Abstract
VP22, encoded by the UL49 gene, is one of the most abundant proteins of the herpes simplex virus 1 (HSV-1) tegument. In the present study we show VP22 is required for optimal protein synthesis at late times in infection. Specifically, in the absence of VP22, viral proteins accumulated to wild-type levels until ∼6 h postinfection. At that time, ongoing synthesis of most viral proteins dramatically decreased in the absence of VP22, whereas protein stability was not affected. Of the individual proteins we assayed, VP22 was required for optimal synthesis of the late viral proteins gE and gD and the immediate-early protein ICP0 but did not have discernible effects on accumulation of the immediate-early proteins ICP4 or ICP27. In addition, we found VP22 is required for the accumulation of a subset of mRNAs to wild-type levels at early, but not late, times in infection. Specifically, the presence of VP22 enhanced the accumulation of gE and gD mRNAs until ∼9 h postinfection, but it had no discernible effect at later times in infection. Also, VP22 did not significantly affect ICP0 mRNA at any time in infection. Thus, the protein synthesis and mRNA phenotypes observed with the UL49-null virus are separable with regard to both timing during infection and the genes affected and suggest separate roles for VP22 in enhancing the accumulation of viral proteins and mRNAs. Finally, we show that VP22's effects on protein synthesis and mRNA accumulation occur independently of mutations in genes encoding the VP22-interacting partners VP16 and vhs.
Herpes simplex virus 1 (HSV-1) virions are composed of a nucleocapsid enclosing the double-stranded linear DNA genome, a proteinaceous layer termed the tegument that surrounds the nucleocapsid, and a host-derived lipid membrane envelope that contains viral glycoproteins. The tegument is unique to herpesviruses and is composed of at least 20 different viral proteins of varied stoichiometries. Tegument proteins have been shown to play a variety of roles in infection, including the regulation of viral and host gene expression and the promotion of virus assembly and egress (1, 16, 34, 38). Tegument proteins enter the cell upon fusion of the viral envelope with the host cell membrane during infection initiation. Thus, these proteins provide a potential means to modulate the host cell and advance viral infection at both very early times in infection upon tegument delivery and at late times in infection, when the tegument proteins are produced in high amounts.
VP22, encoded by the UL49 gene, is one of the most abundant HSV-1 tegument proteins, with an average stoichiometry of 2,000 copies per virion. A number of functions have been attributed to VP22, including association with and reorganization of microtubules in infected and uninfected cells (10, 22) and incorporation of RNA into the virion (42). In previous studies, UL49 truncation or deletion mutants propagated on VP22-expressing cell lines were shown to be significantly debilitated in production of infectious virus on noncomplementing cells at low multiplicities of infection and produced plaques of significantly smaller size (6, 35). Further characterization revealed significant defects in release of infectious virus into the medium in the absence of VP22, thereby decreasing viral spread (6). Virions produced by this UL49− virus upon infection of noncomplementing cells contained decreased levels of ICP0, gD, and gE. These observations are potentially consistent with reports that VP22 interacts with the cytoplasmic tails of gD and gE (4, 14, 33) and is necessary for localization of ICP0 to putative sites of viral assembly within the cytoplasm (8). In contrast to the small plaque phenotype displayed by the UL49− virus on noncomplementing cells after propagation on complementing cells, passage of the UL49− virus on noncomplementing cells rapidly and consistently caused the normally defective virus to produce plaques of wild-type size (5). These data suggest a strong selection for a secondary compensatory mutation(s) that rescues the UL49− small plaque phenotype.
Also pertinent to this report is the observation that VP22 interacts with VP16 (9). VP16 was one of the first transcription factors described and binds host octamer transcription factor 1 and other host factors to mediate transcription from viral immediate-early promoters expressed in the absence of viral protein production (23-26, 40). Thus, VP16 within the tegument is delivered to the nucleus upon initiation of infection to mediate preferential transcription of viral mRNAs over those of the host.
Previous studies indicated that VP16 and the VP16/VP22 complex can interact with the virion host shutoff protein (vhs) (46). vhs, encoded by the UL41 gene, is a riboendonuclease that specifically cleaves host and viral mRNAs (7, 13, 27, 28, 38, 45, 48). The RNase activity of vhs is believed to promote expression of viral proteins early in infection by degrading host mRNAs. Likewise, vhs is believed to promote the transition from the early phase of viral transcription to the late phase by degrading early viral mRNAs at the appropriate time in infection. In contrast, the production of VP16 late in infection suppresses the RNase activity of vhs (29, 41, 44), suggesting a mechanism to ensure appropriate accumulation of late transcripts. Interestingly, a previously described UL49-null mutant derived from an HSV-1 bacterial artificial chromosome accumulated deletion and frameshift mutations in UL41 when propagated on noncomplementing cells (43), suggesting the presence of vhs was detrimental in the absence of VP22.
The present studies were initiated to further characterize the phenotype of the UL49− virus we described previously (6). The data presented herein show VP22 is required for optimal protein synthesis at late times in infection and the accumulation of at least gE, gD, and vhs mRNAs at early times in infection. Interestingly, the protein and mRNA phenotypes are separable with regard to both timing during infection and the genes affected, suggesting separate roles for VP22 in both protein and mRNA accumulation. Moreover, these effects were mediated in the absence of mutations in VP16 or vhs.
MATERIALS AND METHODS
Viruses and cells.
Viral stocks of wild-type HSV-1(F) (wild type [WT]), the UL49 deletion virus (UL49−), and the UL49 repair viruses (UL49R) described previously (6) were propagated exclusively on V49 cells, a Vero-derived cell line that constitutively expresses VP22 (35). Vero and V49 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 4.0 mM l-glutamine, 4.5 g/liter glucose, 125 units/ml penicillin, 0.125 mg/ml streptomycin, and either 10% newborn calf serum or 10% fetal bovine serum, respectively.
Time course of protein synthesis.
