Abstract
The human cytidine deaminase APOBEC3G (A3G) is a part of a cellular defense system against human immunodeficiency virus type 1 (HIV-1) and other retroviruses. Antiretroviral activity of A3G can be severely blunted in the presence of the HIV-1 protein Vif. However, in some cells expressing the enzymatically active low-molecular-mass form of A3G, HIV-1 replication is restricted at preintegration steps, before accumulation of Vif. Here, we show that A3G can be secreted by cells in exosomes that confer resistance to both vif-defective and wild-type HIV-1 in exosome recipient cells. Our results also suggest that A3G is the major exosomal component responsible for the anti-HIV-1 activity of exosomes. However, enzymatic activity of encapsidated A3G does not correlate with the observed limited cytidine deamination in HIV-1 DNA, suggesting that A3G-laden exosomes restrict HIV-1 through a nonenzymatic mechanism. Real-time PCR quantitation demonstrated that A3G exosomes reduce accumulation of HIV-1 reverse transcription products and steady-state levels of HIV-1 Gag and Vif proteins. Our findings suggest that A3G exosomes could be developed into a novel class of anti-HIV-1 therapeutics.
APOBEC3 (A3) proteins belong to a family of cellular cytidine deaminases that restrict replication of a variety of exogenous retroviruses and endogenous retroelements (10, 12, 17, 24, 30, 45, 57, 60). The most prominent member of this family, APOBEC3G (A3G), has a potent activity against vif-deficient human immunodeficiency virus type 1 (HIV-1) (63) but can also function as a postentry restriction factor against wild-type viruses (11, 48, 77). An antiretroviral function of A3G has been linked with its DNA-editing activity (40, 59); however, recent studies point to the existence of additional, editing-independent activities of A3G that may contribute to its antiviral function (21, 43, 71). Anti-HIV-1 activity of A3G usually requires its encapsidation into budding virions, a process mediated by the viral nucleocapsid protein (1, 6, 16, 35, 58) and/or cellular or viral RNAs (28, 72, 78, 83). However, encapsidation of A3G into wild-type HIV-1 virions is severely impaired, reflecting, at least partly, the action of the viral protein Vif, which targets A3G for polyubiquitination and degradation by the 26S proteasome (13, 27, 38, 64, 70, 82).
Identification of numerous cellular proteins commonly found in HIV-1 virions and exosomes (9) and the evidence that HIV-1 budding may occur, at least in some cells, through the exosome release pathway (44) prompted us to investigate whether A3G could be secreted by cells in association with exosomes. In line with this supposition, we have observed that a fraction of cellular A3G associates with multivesicular bodies/late endosomes (1), organelles that release intraluminal vesicles as exosomes after fusion with the plasma membrane (14, 69, 74).
The biological role of exosomes is poorly understood. Exosomes were originally described as vesicles removing unnecessary proteins from maturating reticulocytes (26). However, secretion of morphogenic proteins (33) or adhesion and antigen presentation molecules in association with exosomes (42, 52, 61, 62) suggested that these vesicles may play a role during embryogenesis and in modulation of immune responses (39, 46, 65, 66, 68). In addition, exosomes concentrate several cellular mRNAs and microRNAs (75) that may epigenetically reprogram the recipient cells (15, 53, 54). Exosomes’ role in pathogenesis may include tumor metastasis (20) and transmission of infectious agents including prions (19, 51, 76) and HIV-1 (18, 79).
In the present study, we tested the hypothesis that A3G secreted in exosomes can confer an antiviral phenotype to target cells. We demonstrate that both endogenous and exogenous A3G can be secreted by cells in exosomes. Furthermore, A3G-laden exosomes potently restrict replication of vif-deficient and wild-type HIV-1 in recipient cells, which leads us to speculate that exosomes may contribute to antiretroviral defenses in vivo by exchanging A3G between cells.
MATERIALS AND METHODS
Cells.
Jurkat, SupT1, and H9 cells and peripheral blood mononuclear cells (PBMCs) were propagated in RPMI supplemented with 10% fetal calf serum (FCS; Gemini Bio-Products) and gentamicin (50 μg/ml). HeLa and 293T cells were cultivated in Dulbecco modified Eagle medium with 10% FCS and gentamicin. PBMCs were obtained by Ficoll-Paque (Amersham) density centrifugation from several healthy blood donors (New York Blood Center). PBMCs were depleted of monocytes by plastic adherence and stimulated for 3 days with phytohemagglutinin (PHA) L (2 μg/ml; Roche Applied Science) in RPMI supplemented with 10% FCS and gentamicin. Activated PBMCs were washed and resuspended in fresh medium with interleukin-2 (IL-2; 20 U/ml; Roche Applied Science). After 48 h, the cells were used in experiments.
Virus preparation.
Virus stocks were prepared by transfecting 293T cells with HIV-1 plasmid DNA by using PolyFect reagent (Qiagen). Culture supernatants were collected after 48 h, centrifuged to remove cell debris, filtered through an 0.45-μm filter, and concentrated by ultracentrifugation through a cushion of 20% sucrose in phosphate-buffered saline (PBS). The pelleted virus was resuspended in RPMI with 10% FCS, aliquoted, and stored at −80°C. Virus titer was determined by the reverse transcriptase (RT) activity assay (50). For PCR analysis, virus preparations were treated with DNase I (200 U/ml) for 1 h at room temperature.
Virus entry assay.
The virus entry assay was performed as described previously (31, 56). SupT1 cells (1 × 106) were incubated at 37°C for 90 min with HIV-1 NL4-3 (5 RT cpm/cell) with encapsidated Nef-luciferase fusion protein. After being washed three times with ice-cold PBS, the cells were resuspended in 0.1 ml of luciferase substrate (Promega), and luciferase activity was measured in live cells after 1 min using a Fluostar Optima plate reader. The effect of fusion inhibitor was tested in cells pretreated for 1 h with T20 (1 μg/ml; NIH AIDS Research and Reference Reagent Program).
Viral infectivity assay.
Viral supernatants were harvested on day 3 posttransfection, centrifuged to remove any cellular debris, and kept at −80°C. The assay was performed using HeLa-CD4-long terminal repeat (LTR)-β-galactosidase (β-Gal) cells (29) according to the protocol (NIH AIDS Research and Reference Reagent Program; catalog no. 1470). Viral inoculum was standardized by RT assay.
Apoptosis assay.
Early apoptotic events in cells exposed to exosomes were investigated using the Image-iT live green polycaspases detection kit (Invitrogen). An assay was performed according to the manufacturer's specifications (Invitrogen).
Exosome isolation protocol.
Cells were grown in culture medium supplemented with 10% FCS and depleted of endogenous exosomes. All procedures were performed at 4°C. Exosome-depleted media were prepared by 16 h of ultracentrifugation of culture medium supplemented with 50% FCS at 100,000 × g and filtration through a 0.2-μm filter. Culture supernatants were collected every 24 h and were first centrifuged at 300 × g for 10 min to pellet cells. Cell debris was removed by centrifugation at 2,000 × g for 15 min followed by 30 min of centrifugation at 10,000 × g. The clarified supernatant was filtered through an 0.2-μm filter and ultracentrifuged at 100,000 × g using an SW32 rotor to pellet exosomes. Exosomes were washed by resuspension in 10 ml of PBS and pelleting for 1 h at 100,000 × g using a SW41 rotor. The final pellet was resuspended in 30 to 50 μl of PBS and frozen at −80°C. For further purification, exosomes were mixed with 2 ml of 2.5 M sucrose in PBS and placed on the bottom of a SW41 centrifuge tube, overlaid with 6 ml 2 M sucrose and 3 ml 0.25 M sucrose, and ultracentrifuged for 16 h at 100,000 × g. The purified exosomes accumulating at the 2 M/0.25 M interface were collected, diluted in PBS, and pelleted by ultracentrifugation at 100,000 × g for 30 min. Pelleted exosomes were resuspended in PBS and used immediately or kept at −80°C. Protein concentrations of exosome preparations were determined using the micro-bicinchoninic acid protein assay (Thermo Scientific). On average, we have recovered about 0.32 μg of total proteins in exosomes secreted by 106 cells during a 24-h period.
Exosome treatment and cell infections.
