Abstract
The packaging signal (ψ) of human immunodeficiency virus type 2 (HIV-2) is present in the 5′ noncoding region of RNA and contains a 10-nucleotide palindrome (pal; 5′-392-GGAGUGCUCC) located upstream of the dimerization signal stem-loop 1 (SL1). pal has been shown to be functionally important in vitro and in vivo. We previously showed that the 3′ side of pal (GCUCC-3′) is involved in base-pairing interactions with a sequence downstream of SL1 to make an extended SL1, which is important for replication in vivo and the regulation of dimerization in vitro. However, the role of the 5′ side of pal (5′-GGAGU) was less clear. Here, we characterized this role using an in vivo SELEX approach. We produced a population of HIV-2 DNA genomes with random sequences within the 5′ side of pal and transfected these into COS-7 cells. Viruses from COS-7 cells were used to infect C8166 permissive cells. After several weeks of serial passage in C8166 cells, surviving viruses were sequenced. On the 5′ side of pal there was a striking convergence toward a GGRGN consensus sequence. Individual clones with consensus and nonconsensus sequences were tested in infectivity and packaging assays. Analysis of individuals that diverged from the consensus sequence showed normal viral RNA and protein synthesis but had replication defects and impaired RNA packaging. These findings clearly indicate that the GGRG motif is essential for viral replication and genomic RNA packaging.
The 5′ leader RNA of retroviruses is involved in the regulation of many essential steps of the retroviral replication cycle. These steps include transcription, splicing, translation, genomic RNA dimerization, packaging, and reverse transcription (RT) (1, 8, 12, 27, 34-36, 38, 40). RNA packaging is a critical step during which two molecules of full-length genomic RNA are encapsidated into the budding virion in the form of a dimer (for a review, see reference 42). Furthermore, packaging is a highly selective and specific process in which unspliced genomic RNA is preferentially incorporated in viral particles over a vast excess of cellular mRNAs and spliced viral RNAs (31, 32). This selectivity occurs through the interaction between trans-acting Gag polyprotein and cis-acting elements in the genomic RNA. The cis elements are known as packaging signals (psi or Ψ) and have been mapped in the 5′ leader region of unspliced genomic RNA in many retroviruses (22, 31, 42).
The packaging signal of human immunodeficiency virus type 1 (HIV-1) has been extensively studied. Deletion analyses have found that the most important packaging signal (Ψ core) of HIV-1 genomic RNA lies between the major splice donor site (SD) and the gag initiation codon in the leader region (2, 10, 29). In vitro studies have suggested that the HIV-1 packaging signal is composed of at least four stem-loops located both upstream and downstream of SD (6, 11). Although the HIV-1 Ψ core is located downstream of SD in the leader region, other packaging signals have been characterized either upstream of SD (24) or even outside the 5′ leader region (7). Thus, the full HIV-1 packaging signal is a complex, compound structure.
The HIV-2 packaging signal has been less extensively studied and conclusions about the exact location of the Ψ core have been more controversial (20, 21, 35, 41). Several studies demonstrated the importance of sequences located downstream of SD (20, 41). However, other studies showed that deletion of a 28-nucleotide (nt) sequence located upstream of SD caused a severe packaging defect for HIV-2 genomic RNA packaging, whereas mutation of sequences downstream of SD had mild effects on packaging (21, 35). The unusual localization of the HIV-2 Ψ core upstream of SD suggested that, unlike HIV-1, the strongest HIV-2 packaging signal is present in both unspliced and spliced viral RNAs (35). In addition, a 10-nt palindrome sequence (pal; 5′-392GGAGUGCUCC401) was described within the 28-nt Ψ core sequence, just upstream of the SL1 dimerization signal (see Fig. 1B) (25). Mutations of the pal element revealed its involvement in RNA dimerization both in vitro and in vivo (3, 25, 26, 30).
FIG. 1.
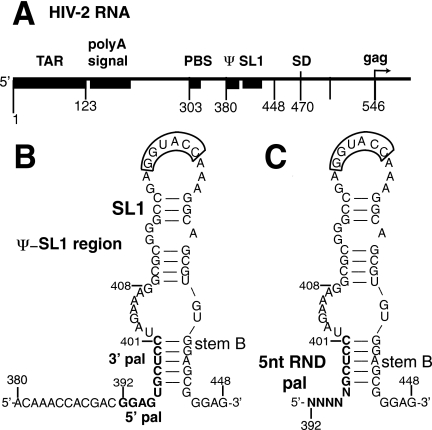
Schematic of the 5′ untranslated leader region of HIV-2 genomic RNA. (A) The 5′ leader region of HIV-2 RNA is represented with boxes and numbers to indicate the landmark sequences with their names indicated above. TAR, poly(A) signal, PBS, ψ, SL1, SD, and gag represent the transactivation region, the poly (A) signal domain, the primer binding site, the packaging signal, the stem-loop 1, the major splice donor site, and the 5′ end of the Gag protein coding region, respectively. (B) The secondary structure of Ψ-SL1 region (nt 380 to 448) (3, 15, 35). The pal element is a 10-nt palindrome sequence (nt 392 to 401) within Ψ, located immediately upstream of SL1 and is represented by boldface letters. nt 392 to 396 and 397 to 401 represent the 5′ and 3′ sides of pal, respectively. The 3′ pal sequence is involved in a base-pairing interaction with a sequence downstream of SL1 and creates stem B. The 5′ side of pal is a purine-rich motif upstream of 3′ pal pyrimidine-rich sequence. The 6-nt autocomplementary sequence essential for SL1-mediated dimerization in the apical loop of SL1 is outlined. (C) Five nucleotides at the 5′ side of pal (positions 392 to 396) were randomized in the insertion segment and inserted into the full-length proviral DNA as described in the text. The last 5 nt of pal (nt 397 to 401) were kept wild type so as not to disrupt stem B.
Since the Pal motif is present in the HIV-2 packaging signal and can interfere with HIV-2 leader RNA dimerization in vitro, its role during viral replication was investigated. Deletion and substitution studies indicated that pal is required for efficient genome packaging and viral replication (26). Reversion analysis of pal mutant viruses in long-term culture suggested that the 3′ side of pal (GCUCC-3′) interacts with a downstream sequence and forms an extension of SL1 called stem B that is required for viral replication and genomic encapsidation (26). In parallel, an in vitro SELEX study found the 3′ side of pal to be involved in the stem B structure that regulates SL1-mediated HIV-2 RNA dimerization in vitro (3). Finally, mutations in the pal/SL1 region showed pal to be the most important determinant for genomic RNA packaging and dimerization (30).