Six-well plates of Vero cells were infected with WT, UL49−, or UL49R viruses in medium 199V at a multiplicity of infection (MOI) of 10 at 4°C for 1 hour to allow viral attachment. Virus-containing medium was then replaced with fresh medium 199 supplemented with 1% newborn calf serum (medium 199V), and the plates were shifted to 37°C to allow synchronization of infection. At various times after the 37°C shift, the medium overlying each well was replaced with DMEM (high glucose, with pyridoxine hydrochloride, no l-glutamine, no l-methionine, no l-cysteine) supplemented with 20 μCi/ml Trans35S-label ([35S]methionine/cysteine; MP Biomedicals, Solon, OH). Cells were labeled for the time periods shown in the figures and figure legends. At the end of each labeling period the media were removed and the cells were collected, washed with phosphate-buffered saline (PBS), resuspended in 150 μl sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (50 mM Tris-Cl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol), boiled for 10 min, and sonicated briefly. Radiolabeled lysates were separated by SDS-12% PAGE and transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA). Labeled proteins were visualized by autoradiography on Pierce CL-X Posure film (Pierce Biotechnology, Rockford, IL). All time course experiments were performed in duplicate. A 10-μl aliquot of each 35S-labeled cell lysate was added to 5 ml scintillation fluid (Ecoscint; National Diagnostics, Atlanta, GA), and the amount of radiolabeled methionine and cysteine in the samples was determined with a Beckman scintillation counter.
Pulse-chase analyses.
Six-well plates of Vero cells were infected with WT, UL49−, or UL49R viruses in medium 199V at an MOI of 10 at 4°C for 1 hour to allow viral attachment. Virus-containing media were then replaced with fresh medium 199V and the plates were shifted to 37°C to allow synchronization of infection. Eight hours after the 37°C shift, the medium overlying each well was replaced with warm DMEM (high glucose, with pyridoxine hydrochloride, no l-glutamine, no l-methionine, no l-cysteine) supplemented with 20 μCi/ml Trans35S-label ([35S]methionine/cysteine). Cells were labeled for 15 min, the 35S-containing medium was removed, and the cells were overlaid with fresh, warm medium 199V. At 0, 15, 30, 60, and 120 min postpulse, the cells were collected, washed with PBS, resuspended in 150 μl SDS-PAGE sample buffer, boiled for 10 min, and sonicated briefly. Radiolabeled lysates were separated by SDS-12% PAGE and transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA). Labeled proteins were visualized by autoradiography on Pierce CL-X Posure film (Pierce Biotechnology, Rockford, IL). All pulse-chase analyses were performed in duplicate. A 10-μl aliquot of each 35S-labeled cell lysate was added to 5 ml scintillation fluid (Ecoscint; National Diagnostics, Atlanta, GA) and analyzed on a Beckman scintillation counter to determine the amount of radiolabeled methionine and cysteine incorporated.
Immunoblot analyses.
The 35S-labeled cell lysates prepared above for the protein synthesis time course experiments were used in the immunoblot analyses. Cell lysates were separated by SDS-12% PAGE and the proteins were transferred to nitrocellulose or polyvinylidene difluoride membranes. Free binding sites on the membranes were blocked by 2% bovine serum albumin in PBS before addition of either anti-ICP0 (1:5,000; Abcam, Cambridge, MA), anti-ICP4 (1:500; Rumbaugh-Goodwin Institute for Cancer Research, Plantation, FL), anti-gE (20) (diluted 1:2,500), anti-gD (1:1,000; Rumbaugh-Goodwin Institute for Cancer Research, Plantation, FL), anti-β-actin (0.5 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA), anti-poly(ADP-ribose) polymerase (PARP; 1:1,000; Cell Signaling Technology, Danvers, MA), anti-ICP27 (1:5,000; Virusys Corporation, Sykesville, MD), anti-VP16 (1:500; sc-17547; Santa Cruz Biotechnology, Santa Cruz, CA), anti-α subunit of eukaryotic initiation factor 2 (eIF2α) phosphor S51 (1:500; Abcam Incorporated, Cambridge, MA), or anti-vhs (38) (diluted 1:500) antibodies. After extensive washing to remove unbound antibody, bound antibodies were detected with anti-rabbit (Amersham Biosciences, Piscataway, NJ) or anti-goat (Jackson ImmunoResearch Laboratories, West Grove, PA) immunoglobulin G conjugated to horseradish peroxidase and visualized by enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ) autoradiography on Pierce CL-X Posure film (Pierce Biotechnology, Rockford, IL).
Quantitative real-time RT-PCR.
Vero cells were infected with WT, UL49−, or UL49R viruses in medium 199V at an MOI of 10 at 4°C for 1 hour to allow viral attachment. Virus-containing medium was then replaced with fresh medium 199V and the plates were shifted to 37°C to allow synchronization of infection. Total RNA was extracted from the infected cells at 3, 6, 9, and 15 h postinfection using the RNAqueous RNA isolation kit (Ambion, Inc., Austin, TX). Residual DNA was removed by DNase I treatment and DNase I was then removed using DNase inactivation reagent (Ambion, Inc.). Before proceeding to the reverse transcription (RT) reactions, RNA samples were tested for contaminating DNA through trial quantitative PCRs (qPCRs) using primers to both 18S and gE and Maxima Sybr green qPCR master mix (Fermentas Inc., Glen Burnie, MD). All RNA samples were found to be free of amplifiable DNA, as PCR product did not accumulate above the level detected in no-template control reaction mixtures.
Samples (0.5 μg) of DNase I-treated total RNA were used as the templates for single-stranded cDNA synthesis reactions primed with random decamers using the RETRO-script kit (Ambion, Inc.). cDNAs were tested for template quality by constructing standard curves using serial dilutions of each cDNA as templates in qRT-PCRs. All cDNA templates gave an assay efficiency of 90 to 105%, and the R2 of all standard curves was >0.98.
Relative, quantitative real-time RT-PCR was performed to quantify ICP0, gE, gD, and vhs mRNA levels over time using 18S rRNA internal controls as follows. Equal amounts of cDNAs were used in qRT-PCRs containing primers to the gene of interest and, in separate reactions, primers to 18S rRNA as a control to normalize template input. All qRT-PCRs were performed in triplicate using Maxima SYBR green qPCR master mix (Fermentas Inc., Glen Burnie, MD) in a MyiQ real-time PCR detection system (Bio-Rad Laboratories, Inc., Hercules, CA). Primer pair sequences were as follows: gD, 5′-AGGTCTGCGGGGTTGGGATGG-3′ and 5′-GGTGCGACAGGCGGAACAGG-3′; gE, 5′-GCCTCGGTGGTCCTGGTGGTG-3′ and 5′-CTCGTCATTGTCATCCTCGTCGTAATCG-3′; ICP0, 5′-CCTCTCCGCATCACCACAGAAGCC-3′ and 5′-CAGGTCTCGGTCGCAGGGAAACAC-3′; vhs, 5′-ATCCAACACAATATCACAGCCCATCAACAG-3′ and 5′-CGCCAACCTCTATCACACCAACACG-3′; 18S rRNA, 5′-CCAGTAAGTGCGGGTCATAAGC-3′ and 5′-GCCTCACTAAACCATCCAATCGG-3′ (Integrated DNA Technologies, Coralville, IA). SYBR green fluorescence was measured over the course of 40 amplification cycles. For each template quantified, the mean cycle number at which product accumulation entered the linear range (CT) was calculated. Replicate CT values were within 0.4 cycles of each other. The 18S rRNA CT value for a given template was subtracted from the CT obtained for each mRNA of interest (gD, gE, ICP0, and vhs) from the same template to obtain the normalized CT value (ΔCT) for each template. The change was then calculated using the ΔΔCT method relative to wild type.
