Abstract
The cystoviruses have genomes of three double-stranded RNA segments. The genes of the L transcript are expressed early in infection, while those of M and S are expressed late. In all cystovirus groups but one, the quantity of the L transcript late in infection is lower than those of the other two because of transcriptional control. In bacteriophage Φ8 and its close relatives, transcription of L is not controlled; instead, the L transcript is turned over rapidly late in infection. The three messages are produced in approximately equal amounts early in infection, but the amount of L is less than 10% of the amounts of the others late in infection. The decay of the Φ8 L message depends upon the production of protein Hb, which is encoded in segment L. It also depends upon a target site within the H gene region. Phage mutants lacking either the Hb gene or the target region do not show the late control of L message quantity. In addition to having a role as a negative regulator, Hb functions to neutralize the activity of protein J, encoded by segment S, which causes the degradation of all viral transcripts.
Bacteriophage Φ8 is a member of the Cystoviridae, a family of phages that have genomes of three double-stranded RNA (dsRNA) molecules packaged within a polyhedral capsid structure (11). The first-discovered isolate of this family was bacteriophage Φ6 (26). The life cycle of Φ6 involves attachment to a pilus, which retracts to place the virus at the outer membrane of its host, Pseudomonas syringae. The external membrane of Φ6 fuses with the outer membrane of the host, which results in the entry of the viral nucleocapsid into the periplasmic space (21). A viral muramidase digests the cell wall, and the viral nucleocapsid enters the cell (1). The nucleocapsid loses the shell of protein 8 (P8). The core particle is composed of four proteins: P1, the major structural protein; P2, the RNA polymerase; P4, a hexameric NTPase essential for genomic packaging; and P7, an auxiliary protein involved in RNA synthesis and packaging (14). The core particle, once inside the cell, transcribes the three genomic dsRNA molecules, L, M, and S, to produce messages l, m, and s, which are extruded from the particle. In the case of Φ6, the concentrations of the early transcripts are almost equimolar, but translation of L predominates (5). At later times, the level of the L transcript is about 5 to 10% of those of M and S, and translation of their genes predominates. The plus strands of the three segments have an 18-base consensus sequence at the 5′ end. There is a 1-base difference between the sequence of L and those of M and S. The L sequence begins with GU, while those of M and S begin with GG (16). In vitro transcription by nucleocapsids results in the production of transcripts of M and S in buffers that do not contain manganese ions. In vitro studies showed that the sequence difference is responsible for the differences between both late transcription and transcription from nucleocapsids and early transcription in infected cells (8). The control of Φ6 transcription is dependent upon the activity of a host protein. Φ8 transcription is independent of this protein (unpublished results).
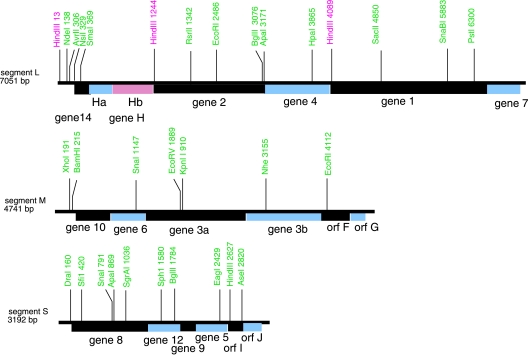
Φ8 is somewhat similar to Φ6 in structure and genetic makeup but has almost no sequence similarity to Φ6 (10). Φ8 plus strands have a shorter 5′ consensus sequence, but the first 7 nucleotides are identical in all three plus strands (10). Φ8 is the only member of the Cystoviridae whose second nucleotide of L is identical to those of S and M. In vitro transcription from inner core particles produces approximately equal amounts of the three plus strands, yet there is little L transcript evident late in the infection cycle (this study). Genetic studies showed that Φ8 differs from Φ6 in several ways. A striking difference is manifested in the arrangement of genes on genomic segment L (Fig. 1). In all other members of the Cystoviridae, the order of genes is 14-7-2-4-1. However, in the case of Φ8, the order is 14-H-2-4-1-7, where gene 7 has been moved out from its usual position and a pair of genes, Ha and Hb, now precedes gene 2 (25). Deletion of Hb prevents normal phage development, but the deletion can be partially complemented by cloned gene Hb (this study). In this report, we will show that gene Hb is involved in the temporal control of the abundance of the L transcript by regulating the L transcript's turnover.