A total of 1 × 106 cells were treated with exosomes (10 μg/ml unless otherwise stated) for 16 h, washed, and infected for 3 h with an RT-standardized amount of HIV-1 (5 RT cpm/cell) in 1 ml of RPMI medium supplemented with 10% FCS. After two washes to remove unbound virus, exosomes (10 μg/ml) were added and cells were cultured for the indicated period of time. Virus replication was monitored by virion-associated RT activity released to the medium (50). Fifty microliters of culture medium was collected daily for RT assay and replaced with fresh culture medium.
Exosome deaminase activity assay.
Exosomes (20 μg protein) were suspended in 25 μl of deaminase buffer (40 mM Tris, pH 8.0, 40 mM KCl, 50 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol, 2% glycerol, 0.1% Triton X-100) with a 5′-biotin-labeled oligonucleotide substrate (50 ng) and incubated at 37°C for 5 h. The reactions were terminated by heating the reaction mixtures to 90°C for 5 min followed by adding 25 μl of 60 mM Tris, pH 8.0, with 1 mM dithiothreitol and then incubating them with uracil DNA glycosylase (0.5 U; Roche Applied Science) for 30 min at 37°C. Subsequently, the samples were treated with 0.15 M NaOH (by adding 0.75 μl of 10 M NaOH) at 37°C for 30 min. Products were neutralized with 0.15 M HCl (by adding 0.7 μl concentrated HCl) and mixed with 2× gel loading buffer, and 40 μl of the mixture was separated on a 15% Tris-buffered EDTA-urea polyacrylamide gel, transferred to a zeta-probe membrane (Bio-Rad), and detected by chemiluminescence with streptavidin-horseradish peroxidase (1:50,000 dilution; Sigma). 5′-Biotin-labeled oligonucleotides (Invitrogen) used in this assay were as described previously (7): 60-nucleotide (nt) CCC, GAG GAA GGG AAG AAA GAG AAA GGG AGA CCC AAA GAG GAA AGG TGA GGA GGT TAA TTT GTG (A3G target site is underlined), and 60-nt TAA (negative control); in this oligonucleotide, the A3G target sequence CCC is changed into TAA.
Exosome labeling, internalization, and confocal microscopy.
Exosomes were labeled at room temperature in a SW41 centrifuge tube using the PKH67 kit (Sigma-Aldrich) according to the manufacturer's instructions. Briefly, 10 μg of exosomes in PBS was resuspended in 1 ml of diluent C, mixed with freshly prepared PKH67 in diluent C at a final concentration of 5 × 10−6 M, and incubated for 3 min. Labeling was stopped by addition of an equal volume of exosome-free FCS for 1 min, followed by the addition of exosome-free RPMI-10% FCS to fill up the centrifuge tube and ultracentrifugation for 30 min at 100,000 × g. After two additional washes in exosome-free RPMI-10% FCS by ultracentrifugation, the exosomes were resuspended in 100 μl of RPMI with 10% FCS. Alternatively, exosomes containing green fluorescent protein (GFP)-A3G were purified from culture supernatants of 293T cells transiently transfected with GFP-A3G expression vector. Exosomes labeled with fluorescent PKH67 or GFP-A3G (10 μg/ml) were added to HeLa cells growing on two-well Lab-Tek II chamber microscope slides (Nalgene Nunc International) and incubated at 37°C for 2 h. The medium with exosomes was replaced with medium supplemented with transferrin-Alexa 594 (5 μg/ml) or LysoTracker (50 nM) (Invitrogen) for 30 min. The cells were washed three times with PBS, fixed in 4% paraformaldehyde for 15 min at room temperature, washed three times with PBS, mounted with ProLong antifade reagent (Invitrogen), and observed under a laser scanning confocal microscope (Nikon TE2000).
Electron microscopy.
Exosomes purified on a sucrose gradient (10 μl in PBS) were dropped onto a sheet of Parafilm, and Formvar-carbon coated grids were floated for 1 min at room temperature to adsorb exosomes, washed in PBS, fixed with 2% paraformaldehyde, washed with PBS, negatively stained with 1% phosphotungstic acid for 30 s, and viewed using a JEOL transmission electron microscope at 80 kV. For immunogold labeling, grids with adsorbed fixed exosomes were blocked with 4% bovine serum albumin or 1% cold-water fish skin gelatin in PBS for 10 min, permeabilized with 0.02% Triton for 5 min, washed, and incubated with primary antibody (1:50) in blocking solution for 30 min. After extensive washing with blocking solution, the grids were incubated for 10 min with protein A-gold conjugates (1:20), washed with PBS, fixed with 2.5% glutaraldehyde for 30 s, washed with PBS and water, stained for 30 s with 1% phosphotungstic acid for 30 s, and observed under an electron microscope as described above.
Detection of hypermutation in viral DNA.
SupT1 cells (1 × 106 cells/ml) were pretreated for 16 h with H9 exosomes (10 μg/ml) or were left untreated. The cells were infected with DNase I-treated vif-negative HIV-1 (5 RT cpm/cell) for 4 h, washed, and incubated with or without exosomes for 72 h. The cells were washed with PBS and treated with DNase I (100 U/ml) at room temperature for 1 h, and total DNA was isolated using the DNeasy kit (Qiagen). A 650-bp DNA fragment encompassing part of the nef/3′-LTR region of HIV-1 was amplified with Platinum Taq DNA polymerase (Invitrogen) using the primers HIV-1 F (5′-AGGCAGCTGTAGATATTAGCCAC) and HIV-1 R (5′-GTATGAGGGATCTCTAGCTACCA) (36). The PCR products were cloned into the TOPO TA cloning vector pCR4 (Invitrogen). Plasmid DNA isolated from transformed bacteria was sequenced and analyzed for mutations.
Quantitative analysis of HIV-1 reverse transcription products.
SupT1 cells (untreated or pretreated with H9 or SupT1 exosomes for 16 h) were washed and infected for 3 h with 5 RT cpm/cell of DNase-treated NL4-3 ΔE-EGFP pseudotyped with vesicular stomatitis virus glycoprotein (VSV-G) (84) or with wild-type NL4-3 in the presence of protease inhibitors amprenavir and nelfinavir (each 5 μM) to limit infection to a single cycle (5). After incubation, the cells were washed and resuspended in RPMI-10% FCS at 1 × 106 cells/ml in 12-well plates. At 0, 8, 24, and 48 h postinfection, equal aliquots of cells were collected and washed with PBS, and total cellular DNA was isolated from the cells using a DNeasy DNA isolation kit (Qiagen). The primers for the detection of early reverse transcription (ERT), late reverse transcription (LRT), two-LTR circle DNA, and glyceraldehyde-3-phosphate dehydrogenase have been described elsewhere (4, 32, 55). Real-time PCR assays were performed on iCycler using iQ Sybr Green Supermix (Bio-Rad). Reaction mixtures contained 250 nM (each) forward and reverse primers with 60 to 100 ng template DNA in a final volume of 25 μl. After initial incubation at 50°C for 2 min and 95°C for 10 min, amplifications (40 cycles) were performed at 95°C for 15 s and 60°C for 1 min. All analyses were performed at least three times with triplicate samples in each experiment.
Construction of vector expressing Nef-luciferase fusion protein.
Nef coding sequences were amplified from p96ZM651nef-opt by using primers KpnI-nef (5′-AGC TCG GTA CCA TGG GCG GCA AGT GGA GCA AG-3′) and HindIII-nef (5′-TGT CAA AGC TTG CAG TCC TTG TAG TAC TCG GG-3′). The amplicon was cloned in frame in the N terminus of the firefly luciferase gene in pSP luc+ NF fusion vector (Promega) in its KpnI and HindIII restriction sites. The Nef-Luc insert was cut out from the vector by KpnI and EcoRI restriction enzymes and subsequently recloned into pcDNA 3.1(+) expression vector (Invitrogen).
Western blot analysis, antibodies, and expression vectors.
Proteins were separated on sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and analyzed as described previously (49). Antibodies for Western blotting were obtained as follows: rabbit anti-Alix from H. Stenmark (Oslo, Norway) and rabbit anti-A3G from K. Strebel (NIH). Antibodies against Tsg101, CD71, CXCR4, HSP90, calnexin, VDAC1, and β-actin were from Santa Cruz Biotechnology. Monoclonal antibodies against HIV-1 p24, Vif, and Nef and Nef expression vector p96ZM651nef-opt were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Plasmids expressing vif-negative NL4-3 and A3G-HA were obtained from K. Strebel (NIH), and vectors pDON/EGFP (coding for GFP) and pDON/EGFP-CEM15 (coding for GFP-A3G) were obtained from A. Takaori-Kondo (Kyoto, Japan).
