplasmic replication of NDV is that it overcomes the inefficient transcription and export from the nucleus of some HIV mRNA (including Gag mRNA) due to the presence of Rev response elements, a problem widely reported for the production of DNA vaccines against HIV (47). Finally, genetic recombination is rare or does not occur in negative-strand RNA viruses, alleviating concerns for vector stability.
Safety is always important for vaccine development and is most critical in an HIV vaccine. NDV is nonpathogenic in humans, with only a few reported cases of naturally occurring human infections. The virus has been reported to cause benign conjunctivitis in people in close contact with infected birds (3). There is no evidence of human-human transmission. NDV has been utilized, due to its oncolytic properties, in human studies over several decades and has been shown to be safe even in immunosuppressed patients (5, 21). In recent studies, the intravenous serial administration of high doses of NDV has led to mild side effects, including influenza-like symptoms and hematologic effects, consistent with the production of proinflammatory cytokines after administration (34, 45, 61). These side effects disappeared in subsequent administrations even with the use of amounts five times higher than the first dose, confirming the safety of NDV in humans.
In contrast to the case with other viruses used in the design of HIV vaccines, such as adenoviruses (Ad5) or human paramyxoviruses (measles virus and mumps virus), most humans do not have preexisting immunity to NDV, an avian virus. NDV replication is limited in mammalian cells; however, it has been reported that NDV is a potent inducer of humoral and cellular responses in mice against foreign antigens, including the influenza HA, simian immunodeficiency virus (SIV) Gag, or respiratory syncytial virus F protein (48, 51, 52). Recently it has been shown that NDV is also highly immunogenic in primates (12, 16). NDV has been studied as an inducer of type I IFN synthesis in vitro and in vivo (18, 41, 57). The ability of NDV to induce high levels of type I IFN locally might correlate with its high immunogenic properties, since type I IFN is known to be an adjuvant for the induction of humoral and cellular immune responses (9, 46, 68, 69). In addition, it has been reported that NDV induces activation of dendritic cells (DCs). DCs are antigen-presenting cells that play an important role in the induction of T-cell responses in vivo. This activation of DCs is at least partially related to NDV's ability to induce IFN (48, 69, 72). It has been found in hu-PBL-SCID mice that DCs activation by IFN followed by stimulation with inactivated HIV are able to generate higher HIV-specific CD8+ T-cell and humoral responses than DCs pretreated with other inducers of maturation, like IL-4. Moreover, the immune response correlates with protection from HIV challenge in these mice (43, 44). The potential of NDV to induce activation of DCs and IFN production in vivo may allow this viral vector to act as a potent adjuvant in the generation of the immune response against NDV-encoded antigens.
The potential importance of Gag as an immunogen has been suggested in numerous studies for several reasons. First, Gag is a highly conserved protein among different HIV strains compared with other potential antigens, such as the Env protein. Second, gag is one of the most immunodominant genes of HIV. The relationship between the plasma viral load and the host cellular immune response has remained controversial, since some studies have established a correlation between low viral loads and cellular responses against Gag, Pol, or Nef while other studies found no correlation or even an inverse correlation (1, 55, 56). Recent studies provide evidences that Gag-specific CD8+ T-cell responses could be effective in the control of chronic HIV infection and the maintenance of immune responses in HIV-infected individuals in comparison with other potential HIV antigens (23, 38). Thus, we selected Gag as the antigen in our study. Nevertheless, NDV-HIVGag could be used in combination with NDVs expressing other HIV antigens, like Nef, Pol, or Env, in an HIV vaccine approach.
The limited replication of NDV in mammalian species is a guarantee of virus safety but at the same time limits the production of viral and foreign proteins. These characteristics of NDV require the optimization of the conditions for administration of the vaccine and expression of the antigen. The selection of a route of administration was based on a previous study comparing different administration routes for NDV expressing SIV Gag in a mouse model (intravenous, intraperitoneal, and intranasal). Every route was able to generate an immune response in mice, but the best results were obtained with intranasal immunizations (52). In addition, intranasal or respiratory tract routes have been tested in nonhuman primates in immunizations with NDV expressing influenza HA, human parainfluenza virus 3 HN, or severe acute respiratory syndrome virus Spike protein (12, 15, 16, 52). The safety profile of NDV allows the administration of high doses of NDV with few undesirable effects.