FIG. 1.
Restriction maps of Φ8 genomic segment cDNA copies.
MATERIALS AND METHODS
Bacterial strains, phage, and plasmids.
Cells and plasmids used in this study are listed in Table 1. Phages are listed in Table 2. LM2489 is a rough derivative of P. syringae pv. phaseolicola HB10Y (26) and was used as the primary host for plating Φ8 (17). LM128 was also used, and LM2691 is LM128 carrying plasmid pLM1086, which is a derivative of pRK290 (7) and pAR1219 (6) and expresses T7 RNA polymerase in pseudomonads. Plasmids pLM2622, pLM2638, and pLM2743 are derivatives of pT7T319U with T7 polymerase promoters and cDNA copies of the Φ8 segments L, M, and S, respectively. Details of the construction of the plasmids are available from the authors. Plasmid transformation into Escherichia coli used strain JM109 (28) or Stratagene supercompetent XL1-Blue cells {recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)]}. Complementation of phage mutants and deletions was accomplished by inserting the relevant genes from cDNA plasmids of Φ8 into the shuttle vector pLM254, which is a derivative of plasmids pUC8 and RSF1010 (15).
TABLE 1.
Bacteriophages used in this study
| Phage | Relevant characteristic(s)a | Figure(s) | Reference or source |
|---|---|---|---|
| Φ8 wild type | 1, 2, 3, 4, 5 | 10 | |
| Φ2797 | Δ14 ΔH, deletion of nucleotides 329 to 1152 | 3, 5, 6, 7 | This study |
| Φ2833 | Δ14 ΔH mJ | 4 | This study |
| Φ2834 | Δ14 ΔH ΔJ | 3, 4, 5 | This study |
| Φ2825 | Δ14 ΔH Δ8 | This study | |
| Φ2835 | Δ14 ΔH m8 | 4 | This study |
| Φ2806 | Δ14 ΔH m8 mJ | 2, 3, 4 | This study |
| Φ2821 | Δ14 ΔH m8 ΔJ | 4 | This study |
| Φ2826 | Δ8 | 4 | This study |
| Φ6 wild type | 5′ GU in L | 2 | 26 |
| Φ2807 | 5′ GG in L | 2 | This study |
| Φ2837 | Δ14 ΔH in L, H insertion in gene 3a of M | This study | |
| Φ2781 | Deletion of nucleotides 329 to 11526 | 6 | This study |
| Φ2895 | Deletion of nucleotides 329 to 781 | 6 | This study |
| Φ2896 | Deletion of nucleotides 329 to 841 | 6 | This study |
| Φ2897 | Deletion of nucleotides 329 to 901 | 6 | This study |
| Φ2916 | Deletion of nucleotides 838 to 1152 | 6 | This study |
mJ, mutation in gene J; m8, mutation in gene 8.
TABLE 2.
Cells and plasmids used in this study
| Cells | Plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|---|
| LM2333 | pLM746 | Rough LPS, LacΩ in pLM254 | 17 |
| LM2489 | Rough LPS | 17 | |
| LM3827 | Tn5 insert in rnr of LM2489 | This study | |
| LM3865 | pLM3200 | rnr in pLM254 in LM3827 | This study |
| LM3863 | pLM3200 | rnr in pLM254 in LM2489 | This study |
| pLM3099 | Hb (olm776 to HindIII) in pET15b | This study | |
| pLM254 | Shuttle expression vector for pseudomonads | 15 | |
| LM3671 | pLM2934 | 14, Ha, and Hb in pLM254 (SmaI/HindIII) | This study |
| LM3722 | pLM3102 | Hb in pLM254 (XbaI/HindIII from pLM3099 in SmaI/HindIII) | This study |
LPS, lipopolysaccharide.
Media.