Statistical analysis.
All experiments were repeated at least three times with similar results. The variation in one experiment was expressed by calculating standard deviation (SD) from the triplicates and presented as the mean value ± SD.
RESULTS
A3G is released by cells in association with exosomes.
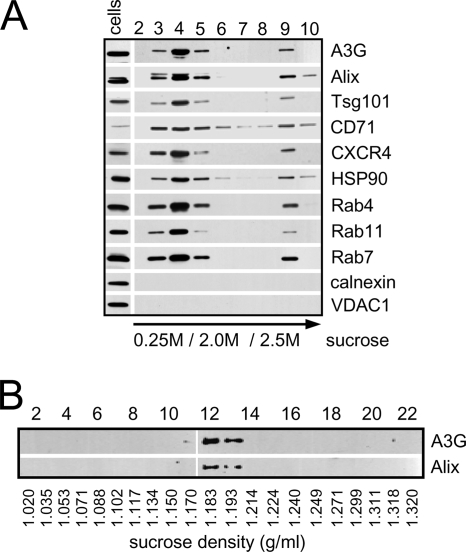
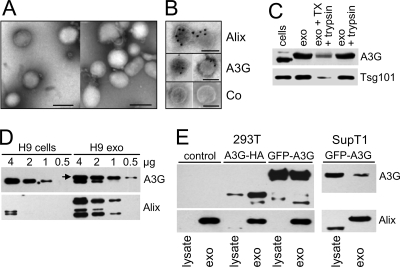
Exosomes prepared from culture supernatants of A3G-expressing H9 CD4+ T cells were subjected to sucrose gradient ultracentrifugation, and the protein composition of collected fractions was analyzed by Western blotting (Fig. 1A). Most of A3G floated in a sucrose gradient and accumulated at the 2.0 M/0.25 M sucrose interface together with markers specific for exosomes including Alix, Tsg101, CD71, HSP90, and Rab proteins. Markers for endoplasmic reticulum (calnexin) or mitochondria (VDAC1) were undetectable in exosomes but evident in cell lysates, suggesting that exosome preparations were not significantly contaminated with membranes from apoptotic or dead cells. An equilibrium sucrose gradient ultracentrifugation of combined fractions 3 to 5 showed that A3G colocalized with Alix (Fig. 1B) at a density of 1.183 to 1.193 g/ml characteristic for exosomes (19). The electron micrographs of exosomes showed vesicles ranging in size from 80 to 150 nm (Fig. 2A) with A3G and Alix localized inside exosomes (Fig. 2B). The intraexosomal localization of A3G and Tsg101 (73) was confirmed by their resistance to trypsin in the absence but not in the presence of detergent (Fig. 2C). Interestingly, the major form of A3G detected in H9 exosomes migrated in an SDS-polyacrylamide gel as a protein with an apparent molecular mass higher by about 2 kDa than that of cellular A3G (Fig. 2D). This result cannot be explained by monoubiquitination, which would result in an increase of molecular mass of A3G of about 8.5 kDa. In addition, distinct-molecular-mass forms of Alix were differentially expressed in exosomes and cells; a lower-molecular-mass form of Alix (70 to 75 kDa) was detected in H9 cells and exosomes and a higher-molecular-mass form of Alix (95 kDa) was detected only in H9 exosomes (Fig. 2D). The reason for the preferential localization of 95-kDa Alix in exosomes is presently unknown. In contrast, gel migration of exosomal Tsg101 was unchanged in comparison with that of cellular Tsg101.
FIG. 1.
A3G is released by H9 cells in association with exosomes. (A) Discontinuous (0.25 M/2 M/2.5 M) sucrose gradient analysis of exosomes secreted by CD4+ H9 cells. Cell lysates (30 μg) and fractions collected from the top of sucrose gradient (50 μl) were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting for the presence of A3G and exosome markers Alix, Tsg101, CD71, HSP90, and Rab proteins. Nonexosomal proteins calnexin (endoplasmic reticulum) and VDAC1 (mitochondrion) are detected in cell lysates. (B) Analysis of exosome density in a sucrose gradient. Fractions 3 to 5 from panel A were combined and subjected to an equilibrium ultracentrifugation in a continuous sucrose gradient. Equal fraction volumes (50 μl) were analyzed by Western blotting for the presence of A3G and Alix. Fraction numbers and densities (g/ml) are indicated.
FIG. 2.
A3G is localized inside exosomes. (A) Exosomes purified on a sucrose gradient were negatively stained with 1% phosphotungstic acid and observed under an electron microscope. (B) Immunogold labeling: grids with adsorbed fixed exosomes were permeabilized with 0.02% Triton and labeled with rabbit anti-Alix, anti-A3G, or control (Co) rabbit serum. Bound antibodies were detected with protein A-gold conjugates. Permeabilized exosomes were stained with 1% phosphotungstic acid. Bars, 100 nm. (C) Sucrose gradient-purified exosomes (10 μg) were left untreated (exo), incubated with trypsin (50 μg/ml) for 30 min at room temperature (exo + trypsin), or incubated with 0.1% Triton X-100 (TX) for 5 min followed by trypsin treatment for 30 min (exo + TX + trypsin). A3G and Tsg101 expression was analyzed by Western blotting. “Cells” indicates lysates prepared from untreated cells (10 μg). (D) HMM forms of A3G and Alix in H9 exosomes. Cellular and exosomal lysates were analyzed by Western blotting for the presence of A3G and Alix. The arrow shows a slower-migrating form of A3G in exosomes. In addition to the 70- to 75-kDa form of Alix detected in cells and exosomes, a 95-kDa form of Alix predominantly accumulates in exosomes. (E) A3G-negative 293T and SupT1 cells transfected with A3G expression vectors secrete exosomes with A3G. Exosomes were isolated from culture medium of transiently transfected 293T cells or from G418-resistant SupT1/GFP-A3G cells. Western blot analysis (5 μg protein) shows 95-kDa Alix predominantly concentrated in exosomes. The 75-kDa Alix is detected in SupT1 cells and, after a longer film exposure (not shown), in 293T cells.
Secretion of A3G in exosomes is not limited to H9 cells expressing endogenous A3G and can be detected in exosomes secreted by 293T cells transiently transfected with A3G expression vectors or in exosomes released by CD4+ SupT1 cells stably expressing GFP-A3G (Fig. 2E). In contrast to exosomal A3G secreted by H9 cells, the gel mobility of A3G released by transfected cells does not noticeably differ from that of cellular A3G. The A3G-HA band detected in 293T cell-derived exosomes corresponded to the upper band of A3G in H9 exosomes (Fig. 2E).
A3G in exosomes is catalytically active.
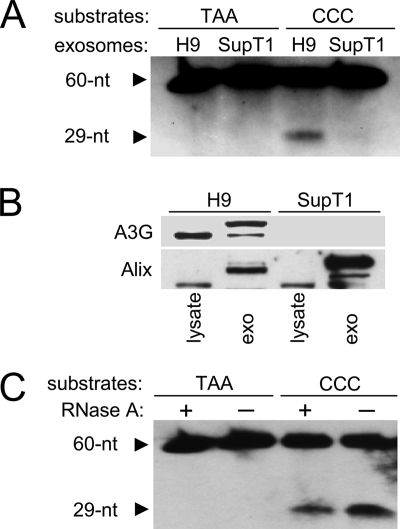
To investigate whether exosomal A3G retains its enzymatic activity, H9 exosome lysates were incubated with a substrate DNA containing a CCC motif specifically targeted by A3G (7). Formation of a biotinylated 29-nt product indicates the presence of cytidine deaminase activity in H9 exosomes (Fig. 3A). In contrast, deaminase activity was not detected in exosomes released by SupT1 cells (Fig. 3A) that do not express A3G (Fig. 3B). Incubation of the SupT1 and H9 exosome lysates with an oligonucleotide mutant TAA did not result in detectable deamination, suggesting that formation of the 29-nt product was specific for A3G present in H9 exosomes. Thus, despite the fact that A3G exists in H9 cells mainly in high-molecular-mass (HMM) ribonucleoprotein complexes with no detectable deaminase activity (11), A3G in exosomes secreted by the cells is enzymatically active. In addition, the presence of RNase A in a deamination reaction slightly decreased rather than increased the enzymatic activity of A3G (Fig. 3C), suggesting that cellular RNA, if present in H9 exosomes (75), does not block the enzymatic activity of A3G.