Viral DNA purification and UL41 and UL48 sequencing.
Viral DNA was purified from plaques produced on Vero cells by the WT, UL49−, and UL49R viruses as well as from two independent large plaques derived from the UL49− virus after two passages on noncomplementing Vero cells. Plaques were picked by Pasteur pipette aspiration through an agarose overlay into 1 ml of medium 199V. Viral DNA was isolated by boiling the plaques for 10 min followed by two phenol-chloroform-isoamyl alcohol (25:24:1) extractions, one chloroform-isoamyl alcohol (24:1) extraction, and ethanol precipitation. Isolated DNAs from individual plaques were used as templates to amplify the upstream, coding, and downstream regions of the UL41 and UL48 genes by PCR. The primers used were 5′-GCAAAAAGCAGTAACCAGGTCCGTCCAGATTCTGC-3′ and 5′-GGAATCCGTCATCCCAACGCGGG-3′, which amplified HSV-1 bp 91061 to 93124 (the UL41 gene runs in reverse orientation from HSV-1 bp 92637 to bp 91170) and 5′-CCACCACCAAATAATCAGACGACAACCGCAGG-3′ and 5′-CGTCGAGTGAAAACTTCCGTACCCAGACAATAAAGCACC-3′, which amplified HSV-1 bp 103482 to 105495 (the UL48 gene runs in reverse orientation from HSV-1 bp 105079 to 103609) (30). The amplified sequences were cloned into a TA cloning vector (Invitrogen, Carlsbad, CA). Clonal plasmid DNA was isolated and used in sequencing reactions with the following primers: bp 91061 For, 5′-GCAAAAAGCAGTAACCAGGTCCGTCCAGATTCTGC; bp 91460 For, 5′-CGTCGTGGATGACGTGGCGTCG; bp 91868 For, 5′-GGACAAAGCGGGCCAGGAACTGG; bp 92247 For, 5′-GCTGGGTCTGGAGTCGGTGATGG; bp 92642 For, 5′-GGTCAATTGTAACTGCGGATCGGCCTAACTAAGG; bp 93123 Rev, 5′-GGAATCCGTCATCCCAACGCGGG; bp 103482 For, 5′-CCACCACCAAATAATCAGACGACAACCGCAGG; bp 103872 For, 5′-GGAGAAAGGACAGGCGCGGAGC; bp 104265 For, 5′-GCTGGAACAGACCCGCCAACTGAC; bp 104693 For, 5′-CGAATGTCGATTTGGGTGCGTTCGG; bp 105050 For, 5′-GCAAACAGCTCGTCGACCAAGAGGTCC; bp 105495 Rev, 5′-CGTCGAGTGAAAACTTCCGTACCCAGACAATAAAGCACC.
In addition, viral DNA was isolated from all viral stocks used throughout this study by three phenol-chloroform-isoamyl alcohol (25:24:1) extractions, one chloroform-isoamyl alcohol (24:1) extraction, and ethanol precipitation. Isolated DNAs from viral stocks were used as templates to amplify and sequence the UL41 gene.
RESULTS
Steady-state levels of ICP0, gE, and gD are decreased at late, but not early, times in UL49−-infected cells.
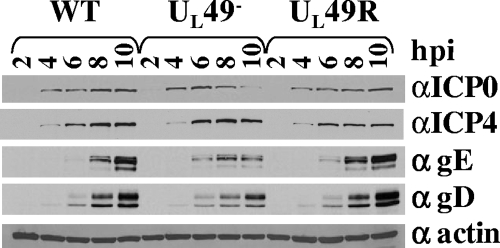
Previous studies showed that virions produced by various HSV-1 UL49− viruses contained decreased amounts of ICP0, ICP4, gE, and gD compared to WT and UL49R virions but contained WT amounts of β-actin (6, 8, 37). One possible reason for this aspect of the UL49-null phenotype is that VP22 is required for physical incorporation of ICP0, ICP4, gE, and gD proteins into virions. Another possibility is that these proteins are not produced to WT levels in the absence of VP22 and therefore cannot be efficiently incorporated into virions. To test the latter possibility, lysates of equal numbers of Vero cells infected with the WT, UL49−, and UL49R viruses for 2, 4, 6, 8, and 10 h were subjected to immunoblotting using antibodies against ICP0, gE, gD, and β-actin. As shown in Fig. 1, steady-state levels of ICP0, ICP4, gE, gD, and β-actin were very similar in WT-, UL49−-, and UL49R-infected cells at 2, 4, and 6 h postinfection. However, at 8 and 10 h postinfection, UL49−-infected cells contained decreased levels of ICP0, gE, and gD but WT levels of ICP4 and β-actin. Thus, VP22 is required for accumulation of ICP0, gE, and gD to WT levels at late, but not early, times in infection.
FIG. 1.
Immunoblot analysis of steady-state ICP0, ICP4, gE, gD, and β-actin protein levels throughout WT, UL49−, and UL49R infections. Lysates of Vero cells infected with the WT, UL49−, or UL49R virus for 2, 4, 6, 8, or 10 h were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed for the presence of ICP0 (top row), gE (second row), gD (third row), and β-actin (bottom row).
VP22 affects global protein synthesis at late times in infection.