The media used were LC and M8 (24). Ampicillin plates contained 200 μg of ampicillin per ml in LC agar. Kanamycin was used at 40 μg per ml.
Reverse genetics of Φ8.
Mutant forms of the phage were prepared by constructing deletions or modifications in plasmids containing cDNA copies of the genomic segments. These were then electroporated into host cells containing plasmids expressing T7 RNA polymerase. In general, thousands of plaques were produced, samples were purified, and their RNA was analyzed by gel behavior and by the preparation of DNA copies by reverse transcription-PCR and subsequently checked by sequence analysis at the DNA sequencing core facility of UMDNJ.
Labeling of RNA during phage infection.
Host cells were diluted from an overnight culture in synthetic M8 medium and grown to 5 × 108 cells per ml. One-milliliter aliquots were placed on ice, and phage was added at a multiplicity of infection of 10. The cells were left on ice for 30 minutes and then transferred to 28°C. [5,6-3H]uracil (10 μCi) was added at various times. The cultures were spun at 6,000 × g 10 minutes later, and the cells were resuspended in 200 μl lysis buffer (20 mM Tris-HCl, pH 8, 1% sodium dodecyl sulfate, 5 mM EDTA) with 2 M sodium acetate, pH 5.4. The mixture was then frozen at −20°C for 20 min and then spun at 8,000 × g for 10 min at room temperature. The supernatant liquid was extracted twice with phenol-chloroform and precipitated with ethanol. The precipitate was resuspended in 20 μl DNA buffer and analyzed on 0.8% SeaKem GTG agarose (FMC BioProducts) in Tris-borate-EDTA with 2 μg/ml ethidium bromide. The bands were visualized with UV light and subjected to autoradiography.
Transposon mutagenesis.
EZ::TN (Epicentre), which is a complex of transposase and transposon (9), was electroporated into cells of LM2489, which were then plated on LB agar containing kanamycin. Colonies were tested by cross-streaking for sensitivity to bacteriophages Φ8, Φ6, and Φ13. Candidates that showed resistance to Φ8 were tested further by plating dilutions of phage on lawns. The sites of insertions were determined by preparation of EcoRI restriction digests of chromosomal DNA, ligating the DNA to form circles, and then amplifying inserts by PCR using forward and reverse primers supplied by Epicentre. The PCR products were then sequenced using the forward primer supplied by Epicentre, and the sequences were compared with those in the NIH data bank using the BLAST search facility. All insert sequences showed high identity to sequences of P. syringae pv. phaseolicola 1448A (GenBank accession number CP000058).
Cloning rnr of P. syringae pv. phaseolicola.
The rnr (vacB) gene was copied from chromosomal DNA using oligonucleotides olm824 (CCCGTCGACGGAGTTGACAAATGGCCGATTGGCA) for the 5′ end and olm829 (TGACGGCCGTAGATTTTTTCCAGC) for the 3′ end. The PCR product was cut with restriction enzymes SalI and EagI and inserted into the shuttle vector pLM254. The base sequence of this rnr gene is 99% identical to that of Pseudomonas syringae pv. phaseolicola 1448A (locus tag AAZ33054), and the amino acid sequence has 99% identity.
Nucleotide sequence accession numbers.
Open reading frames (ORFs) Ha and Hb in segment L identified in this study have been deposited in GenBank as accession number AF226851. ORF J in segment S is GenBank accession number AF226853.
RESULTS
Temporal control of plus-strand abundance in Φ6 and Φ8 infections.
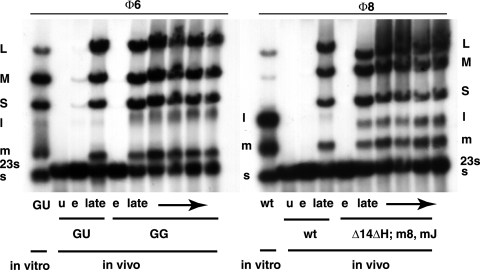
In vitro transcription using nucleocapsids of Φ6 results in unbalanced synthesis, in that the transcripts of segments S and M are more than 10 times as abundant as those of L (19) (Fig. 2). This behavior is dependent upon the difference in the second nucleotide of the L plus strand from that of S or M (8). The sequence at the 5′ end of L is GU, while it is GG for S and M. Early in infection, there is a higher level of the L transcript than seen in the in vitro transcription, but late in the infection period, the amount of L transcript diminishes drastically. We have prepared Φ6 phage with GG as the first 2 nucleotides of L and demonstrate that production of the L transcript is much higher than normal in late infection (Fig. 2). The elucidation of the control of Φ6 transcription is the subject of a separate publication.