FIG. 3.
A3G in exosomes is catalytically active. (A) Cytidine deaminase activity in exosomes from A3G-expressing H9 and A3G-negative SupT1 cells (B) was measured with 5′ biotin-labeled deoxyoligonucleotides containing the A3G-targeted sequence CCC and a mutant sequence in which CCC was replaced with TAA. 5′-biotinylated 60-nt substrate and deaminated 29-nt product were separated on a 15% Tris-buffered EDTA-urea polyacrylamide gel and detected with streptavidin-horseradish peroxidase. (B) SupT1 cells release exosomes without A3G. Cell lysates (10 μg) and sucrose gradient-purified exosomes (5 μg) released by H9 and SupT1 cells were analyzed by Western blotting for the expression of A3G and Alix. Faster-migrating protein reacting with anti-Alix is detected in cell lysates. (C) Enzymatic activity of exosomal A3G is not inhibited by RNA. Cytidine deaminase activity of H9 exosomes was measured in the absence or presence of RNase A (1 μg) in the enzymatic reaction. Biotinylated deoxyoligonucleotide substrates and deaminated products were detected as described for panel A.
A3G-laden exosomes are internalized by cells and inhibit HIV-1 replication.
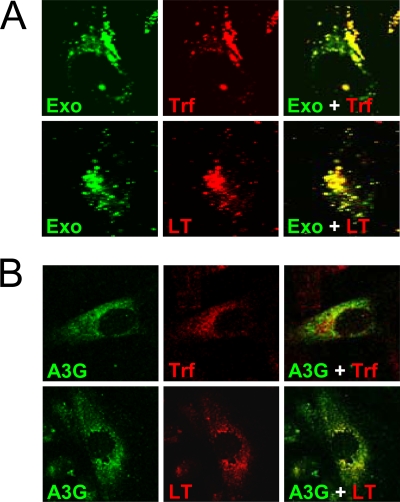
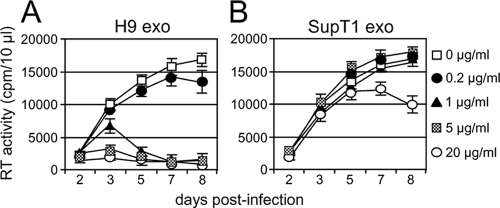
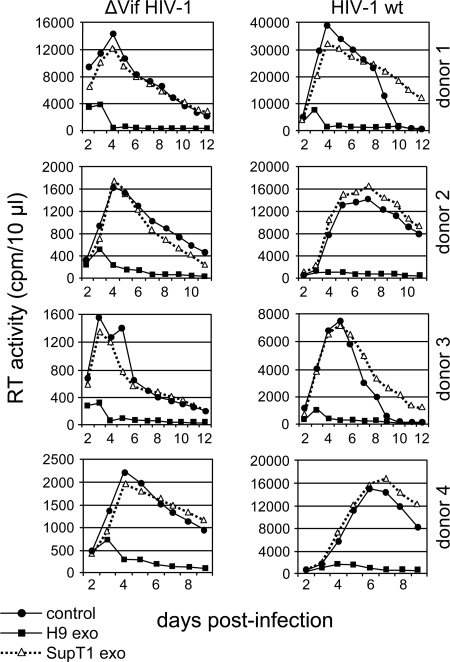
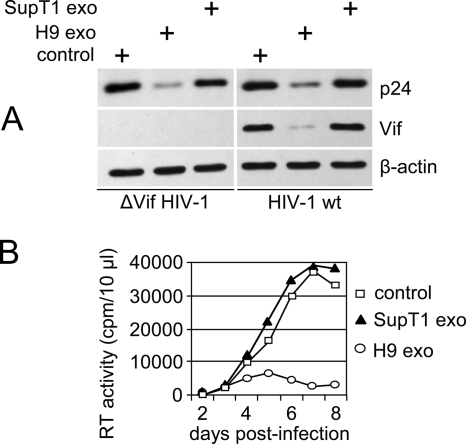
To investigate exosome internalization, H9 exosomal membranes were labeled with green fluorescent PKH67 (34, 41). PKH67-labeled exosomes were rapidly endocytosed by HeLa cells and substantially colocalized with markers specific for early endosomes (transferrin) and late endosomes/lysosomes (LysoTracker) (Fig. 4A). Exosomes fluorescently labeled with encapsidated GFP-A3G showed less-extensive colocalization with late endosomes/lysosomes (Fig. 4B). Although the mechanism of A3G release from internalized exosomes into the cytoplasm is unknown, one possibility is that exosomal A3G escapes degradation by fusion of the endocytosed exosome membrane with the limiting membrane of endosomes and release of A3G into the cytoplasm. Consequently, A3G delivery into the cytoplasm could confer resistance to HIV-1 infection. To investigate this possibility, we analyzed replication of vif-negative HIV-1 in Jurkat T cells preexposed to exosomes. Virus replication was potently blocked by H9 exosomes in a manner depending on exosome protein concentration (Fig. 5A). In contrast, exosomes secreted by A3G-negative SupT1 cells inhibited virus replication only moderately at the highest dose used (Fig. 5B). The inhibitory effect of H9 exosomes on vif-negative HIV-1 replication was not limited to CD4+ T-cell lines but was evident in PHA/IL-2-activated PBMCs obtained from four different blood donors (Fig. 6). Strikingly, replication of wild-type virus was also potently restricted by H9 exosomes in activated PBMCs, suggesting that inhibition could take place early in the virus replication cycle, likely before Vif accumulation. Accordingly, low levels of Vif were detected in H9 exosome-pretreated SupT1 cells infected with wild-type HIV-1 (Fig. 7A). Inhibition of virus replication was confirmed by reduced accumulation of cellular HIV-1 Gag p24 and low levels of released RT (Fig. 7B). In contrast, virus replication in cells pretreated with A3G-negative SupT1 exosomes was slightly enhanced in comparison with that in untreated control cells (Fig. 7B).
FIG. 4.
A3G exosomes are internalized by HeLa cells. (A) PKH67-labeled H9 exosomes (Exo) were incubated with HeLa cells for 2 h followed by incubation for 30 min with transferrin-Alexa Fluor 594 (Trf) or LysoTracker Red (LT). Separate and merged confocal microscopy images are shown. (B) Confocal images of GFP-A3G exosomes internalized by HeLa cells. GFP-A3G exosomes were produced by 293T cells transfected with GFP-A3G expression vector. Exosome incubation with HeLa cells was performed as described for panel A.
FIG. 5.
A3G exosomes inhibit replication of vif-negative HIV-1 in Jurkat T cells. Cells (1 × 106/ml) were preincubated for 16 h with indicated doses of H9 exosomes (A) or SupT1 exosomes (B), followed by 3 h of infection with vif-negative HIV-1 (5 RT cpm/ml). After cell washing, additional indicated doses of exosomes were added to the cell culture. Virus replication was monitored by RT activity released into the culture supernatants. Error bars, mean values ± SDs of triplicate samples.
FIG. 6.
H9 exosomes inhibit HIV-1 replication in PBMCs. PHA/IL-2-activated PBMCs from four different donors were preincubated for 16 h with H9 or SupT1 exosomes, washed, and infected for 3 h with vif-negative (ΔVif HIV-1) or wild-type NL4-3 (HIV-1 wt). Additional exosome doses (10 μg/ml) were added to cell culture on day 2 and day 3 postinfection. Virus replication was monitored by RT activity in the culture supernatants. Control, no exosomes were added.
FIG. 7.
H9 exosomes inhibit HIV-1 replication in SupT1 cells. SupT1 cells were pretreated with A3G-positive (H9 exo) or A3G-negative (SupT1 exo) exosomes or left untreated (control) and infected with vif-negative (ΔVif HIV-1) or wild-type NL4-3 (HIV-1 wt). On day 2 and day 3 postinfection, two additional doses of exosomes were added. (A) Cell lysates were prepared on day 5 postinfection and analyzed by Western blotting for the expression of HIV-1 Gag p24, Vif, and β-actin. (B) Replication of wild-type HIV-1 in untreated or exosome-treated cells was monitored by RT activity released into the culture supernatants.