These studies have clearly suggested a structural and functional role for the 3′ side of pal in vitro and in vivo. To understand the role of the 5′ side of pal, we used an in vivo randomization and selection methodology similar to one first used by Berkhout and Klaver (4) to study the role of the bulge and loop of the TAR element during viral replication in eukaryotic cells. Using this technique, we selected for replication-competent viruses from a population of viruses that were randomized in pal. We constructed a library of full-length HIV-2 proviral DNA genomes with random sequences in the 5′ side of pal and selected viable viruses after transfection and subsequent infection of permissive cells. Our sequencing data showed that 5′ pal nucleotides of the selected viable (“winner”) viruses converged to the consensus sequence 5′-GGRGN. Analysis of individual clones that diverged from this sequence showed replication defects and impaired RNA packaging, whereas sequences of the type 5′-GGRGN restored these features. These data strongly support the hypothesis that pal is a critical packaging signal during the HIV-2 replication cycle.
MATERIALS AND METHODS
Construction of plasmids for generation of randomized proviral DNA libraries.
To rule out the chance of contamination by wild-type proviral DNA plasmid in the pal-randomized libraries, we used a parent plasmid called pSCR2, which harbors deleterious mutations in the pal sequence to construct our random library. pSCR2 was derived from modified pROD10 (a full-length plasmid containing infectious HIV-2 DNA sequence) and remained noninfectious even after several months in culture (26).
To create a vector for generating the randomized libraries, a derivative of pSCR2 plasmid called pSCR2AleIΔ(396-2030) was constructed, in which a fragment encompassing nt 396 to 2030 of the viral genome, including part of the noncoding region, and most of the Gag coding region up to the XhoI site was deleted, and a nucleotide was substituted (A394T) to introduce a unique AleI restriction site. All nucleotide numbering in the present study is referenced to the RNA sequence of HIV-2 (ROD isolate, GenBank no. M15390). The mutated vector sequence is as follows: 5′-380ACAAACCACGACGGtG395//2031CTCGAG-3′, where AleI and XhoI sites are underlined and the lowercase “t” indicates the changed nucleotide.
pSCR2AleIΔ(396-2030) was constructed as follows. A fragment containing the long terminal repeat (LTR) and the sequence up to nt 395 with the A394T substitution, along with the 6-nt subsequent sequence of the XhoI site, was amplified, using a sense primer [M13 forward (−41) binding upstream of a unique AatII site; Table 1] and an antisense primer (asXhoI-AleI, containing the XhoI and AleI sites; Table 1). The amplified product was digested with AatII/XhoI and ligated into the pSCR2 plasmid vector missing the original AatII-XhoI fragment. The ligated product was transformed in Escherichia coli DH5α cells, and ampicillin-resistant colonies were selected, followed by plasmid DNA purification.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| M13 forward (−41) | CGC CAG GGT TTT CCC AGT CAC GAC |
| asXhoI-AleI | AAC TCG AGC ACC GTC GTG GTT TGT TCC TG |
| sPal-random-5 | GGG CGG CAG GAA CAA ACC ACG ACN NNN NGC TCC TAG AAA GGC GCG GGC CGA GG |
| asROD2050 | GGT GTC TCC CCC TGC TC |
| sBamT7R | TAG GAT CCT AAT ACG ACT CAC TAT AGG TCG CTC TGC GGA GAG |
| asEco561 | AAG AAT TCA GTT TCT CGC GCC CAT CTC CC |
| sPal-SM2 | GG CGG CAG GAA CAA ACC ACG ACC CTC TGC TCC TAG AAA GGC GCG G |
| sPal-TTB-16 | GGG CGG CAG GAA CAA ACC ACG ACA CCT GGC TCC TAG AAA GGC GCG G |
| sPal-random-2 | GGG CGG CAG GAA CAA ACC ACG ACG GNG NGC TCC TAG AAA GGC GCG G |
Generation of pal-randomized proviral DNA library.
To generate the proviral DNA library that is randomized at the 5′ side of pal (5-nt RND pal), we used the In-Fusion (Clontech) in vitro DNA recombination protocol, which obviates the need of a DNA ligase step and combines the insert and vector due to the homologous regions at both the 5′ and 3′ ends of the insert and digested vector. We constructed our vector and randomized insert with these homologous regions by the use of long primers according to the manufacturer's protocol.
To create the vector for use in the In-Fusion reaction, the pSCR2AleIΔ(396-2030) plasmid was digested with AleI and XhoI and purified by agarose gel electrophoresis, followed by gel extraction (5PRIME) prior to combining with the randomized insert.
To construct the randomized insert, a PCR product (nt 369 to 2050) was generated, using a mutagenic sense primer (sPal-random-5; Table 1) with degenerate nucleotides at positions 392 to 396 and an antisense primer (asROD2050; Table 1) and using pSCR2 as a template. The PCR mixture contained 0.4 ng of pSCR2, 0.2 μM concentrations of the sense and antisense primers, a 0.2 mM deoxynucleoside triphosphate mix, 1× Taq standard buffer, and 0.5 μl of Taq polymerase enzyme (New England Biolabs) in a final reaction volume of 50 μl. A 40-cycle PCR protocol with a 55°C annealing temperature was used. The PCR product (randomized insert) was purified by agarose gel electrophoresis.
The randomized insert was combined with the digested pSCR2AleIΔ(396-2030) vector at a 2:1 ratio and transferred to an In-Fusion reaction tube. The resulting recombined product was transformed into E. coli DH5α, and DNA was extracted from bulk transformation reactions to obtain the randomized proviral DNA library. The degeneracy of pal region in the plasmids of this library was checked by sequencing (see Table S1 in the supplemental material). Although there was a bias in oligonucleotide synthesis that favored the incorporation of purine samples somewhat over pyrimidines, each nucleotide was sufficiently represented to ensure adequate mutagenesis at each position in the reconstructed proviral DNA library.
Cell culture and transfections of 5-nt RND pal library.
COS-7 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, penicillin, and streptomycin (Invitrogen). COS-7 cells were transfected with randomized library (5-nt RND pal plasmids) by using a Trans-IT-COS transfection kit (Mirus). At 2 days posttransfection, the cells and media were harvested, and the presence of virus in the medium was monitored by using a p27-specific enzyme-linked immunosorbent assay (ELISA; SIV p27 ELISA; Zeptometrix).
Cell culture and infections of RND viruses.