FIG. 2.
Autoradiogram of an agarose gel with [3H]uracil-labeled infected cells of LM2489. Infection was with Φ6 or Φ8. In vitro columns show products of transcription reactions of inner cores of purified virions. Note that, in vitro, Φ6 shows very little L transcript but that Φ8 has abundant L transcript. Also note that late transcripts in Φ6-infected cells show little L transcript but that the GG mutant has a significant amount. In the case of Φ8-infected cells, there is also little L transcript unless gene H is missing (deletion of positions 329 to 1152). wt, wild type; u, uninfected cells; e, early labeling (15 min); late, labeling at 60, 90, 120, 165, and 210 min. ΔH, m8, and mJ refer to phage Φ2806. m8, mutation in gene 8; mJ, mutation in gene J.
In vitro transcription using nucleocapsids of Φ8 results in approximately equimolar synthesis of the three transcripts (Fig. 2). Yet, late in infection there is a dramatic reduction in the relative amount of the L transcript. This reduction is not the result of differential transcription but is due to turnover of the L transcript (Fig. 3). The amount of L transcript is seen to be low even after very short pulses of labeling. However, transcription in Φ8 involves displacement of the plus strand from the double-stranded template by the newly synthesized transcript. Therefore, the plus strand of the dsRNA is a measure of the amount of transcription taking place. We observe that labeling of the dsRNA for L is commensurate with that for S and M. Denaturation of the genomic dsRNA shows that plus strands of L are being synthesized at rates comparable to those of S and M (Fig. 3).
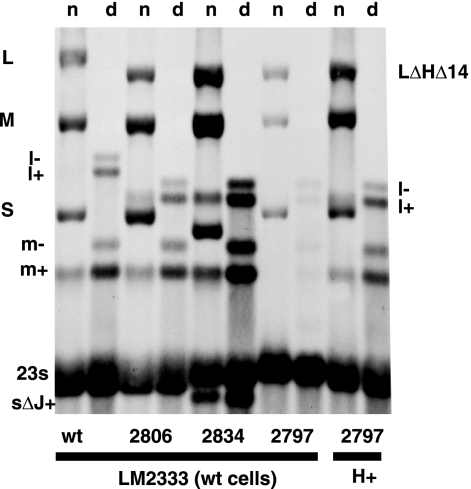
FIG. 3.
Autoradiogram showing the synthesis of plus strands of L despite their absence in single-stranded RNA. Note that in wild-type (wt) infection, there is visible M transcript (m+) but not L transcript (l+) in the nondenatured lanes (lanes n); however, in the lanes with denatured RNA (lanes d) (18), the L transcript is apparent. Phage with H deleted (deletion of positions 329 to 1152) but with genes 8 and J mutated (Φ2806) or J deleted (Φ2834) show L transcript in the nondenatured lanes. Phage with H deleted (Φ2797) shows very low levels of RNA synthesis in wild-type cells but increased levels in cells producing protein H in trans. Note that the L transcript is visible because trans complementation of H does not restore degradation of l+ because Hb is missing from genomic segment L.
Deletion of the H region.