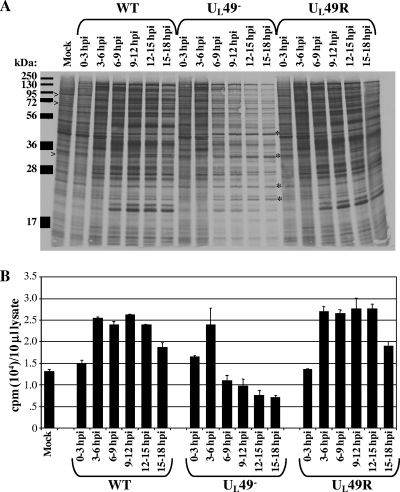
Given the disparate effects on expression of different genes, we next asked whether VP22 affects protein production on a more global scale. Vero cells were mock infected or infected with the WT, UL49−, or UL49R virus for 0, 3, 6, 9, or 12 h. At these time points, infected cells were incubated in medium containing [35S]methionine/cysteine for 3 hours. Following each 35S-labeling period, the medium was removed, cells were washed and lysed, and the labeled proteins were separated by SDS-PAGE followed by electrophoretic transfer and autoradiography to visualize individual proteins. Aliquots of labeled cell lysates were also analyzed by scintillation counting to determine total protein production and accumulation during each labeling period.
As shown in Fig. 2, protein production in WT-, UL49−-, and UL49R-infected cells was very similar from 0 to 3 and 3 to 6 h postinfection with regard to both the individual proteins produced (Fig. 2A) and total protein accumulation (Fig. 2B). By comparing the electrophoretic profiles of labeled polypeptides from WT-, UL49−-, and UL49R-infected cells to those of mock-infected cells, it was also clear that all three viruses were able to mediate shutoff of host protein synthesis (Fig. 2A). These data indicate that the absence of VP22 has virtually no effect on viral protein synthesis at early times in infection or on the onset of host shutoff.
FIG. 2.
Analysis of global protein synthesis and accumulation throughout WT, UL49−, and UL49R infections. Vero cells synchronously infected with the WT, UL49−, and UL49R viruses were incubated in the presence of [35S]methionine/cysteine from 0 to 3, 3 to 6, 6 to 9, 9 to 12, 12 to 15, or 15 to 18 h postinfection (hpi). Following cell washing and lysis, labeled proteins were detected by either autoradiography following SDS-PAGE separation (A) or scintillation counting (B). Example cellular proteins that undergo shutoff in WT-, UL49−-, and UL49R-infected cells are indicated by >, and example proteins whose synthesis does not decrease at late times during UL49− infection are indicated by *.
In contrast to observations at early times, protein production dramatically decreased in UL49−-infected compared to WT- and UL49R-infected cells starting at ∼6 h postinfection. Total 35S incorporation in UL49−-infected cells was 110% and 94% of WT incorporation from 0 to 3 and 3 to 6 h postinfection, respectively, but dropped to 46% from 6 to 9 h postinfection, 37% from 9 to 12 h postinfection, 32% from 12 to 15 h postinfection, and 38% from 15 to 18 h postinfection. Figure 2A shows the majority of individual proteins in UL49−-infected cells were produced in decreased amounts starting at ∼6 h postinfection, corresponding with the drop in total 35S incorporation measured by scintillation counting. Interestingly, production of a few proteins was unaffected by the absence of VP22 (Fig. 2A). These data show VP22 is necessary for accumulation of most proteins to WT levels at late times in infection.
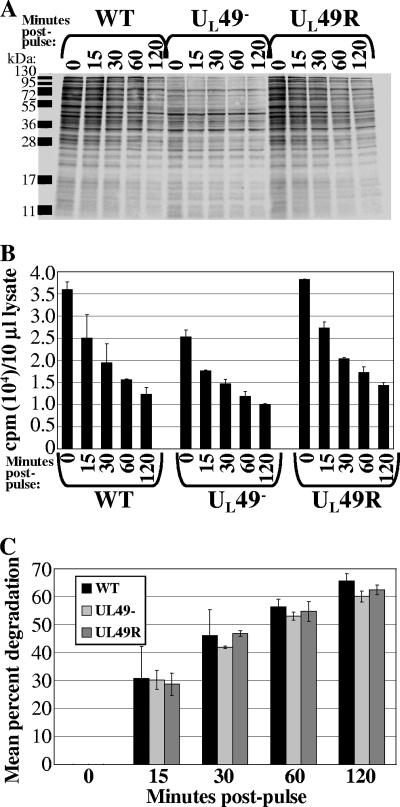
To determine whether the decreased accumulation of proteins at late times during UL49− infection was a consequence of decreased protein synthesis, decreased protein stability, or both, we performed pulse-chase assays. Vero cells infected with the WT, UL49−, and UL49R viruses for 8 h (a time in infection corresponding to the observed decrease in protein accumulation) were pulse-labeled with [35S]methionine/cysteine for 15 min at 37°C. Following the pulse, the 35S-containing medium was replaced with medium 199V and the infected, labeled cells were incubated at 37°C. At 0, 15, 30, 60, or 120 min after pulse-labeling the cells were washed and lysed. Labeled polypeptides were separated by SDS-PAGE, electrophoretically transferred, and visualized by autoradiography (Fig. 3A). Aliquots of pulse-labeled cell lysates were also analyzed by scintillation counting to determine total protein degradation in cells infected by each virus over time (Fig. 3B and C).
FIG. 3.
Pulse-chase analysis of protein degradation during WT, UL49−, and UL49R infections. Vero cells synchronously infected for 8 h with the WT, UL49−, and UL49R viruses were incubated in the presence of [35S]methionine/cysteine for 15 min. Following replacement of 35S-containing medium with non-35S-containing medium, cells were incubated for a further 0, 15, 30, 60, or 120 min. Following cell washing and lysis, labeled proteins were detected by either autoradiography following SDS-PAGE separation (A) or by scintillation counting (B). (C) Mean percent protein degradation during WT, UL49−, and UL49R infections. To account for differences in protein synthesis between the WT, UL49−, and UL49R infections during the 15-min labeling period, 35S incorporation following the 15-, 30-, 60-, and 120-min chases, as measured for panel B, were normalized to each virus's 35S incorporation following the initial labeling period (0 min chase). The normalized 35S incorporation values corresponding to the 15-, 30-, 60-, and 120-min chases were then divided by the normalized 0-min chase 35S incorporation values and multiplied by 100 to determine the relative percent protein degradation occurring over time in WT, UL49−, and UL49R infections.
Figure 3B shows there was less total 35S incorporation in UL49−-infected cells collected immediately after the short 15-min labeling (0 min postpulse) compared to WT- and UL49R-infected cells, indicating protein synthesis is decreased in UL49−-infected cells at 8 h postinfection. In Fig. 3C, the amount of 35S incorporation measured following the chases at 15, 30, 60, and 120 min was normalized to each virus's amount of incorporation following the initial labeling (0 min postpulse) and expressed as the percent degradation. These data show there is not a global increase in protein degradation in UL49−-infected cells compared to WT- and UL49R-infected cells at 8 h postinfection. Figure 3A shows the degradation of individual proteins over time. Overall, the proteins that appeared most stable in WT- and UL49R-infected cells were also most stable in UL49−-infected cells. Likewise, the most unstable proteins in WT- and UL49R-infected cells were relatively unstable in UL49−-infected cells. We conclude from the above data that VP22 is required for WT levels of protein synthesis, but not protein stability, at late times in infection.