C8166 cells (NIH AIDS reagent 404) were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, penicillin, streptomycin, and glutamine (Invitrogen). The media of transfected cells containing 5-nt pal viruses was filtered through 0.22-μm-pore-size filters. Viruses were collected from the media by centrifugation for 2 h at 4°C and at 8,500 rpm (10,015 relative centrifugal force [RCF]). The resulting viral pellet was resuspended in 2.0 ml of RPMI 1640. Subsequently, 1.5 ml was used for infection in 105 C8166 cells. This 1.5 ml corresponded to 250 ng of HIV-2 CAp27 for 5-nt RND pal viruses, as determined by ELISA. An aliquot of 0.5 ml was saved at −80°C for RNA extraction. After 12 h at 37°C, cells were washed and resuspended in fresh RPMI medium-fetal calf serum. Every 7 to 10 days, the viruses produced in the media were filtered through 0.22-μm-pore-size syringe filters and used to reinfect 105 fresh uninfected C8166 cells.
Virus propagation for the 5-nt RND pal library was monitored by quantifying the concentration of CAp27 protein (determined by ELISA; Zeptometrix) in the medium at different days postinfection (data not shown). In addition, aliquots of medium containing filtered viruses were saved at 4, 9, 15, and 41 days postinfection (dpi) for RNA isolation and analysis.
RNA isolation and analysis of nonselected and selected sequences from the 5-nt RND pal library.
The total COS-7 intracellular vRNA and fractions of COS-7 and C8166 (19 and 41 dpi) extracellular vRNAs from 5-nt RND pal viruses were purified, using a Stratagene Absolutely RNA microprep kit. To pellet the virus particles, saved aliquots were centrifuged for 2 h at 4°C and 15,000 rpm (21,882 RCF). Purified RNAs were used for RT-PCR (AccuScript High-Fidelity RT-PCR system; Stratagene) using sBamT7R and asEco561 primers (Table 1). An aliquot of each RT-PCR was sequenced directly as a pool, while the rest of the reaction was digested with BamHI/EcoRI to clone individual sequences. The digested products were ligated with the BamHI and EcoRI sites of pUC18 vector and transformed in competent DH5α E. coli. Individual DNA plasmids were sequenced to examine the in vivo selection of the viable sequences and for further characterization.
Construction of individual plasmids with winner and loser sequences in the 5′ pal region.
To construct full-length plasmids with the selected and nonselected sequences in the 5′ pal region, we used the In-Fusion strategy described above with the same vector but with different inserts that contained the desired mutations in the 5′ pal region. The inserts with nonselected “loser” (SM2 and TTB-16) and selected “winner” (TTB-78) sequences were constructed, using sense primers (sPal-SM2, sPal-TTB-16, and sPal-random-2, respectively) and the antisense primer asROD2050 in separate PCRs. Since the wild-type sequence was one of the selected sequences (TTB-61 and TTB-76, see Table 3), we used the modified pROD10 plasmid (26).
TABLE 3.
S′ Pal sequences of viruses that were selected after several rounds of infection in C8166 cellsa
| Time postinfection and clone no. | Nucleotide at position:
|
||||
|---|---|---|---|---|---|
| 392 | 393 | 394 | 395 | 396 | |
| 9 dpi | |||||
| Wild type | G | G | A | G | U |
| TTB-47/48 | G | G | A | A | A |
| TTB-49/50 | G | G | A | G | G |
| TTB-51/52 | G | G | G | G | G |
| TTB-53/54 | G | G | A | G | A |
| TTB-55 | G | G | G | U | A |
| TTB-56 | G | G | G | C | G |
| TTB-57/58 | G | G | G | C | A |
| TTB-59 | G | G | U | A | G |
| TTB-60 | G | G | U | A | A |
| TTB-61 | G | G | A | G | U |
| TTB-62/65 | G | A | G | G | G |
| 41 dpi | |||||
| Wild type | G | G | A | G | U |
| TTB-66/71 | G | G | A | G | A |
| TTB-72/75 | G | G | A | G | G |
| TTB-76 | G | G | A | G | U |
| TTB-77 | G | G | A | A | G |
| TTB-78/80 | G | G | G | G | U |
| TTB-81 | G | G | G | U | A |
| TTB-82 | G | G | G | U | G |
vRNAs extracted from viruses at 9 and 41 dpi were subjected to RT-PCR and cloning as described in Materials and Methods. TTB-47 to TTB-65 (9 dpi) and TTB-66 to TTB-82 (41 dpi) clones were sequenced in the 5′ pal region. Sequences that appeared two or more times are shown in only one row (e.g., TTB-66/71 for TTB-66 to TTB-71). The individual clones TTB-76 (wild type) and TTB-78 were tested for infectivity and packaging.
Transfections of nonselected and selected individual plasmids.
Full-length plasmids with loser and winner sequences were transfected in COS-7 cells individually as described above.
Replication assay of nonselected and selected clones.
A total of 200 μl of medium from transfected COS-7 cells containing 0.6 ng of HIV-2 p27 capsid protein, as determined by ELISA, was used to infect 105 C8166 cells. After 12 h, cells were washed twice and resuspended in fresh RPMI medium-fetal calf serum. Viral replication was monitored for up to 16 days for the selected and nonselected individuals by quantifying the concentration of CAp27 protein (determined by ELISA; Zeptometrix) in the medium at different days postinfection.
Single-round infectivity assay of nonselected and selected individuals.
The infectivity of viruses in the medium of COS-7 cells was quantified by using CMMT-CD4-LTR-β-Gal indicator cells. This cell line expresses the CD4 receptor and contains a β-galactosidase gene fused to the HIV LTR, thus detecting synthesis of the transactivating (Tat) protein after productive infection of the cells by the input virus (9). After 2 days of culture postinfection, the cells were stained for β-galactosidase activity, and the blue infected cells were counted. The numbers were normalized first to the amounts of input viruses, as determined by ELISA, and second to the level of infection by wild-type virus to give the infectivities of viruses relative to the wild type.
RNA isolation and RNase protection assay for nonselected and selected individuals.
The COS-7 intracellular vRNAs were purified by using a Stratagene Absolutely RNA miniprep kit. For extracellular vRNAs, a fraction (1 ml) of the medium was centrifuged for 2 h at 4°C and 15,000 rpm (21,882 RCF) to pellet the viral particles, followed by lysis and RNA purification. Purified RNAs were used separately for RNase protection assays (RPA III; Ambion), as described by the manufacturer. The RNase protection assay was carried out using a 32P-labeled antisense RNA probe complementary to positions 401 to 562 of the HIV-2 RNA ROD isolate as described in reference 26). The antisense region was cloned into the Promega pGEM 7Zf(+) vector (Novagen) so that the T7 transcript had approximately 45 nucleotides of vector at its 5′ end. The non-HIV-2 tail of the probe was used as a marker of the RNase digestion efficiency during the experiment.
RESULTS
Generation and selection of 5-nt RND pal library.