A3G-containing exosomes do not block virus entry or induce apoptosis.
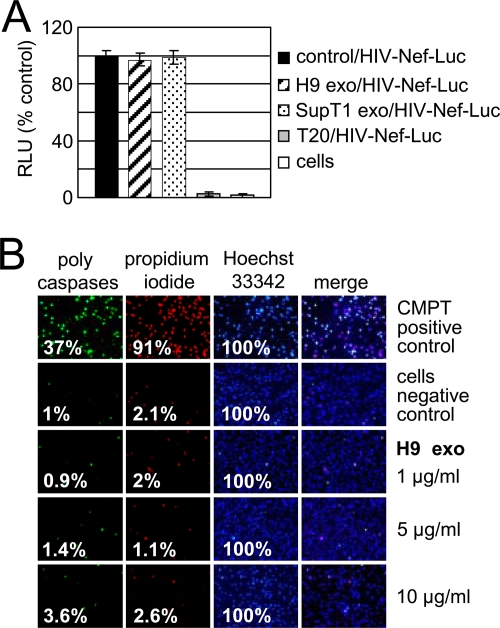
Virus entry was monitored by luciferase activity of a Nef-luciferase fusion protein encapsidated into HIV-1 virions (HIV-Nef-Luc) and released into cytoplasm after virus-cell fusion occurred (31, 56). Infected cells were subsequently exposed to a plasma membrane-permeable luciferase substrate, and luciferase activity was measured in live cells. Exosome pretreatment did not affect virus entry into SupT1 cells; however, in the presence of a fusion inhibitor, T20, HIV-1 entry was almost completely blocked (Fig. 8A). To investigate whether exosomes stimulate apoptosis and cell death, we analyzed activation of caspases and plasma membrane integrity in SupT1 cells cultivated for 72 h with different doses of H9 exosomes (Fig. 8B). Using a fluorescently labeled inhibitor of caspases that binds covalently to activated caspases 1 and 3 to 9, we detected activation of caspases in 3.6% of cells incubated with 10 μg/ml exosomes and in 1% of control cells. Cell death was observed at the levels of 2.6% and 2.1% in exosome-treated and untreated cells, respectively. Thus, H9 exosome treatment did not significantly affect cell viability and thus cannot account for the observed severe inhibition of HIV-1 replication.
FIG. 8.
H9 exosomes do not inhibit HIV-1 entry or induce apoptosis. (A) Entry of HIV-1 into SupT1 cells was measured after 90 min of incubation of Nef-luciferase virus (HIV-Nef-Luc) with 1 × 106 SupT1 cells preincubated for 16 h with exosomes or pretreated for 1 h with HIV-1 entry inhibitor T20 (1 μg/ml). Following incubation, cells were washed and incubated with cell-permeable luciferase substrate, and luciferase activity was measured in live cells. Luciferase activity in treated cells was normalized to luciferase activity of untreated cells (control) exposed to HIV-Nef-Luc (100%). RLU, relative light units; “cells” indicates untreated and uninfected cells. Error bars, mean values ± SDs of triplicate samples. (B) Activation of caspases and cell death in SupT1 cells exposed to H9 exosomes. Cells were left untreated (negative control) or were treated for 24 h with 10 μM camptothecin (CMPT) to induce apoptosis and cell death (positive control). Cells were treated with different doses of exosomes for 72 h followed by staining with FLICA (fluorescently labeled inhibitor of caspases) reagent (caspases) and subsequent staining with 1 μM Hoechst 33342 (nuclei) and 5 μM propidium iodide (dead cells). Fixed cells were observed under a fluorescence microscope using standard filter sets. Percentages of positively stained cells are shown.
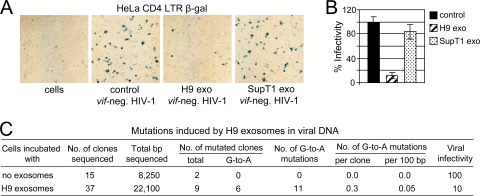
A3G exosomes reduce HIV-1 infectivity without extensive deamination of cytidine in viral DNA.
Using HeLa-CD4-LTR-β-Gal reporter cells, we demonstrated that virus released from SupT1 cells treated with H9 exosomes was approximately 10- and 8-fold less infectious than virus produced by untreated cells or cells exposed to SupT1 exosomes, respectively (Fig. 9A and B). To determine whether the reduced HIV-1 infectivity correlates with deoxycytidine deaminase activity of A3G exosomes, we sequenced DNA fragments spanning a 650-bp nef/3′-LTR region in HIV-1 DNA produced after infection of untreated SupT1 cells or cells pretreated with H9 exosomes. No G-to-A mutations were detected in an HIV-1 DNA fragment from control cells infected with vif-negative HIV-1 (Fig. 9C). In cells exposed to H9 exosomes, only six clones out of 37 tested were carrying a total of 11 G-to-A mutations in the analyzed fragment, which gives an average mutational rate of 0.05%. However, the infectivity of the virus released from SupT1 cells exposed to A3G exosomes was reduced about 10-fold, suggesting that deaminase activity of the encapsidated A3G cannot be the major factor responsible for the reduced infectivity of the progeny virus.
FIG. 9.
H9 exosomes reduce HIV-1 infectivity without extensive cytidine deamination in viral DNA. (A) SupT1 cells pretreated with H9 or SupT1 exosomes or not treated (control) were infected with vif-negative HIV-1, and viral supernatants were harvested on day 3 posttransfection. The same amount of viral inoculum (RT assay standardized) was added in triplicate to HeLa-CD4-LTR-β-Gal indicator cells in a 12-well plate. Three dilutions of viral inoculum were applied: 2 × 105, 2 × 104, and 2 × 103 RT cpm. After 2 days, cells were fixed and stained for β-Gal and positive syncytia (blue) were counted (B). The highest viral inoculum (2 × 105 RT cpm) applied to indicator cells is shown. Percent infectivity is presented relative to infectivity of virus produced by SupT1 cells in the absence of exosomes (100%; control). Results representative of three experiments are shown. Error bars, means ± SDs of triplicate samples. (C) Mutations induced by H9 exosomes in a 650-bp nef/3′-LTR HIV-1 DNA fragment were analyzed as described in Materials and Methods.
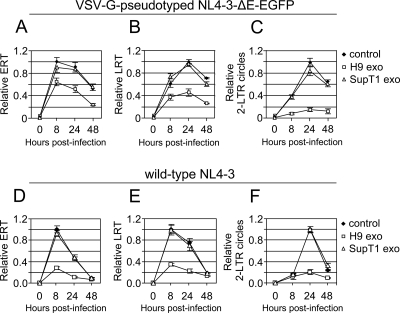
H9 exosomes reduce the levels of HIV-1 reverse transcripts and inhibit accumulation of two-LTR circles.
To explore the mechanism of exosome-mediated HIV-1 inhibition, we investigated accumulation of reverse transcription products in cells treated with exosomes. To limit infection to a single round, SupT1 cells were infected with VSV-G-pseudotyped NL4-3-ΔE-EGFP (84) or with a wild-type NL4-3 virus in the presence of protease inhibitors amprenavir and nelfinavir (5). Accumulation of viral reverse transcription products was monitored by quantitative real-time PCR. In cells infected with VSV-G-pseudotyped virus and treated with H9 exosomes, ERT and LRT decreased by 38% and 57%, respectively, at their peak accumulation (Fig. 10A and B). Even more pronounced inhibition by H9 exosomes was observed in cells infected with wild-type NL4-3 virus. Accumulation of ERT and LRT was decreased by about 70% and 63%, respectively (Fig. 10D and E). In contrast, treatment with SupT1 exosomes did not considerably affect relative levels of ERT and LRT. The highest inhibition was observed in the accumulation of two-LTR circles; their levels were reduced by about 80% at 24 h postinfection (Fig. 10C and F). Since two-LTR circles are formed exclusively in the nucleus, these results suggest that H9 exosomes may inhibit nuclear transport of HIV-1 DNA or formation/stability of two-LTR circles in the nucleus.
FIG. 10.