The optimization of protein expression is another important issue. One would predict that the higher the protein expression achieved by the vaccine vector, the higher the immune response elicited against the expressed protein. We have investigated the best location of the gag gene in the NDV genome for higher levels of the Gag protein in NDV-infected cells. The NDV genome contains six transcriptional units, NP-P-M-F-HN-L. It is known that paramyxovirus polymerases gain access to the viral genes through the 3′ end of the viral genome. NP is the first transcriptional unit to be transcribed. Then, the polymerase skips the intergenic region and reinitiates mRNA synthesis of the second transcriptional unit at the start site. This sequential mechanism continues across the viral genome. The jump between transcriptional units is not always successful, and this imperfect reinitiation leads to a gradient in the expression of NDV genes according to the distance from the genome 3′ end (37). According to this strategy, the optimal place to insert an antigen in the NDV genome would appear to be the closest to the 3′end. On the other hand, the insertion of a foreign gene should affect the transcription of the genes located after the insertion place, and these changes could affect virus fitness and reduce viral replication, resulting in lower levels of antigen expression. We inserted HIV gag into every transcription unit throughout the NDV genome except behind the L gene, due to the low expression of protein associated with this position (76).
We rescued recombinant viruses expressing HIV Gag from every position except in front of NP. In previous studies, Huang and collaborators described the rescue of the NDV LaSota strain expressing two different genes in front of NP with only a slight effect on virus growth (35, 36). In our case, the inability to rescue a virus with HIV gag in this position might be related to the insert size (1.5 kb, larger than previous genes introduced at this position). Moreover, we are using a different strain of NDV, Hitchner B1, and specific characteristics of the strain may explain the inability to rescue the virus.
The rescued recombinant viruses grew to similar levels in embryonated eggs or Vero cells and expressed HIV Gag in a stable manner. A gradient in the expression of HIV Gag was detected by Western blotting in Vero cells infected with the different recombinant viruses. The highest level of expression of HIV Gag was obtained after insertion of HIV gag between the P and M genes. The level of expression was inversely correlated with the proximity to the 5′ end of NDV genome, except that very low levels of HIV Gag expression were detected after insertion of the gene between the NP and P genes. At late time points, differences in HIV Gag expression are less remarkable, probably due to the accumulation of the protein, making it difficult to establish differences among the viruses (Fig. 4B). The delay in HIV Gag production observed in NDV-HIVGag-SacII-infected cells at early points together with limited replication of the NDV in mice might be the cause of the poor immune responses generated by this virus. The insertion of a foreign gene in the NDV genome could alter the stoichiometry of expression of viral proteins and affect NDV replication. It has been shown in vesicular stomatitis virus that the ratio between these proteins may be critical for optimal replication of the virus (60, 75). In addition, Zhao and Peeters have shown low expression of alkaline phosphatase when the gene is inserted between the NP and P genes compared with results for other locations (76). These earlier data together with our results would confirm that insertion of a foreign gene in this position may not be the optimal place to achieve high levels of expression. Since we did not observe differences in growth levels in embryonated eggs or Vero cells between the different NDV-HIVGag viruses, it is possible that there is an inefficient use of the gene start and gene end signals for expression of Gag mRNA when the gene is located between the NP and P genes.
Previous results in our laboratory showed the ability of NDV to generate specific cellular responses against SIV Gag in mice immunized with NDV-SIVGag. This immune response was improved in priming/boosting combinations with an influenza virus expressing SIV Gag (52). Priming/boosting combinations of different vectors appear to offer an efficient way to increase the immune responses in a vaccinee (52, 53). We now describe the optimal location of the Gag antigen gene in the NDV genome in order to achieve a high level of immune response. Virus expressing HIV gag between P and M generated the best cellular immune response, measured by the production of IFN-γ by specific HIV Gag CD8+ T cells and by protection after challenge with Vac-HIVGag. We found a correlation between the levels of protein expression and the induction of cellular protective responses. We also found that Gag-specific antiviral immunity induced by NDV-HIVGag was detected 7 weeks after the last immunization, especially in NDV-HIVGag-XbaI-immunized mice. These results suggest that the immunity induced by NDV may last for a long time in mammals.