VP22 enhances mRNA accumulation in a manner separate from its effects on protein synthesis.
To determine whether VP22's role in protein synthesis reflected a role in accumulation of mRNAs, we performed quantitative real-time RT-PCR to quantify ICP0, gE, gD, and β-actin mRNA levels over time. Total RNA was collected from cells infected with the WT, UL49−, and UL49R viruses for 3, 6, 9, and 15 h. The RNAs were treated to remove contaminating DNA and then used as templates to synthesize cDNA using random primers. Equal amounts of cDNA were used in qRT-PCRs containing primers to the gene of interest and Sybr green for product quantification. Parallel reaction mixtures contained primers to 18S rRNA as a control to normalize template input.
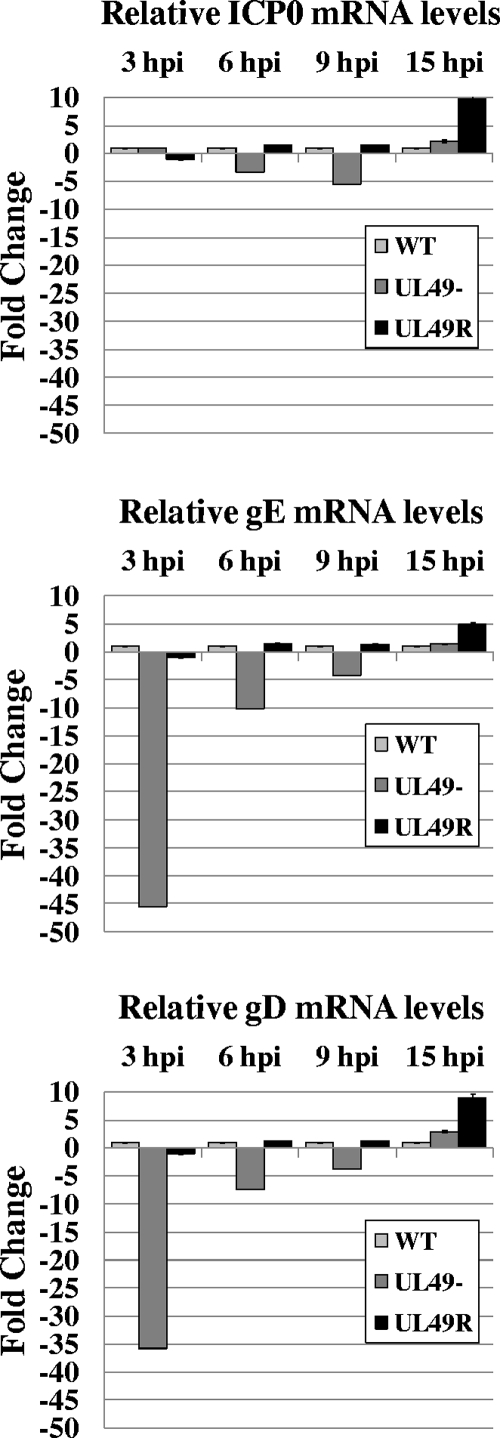
As shown in Fig. 4, ICP0 mRNA levels were very similar among the three viruses throughout the course of infections even though ICP0 protein levels were decreased at late times in UL49− infections compared to WT and UL49R infections (Fig. 1). In contrast, levels of gE and gD mRNAs were greatly reduced in UL49−-infected cells compared to WT- and UL49R-infected cells at 3 hours postinfection and then rose quickly and were present at wild-type levels later in infection. Specifically, gE mRNA levels were reduced 45-fold at 3 h postinfection, 10-fold at 6 h postinfection, and 4-fold at 9 h postinfection and were increased 1.4-fold at 15 h postinfection in UL49−-infected cells compared to WT-infected cells. Likewise, gD mRNA levels were reduced 36-fold at 3 h postinfection, 7-fold at 6 h postinfection, and 4-fold at 9 h postinfection and were increased 3-fold at 15 h postinfection in UL49−-infected cells compared to WT-infected cells. As an additional control, Northern blot analysis was performed and indicated that, once detectable, the size of gE mRNA did not change significantly during the course of infection (data not shown). Thus, the quantitative PCR results were a reliable indicator of mRNA accumulation rather than spurious transcription. Interestingly, gE and gD UL49− mRNA levels did not correlate with gE and gD protein levels in UL49−-infected cells; while gE and gD protein levels were unaffected at early times in infection and dropped off starting at ∼8 h postinfection (Fig. 1), levels of gE and gD mRNAs were reduced in UL49−-infected cells at early times in infection and present at wild-type levels later in infection.
FIG. 4.
Relative quantitative real-time RT-PCR analysis of ICP0, gE, and gD mRNA levels throughout WT, UL49−, and UL49R infections. Total RNA was collected from Vero cells infected with the WT, UL49−, and UL49R viruses for 3, 6, 9, or 15 h. Following contaminating DNA removal and reverse transcriptase reactions, equal amounts of cDNAs were used in qRT-PCRs containing primers to the gene of interest.
The above data show (i) a subset of mRNAs (e.g., gD and gE, but not ICP0) are present in reduced levels early in UL49− infections, and (ii) the protein synthesis shutdown observed at late times in UL49− infections does not appear to occur at an mRNA regulatory level. Together, the above data indicate VP22 plays roles in both protein synthesis and mRNA accumulation. Interestingly, these roles are distinct and separable with regard to both timing during infection and the genes affected.
Effects of VP22 on protein synthesis and mRNA accumulation are not mediated by antiapoptotic activities.
HSV-1 has been shown to induce apoptosis at early times in infection and later block the apoptotic pathway through the production of antiapoptotic viral proteins (31). To determine whether VP22 promotes protein synthesis and mRNA accumulation through an antiapoptotic role during HSV-1 infection, we used immunoblot assays to study PARP accumulation and cleavage over time in cells infected with the WT, UL49−, and UL49R viruses. PARP is a substrate for caspase-3, a protease that is activated during apoptosis, and PARP cleavage is an indicator of unblocked apoptosis. In Vero cells infected with HSV-1 and treated with cycloheximide, PARP cleavage is seen at 24 h postinfection (32). Figure 5A shows that UL49−-infected cells did not show either decreased PARP accumulation or increased PARP cleavage compared to WT- or UL49R-infected cells over the course of infection. We therefore conclude that VP22 does not promote protein synthesis and mRNA accumulation through antiapoptotic activities.