We previously showed that the pal sequence (5′-GGAGUGCUCC-3′), located at the 3′ end of Ψ, was important for regulating SL1-mediated HIV-2 RNA dimerization in vitro (25). Using SELEX methodology, we showed that this regulation is mediated by the formation of an extended SL1 structure created by the interaction of the 3′ side of pal (GCUCC-3′) with an immediate downstream sequence of SL1 (3). Moreover, formation of the extended SL1 structure is necessary during viral replication (26). Although both studies demonstrated the importance of the 3′ side of pal, the role of the 5′ side of pal (5′-GGAGU) was not well understood. Therefore, we investigated the role of 5′ side of pal in HIV-2 replication by subjecting this region to randomization and subsequent in vivo selection.
The in vivo SELEX method starts with the generation of a pool of viral genomes containing many possible mutations at randomized sequences in a particular region (4). We synthesized a pool of HIV-2 proviral DNA genomes with five randomized nucleotides of pal at positions 392 to 396 so that the DNA library harbored 392NNNNN396 sequence (where N has an equal probability of being A, C, G, or T; Fig. 1C).
The recombinagenic method used for the generation of the 5-nt RND pal library is detailed in Materials and Methods. Briefly, a pSCR2AleIΔ(396-2030) vector and an insert (randomized at 5′ pal) were assembled in an In-Fusion (Clontech) recombinagenic reaction by virtue of the engineered homologous regions at their 5′ and 3′ ends. The recombined product was transformed in E. coli to produce a library of full-length HIV-2 proviral DNA plasmids with up to 1,024 (i.e., 45) possible sequence variations at the 5′ side of pal. To verify the degeneracy of the library, a fraction of the bacteria transformed with the recombinagenic PCR products were counted and their plasmids sequenced. Sequencing of 38 individual clones (see Table S1 in the supplemental material) revealed the presence of all four nucleotides at each position, although some purine bias in the synthesized oligonucleotides was noted. The diversity of the library allowed adequate interrogation of each position in the 392-396 pal region.
To initiate selection, the purified plasmid DNA library was first transfected into COS-7 cells to produce the library of 5-nt RND pal viruses. To monitor the sequence diversity and to confirm that no individual sequence was dominating in the 5-nt RND pal library at the transfection stage, we analyzed the leader sequences of intracellular and packaged genomic RNAs after RT-PCR. Sequence analysis of the RT-PCR product as a pool showed that COS-7 intracellular and packaged vRNAs contained random sequences at the 5′ side of pal (data not shown). In addition, RT-PCR products were subcloned to obtain the sequence of individual clones. Sequence analysis of 25 and 21 individual clones from COS-7 intracellular and packaged (extracellular) vRNAs, respectively, confirmed that no single sequence was predominating in the library at this stage (Table 2). However, a bias toward purine samples at nt 392 to 396 was already evident in the viral RNAs that were packaged and exported as viral particles into the media of the transfected COS-7 cells.
TABLE 2.
Sequence of 5-nt RND pal virus library inside and outside of COS-7 cells after transfectiona
| Category and clone no. | Nucleotide at position:
|
||||
|---|---|---|---|---|---|
| 392 | 393 | 394 | 395 | 396 | |
| Intracellular vRNA | |||||
| Wild type | G | G | A | G | U |
| TTB-1 | A | G | C | U | U |
| TTB-2 | A | A | U | U | U |
| TTB-3 | U | C | U | A | A |
| TTB-4 | A | G | G | A | A |
| TTB-5 | G | G | G | A | G |
| TTB-6 | C | A | G | G | G |
| TTB-7 | G | G | C | C | G |
| TTB-8 | A | U | G | C | U |
| TTB-9 | A | A | G | U | C |
| TTB-10 | A | A | A | A | A |
| TTB-11 | A | G | G | A | A |
| TTB-12 | A | G | A | A | C |
| TTB-13 | U | A | A | C | G |
| TTB-14 | G | G | C | A | G |
| TTB-15 | U | A | G | G | U |
| TTB-16 | A | C | C | U | G |
| TTB-17 | U | U | A | G | U |
| TTB-18 | G | U | G | G | C |
| TTB-19 | G | U | U | U | U |
| TTB-20 | A | A | G | G | C |
| TTB-21 | G | G | C | A | G |
| TTB-22 | A | C | G | G | G |
| TTB-23 | A | C | U | G | G |
| TTB-24 | U | A | C | A | G |
| TTB-25 | G | G | A | A | G |
| Extracellular vRNA | |||||
| Wild type | G | G | A | G | U |
| TTB-26 | G | U | G | A | G |
| TTB-27 | A | G | A | A | C |
| TTB-28 | G | G | A | A | A |
| TTB-29 | A | A | G | G | G |
| TTB-30 | G | U | A | A | G |
| TTB-31 | C | A | U | A | G |
| TTB-32 | A | A | G | G | A |
| TTB-33 | G | G | G | G | G |
| TTB-34 | U | U | A | A | C |
| TTB-35 | G | G | G | U | A |
| TTB-36 | G | A | A | G | U |
| TTB-37 | G | G | G | U | U |
| TTB-38 | A | G | A | G | G |
| TTB-39 | A | G | G | A | U |
| TTB-40 | G | U | A | G | G |
| TTB-41 | G | C | U | U | U |
| TTB-42 | G | G | G | G | C |
| TTB-43 | G | G | C | U | G |
| TTB-44 | G | G | A | A | A |
| TTB-45 | G | G | G | U | G |
| TTB-46 | A | U | G | C | G |
vRNAs extracted from COS-7 cells (intracellular) and media (extracellular) were subjected to RT-PCR, cloning, and sequencing as described in Materials and Methods. TTB-1 to TTB-25 represent RNAs that were expressed intracellularly and may or may not be packageable. TTB-26 to TTB-46 represent vRNAs that were exported as viral particles into the media of COS-7 cells and thus were at least marginally competent for packaging. TTB-16 was tested for infectivity and packaging.
To study the selection of 5′ pal sequences during viral replication and infection cycles, media from COS-7 transfected cells were used to infect permissive C8166 cells. Virus production was detected within a few days of infection in the media of C8166 cells by p27 capsid ELISA. Therefore, we harvested virus at 4 dpi, followed by vRNA purification, RT-PCR, and sequencing. Sequence analysis of the RT-PCR products showed selection at 4 dpi for G's at positions 392 and 393 (data not shown). Sequencing viral RNA from 9 and 15 dpi showed that the selection was complete at position 392 and nearly complete for positions 393 and 395 (Table 3). By 41 dpi, selection was obvious for the entire RND region: viral RNAs had converged on the consensus sequence GGRGN (Table 3), where N represents T, G, or A at position 396 (no C's were observed). The absence of C396 in the recovered viral RNAs may have been a result of negative selection, a sampling artifact, or because of the relative underrepresentation of C in the library. C396 viruses were present as packageable viruses in the posttransfection sampling, so it appears that they are viable but are likely overrun during longer infection schemes (Table 2).