Another strategy we used to optimize the expression of the antigen by NDV was to increase its translational efficiency using a mammalian codon-optimized version of HIV gag. Optimized codon usage has been an effective method to increase the antigen production and immunogenicity of DNA vaccines (22, 25, 65). After the insertion of codon-optimized HIV gag in NDV (NDV-HIVGag-opt), we obtained a further increase in the expression of HIV Gag in NDV-infected cells. This correlated with a higher level of protection in immunized mice after challenge with vaccinia virus expressing HIV Gag. Increases in the cellular immune responses were also detected. Moreover, 8 days after immunization with NDV-HIVGag-opt, we observed a specific anti-Gag cytotoxic activity in vivo, while we could not detect this in animals immunized with NDV-HIVGag-XbaI even if we were able to see some Gag-specific CD8+ activity in vitro 10 days after priming by ELISPOT (Fig. 9B). That could be due to different sensitivities between the two techniques.
Finally, we studied the kinetic of generation of the cellular responses induced after vaccination with four viruses NDV-HIVGag-SacII, NDV-HIVGag-XbaI, NDV-HIVGag-opt, and NDV-GFP, used as a control. In response to immunization, antigen-specific CD8+ T cells proliferated, and we observed an increase in the number of the cells 10 days after the prime, 7 days after the boost, and 5 days after challenge with vaccinia. The magnitude of the response was increase after the boost and later on by the vaccinia HIV Gag challenge, most likely due to the recall effect after restimulation with the antigen. After boosting, the numbers of antigen-specific CD8+ T cells underwent a decline during the “cell death” or “contraction” phase, which eliminated a big percentage of the antigen-specific CD8+ T cells. The elimination of the cells was not complete, and a small number of the cells stayed as memory CD8+ T cells, a pattern observed at 14 and 28 days after the boost. The amount of the CD8 response generated by immunization with NDV-HIVGag-opt was substantially bigger than those with immunization with NDV-HIVGag-XbaI and NDV-HIVGag-SacII.
In summary, we have described the successful recovery of NDV viruses expressing HIV Gag at different intergenic locations within the virus genome. We showed that these viruses are safe and immunogenic in mice and determined the optimal location for insertion of the Gag antigen gene for the maximal cellular immune response. Moreover, we have described how the use of a mammalian codon-optimized gene in the optimal location can increase considerably the immune response generated against the antigen HIV Gag. These results support the use of NDV as a vaccine vector for further studies aimed at the development of an effective HIV vaccine.
Acknowledgments
We thank Richard Cadagan for expert technical assistance and Sharon Czelusniak for expertise in animal care. We also thank Peter Palese, Eric Bortz, Donna Tscherne, and Alan Belicha for helpful discussions and proofreading of the manuscript. We thank members of the P. Heeger lab and specially Denise Richards for using the ELISPOT reader and M. J. Schnell and Y. Huang for reagents used in our study. The monoclonal antibody to HIV-1 p24 (71-31) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Susan Zolla-Pazner. The HIV-1 p24 Gag monoclonal antibody (no. 24-4) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Michael H. Malim.
This study was partially supported by a Fundacion Ramon Areces postdoctoral fellowship to E.C. and by a grant from the Bill and Melinda Gates Foundation (to A.G.-S. and T.M.).
Footnotes
Published ahead of print on 12 November 2008.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 772081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, D. 1997. Newscastle disease and other avian Paramyxoviridae infections, p. 541-569. In B. W. Calnek et al. (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames.