FIG. 5.
Immunoblot analyses of steady-state PARP, ICP27, VP16, and vhs protein levels throughout WT, UL49−, and UL49R infections. (A) Immunoblot analysis of PARP protein levels and cleavage. Lysates of Vero cells infected with the WT, UL49−, and UL49R viruses for 12, 18, 24, or 30 h were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed for the presence of PARP. The upper band, present at all time points, corresponds to full-length PARP, while the lower band present at 24 and 30 h postinfection (hpi) corresponds to cleaved PARP. (B) Immunoblot analysis of ICP27, VP16, and vhs levels. Lysates of Vero cells infected with the WT, UL49−, and UL49R viruses for 3, 6, 9, 12, 15, or 18 h were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed for the presence of ICP27 (top row), VP16 (second row), and vhs (bottom row).
Effects of VP22 on protein synthesis occur independently of ICP27, VP16, vhs, or phosphorylated eIF2α protein levels.
ICP27 is an immediate-early protein that contributes to HSV-1 infection through roles in transcriptional regulation of HSV-1 early and late genes (19, 47), translation initiation (12, 15), and mRNA export (2, 3, 21). VP22 could contribute to global protein synthesis at late times and/or the accumulation of viral mRNAs throughout the viral life cycle by increasing the amount of ICP27 available to facilitate RNA export and/or transcriptional activation. To determine whether the presence of VP22 affects ICP27 accumulation, we examined steady-state levels of ICP27 protein over time in Vero cells infected with the WT, UL49−, and UL49R viruses. Figure 5B shows ICP27 protein levels were unaffected throughout the course of infection by the absence of UL49. Thus, VP22's role in protein synthesis does not appear to take place through an effect on ICP27 protein levels.
We next asked whether VP22 promotes mRNA accumulation through an effect on VP16 accumulation. We previously showed VP16 is incorporated into virions at near-WT levels (6). Therefore, VP16's role in immediate-early mRNA synthesis should not be affected by a lack of VP22, consistent with the WT levels of ICP0 mRNA (Fig. 4) and ICP4 and ICP27 proteins (Fig. 1 and 5B) we observed throughout UL49− infection. At late times in infection, VP16 suppresses the RNA degradative activity of vhs, suggesting a mechanism that ensures appropriate accumulation of late transcripts (29). Given its potential relevance to the VP22 phenotypes shown herein, we examined the accumulation of VP16 in WT-, UL49−-, and UL49R-infected cells over time. Like the majority of other proteins studied herein, we found VP16 accumulated to WT levels in UL49−-infected cells at early times in infection (Fig. 5B, 3 and 6 h postinfection), but relative VP16 levels decreased in UL49−-infected cells at ∼9 h postinfection and beyond.
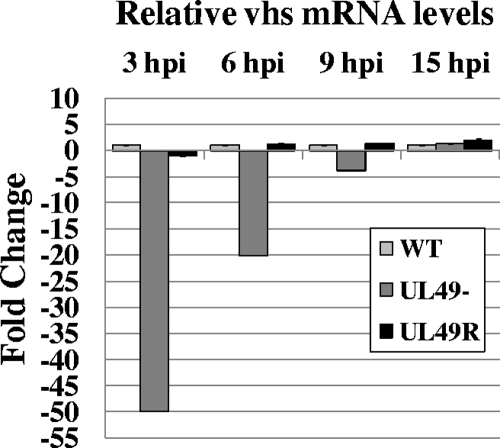
FIG. 6.
Relative quantitative real-time RT-PCR analysis of vhs mRNA levels throughout WT, UL49−, and UL49R infections. Total RNA was collected from Vero cells infected with the WT, UL49−, and UL49R viruses for 3, 6, 9, or 15 h. Following contaminating DNA removal and reverse transcriptase reactions, equal amounts of cDNAs were used in qRT-PCRs containing primers to UL41.
Because vhs both interacts with and is modulated by VP16 (29, 41, 44), the decreased levels of VP16 in UL49−-infected cells could affect mRNA accumulation and/or protein synthesis if vhs were present at WT levels in UL49− cells. Therefore, we next examined vhs protein levels in WT-, UL49−-, and UL49R-infected cells over the course of infection. We found very little vhs protein present in UL49−-infected cells (Fig. 5B). As vhs was first detected in WT- and UL49R-infected cells at 9 h postinfection, a time at which shutdown of protein synthesis has already begun in UL49−-infected cells, it is reasonable that the lack of vhs observed in UL49−-infected cells is due to this UL49− phenotype. To ensure the decreased vhs protein levels found in UL49−-infected cells were not due to a disruption in vhs transcription, we performed qRT-PCR using primers against vhs and 18S (Fig. 6). As with gE and gD, we found vhs mRNA levels were greatly reduced at early, but not late, times in UL49− infection. Specifically, vhs mRNA levels were reduced 50-fold at 3 h postinfection, 20-fold at 6 h, and 3.9-fold at 9 h and were increased 1.3-fold at 15 h postinfection in UL49−-infected cells compared to WT-infected cells. Together, the above data indicate that the decreased protein synthesis and mRNA accumulation observed in UL49−-infected cells is not due to an increase in the ratio of vhs versus VP16 protein levels and that the absence of VP22 affects vhs, gD, and gE mRNA and protein levels in a very similar manner.
Finally, we asked whether VP22 promotes protein synthesis late in infection by modulating, directly or indirectly, the phosphorylation status of the translation initiation factor eIF-2α. During the normal course of infection, double-stranded RNA produced through complementary transcript annealing leads to the activation of protein kinase R, the subsequent phosphorylation of eIF-2α at serine-51, and a consequent shutdown in protein synthesis (17, 39). The HSV-1 ICP34.5 protein acts to preclude the shutoff of protein synthesis by directing the dephosphorylation of eIF-2α by the host protein phosphatase 1α (5, 18). It is possible that VP22 increases protein synthesis late in infection through an ICP34.5-dependent or -independent role in eIF-2α dephosphorylation. To determine whether UL49−-infected cells contained increased levels of phosphorylated eIF-2α, we performed immunoblot assays with lysates from cells infected for 3, 6, 9, 12, 15, and 18 h with the WT, UL49−, and UL49R viruses using an antibody specific to the phosphorylated form of eIF-2α's serine-51. We found approximately equal and barely detectable levels of phosphorylated eIF-2α in cells infected with all three viruses (data not shown). Therefore, we conclude VP22 does not promote protein synthesis at late times in infection by modulating eIF-2α's phosphorylation status.