H9 exosomes reduce accumulation of HIV-1 reverse transcripts and two-LTR circles. SupT1 cells were preincubated for 16 h with H9 or SupT1 exosomes or left untreated (control). Cells were infected for 3 h either with VSV-G-pseudotyped NL4-3-ΔE-EGFP (A to C) or with wild-type NL4-3 in the presence of protease inhibitors amprenavir and nelfinavir, each at 5 μM (D to F), washed, and cultured without (control) or with an additional dose of exosomes. Total DNA was isolated at indicated times postinfection. Relative levels of HIV-1 ERT (A and D), LRT (B and E), and two-LTR circles (C and F) were quantified by real-time PCR. Error bars, means ± SDs of triplicate samples from a representative experiment.
A3G contributes to the antiretroviral activity of exosomes.
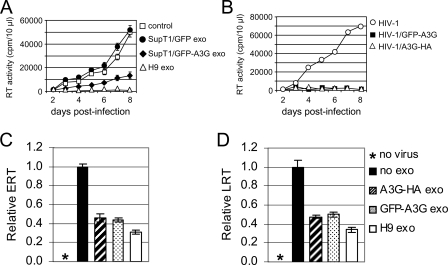
A3G short interfering RNA approaches to considerably reduce A3G levels in H9 cells were unsuccessful (data not shown). Thus, to investigate the contribution of A3G to anti-HIV-1 activity of exosomes, we transfected A3G-negative SupT1 cells with GFP-A3G or GFP expression vectors. Exosomes produced by transfected SupT1/GFP cells did not inhibit replication of vif-negative HIV-1; however, exosomes secreted by SupT1/GFP-A3G cells significantly reduced virus replication, although less effectively than did H9 exosomes (Fig. 11A). Both GFP-A3G and A3G-HA were fully functional when encapsidated into vif-negative virions and severely inhibited virus replication (Fig. 11B). We also tested the effect of exosomes secreted by 293T cells transfected transiently with A3G expression vectors on the accumulation of ERT and LRT upon infection with wild-type NL4-3. Results show that 293T exosomes inhibited accumulation of ERT and LRT in SupT1 cells, although not as effectively as H9 exosomes (Fig. 11C and D).
FIG. 11.
Exosomal A3G contributes to inhibition of HIV-1 replication. (A) Exosomes were isolated from culture medium of SupT1 cells transfected with GFP-A3G or GFP expression vectors. SupT1 cells were incubated without (control) or with indicated exosomes and then infected with vif-negative HIV-1. Additional doses of exosomes (10 μg/ml) were added 2, 3, and 5 days postinfection. Virus replication was monitored by RT activity released into the culture supernatants. (B) A3G tagged with HA or GFP retains full anti-HIV-1 activity when encapsidated into vif-negative virions. HIV-1 virions were produced by 293T cells transfected with vif-negative pNL4-3 plasmid DNA alone or in combination with expression vectors coding for GFP-A3G or A3G-HA. SupT1 cells were infected with equivalent amounts of viruses, and virus replication was monitored daily by RT activity released into the culture supernatants. (C and D) Exosomes secreted by 293T cells expressing A3G reduce accumulation of HIV-1 ERT (C) and LRT (D). SupT1 cells were preincubated for 16 h with 293T-derived exosomes or H9 exosomes or left untreated (no exo). Cells were infected for 2 h with wild-type NL4-3, washed, and cultured with an additional dose of exosomes for additional 6 h. Uninfected cells (no virus) served as a negative control. Relative levels of ERT and LRT were quantified by real-time PCR. Error bars, means ± SDs of triplicate samples.
Together, these data suggest that exosomal A3G significantly contributes to anti-HIV-1 activity of exosomes secreted by various cell types. We cannot, however, exclude the possibility that factors other than A3G may also contribute to H9 exosome-mediated restriction of HIV-1.
DISCUSSION
In this study, we tested the hypothesis that A3G is secreted in exosomes and that A3G-laden exosomes can confer an antiviral phenotype to target cells. We have shown that H9 CD4+ T cells expressing endogenous A3G incorporate this protein into vesicles that are released into the culture medium. Vesicles released by H9 cells meet the criteria for exosome identity; their size, density, and protein composition are similar to those described for exosomes released by other cells (73, 74, 80). We have also demonstrated that exosomal A3G is localized inside vesicles and thus can be protected from degradation by extracellular proteases. Interestingly, the major form of exosomal A3G migrates slower in a polyacrylamide gel than does its cytoplasmic counterpart. The significance of the observation is currently unknown, since A3G in exosomes secreted by 293T or SupT1 cells transfected with A3G constructs does not show aberrant migration in a gel.
Previous results show that the enzymatic activity of A3G in H9 cells is suppressed by its recruitment into HMM ribonucleoprotein complexes from which A3G activity can be rescued by RNase A treatment (11). Similarly, activity of intravirion A3G, blocked by HIV-1 or cellular RNAs, is rescued during reverse transcription by viral RNase H (67). In contrast, exosomal A3G remains enzymatically active. Since exosomes may incorporate cellular RNAs (75), our experiments with RNase A suggest that cellular RNAs, if present in H9 exosomes, do not suppress enzymatic activity of A3G.
Exosome internalization is required to affect at least some target cell functions (34, 41, 81). Although we cannot exclude the possibility that a fraction of exosomes fuse with the plasma membrane and deliver their cargo into the cytosol, we have shown that H9 exosomes labeled with fluorescent lipid PKH67 are efficiently internalized by the cells and colocalize with markers specific for early and late endosomes or lysosomes. Since PKH67 labels only exosomal membrane, a fluorescent signal found in endosomal compartments may not accurately trace localization of intraexosomal proteins. For instance, intraexosomal proteins may escape degradation in lysosomes when endocytosed exosomes fuse with a limiting membrane of endocytic vesicles and release them into the cytosol, before endocytic vesicles reach a degradation compartment. This possibility, although not directly proved, seems to be supported by reduced colocalization of exosomal GFP-A3G with late endosomes or lysosomes compared with exosomal membranes. In support of the notion that molecules delivered by exosomes may escape degradation is the observation that some cellular receptors (37), transcription factors (53), or mRNAs (15, 53, 75) transferred by microvesicles or exosomes remain functional in recipient cells. Indeed, supporting this idea, our results show that A3G-positive H9 exosomes, but not exosomes released by A3G-negative SupT1 cells, restrict replication of HIV-1. The inhibitory effect of H9 exosomes was detected both in CD4+ T-cell lines and in stimulated PBMCs from different donors. Using a real-time virus entry assay (31, 56), we have ruled out the possibility that virus entry is affected by exosome pretreatment. Similarly, H9 exosomes do not noticeably stimulate apoptosis or cell death in target cells, and thus, these events are not implicated in exosome-mediated inhibition of virus replication.
Although our experiments suggest that A3G in exosomes is responsible for the inhibition of HIV-1 replication, a direct proof would require obtaining H9 exosomes without A3G. However, using a typical short interfering RNA approach, we were unable to downregulate A3G in H9 cells to levels low enough to observe a sufficient reduction of A3G in exosomes. Nevertheless, we successfully introduced the A3G gene into A3G-negative SupT1 cells. Exosomes secreted by these cells inhibited HIV-1 replication, yet not as potently as H9 exosomes did. Similarly, exosomes secreted by 293T cells transiently transfected with A3G have anti-HIV-1 activity, although not as potent as that of H9 exosomes. Thus, we cannot exclude the possibility that in addition to A3G, other factors present in exosomes may contribute to HIV-1 restriction. Recent reports showing that microvesicles and exosomes shuttle functional mRNAs, microRNAs, and bioactive proteins further support this possibility (22, 23, 53, 54, 75).
An intriguing observation is that H9 exosomes also inhibit replication of wild-type HIV-1. This seems to contradict reports showing that HIV-1 Vif mediates A3G degradation (38, 64, 82). Since A3G in exosomes is enzymatically active and thus most likely present in low-molecular-mass form (11), we speculate that A3G delivered by exosomes to cytosol does not convert into HMM form. Consequently, exosomal A3G would function as a postentry restriction factor as proposed for cellular low-molecular-mass A3G in resting CD4+ T cells (8, 11), monocytes and monocyte-derived dendritic cells (47, 48), or peripheral plasmacytoid dendritic cells (77). This notion is consistent with our observations showing reduced accumulation of HIV-1 reverse transcription products and nuclear two-LTR circles in cells exposed to H9 exosomes. Accordingly, we observed low cellular levels of viral Gag p24 and Vif. Thus, low levels of Vif may inefficiently degrade A3G delivered by exosomes. Although the infectivity of progeny viruses released from H9 exosome-treated cells is reduced by about 10-fold, the low frequency of G-to-A mutations detected in viral cDNA suggests that an editing-independent mechanism may possibly play a role in HIV-1 restriction (11, 21, 25, 43).