- 3.Alexander, D. J. 2000. Newcastle disease and other avian paramyxoviruses. Rev. Sci. Tech. 19443-462. [DOI] [PubMed] [Google Scholar]
- 4.Baba, T. W., V. Liska, A. H. Khimani, N. B. Ray, P. J. Dailey, D. Penninck, R. Bronson, M. F. Greene, H. M. McClure, L. N. Martin, and R. M. Ruprecht. 1999. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat. Med. 5194-203. [DOI] [PubMed] [Google Scholar]
- 5.Batliwalla, F. M., B. A. Bateman, D. Serrano, D. Murray, S. Macphail, V. C. Maino, J. C. Ansel, P. K. Gregersen, and C. A. Armstrong. 1998. A 15-year follow-up of AJCC stage III malignant melanoma patients treated postsurgically with Newcastle disease virus (NDV) oncolysate and determination of alterations in the CD8 T cell repertoire. Mol. Med. 4783-794. [PMC free article] [PubMed] [Google Scholar]
- 6.Berkley, S. F., and W. C. Koff. 2007. Scientific and policy challenges to development of an AIDS vaccine. Lancet 37094-101. [DOI] [PubMed] [Google Scholar]
- 7.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 7511983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 1074781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonjardim, C. A. 2005. Interferons (IFNs) are key cytokines in both innate and adaptive antiviral immune responses—and viruses counteract IFN action. Microbes Infect. 7569-578. [DOI] [PubMed] [Google Scholar]
- 10.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 686103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3205-211. [DOI] [PubMed] [Google Scholar]
- 12.Bukreyev, A., Z. Huang, L. Yang, S. Elankumaran, M. St Claire, B. R. Murphy, S. K. Samal, and P. L. Collins. 2005. Recombinant Newcastle disease virus expressing a foreign viral antigen is attenuated and highly immunogenic in primates. J. Virol. 7913275-13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bukreyev, A., M. H. Skiadopoulos, B. R. Murphy, and P. L. Collins. 2006. Nonsegmented negative-strand viruses as vaccine vectors. J. Virol. 8010293-10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Currier, J. R., E. G. Kuta, E. Turk, L. B. Earhart, L. Loomis-Price, S. Janetzki, G. Ferrari, D. L. Birx, and J. H. Cox. 2002. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J. Immunol. Methods 260157-172. [DOI] [PubMed] [Google Scholar]
- 15.DiNapoli, J. M., A. Kotelkin, L. Yang, S. Elankumaran, B. R. Murphy, S. K. Samal, P. L. Collins, and A. Bukreyev. 2007. Newcastle disease virus, a host range-restricted virus, as a vaccine vector for intranasal immunization against emerging pathogens. Proc. Natl. Acad. Sci. USA 1049788-9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiNapoli, J. M., L. Yang, A. Suguitan, Jr., S. Elankumaran, D. W. Dorward, B. R. Murphy, S. K. Samal, P. L. Collins, and A. Bukreyev. 2007. Immunization of primates with a Newcastle disease virus-vectored vaccine via the respiratory tract induces a high titer of serum neutralizing antibodies against highly pathogenic avian influenza virus. J. Virol. 8111560-11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duerr, A., J. N. Wasserheit, and L. Corey. 2006. HIV vaccines: new frontiers in vaccine development. Clin. Infect. Dis. 43500-511. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Sesma, A., S. Marukian, B. J. Ebersole, D. Kaminski, M. S. Park, T. Yuen, S. C. Sealfon, A. Garcia-Sastre, and T. M. Moran. 2006. Influenza virus evades innate and adaptive immunity via the NS1 protein. J. Virol. 806295-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 164531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. StJohn, A. Khatri, K. Davis, J. Mullins, P. J. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 782187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman, A. I., Z. Zakay-Rones, J. M. Gomori, E. Linetsky, L. Rasooly, E. Greenbaum, S. Rozenman-Yair, A. Panet, E. Libson, C. S. Irving, E. Galun, and T. Siegal. 2006. Phase I/II trial of intravenous NDV-HUJ oncolytic virus in recurrent glioblastoma multiforme. Mol. Ther. 13221-228. [DOI] [PubMed] [Google Scholar]
- 22.Frelin, L., G. Ahlen, M. Alheim, O. Weiland, C. Barnfield, P. Liljestrom, and M. Sallberg. 2004. Codon optimization and mRNA amplification effectively enhances the immunogenicity of the hepatitis C virus nonstructural 3/4A gene. Gene Ther. 11522-533. [DOI] [PubMed] [Google Scholar]