Small and large plaque variants of the UL49− virus do not possess secondary mutations in UL41 or UL48.
In WT- and UL49R-infected cells, vhs is produced starting at ∼9 h postinfection (Fig. 5B), the time corresponding to the global dropoff in protein synthesis observed in UL49−-infected cells. Therefore, the lack of vhs observed in UL49−-infected cells (Fig. 5B) was not surprising. However, a recent study by Sciortino et al. showed that three independently derived UL49 deletion viruses contained secondary mutations that led to either the truncation or deletion of vhs (43). To ensure the lack of vhs expression observed during infection with our UL49− virus was due to the lack of VP22 and not to secondary mutations in UL41 encoding vhs, and to ensure the vhs protein encoded by our UL49− virus did not contain mutations that could preclude interaction with or modulation by VP16, we sequenced the UL41 coding region from WT, UL49−, and UL49R viral DNA obtained from individual plaques and from the virus stocks used in the above studies. The sequenced region started 487 bp upstream of the UL41 start codon and extended through the UL41 promoter, the UL41 open reading frame, and downstream of the stop codon through the UL41 poly(A) tract. There were no DNA sequence differences between WT, UL49−, and UL49R viral DNAs for the entire UL41 coding region of the genome (data not shown).
We previously reported that transfection of UL49− BAC DNA into or passage of the UL49− virus on noncomplementing cells reproducibly led to the production of UL49− large plaque variants, presumably due to the acquisition of secondary compensatory mutations. The UL49 deletion viruses carrying secondary mutations in the UL41 gene constructed by Sciortino et al. were generated through transfection of noncomplementing cells with UL49− BAC DNA (43). To determine whether large plaque variants produced by our UL49− virus after two passages on noncomplementing cells had acquired secondary mutations in the UL41 gene, we sequenced the above UL41 coding regions of viral DNA obtained from two independent UL49− large plaque variants. We found no sequence changes within the entire UL41 coding region for either of the large plaque variants. We therefore conclude that passage of the UL49− virus on noncomplementing cells can lead to the production of UL49− large plaque variants by means other than the acquisition of secondary mutations in the UL41 region of the genome.
Secondary mutations in the UL48 gene encoding VP16 could affect the ability of VP16 to modulate vhs, rendering the trace amounts of vhs present in UL49−-infected cells very active in mRNA degradation. Therefore, we also sequenced the UL48 coding region, starting 416 bp upstream of the UL48 start codon and extending through the UL48 promoter, the UL48 open reading frame, and downstream of the stop codon through the UL48 poly(A) tract, from the above DNAs obtained from WT, UL49−, and UL49R plaques. There were no DNA sequence differences between WT, UL49−, and UL49R viral DNA for the entire UL48 coding region of the genome (data not shown).
DISCUSSION
In previous studies of a UL49 deletion mutant we showed VP22 greatly enhanced plaque size and viral replication at low multiplicities of infection (6). We also showed UL49− virions contained decreased amounts of ICP0, gE, and gD. In the current study we aimed to determine whether the latter phenomenon was due to decreased virion incorporation or decreased synthesis of ICP0, gE, and gD in the absence of VP22. We found that at late times in infection, VP22 is required for WT levels of protein synthesis on a near-global scale. We also found VP22 contributes to the accumulation of some viral mRNAs at early, but not late, times in infection. Interestingly, VP22's effects on protein and mRNA levels are distinct and separable with regard to both timing during infection and the genes affected. Specifically, (i) gE and gD mRNA levels were reduced in UL49−-infected cells at early times in infection whereas gE and gD protein levels were reduced at late times in infection, and (ii) gE, gD, and ICP0 protein levels were all reduced late in UL49− infections whereas only gE and gD mRNA levels were substantially reduced in UL49−-infected cells relative to WT- and UL49R-infected cells.
Role of VP22 in protein synthesis at late times in infection.
Immunoblot analyses of steady-state ICP0, ICP4, ICP27, gE, gD, and β-actin levels in WT-, UL49−-, and UL49R-infected cells at various times in infection showed these proteins were present in similar amounts regardless of the infecting virus at early times in infection. However, beginning at ∼6 to 8 h postinfection ICP0, gE, and gD levels dropped dramatically in UL49−-infected cells, whereas ICP4, ICP27, and β-actin levels were not affected. Using [35S]methionine/cysteine labeling, we found the dropoff in protein synthesis during UL49− infection took place on a near-global scale and decreased total steady-state protein levels by up to 68%. Using pulse-chase analyses, we determined the defect was in protein synthesis, rather than protein stability. We also determined that this drop-off in protein synthesis during UL49− infections did not reflect an aberrant induction of apoptosis or an increase in eIF-2α phosphorylation, either of which might account for a general failure of protein synthesis.
VP22's function in protein synthesis at late times in infection appears to involve direct and/or indirect roles in mRNA export and/or mRNA translation. VP22 has been shown to interact with mRNA (42) and is found in both the nucleus and cytoplasm (11, 36), pointing to a possible direct role for VP22 in mRNA export and/or translation. Alternatively, VP22 may aid another viral protein in these activities. Although we found no difference in ICP27 protein levels in UL49−-infected cells compared to cells infected with wild-type virus, it is possible that VP22 serves to activate ICP27 at late times in infection, boosting ICP27's mRNA export and translation initiation activities.
Role of VP22 in mRNA accumulation at early times in infection.
We have shown that whereas ICP0 mRNAs were present at WT levels throughout the course of UL49− infection, gE and gD mRNA levels were significantly reduced early in infection and rose to WT levels by late times in infection. VP22 may act to increase steady-state mRNA levels early in infection via a direct or indirect route. For example, VP22's mRNA binding activity may serve to directly protect viral mRNAs from degradation. Alternatively, VP22 may contribute to mRNA accumulation by modulating the RNase activity of vhs. VP22, VP16, and vhs are known to form a complex (46) and VP16 is known to modulate the vhs RNase activity (29). Although the amount of vhs present in UL49−-infected cells is severely decreased relative to WT-infected cells, we cannot exclude the hypothesis that VP22 may contribute to VP16's modulation of vhs, and thus, in the absence of VP22 the small amount of vhs present may be extremely active.