In conclusion, findings reported here suggest that A3G is packaged into exosomes that potently restrict replication of HIV-1 in recipient cells in experimental conditions in vitro. One of the reasons that exosomes in vivo do not inhibit HIV-1 may be due to the fact that only a small population of exosomes, produced by A3G-expressing cells, carry A3G and that HIV-1 infects cells that are primary producers of A3G exosomes (activated T cells), depleting A3G from the cells and consequently from exosomes.
The functional relevance of exosomes in vivo is still debated; however, accumulating data suggest that exosomes may play a role in modulation of the immune system. In this regard, the presence of A3G in exosomes may be “intended” or may just represent a way of disposing of excessive amounts of APOBECs that might otherwise cause genomic instability (2). It may also be possible that exosomes carrying A3G are addressed in vivo to specific cells (e.g., A3G-negative cells) with a task different than inhibition of HIV-1. For instance, A3G-rich exosomes could inhibit mobility of retrotransposons and endogenous retroviruses (12, 17, 24, 60) in cells that do not normally express A3G. Moreover, recent findings show that A3G prevents the decay of microRNA-targeted mRNA and prevents inhibition of protein synthesis (22). Together with the finding that A3 is expressed in mouse germ cells (3), these results suggest that A3G delivered by exosomes may play a role during embryogenesis.
Exosomes, whether natural or engineered, may serve as intercellular vehicles for the exchange of powerful antiretroviral factors. This could lead to the development of novel treatments for AIDS and other infectious diseases.
Acknowledgments
This work was supported by the Meharry Center for AIDS Health Disparities Research (NCRR grant U54RR019192) and the Meharry-Vanderbilt Center for AIDS Research (NIAID grant P30AI054999) and in part by the Research Centers in Minority Institutes program (G12RR003032).
We thank A. Takaori-Kondo, H. Stenmark, and K. Strebel for providing reagents. Fusion inhibitor T20 and HeLa-CD4—LTR-β-Gal cells were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Confocal microscopy experiments were performed in the MMC Morphology Core, supported in part by NIH grants U54NS041071, G12RR03032, and U54CA91408. We thank Jared Elzey for his assistance in manuscript editing and preparation.
Footnotes
Published ahead of print on 5 November 2008.
REFERENCES
- 1.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 27934083-34086. [DOI] [PubMed] [Google Scholar]
- 2.Anant, S., and N. O. Davidson. 2003. Hydrolytic nucleoside and nucleotide deamination, and genetic instability: a possible link between RNA-editing enzymes and cancer? Trends Mol. Med. 9147-152. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya, C., S. Aggarwal, M. Kumar, A. Ali, and A. Matin. 2008. Mouse apolipoprotein B editing complex 3 (APOBEC3) is expressed in germ cells and interacts with dead-end (DND1). PLoS ONE 3e2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7631-634. [DOI] [PubMed] [Google Scholar]
- 5.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 763739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 27933177-33184. [DOI] [PubMed] [Google Scholar]
- 7.Chelico, L., P. Pham, P. Calabrese, and M. F. Goodman. 2006. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 13392-399. [DOI] [PubMed] [Google Scholar]
- 8.Chen, K., J. Huang, C. Zhang, S. Huang, G. Nunnari, F. X. Wang, X. Tong, L. Gao, K. Nikisher, and H. Zhang. 2006. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J. Virol. 807645-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chertova, E., O. Chertov, L. V. Coren, J. D. Roser, C. M. Trubey, J. W. Bess, Jr., R. C. Sowder II, E. Barsov, B. L. Hood, R. J. Fisher, K. Nagashima, T. P. Conrads, T. D. Veenstra, J. D. Lifson, and D. E. Ott. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 809039-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26317-353. [DOI] [PubMed] [Google Scholar]
- 11.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435108-114. [DOI] [PubMed] [Google Scholar]
- 12.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 10315588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 132009-2013. [DOI] [PubMed] [Google Scholar]
- 14.de Gassart, A., C. Geminard, D. Hoekstra, and M. Vidal. 2004. Exosome secretion: the art of reutilizing nonrecycled proteins? Traffic 5896-903. [DOI] [PubMed] [Google Scholar]
- 15.Deregibus, M. C., V. Cantaluppi, R. Calogero, M. Lo Iacono, C. Tetta, L. Biancone, S. Bruno, B. Bussolati, and G. Camussi. 2007. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 1102440-2448. [DOI] [PubMed] [Google Scholar]
- 16.Douaisi, M., S. Dussart, M. Courcoul, G. Bessou, R. Vigne, and E. Decroly. 2004. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem. Biophys. Res. Commun. 321566-573. [DOI] [PubMed] [Google Scholar]
- 17.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433430-433. [DOI] [PubMed] [Google Scholar]
- 18.Fang, Y., N. Wu, X. Gan, W. Yan, J. C. Morrell, and S. J. Gould. 2007. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 5e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fevrier, B., D. Vilette, F. Archer, D. Loew, W. Faigle, M. Vidal, H. Laude, and G. Raposo. 2004. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA 1019683-9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao, S., Z. Ye, F. Li, Q. Meng, M. Qureshi, J. Yang, and J. Xiang. 2006. Epigenetic transfer of metastatic activity by uptake of highly metastatic B16 melanoma cell-released exosomes. Exp. Oncol. 28126-131. [PubMed] [Google Scholar]
- 21.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 2822587-2595. [DOI] [PubMed] [Google Scholar]
- 22.Huang, J., Z. Liang, B. Yang, H. Tian, J. Ma, and H. Zhang. 2007. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J. Biol. Chem. 28233632-33640. [DOI] [PubMed] [Google Scholar]
- 23.Huang, J., F. Wang, E. Argyris, K. Chen, Z. Liang, H. Tian, W. Huang, K. Squires, G. Verlinghieri, and H. Zhang. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 131241-1247. [DOI] [PubMed] [Google Scholar]
- 24.Hulme, A. E., H. P. Bogerd, B. R. Cullen, and J. V. Moran. 2007. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 390199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwatani, Y., D. S. Chan, F. Wang, K. S. Maynard, W. Sugiura, A. M. Gronenborn, I. Rouzina, M. C. Williams, K. Musier-Forsyth, and J. G. Levin. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 357096-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnstone, R. M., A. Mathew, A. B. Mason, and K. Teng. 1991. Exosome formation during maturation of mammalian and avian reticulocytes: evidence that exosome release is a major route for externalization of obsolete membrane proteins. J. Cell. Physiol. 14727-36. [DOI] [PubMed] [Google Scholar]
- 27.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 7711398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan, M. A., S. Kao, E. Miyagi, H. Takeuchi, R. Goila-Gaur, S. Opi, C. L. Gipson, T. G. Parslow, H. Ly, and K. Strebel. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 795870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 662232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinomoto, M., T. Kanno, M. Shimura, Y. Ishizaka, A. Kojima, T. Kurata, T. Sata, and K. Tokunaga. 2007. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 352955-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolokoltsov, A. A., and R. A. Davey. 2004. Rapid and sensitive detection of retrovirus entry by using a novel luciferase-based content-mixing assay. J. Virol. 785124-5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, X. Y., F. Guo, L. Zhang, L. Kleiman, and S. Cen. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 28232065-32074. [DOI] [PubMed] [Google Scholar]
- 33.Liegeois, S., A. Benedetto, J. M. Garnier, Y. Schwab, and M. Labouesse. 2006. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol. 173949-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, C., S. Yu, K. Zinn, J. Wang, L. Zhang, Y. Jia, J. C. Kappes, S. Barnes, R. P. Kimberly, W. E. Grizzle, and H. G. Zhang. 2006. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J. Immunol. 1761375-1385. [DOI] [PubMed] [Google Scholar]
- 35.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X. F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 7811841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X. F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 817238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mack, M., A. Kleinschmidt, H. Bruhl, C. Klier, P. J. Nelson, J. Cihak, J. Plachy, M. Stangassinger, V. Erfle, and D. Schlondorff. 2000. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat. Med. 6769-775. [DOI] [PubMed] [Google Scholar]