- 23.Fu, T. M., S. A. Dubey, D. V. Mehrotra, D. C. Freed, W. L. Trigona, L. Adams-Muhler, J. H. Clair, T. G. Evans, R. Steigbigel, J. M. Jacobson, P. A. Goepfert, M. J. Mulligan, S. A. Kalams, C. Rinaldo, L. Zhu, K. S. Cox, L. Guan, R. Long, N. Persaud, M. J. Caulfield, J. C. Sadoff, E. A. Emini, S. Thaler, and J. W. Shiver. 2007. Evaluation of cellular immune responses in subjects chronically infected with HIV type 1. AIDS Res. Hum. Retrovir. 2367-76. [DOI] [PubMed] [Google Scholar]
- 24.Gallili, G. E., and D. Ben-Nathan. 1998. Newcastle disease vaccines. Biotechnol. Adv. 16343-366. [DOI] [PubMed] [Google Scholar]
- 25.Gao, F., Y. Li, J. M. Decker, F. W. Peyerl, F. Bibollet-Ruche, C. M. Rodenburg, Y. Chen, D. R. Shaw, S. Allen, R. Musonda, G. M. Shaw, A. J. Zajac, N. Letvin, and B. H. Hahn. 2003. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retrovir. 19817-823. [DOI] [PubMed] [Google Scholar]
- 26.Gao, Q., M. S. Park, and P. Palese. 2008. Expression of transgenes from newcastle disease virus with a segmented genome. J. Virol. 822692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garber, D. A., and M. B. Feinberg. 2003. AIDS vaccine development: the long and winding road. AIDS Rev. 5131-139. [PubMed] [Google Scholar]
- 28.Ge, J., G. Deng, Z. Wen, G. Tian, Y. Wang, J. Shi, X. Wang, Y. Li, S. Hu, Y. Jiang, C. Yang, K. Yu, Z. Bu, and H. Chen. 2007. Newcastle disease virus-based live attenuated vaccine completely protects chickens and mice from lethal challenge of homologous and heterologous H5N1 avian influenza viruses. J. Virol. 81150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorny, M. K., V. Gianakakos, S. Sharpe, and S. Zolla-Pazner. 1989. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 861624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goulder, P. J., M. A. Altfeld, E. S. Rosenberg, T. Nguyen, Y. Tang, R. L. Eldridge, M. M. Addo, S. He, J. S. Mukherjee, M. N. Phillips, M. Bunce, S. A. Kalams, R. P. Sekaly, B. D. Walker, and C. Brander. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3212-217. [DOI] [PubMed] [Google Scholar]
- 32.Harty, J. T., and V. P. Badovinac. 2008. Shaping and reshaping CD8+ T-cell memory. Nat. Rev. Immunol. 8107-119. [DOI] [PubMed] [Google Scholar]
- 33.Hendel, H., S. Caillat-Zucman, H. Lebuanec, M. Carrington, S. O'Brien, J. M. Andrieu, F. Schachter, D. Zagury, J. Rappaport, C. Winkler, G. W. Nelson, and J. F. Zagury. 1999. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J. Immunol. 1626942-6946. [PubMed] [Google Scholar]
- 34.Hotte, S. J., R. M. Lorence, H. W. Hirte, S. R. Polawski, M. K. Bamat, J. D. O'Neil, M. S. Roberts, W. S. Groene, and P. P. Major. 2007. An optimized clinical regimen for the oncolytic virus PV701. Clin. Cancer Res. 13977-985. [DOI] [PubMed] [Google Scholar]
- 35.Huang, Z., S. Elankumaran, A. S. Yunus, and S. K. Samal. 2004. A recombinant Newcastle disease virus (NDV) expressing VP2 protein of infectious bursal disease virus (IBDV) protects against NDV and IBDV. J. Virol. 7810054-10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, Z., S. Krishnamurthy, A. Panda, and S. K. Samal. 2001. High-level expression of a foreign gene from the most 3′-proximal locus of a recombinant Newcastle disease virus. J. Gen. Virol. 821729-1736. [DOI] [PubMed] [Google Scholar]
- 37.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23477-484. [DOI] [PubMed] [Google Scholar]
- 38.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 1346-53. [DOI] [PubMed] [Google Scholar]
- 39.Kotsopoulou, E., V. N. Kim, A. J. Kingsman, S. M. Kingsman, and K. A. Mitrophanous. 2000. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 744839-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 684650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumagai, Y., O. Takeuchi, H. Kato, H. Kumar, K. Matsui, E. Morii, K. Aozasa, T. Kawai, and S. Akira. 2007. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27240-252. [DOI] [PubMed] [Google Scholar]
- 42.Lamb, R., and G. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1450-1496. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott-Raven, Philadelphia, PA.