A purine-rich sequence on the 5′ side of pal is important for viral replication.
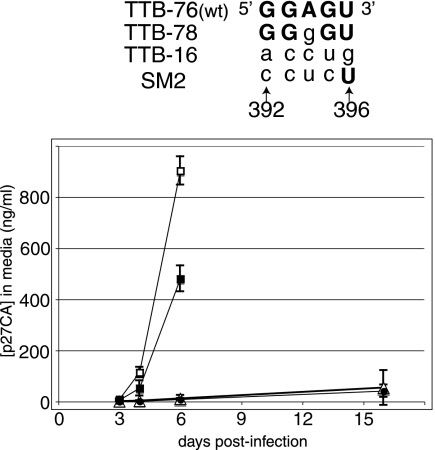
In wild-type HIV-2 the 5′ side of pal contains a purine-rich sequence (5′-GGAGU) and the in vivo 5-nt RND pal selection demonstrated the importance of the 5′-GGRGN motif. We therefore compared the viral replication properties of pyrimidine-rich nonconsensus and purine-rich consensus clonal viruses. The pyrimidine-rich clones used in this experiment were TTB-16, a nonsurviving intracellular COS-7 sequence (5′-accug, where lowercase letters signify nucleotides different from the wild-type sequence; Table 2), and SM2 (5′-cuccU; Fig. 2), a replication-deficient 5′ pal pyrimidine-rich mutant previously described by Lever and coworkers (30). The winner viruses were the purine-rich clones TTB-76 (5′-GGAGU [wild-type sequence]; Table 3) and TTB-78 (GGgGU; Table 3).
FIG. 2.
Mutant HIV-2 viruses harboring pyrimidines at the 5′ side of pal exhibit replication defects. (Top) Sequence at the 5′ side of pal (nt 392 to 396) in wild-type and mutant individuals used in this experiment. TTB-76 (wild type) and TTB-78 contain the pal sequence derived from viral clones that were winners after extended serial passage (Table 3). TTB-16 represents a sequence that was observed intracellularly after transfection but that did not survive during serial infections in C8166 cells (Table 2). SM2 is a packaging-deficient mutant previously characterized by the Lever group that was used here for comparison with our own clones containing pyrimidines in the 5′ side of pal (30). The mutated residues are represented by lowercase letters. (Bottom) Replication kinetics of wild-type (▪), TTB-78 (□), TTB-16 (○), and SM2 (▵) viruses in C8166 cells. Equal amounts of viral particles (0.6 ng of p27 capsid, as determined by ELISA), produced in the media by transfection of DNA plasmids in COS-7 cells, were used to infect permissive C8166 cells. Virus production from these cells was assayed for the production of p27 capsid at various intervals postinfection. The results are based on infections of separate plates of permissive cells using virus harvested from two independent transfections for each clone. Error bars represent one standard deviation for a representative experiment.
Duplicate samples of these four DNA constructs were transfected into COS-7 cells. At 48 h after transfection, viruses were harvested and used to infect C8166 cells with an equivalent amount of p27 capsid, as measured by ELISA. Virus spread was followed by measuring p27 capsid in the C8166 media by ELISA. Infections with purine-rich TTB-76 and TTB-78 viruses led to robust virus production, whereas those with pyrimidine-rich SM2 and TTB-16 sequences showed low or no virus production for up to 16 dpi (Fig. 2). These results support our hypothesis that the purine-rich 5′-GGRGN in the 5′ side of pal is essential for viral replication.
Purine-rich sequence (GGRGN) on the 5′ side of pal is important for genomic RNA packaging.
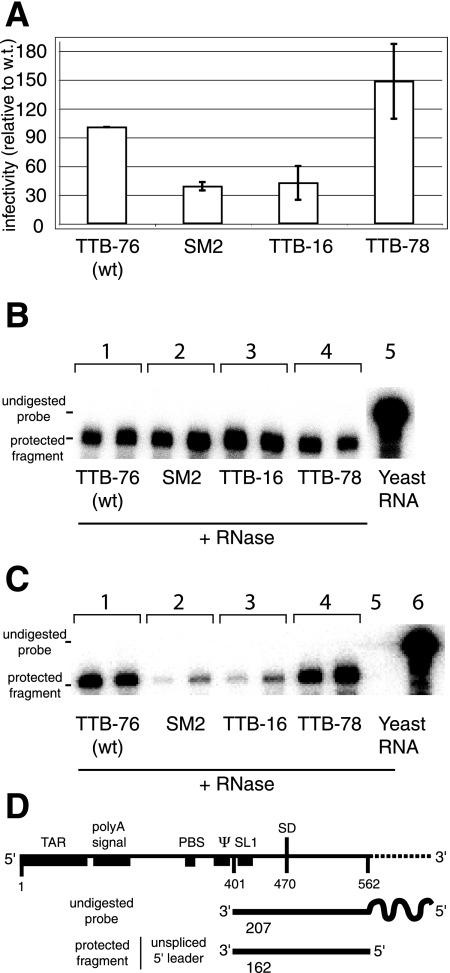
To further identify which steps of HIV-2 viral replication in C8166 cells were affected by non-wild-type substitutions in the 5′ side of pal, viruses produced from transfection of COS-7 cells with SM2, TTB-16, TTB-76 (wild type), and TTB-78 proviral DNAs were assessed for infectivity and/or entry in CMMT-CD4-LTR-β-Gal indicator cells. The infectivity of these cells can be examined by the detection of a Tat-driven reporter gene, whose expression is dependent on HIV entry into the cell (9). As observed in C8166 cells, SM2 and TTB-16 also showed a lower level of infectivity in CMMT-CD4-LTR-β-Gal indicator cells compared to TTB-76 and TTB-78 (Fig. 3 A).
FIG. 3.