- 38.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 91398-1403. [DOI] [PubMed] [Google Scholar]
- 39.Mehta, S. H., J. Astemborski, T. R. Sterling, D. L. Thomas, and D. Vlahov. 2006. Serum albumin as a prognostic indicator for HIV disease progression. AIDS Res. Hum. Retrovir. 2214-21. [DOI] [PubMed] [Google Scholar]
- 40.Miyagi, E., S. Opi, H. Takeuchi, M. Khan, R. Goila-Gaur, S. Kao, and K. Strebel. 2007. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 8113346-13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morelli, A. E., A. T. Larregina, W. J. Shufesky, M. L. Sullivan, D. B. Stolz, G. D. Papworth, A. F. Zahorchak, A. J. Logar, Z. Wang, S. C. Watkins, L. D. Falo, Jr., and A. W. Thomson. 2004. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 1043257-3266. [DOI] [PubMed] [Google Scholar]
- 42.Muntasell, A., A. C. Berger, and P. A. Roche. 2007. T cell-induced secretion of MHC class II-peptide complexes on B cell exosomes. EMBO J. 264263-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15166-170. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 27852347-52354. [DOI] [PubMed] [Google Scholar]
- 45.Niewiadomska, A. M., C. Tian, L. Tan, T. Wang, P. T. Sarkis, and X. F. Yu. 2007. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 819577-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peche, H., K. Renaudin, G. Beriou, E. Merieau, S. Amigorena, and M. C. Cuturi. 2006. Induction of tolerance by exosomes and short-term immunosuppression in a fully MHC-mismatched rat cardiac allograft model. Am. J. Transplant. 61541-1550. [DOI] [PubMed] [Google Scholar]
- 47.Peng, G., T. Greenwell-Wild, S. Nares, W. Jin, K. J. Lei, Z. G. Rangel, P. J. Munson, and S. M. Wahl. 2007. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood 110393-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pion, M., A. Granelli-Piperno, B. Mangeat, R. Stalder, R. Correa, R. M. Steinman, and V. Piguet. 2006. APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J. Exp. Med. 2032887-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popik, W., T. M. Alce, and W. C. Au. 2002. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4+ T cells. J. Virol. 764709-4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popik, W., and P. M. Pitha. 2000. Inhibition of CD3/CD28-mediated activation of the MEK/ERK signaling pathway represses replication of X4 but not R5 human immunodeficiency virus type 1 in peripheral blood CD4+ T lymphocytes. J. Virol. 742558-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porto-Carreiro, I., B. Fevrier, S. Paquet, D. Vilette, and G. Raposo. 2005. Prions and exosomes: from PrPc trafficking to PrPsc propagation. Blood Cells Mol. Dis. 35143-148. [DOI] [PubMed] [Google Scholar]
- 52.Raposo, G., H. W. Nijman, W. Stoorvogel, R. Liejendekker, C. V. Harding, C. J. Melief, and H. J. Geuze. 1996. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1831161-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ratajczak, J., K. Miekus, M. Kucia, J. Zhang, R. Reca, P. Dvorak, and M. Z. Ratajczak. 2006. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20847-856. [DOI] [PubMed] [Google Scholar]
- 54.Ratajczak, J., M. Wysoczynski, F. Hayek, A. Janowska-Wieczorek, and M. Z. Ratajczak. 2006. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia 201487-1495. [DOI] [PubMed] [Google Scholar]
- 55.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2005. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res. Hum. Retrovir. 21611-619. [DOI] [PubMed] [Google Scholar]
- 56.Saeed, M. F., A. A. Kolokoltsov, and R. A. Davey. 2006. Novel, rapid assay for measuring entry of diverse enveloped viruses, including HIV and rabies. J. Virol. Methods 135143-150. [DOI] [PubMed] [Google Scholar]
- 57.Sasada, A., A. Takaori-Kondo, K. Shirakawa, M. Kobayashi, A. Abudu, M. Hishizawa, K. Imada, Y. Tanaka, and T. Uchiyama. 2005. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schafer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328163-168. [DOI] [PubMed] [Google Scholar]
- 59.Schumacher, A. J., G. Hache, D. A. Macduff, W. L. Brown, and R. S. Harris. 2008. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 822652-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 1029854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Segura, E., C. Guerin, N. Hogg, S. Amigorena, and C. Thery. 2007. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J. Immunol. 1791489-1496. [DOI] [PubMed] [Google Scholar]
- 62.Segura, E., C. Nicco, B. Lombard, P. Veron, G. Raposo, F. Batteux, S. Amigorena, and C. Thery. 2005. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood 106216-223. [DOI] [PubMed] [Google Scholar]
- 63.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 64.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 91404-1407. [DOI] [PubMed] [Google Scholar]
- 65.Skokos, D., H. G. Botros, C. Demeure, J. Morin, R. Peronet, G. Birkenmeier, S. Boudaly, and S. Mecheri. 2003. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J. Immunol. 1703037-3045. [DOI] [PubMed] [Google Scholar]
- 66.Skokos, D., S. Le Panse, I. Villa, J. C. Rousselle, R. Peronet, B. David, A. Namane, and S. Mecheri. 2001. Mast cell-dependent B and T lymphocyte activation is mediated by the secretion of immunologically active exosomes. J. Immunol. 166868-876. [DOI] [PubMed] [Google Scholar]
- 67.Soros, V. B., W. Yonemoto, and W. C. Greene. 2007. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 3e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sprent, J. 2005. Direct stimulation of naive T cells by antigen-presenting cell vesicles. Blood Cells Mol. Dis. 3517-20. [DOI] [PubMed] [Google Scholar]
- 69.Stoorvogel, W., M. J. Kleijmeer, H. J. Geuze, and G. Raposo. 2002. The biogenesis and functions of exosomes. Traffic 3321-330. [DOI] [PubMed] [Google Scholar]
- 70.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12591-601. [DOI] [PubMed] [Google Scholar]
- 71.Strebel, K. 2005. APOBEC3G & HTLV-1: inhibition without deamination. Retrovirology 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 27935822-35828. [DOI] [PubMed] [Google Scholar]
- 73.Thery, C., M. Boussac, P. Veron, P. Ricciardi-Castagnoli, G. Raposo, J. Garin, and S. Amigorena. 2001. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 1667309-7318. [DOI] [PubMed] [Google Scholar]
- 74.Thery, C., L. Zitvogel, and S. Amigorena. 2002. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2569-579. [DOI] [PubMed] [Google Scholar]
- 75.Valadi, H., K. Ekstrom, A. Bossios, M. Sjostrand, J. J. Lee, and J. O. Lotvall. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9654-659. [DOI] [PubMed] [Google Scholar]
- 76.Vella, L. J., R. A. Sharples, V. A. Lawson, C. L. Masters, R. Cappai, and A. F. Hill. 2007. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 211582-590. [DOI] [PubMed] [Google Scholar]
- 77.Wang, F. X., J. Huang, H. Zhang, X. Ma, and H. Zhang. 2008. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J. Gen. Virol. 89722-730. [DOI] [PubMed] [Google Scholar]
- 78.Wang, T., C. Tian, W. Zhang, K. Luo, P. T. Sarkis, L. Yu, B. Liu, Y. Yu, and X. F. Yu. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 8113112-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wiley, R. D., and S. Gummuluru. 2006. Immature dendritic cell-derived exosomes can mediate HIV-1 trans infection. Proc. Natl. Acad. Sci. USA 103738-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wubbolts, R., R. S. Leckie, P. T. Veenhuizen, G. Schwarzmann, W. Mobius, J. Hoernschemeyer, J. W. Slot, H. J. Geuze, and W. Stoorvogel. 2003. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 27810963-10972. [DOI] [PubMed] [Google Scholar]
- 81.Yu, S., C. Liu, K. Su, J. Wang, Y. Liu, L. Zhang, C. Li, Y. Cong, R. Kimberly, W. E. Grizzle, C. Falkson, and H. G. Zhang. 2007. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J. Immunol. 1786867-6875. [DOI] [PubMed] [Google Scholar]
- 82.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 3021056-1060. [DOI] [PubMed] [Google Scholar]
- 83.Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 7812058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang, H., Y. Zhou, C. Alcock, T. Kiefer, D. Monie, J. Siliciano, Q. Li, P. Pham, J. Cofrancesco, D. Persaud, and R. F. Siliciano. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 781718-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]