- 43.Lapenta, C., S. M. Santini, M. Logozzi, M. Spada, M. Andreotti, T. Di Pucchio, S. Parlato, and F. Belardelli. 2003. Potent immune response against HIV-1 and protection from virus challenge in hu-PBL-SCID mice immunized with inactivated virus-pulsed dendritic cells generated in the presence of IFN-alpha. J. Exp. Med. 198361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapenta, C., S. M. Santini, M. Spada, S. Donati, F. Urbani, D. Accapezzato, D. Franceschini, M. Andreotti, V. Barnaba, and F. Belardelli. 2006. IFN-alpha-conditioned dendritic cells are highly efficient in inducing cross-priming CD8(+) T cells against exogenous viral antigens. Eur. J. Immunol. 362046-2060. [DOI] [PubMed] [Google Scholar]
- 45.Laurie, S. A., J. C. Bell, H. L. Atkins, J. Roach, M. K. Bamat, J. D. O'Neil, M. S. Roberts, W. S. Groene, and R. M. Lorence. 2006. A phase 1 clinical study of intravenous administration of PV701, an oncolytic virus, using two-step desensitization. Clin. Cancer Res. 122555-2562. [DOI] [PubMed] [Google Scholar]
- 46.Le Bon, A., and D. F. Tough. 2002. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 14432-436. [DOI] [PubMed] [Google Scholar]
- 47.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338254-257. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Sobrido, L., N. Gitiban, A. Fernandez-Sesma, J. Cros, S. E. Mertz, N. A. Jewell, S. Hammond, E. Flano, R. K. Durbin, A. Garcia-Sastre, and J. E. Durbin. 2006. Protection against respiratory syncytial virus by a recombinant Newcastle disease virus vector. J. Virol. 801130-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masemola, A., T. Mashishi, G. Khoury, P. Mohube, P. Mokgotho, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. A. Karim, H. W. Sheppard, and C. M. Gray. 2004. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J. Virol. 783233-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McMichael, A. 2003. Prospects for an AIDS vaccine. Clin. Med. 3269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakaya, T., J. Cros, M. S. Park, Y. Nakaya, H. Zheng, A. Sagrera, E. Villar, A. Garcia-Sastre, and P. Palese. 2001. Recombinant Newcastle disease virus as a vaccine vector. J. Virol. 7511868-11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakaya, Y., T. Nakaya, M. S. Park, J. Cros, J. Imanishi, P. Palese, and A. Garcia-Sastre. 2004. Induction of cellular immune responses to simian immunodeficiency virus Gag by two recombinant negative-strand RNA virus vectors. J. Virol. 789366-9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakaya, Y., H. Zheng, and A. Garcia-Sastre. 2003. Enhanced cellular immune responses to SIV Gag by immunization with influenza and vaccinia virus recombinants. Vaccine 212097-2106. [DOI] [PubMed] [Google Scholar]
- 54.Nemunaitis, J. 2002. Live viruses in cancer treatment. Oncology 161483-1492, 1495-1497. [PubMed] [Google Scholar]
- 55.Novitsky, V., P. Gilbert, T. Peter, M. F. McLane, S. Gaolekwe, N. Rybak, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 2792103-2106. [DOI] [PubMed] [Google Scholar]
- 57.Park, M. S., A. Garcia-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 779522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 771501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park, M. S., J. Steel, A. Garcia-Sastre, D. Swayne, and P. Palese. 2006. Engineered viral vaccine constructs with dual specificity: avian influenza and Newcastle disease. Proc. Natl. Acad. Sci. USA 1038203-8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pattnaik, A. K., and G. W. Wertz. 1990. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J. Virol. 642948-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pecora, A. L., N. Rizvi, G. I. Cohen, N. J. Meropol, D. Sterman, J. L. Marshall, S. Goldberg, P. Gross, J. D. O'Neil, W. S. Groene, M. S. Roberts, H. Rabin, M. K. Bamat, and R. M. Lorence. 2002. Phase I trial of intravenous administration of PV701, an oncolytic virus, in patients with advanced solid cancers. J. Clin. Oncol. 202251-2266. [DOI] [PubMed] [Google Scholar]