HIV-2 virus infectivity and genomic RNA packaging are decreased by the presence of pyrimidines at the 5′ side of pal. (A) Single-round infectivity assay for wild-type and 5′ pal mutant viruses isolated from transfected COS-7 cells using CMMT-CD4-LTR-β-Gal indicator cells (see Materials and Methods). Sequences of wild-type and mutant 5′ pal for the viruses used in this experiment are shown at the top panel of Fig. 2. Virus generated by transfection of COS-7 cells was quantified by p27 ELISA and used to infect the indicator cells. At 48 h postinfection, cells were stained to visualize cellular β-Gal expression. Infectivity was normalized to the infectivity of the wild-type virus, set at 100%. Infectivity was measured in two independent experiments, and results are shown with error bars that represent the standard deviation. (B) Quantification of full-length genomic RNA in the cytoplasmic fraction of COS-7 cells transfected with wild-type and 5′ pal mutants using an RNase protection assay. Cells were harvested from two independent transfections of wild-type and 5′ pal mutants. A 3-μg portion of cytoplasmic RNA was probed with 2 × 106 cpm of labeled riboprobe. Lanes 1, 2, 3, and 4 represent amounts of genomic RNAs from TTB-76 (wild type) (5′-GGAGU), SM2 (5′-ccucU) (30), TTB-16 (5′-accug), and TTB-78 (5′-GGgGU) viruses, respectively. Lanes 5 represents yeast RNA used as a nonspecific target RNA control, incubated without RNases. (C) Quantification of full-length genomic RNA in wild-type and 5′ pal mutant viruses using an RNase protection assay. Viruses were harvested from the media of transfected COS-7 cells from two independent transfections of wild-type and 5′ pal mutants. First, the extracted RNAs from these viruses were normalized to the amount of viral particles (p27 capsid as determined by ELISA), and an equivalent amount of these RNAs was probed with 5 × 105 cpm of labeled riboprobe. Lanes 1, 2, 3, and 4 represent amounts of genomic RNAs from COS-7 cells transfected with TTB-76 (wild type) (5′-GGAGU) and the 5′ pal mutants (SM2 (5′-ccucU) (30), TTB-16 (5′-accug), and TTB-78 (5′-GGgGU), respectively. Lanes 5 and 6 represent yeast RNA used as a nonspecific target RNA control, incubated with or without RNases, respectively. (D) Schematic representation of the radioactive probe used in this experiment and a predicted protected fragment corresponding to the unspliced leader region. wt, wild type.
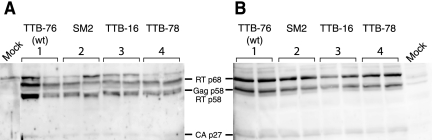
Since the packaging signals in retroviruses contain purine-rich motifs (5) and the 5′ pal element is both purine-rich and located within the HIV-2 packaging signal (21, 26), we investigated the role of 5′ side of pal as an important RNA packaging element. We assessed and compared the packaging of RNAs containing consensus 5′ pal sequence (TTB-76 [wild type] and TTB-78) with packaging of RNAs containing the nonconsensus 5′ pal sequence (TTB-16 and SM2). We performed RNase protection assays (Fig. 3) on TTB-16, SM2, TTB-76 (wild type), and TTB-78 RNAs extracted from the cytoplasm of COS-7 transfected cells and from the COS-7-produced viruses. As seen in Fig. 3, none of the sequences affected the level of genomic RNA in the cytoplasm of the transfected cells relative to the wild type. However, virions produced by these transfected cells showed that the level of genomic RNA packaging was high in the consensus 5-nt RND pal viruses (TTB-76 and TTB-78) but was reduced in the pyrimidine-rich 5′ pal viruses (TTB-16 and SM2) (10 to 15%) compared the wild type (Fig. 3C). Further, to ensure that mutations in 5′ side of pal did not affect viral gene expression, we analyzed viral proteins from intra- and extracellular COS-7 fractions by Western blot analysis. Our data show that mutation of 5′ pal region does not affect viral gene expression (Fig. 4). These data taken together suggest that the replication defect seen in viruses with a pyrimidine-rich 5′ pal sequence is due to a packaging defect.
FIG. 4.
Mutations within the 5′ side of pal do not affect viral gene expression. (A) Western blot analysis of COS-7 extracellular viral proteins. Protein samples from equivalent number of virions, as determined by p27 capsid ELISA, were loaded into each lane. HIV-2 proteins were detected with human serum from an HIV-2-infected patient. Lanes 1, 2, 3, and 4 represent proteins from the TTB-76 (wild type) (5′-GGAGU), SM2 (5′-ccucU) (30), TTB-16 (5′-accug), and TTB-78 (5′-GGgGU) viruses, respectively. (B) Western blot analysis of COS-7 intracellular viral proteins. Wild-type and 5′ pal mutants were tested for viral gene expression inside the COS-7 cells. An equal amount of cell lysate was loaded into each lane. HIV-2 proteins were detected with human serum from an HIV-2-infected patient (HIV-2 reference sera, NIH AIDS Research and Reference Reagent Program 7G 1337 and 6F D150). Lanes 1, 2, 3, and 4 represent proteins from COS-7 cells transfected with TTB-76 (wild type) (5′-GGAGU) and the 5′ pal mutants (SM2 (5′-ccucU) (30), TTB-16 (5′-accug), and TTB-78 (5′-GGgGU), respectively. The positions of the detected proteins with their molecular sizes are shown.
DISCUSSION
Retroviral packaging occurs by means of interactions between the nucleocapsid (NC) domain of the Gag polyprotein and the packaging signal (Ψ) of unspliced genomic RNA (for a review, see reference (17). For HIV-2, Ψ is located upstream of the major splice donor site, SD, in the 5′ untranslated leader region of genomic RNA (Fig. 1). In the present study, we investigated the role of pal, a 10-nt palindrome sequence (5′-392GGAGUGCUCC401) situated within Ψ (Fig. 1). The 3′ side of pal (GCUCC-3′) contributes to the formation of stem B, an extension of SL1 involved in the packaging of HIV-2 RNA, although the function of the 5′ side of pal (5′-GGAGU) was less well defined (26). To ascertain the sequence constraints and role(s) of pal, we used a randomization/in vivo selection methodology to investigate the sequences in pal that are important for packaging and replication. In vivo selection approaches have been used previously to isolate replication-competent viruses from a complex population of viruses during replication in eukaryotic cells (4, 16, 37). This method allows for mutational analysis of an ensemble of variant viruses without making a priori assumptions that might influence the choice of mutations. In contrast to the methods used before, we used a recombinagenic method that enables high-efficiency incorporation of the randomized region into the proviral DNA vector. We were able to select from among a large number of mutant viral sequences only those that were viable.
After in vivo selection of the RND pal viral libraries, sequence analysis showed that nucleotide variation was not well tolerated at positions 392, 393, and 395 of the 5′ pal region during viral replication, and G was strongly selected at each of these positions. For position 394, the dominance of A among the viable winner viruses also reflects its importance for viral replication. This finding is supported by phylogenetic analysis (Fig. 5B) and our previous phenotypic reversion study in which mutant pal viruses harboring an A394G substitution reverted back to A (26). Here, in the 5-nt RND pal selection, a consensus sequence (5′-GGRG) emerged in all viruses surviving the competition after several passages. We interpret this to mean that the 5′-GGRG sequence is essential for optimal HIV-2 replication. In support of this, two nonselected loser viruses with nonconsensus pyrimidine-rich sequences were deficient in infectivity and replication (Fig. 2 and 3).