In summary, we have further characterized an HSV-1 UL49-null virus and have discovered roles for VP22 in both protein synthesis at late times in infection and mRNA accumulation of some viral mRNAs at early times in infection. Future studies will focus on gaining a more comprehensive view of the genes affected by the absence of VP22 and determining VP22's precise role(s) in enhancing protein synthesis and mRNA accumulation.
Acknowledgments
These studies were supported by NIH R01 grants GM 50740 and AI 52341 to J.D.B.
We thank G. Sullivan Read for the gift of the anti-vhs antibody, Rozanne Sandri-Goldin for the gift of the anti-ICP27 antibody, David Johnson for the gift of the anti-gE antibody, and John Blaho for the VP22-expressing cell line.
Footnotes
Published ahead of print on 5 November 2008.
REFERENCES
- 1.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 1801-19. [DOI] [PubMed] [Google Scholar]
- 2.Chen, I. H., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 793949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 7612877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86253-261. [DOI] [PubMed] [Google Scholar]
- 5.Chou, J., J. J. Chen, M. Gross, and B. Roizman. 1995. Association of a Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with γ134.5− mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 9210516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffy, C., J. H. LaVail, A. N. Tauscher, E. G. Wills, J. A. Blaho, and J. D. Baines. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J. Virol. 808664-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 737153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott, G., W. Hafezi, A. Whiteley, and E. Bernard. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 799735-9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 697932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 726448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 734110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellison, K. S., R. A. Maranchuk, K. L. Mottet, and J. R. Smiley. 2005. Control of VP16 translation by the herpes simplex virus type 1 immediate-early protein ICP27. J. Virol. 794120-4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everly, D. N., Jr., P. Feng, I. S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 768560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnsworth, A., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontaine-Rodriguez, E. C., T. J. Taylor, M. Olesky, and D. M. Knipe. 2004. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology 330487-492. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 766729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gale, M., Jr., and M. G. Katze. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 7829-46. [DOI] [PubMed] [Google Scholar]
- 18.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jean, S., K. M. LeVan, B. Song, M. Levine, and D. M. Knipe. 2001. Herpes simplex virus 1 ICP27 is required for transcription of two viral late (gamma 2) genes in infected cells. Virology 283273-284. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, D. C., M. C. Frame, M. W. Ligas, A. m. Cross, and N. D. Stow. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 621347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 205769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotsakis, A., L. E. Pomeranz, A. Blouin, and J. A. Blaho. 2001. Microtubule reorganization during herpes simplex virus type 1 infection facilitates the nuclear localization of VP22, a major virion tegument protein. J. Virol. 758697-8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristie, T. M., J. H. LeBowitz, and P. A. Sharp. 1989. The octamer-binding proteins form multi-protein-DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 84229-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristie, T. M., and B. Roizman. 1987. Host cell proteins bind to the cis-acting site required for virion-mediated induction of herpes simplex virus 1 alpha genes. Proc. Natl. Acad. Sci. USA 8471-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristie, T. M., and P. A. Sharp. 1990. Interactions of the Oct-1 POU subdomains with specific DNA sequences and with the HSV alpha-trans-activator protein. Genes Dev. 42383-2396. [DOI] [PubMed] [Google Scholar]
- 26.Kristie, T. M., and P. A. Sharp. 1993. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus alpha-trans-induction factor (VP16). J. Biol. Chem. 2686525-6534. [PubMed] [Google Scholar]
- 27.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 841926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong, A. D., J. A. Kruper, and N. Frenkel. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 152575-2581. [PMC free article] [PubMed] [Google Scholar]
- 30.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 691531-1574. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen, M. L., and J. A. Blaho. 2007. Apoptosis during herpes simplex virus infection. Adv. Virus Res. 6967-97. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen, M. L., R. M. Kraft, and J. A. Blaho. 2005. African green monkey kidney Vero cells required de novo protein synthesis for efficient herpes simplex virus 1-dependent apoptosis. Virology 336274-290. [DOI] [PubMed] [Google Scholar]
- 33.O'Regan, K. J., M. A. Bucks, M. A. Murphy, J. W. Wills, and R. J. Courtney. 2007. A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358192-200. [DOI] [PubMed] [Google Scholar]
- 34.Pellett, P. E., J. L. McKnight, F. J. Jenkins, and B. Roizman. 1985. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc. Natl. Acad. Sci. USA 825870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 7410041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 736769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potel, C., and G. Elliott. 2005. Phosphorylation of the herpes simplex virus tegument protein VP22 has no effect on incorporation of VP22 into the virus but is involved in optimal expression and virion packaging of ICP0. J. Virol. 7914057-14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 677149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, W. K., A. Hovanessian, R. E. Brown, M. J. Clemens, and I. M. Kerr. 1976. Interferon-mediated protein kinase and low-molecular-weight inhibitor of protein synthesis. Nature 264477-480. [DOI] [PubMed] [Google Scholar]
- 40.Roizman, B., T. Kristie, J. L. McKnight, N. Michael, P. Mavromara Nazos, and D. Spector. 1988. The trans-activation of herpes simplex virus gene expression: comparison of two factors and their cis sites. Biochimie 701031-1043. [DOI] [PubMed] [Google Scholar]
- 41.Schmelter, J., J. Knez, J. R. Smiley, and J. P. Capone. 1996. Identification and characterization of a small modular domain in the herpes simplex virus host shutoff protein sufficient for interaction with VP16. J. Virol. 702124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sciortino, M. T., B. Taddeo, A. P. Poon, A. Mastino, and B. Roizman. 2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 998318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sciortino, M. T., B. Taddeo, M. Giuffre-Cuculletto, M. A. Medici, A. Mastino, and B. Roizman. 2007. Replication-competent herpes simplex virus 1 isolates selected from cells transfected with a bacterial artificial chromosome DNA lacking only the UL49 gene vary with respect to the defect in the UL41 gene encoding host shutoff RNase. J. Virol. 8110924-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 682339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strom, T., and N. Frenkel. 1987. Effects of herpes simplex virus on mRNA stability. J. Virol. 612198-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taddeo, B., M. T. Sciortino, W. Zhang, and B. Roizman. 2007. Interaction of herpes simplex virus RNase with VP16 and VP22 is required for the accumulation of the protein but not for accumulation of mRNA. Proc. Natl. Acad. Sci. USA 10412163-12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uprichard, S. L., and D. M. Knipe. 1996. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J. Virol. 701969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zelus, B. D., R. S. Stewart, and J. Ross. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 702411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]