- 62.Peeters, B. P., O. S. de Leeuw, G. Koch, and A. L. Gielkens. 1999. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J. Virol. 735001-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peeters, B. P., O. S. de Leeuw, I. Verstegen, G. Koch, and A. L. Gielkens. 2001. Generation of a recombinant chimeric Newcastle disease virus vaccine that allows serological differentiation between vaccinated and infected animals. Vaccine 191616-1627. [DOI] [PubMed] [Google Scholar]
- 64.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 941890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramakrishna, L., K. K. Anand, K. M. Mohankumar, and U. Ranga. 2004. Codon optimization of the Tat antigen of human immunodeficiency virus type 1 generates strong immune responses in mice following genetic immunization. J. Virol. 789174-9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Riviere, Y., M. B. McChesney, F. Porrot, F. Tanneau-Salvadori, P. Sansonetti, O. Lopez, G. Pialoux, V. Feuillie, M. Mollereau, S. Chamaret, et al. 1995. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res. Hum Retrovir. 11903-907. [DOI] [PubMed] [Google Scholar]
- 67.Romer-Oberdorfer, A., E. Mundt, T. Mebatsion, U. J. Buchholz, and T. C. Mettenleiter. 1999. Generation of recombinant lentogenic Newcastle disease virus from cDNA. J. Gen. Virol. 802987-2995. [DOI] [PubMed] [Google Scholar]
- 68.Santini, S. M., T. Di Pucchio, C. Lapenta, S. Parlato, M. Logozzi, and F. Belardelli. 2002. The natural alliance between type I interferon and dendritic cells and its role in linking innate and adaptive immunity. J. Interferon Cytokine Res. 221071-1080. [DOI] [PubMed] [Google Scholar]
- 69.Santini, S. M., C. Lapenta, M. Logozzi, S. Parlato, M. Spada, T. Di Pucchio, and F. Belardelli. 2000. Type I interferon as a powerful adjuvant for monocyte-derived dendritic cell development and activity in vitro and in Hu-PBL-SCID mice. J. Exp. Med. 1911777-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneider, R., M. Campbell, G. Nasioulas, B. K. Felber, and G. N. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 714892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simon, J. H., R. A. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 715259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tough, D. F. 2004. Type I interferon as a link between innate and adaptive immunity through dendritic cell stimulation. Leuk. Lymphoma 45257-264. [DOI] [PubMed] [Google Scholar]
- 73.UNAIDS. 2006. “UNAIDS/WHO” AIDS epidemic update. UNAIDS/WHO, Geneva, Switzerland.
- 74.Vigil, A., M. S. Park, O. Martinez, M. A. Chua, S. Xiao, J. F. Cros, L. Martinez-Sobrido, S. L. Woo, and A. Garcia-Sastre. 2007. Use of reverse genetics to enhance the oncolytic properties of Newcastle disease virus. Cancer Res. 678285-8292. [DOI] [PubMed] [Google Scholar]
- 75.Wertz, G. W., R. Moudy, and L. A. Ball. 2002. Adding genes to the RNA genome of vesicular stomatitis virus: positional effects on stability of expression. J. Virol. 767642-7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao, H., and B. P. Peeters. 2003. Recombinant Newcastle disease virus as a viral vector: effect of genomic location of foreign gene on gene expression and virus replication. J. Gen. Virol. 84781-788. [DOI] [PubMed] [Google Scholar]