FIG. 5.
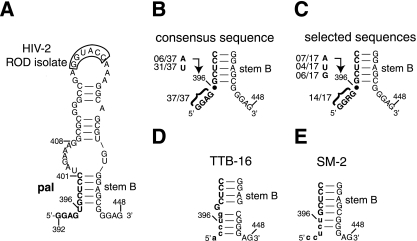
Predicted secondary structures of selected (winner) and nonselected (loser) RNAs. (A) Secondary structure model of pal-SL1 domain in HIV-2 ROD isolate (GenBank accession no. M15390) (3, 26) as predicted by Mfold (44). Stem B represents the interaction between the 5′-GCUCC of pal and the GGAGC-3′ sequence just downstream of SL1. (B and C) Stem B structure in HIV-2/SIV strains (28) and in the selected (winner) sequences derived from serial passage of the 5-nt RND library is indicated. The stem B structure was predicted using two-state hybridization model on the DINAMelt server (http://www.bioinfo.rpi.edu/applications/hybrid/twostate.php) using standard parameters (RNA at 37°C, [Na+] = 1 M, [Mg2+] = 0 M, and [strand] = 0.00001 M) (33). The consensus purine-rich motif and the proportion of bases at position 396 in HIV-2/SIV strains and the winner viruses is shown to the left. (D and E) The GGAG sequence at 3′ end of stem B in TTB-16 and SM2 is sequestered by base pairing with pyrimidines at the 5′ end of pal and potentially makes stem B significantly more stable. The nucleotides different from the wild-type sequence are shown with lowercase letters.
Compared to the strong selective pressure seen for positions 392 to 395, there were no obvious sequence constraints at position 396, except that we did not observe any C's at this position in the final pool. The lack of C's at 396 may indicate that viruses with C396 are somewhat impaired and are successfully outcompeted in this assay, or it may reflect a sampling artifact, since we have observed in previous studies that mutant viruses harboring A or C at position 396 replicated like the wild type during short-time-course infections (J.-M. Lanchy, unpublished data). It should be noted that position 396 exhibits a lower degree of phylogenetic conservation than adjacent residues among HIV-2/SIV isolates (28).
On the basis of previous studies (26, 30) and the present work, we propose that the 5′ pal region comprises an essential component of the core packaging signal of HIV-2 RNA. First, as described above, winning viruses derived from the in vivo selection shared a consensus 5′-GGRG sequence (Table 3). Second, this region is highly conserved among different strains of HIV-2 and SIV, where 37 of 37 published sequences show the 5′-GGAG sequence at this location (Fig. 5) (28). Third, packaging was strongly diminished in mutant viruses harboring sequences other than the 5′-GGRG sequence (Fig. 3C) (30).
Several studies have demonstrated that the interaction between RNA and the packaging proteins Gag/NC is enhanced by the presence of unpaired G residues in the target RNA (13, 39, 43). In the present study, secondary structure predictions of winner sequences suggest that, like that of the wild type, the 5′-GGRG sequence is unpaired (compare Fig. 5A and C), while among the loser viruses the 5′ side of pal was predicted to engage in stable base pairing with sequences downstream of SL1 (Fig. 5D and E). Because of the similarities with known Gag/NC binding preferences (13, 19, 26, 30, 39), the 5′-GGRG consensus is likely to be an essential element for Gag/NC recognition and binding.
In addition to its probable importance for Gag/NC binding, the Ψ-SL1 region of HIV-2 RNA adopts a specific conformation (at least transiently) during replication that is essential for efficient packaging. This conformation involves base pairing between the 3′ side of pal and the sequence downstream of SL1 that results in the formation of stem B (Fig. 1B and 5A). Mutations disrupting stem B negatively affect RNA packaging (21, 26, 35). Our present selection and RNA folding results raise the possibility that RNAs of TTB-16 and SM2 were packaging deficient due to the increased stability of extended SL1. The ΔG values for TTB-16 and SM2 SL1 RNA structures were −9.9 and −11.7 kcal/mol, respectively, while wild-type and winner RNA sequences folded with a predicted ΔG value of −8.3 kcal/mol. Hyperstabilization of stem B may lead to a less favorable interaction of NC with this region (23) or to the sequestration of normally unpaired G residues that may be critical NC binding determinants (13, 19, 39).
Analysis of the predicted extended SL1 structures highlighted the presence of additional conserved 5′-GGAG sequences located downstream of stem B. In particular, packaging models presented by Summers and coworkers suggest that Gag/NC binds poorly to RNAs in which NC-binding elements are sequestered by base-pairing interactions with other regions (14, 18). We suggest that the downstream 5′-GGAG sequence(s) could be a target for NC binding and cooperate with the 5′ GGRG motif to mediate packaging of HIV-2 RNA. This might explain why TTB-16 and SM2, in which the second 5′-GGAG was sequestered in an extended stem B (Fig. 5D and E), were packaging impaired (Fig. 3C).
On a technical note, we found that the use of the recombinagenic method provided a more efficient route to generate the randomized proviral DNA than traditional ligation protocols. In vivo selection has been used to characterize important viral RNA signals previously, but difficulty in generating the library of randomized proviral DNA limited the number of sequences that could be simultaneously examined (4, 16; T. T. Baig and J.-M. Lanchy, unpublished data). This enhanced synthesis of randomized proviral DNA allows a larger degenerate segment to be tested.
A growing body of evidence points to the importance of the pal element within the HIV-2 core packaging signal (21, 26, 30, 35). The palindromic nature of this element has been enigmatic, but the present study demonstrates that pal is really composed of two complementary motifs, one that is required to be single stranded and a likely target for protein recognition, and the other that is crucial for an RNA-RNA interaction to form stem B. Because the major packaging signal (Ψ) in HIV-2 RNA is located upstream of SD and is thus present in both spliced and unspliced RNA species (Fig. 1), it is remarkable that only unspliced RNA is packaged. To gain further insights into the mechanism of HIV-2 packaging and RNA selectivity, it will be interesting to investigate the binding affinities of packaging proteins Gag/NC for spliced and unspliced RNA constructs. These experiments are under way in our laboratory.
Supplementary Material
Acknowledgments
We gratefully acknowledge Christy Strong and Leila Sears for critical reading of the manuscript.
This study was funded by National Institutes of Health grant AI45388 to J.S.L. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institutes of Health (C8166 cells, no. 404) and (HIV-2 Reference Sera, sample ID 7G 1337 and 6F D150). The plasmid pROD10 was provided by the EU program EVA/MRC Centralized Facility for AIDS Reagents, NIBSC, United Kingdom (grant no. QLK2-CT-1999-00609 and GP828102).
Footnotes
Published ahead of print on 29 October 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abbink, T. E., and B. Berkhout. 2008. RNA structure modulates splicing efficiency at the human immunodeficiency virus type 1 major splice donor. J. Virol. 823090-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini, A., and R. A. Young. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 641920-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baig, T. T., J. M. Lanchy, and J. S. Lodmell. 2007. HIV-2 RNA dimerization is regulated by intramolecular interactions in vitro. RNA 131341-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout, B., and B. Klaver. 1993. In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res. 215020-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214177-218. [DOI] [PubMed] [Google Scholar]
- 6.Berkowitz, R. D., and S. P. Goff. 1994. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology 202233-246. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz, R. D., M. L. Hammarskjold, C. Helga-Maria, D. Rekosh, and S. P. Goff. 1995. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology 212718-723. [DOI] [PubMed] [Google Scholar]
- 8.Buck, C. B., X. Shen, M. A. Egan, T. C. Pierson, C. M. Walker, and R. F. Siliciano. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75181-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chackerian, B., N. L. Haigwood, and J. Overbaugh. 1995. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology 213386-394. [DOI] [PubMed] [Google Scholar]
- 10.Clavel, F., and J. M. Orenstein. 1990. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J. Virol. 645230-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever, J., C. Sassetti, and T. G. Parslow. 1995. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 692101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever, J. L., D. A. Eckstein, and T. G. Parslow. 1999. Genetic dissociation of the encapsidation and reverse transcription functions in the 5′ R region of human immunodeficiency virus type 1. J. Virol. 73101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279384-388. [DOI] [PubMed] [Google Scholar]
- 14.Dey, A., D. York, A. Smalls-Mantey, and M. F. Summers. 2005. Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus. Biochemistry 443735-3744. [DOI] [PubMed] [Google Scholar]
- 15.Dirac, A. M., H. Huthoff, J. Kjems, and B. Berkhout. 2001. The dimer initiation site hairpin mediates dimerization of the human immunodeficiency virus, type 2 RNA genome. J. Biol. Chem. 27632345-32352. [DOI] [PubMed] [Google Scholar]
- 16.Doria-Rose, N. A., and V. M. Vogt. 1998. In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J. Virol. 728073-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3643-655. [DOI] [PubMed] [Google Scholar]
- 18.D'Souza, V., and M. F. Summers. 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431586-590. [DOI] [PubMed] [Google Scholar]
- 19.Fisher, R. J., A. Rein, M. Fivash, M. A. Urbaneja, J. R. Casas-Finet, M. Medaglia, and L. E. Henderson. 1998. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 721902-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garzino-Demo, A., R. C. Gallo, and S. K. Arya. 1995. Human immunodeficiency virus type 2 (HIV-2): packaging signal and associated negative regulatory element. Hum. Gene Ther. 6177-184. [DOI] [PubMed] [Google Scholar]
- 21.Griffin, S. D., J. F. Allen, and A. M. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 7512058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz, R. A., R. W. Terry, and A. M. Skalka. 1986. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J. Virol. 59163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerwood, D. J., M. J. Cavaluzzi, and P. N. Borer. 2001. Structure of SL4 RNA from the HIV-1 packaging signal. Biochemistry 4014518-14529. [DOI] [PubMed] [Google Scholar]
- 24.Kim, H. J., K. Lee, and J. J. O'Rear. 1994. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology 198336-340. [DOI] [PubMed] [Google Scholar]
- 25.Lanchy, J. M., J. D. Ivanovitch, and J. S. Lodmell. 2003. A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA. RNA 91007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanchy, J. M., and J. S. Lodmell. 2007. An extended stem-loop 1 is necessary for human immunodeficiency virus type 2 replication and affects genomic RNA encapsidation. J. Virol. 813285-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laughrea, M., L. Jette, J. Mak, L. Kleiman, C. Liang, and M. A. Wainberg. 1997. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J. Virol. 713397-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leitner, T., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber. 2005. HIV sequence compendium 2005. Los Alamos National Laboratory, Los Alamos, NM.
- 29.Lever, A., H. Gottlinger, W. Haseltine, and J. Sodroski. 1989. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 634085-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L'Hernault, A., J. S. Greatorex, R. A. Crowther, and A. M. Lever. 2007. Dimerisation of HIV-2 genomic RNA is linked to efficient RNA packaging, normal particle maturation and viral infectivity. Retrovirology 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linial, M. L., and A. D. Miller. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. Immunol. 157125-152. [DOI] [PubMed] [Google Scholar]
- 32.Luban, J., and S. P. Goff. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 683784-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markham, N. R., and M. Zuker. 2005. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 33W577-W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride, M. S., and A. T. Panganiban. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 702963-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCann, E. M., and A. M. Lever. 1997. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 714133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miele, G., A. Mouland, G. P. Harrison, E. Cohen, and A. M. Lever. 1996. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 70944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris, S., M. Johnson, E. Stavnezer, and J. Leis. 2002. Replication of avian sarcoma virus in vivo requires an interaction between the viral RNA and the TpsiC loop of the tRNA(Trp) primer. J. Virol. 767571-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohlmann, T., M. Lopez-Lastra, and J. L. Darlix. 2000. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 27511899-11906. [DOI] [PubMed] [Google Scholar]
- 39.Paoletti, A. C., M. F. Shubsda, B. S. Hudson, and P. N. Borer. 2002. Affinities of the nucleocapsid protein for variants of SL3 RNA in HIV-1. Biochemistry 4115423-15428. [DOI] [PubMed] [Google Scholar]
- 40.Patel, J., S. W. Wang, E. Izmailova, and A. Aldovini. 2003. The simian immunodeficiency virus 5′ untranslated leader sequence plays a role in intracellular viral protein accumulation and in RNA packaging. J. Virol. 776284-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poeschla, E., J. Gilbert, X. Li, S. Huang, A. Ho, and F. Wong-Staal. 1998. Identification of a human immunodeficiency virus type 2 (HIV-2) encapsidation determinant and transduction of nondividing human cells by HIV-2-based lentivirus vectors. J. Virol. 726527-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rein, A. 1994. Retroviral RNA packaging: a review. Arch. Virol. 1994(Suppl. 9)513-522. [DOI] [PubMed] [Google Scholar]
- 43.South, T. L., and M. F. Summers. 1993. Zinc- and sequence-dependent binding to nucleic acids by the N-terminal zinc finger of the HIV-1 nucleocapsid protein: NMR structure of the complex with the Psi-site analog, dACGCC. Protein Sci. 23-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.