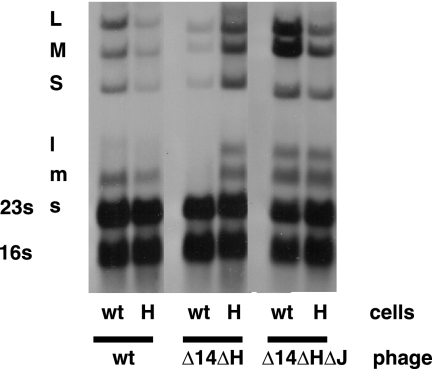
The H region of segment L contains two ORFs, Ha and Hb. A protein product has been observed only for Hb. We prepared virus with a deletion of the H region in order to determine its role in the life cycle of Φ8. This was accomplished by creating an NsiI site at position 1152 of the cDNA copy of segment L and then deleting from NsiI positions 329 to 1152, which eliminates part of gene 14, all of Ha, and most of Hb and is designated the Δ14 ΔH construct. Gene 14 has been found to be dispensable in this study. The resulting plasmid, pLM3007, along with plasmids pLM2743 and pLM3056, which carry the cDNA copies of normal segment S and segment M, respectively, in which Ha and Hb are embedded in gene 3a, was electroporated into host cells containing plasmids expressing T7 RNA polymerase and gene 3a. Plaques were obtained, and the phage was crossed with a phage with normal segment M but with a deletion in gene 4 of segment L. The resulting phage, Φ2797, contained normal segments S and M and the deletion of the H region in segment L. Phage from these plaques did not propagate well on normal host strains. The efficiency of plating was orders of magnitude lower than that found on the strain carrying a plasmid containing the H region. Only a few plaques were obtained, and these were very small. Propagation of these phages led to the production of larger plaques, Φ2806, that were found to be capable of reproduction on normal host cells. Analysis of these phages showed that they indeed carried the deletion of H and that they carried suppressor mutations in segment S. The suppressor mutations were located in gene 8, which encodes a membrane protein in Φ8 but the determinant of a middle shell in Φ6, and in gene J, which is a unique gene, found only in segment S of Φ8 (Fig. 1). Gene J is found between nucleotides 2850 and 3086. The mutation in gene 8 is A738G (K→R), and the mutation in J is T2872C (F→S). P8 contains 366 amino acids, while protein J contains 78. Although the mutant phages with the deletion of H and suppressor mutations in genes 8 and J were able to propagate well, they showed a new pattern of RNA labeling late in infection (Fig. 2, 3, and 4). Whereas the concentration of the transcript of segment L was usually low at late times in normal virus infection, it was close to equimolar with those of segments M and S in the mutant viruses. The phage with the deletion of H but lacking the suppressor mutations showed low levels of transcripts for all three segments (Fig. 3, 4, and 5). It appears that H is necessary for the downregulation of the L transcript late in infection, while it also seems to be necessary for the normal levels of the S and M transcripts. In the absence of the normal gene J product, either because of mutation or deletion, H is not necessary for the normal levels of S and M transcripts. Gene J is not necessary for normal phage reproduction (data not shown), but when J is present, H is required to counter its effects. The mutation in gene 8 is necessary for plaque formation in the absence of H (data not shown), but it does not have a role in the regulation of RNA decay (Fig. 4). The nature of the role of the gene J product will be the subject of another publication.
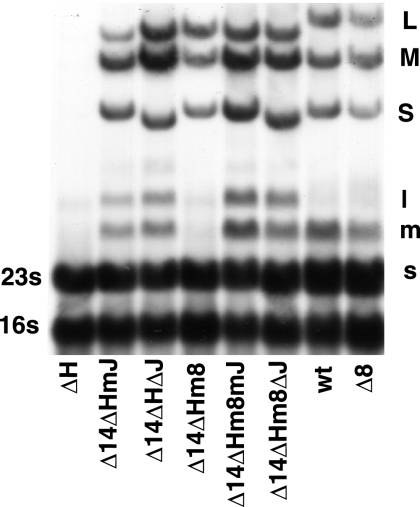
FIG. 4.
Autoradiogram showing that the dsRNA and transcripts of the genomic segments are missing when gene H is deleted (deletion of positions 329 to 1152) but that deletion of gene J (Φ2834) or mutation in J (Φ2833) suffices to restore synthesis and to prevent the decay of the L transcript. Deletion (Φ2825) or mutation (Φ2835) of gene 8 shows little effect despite their requirement for plaque formation when H is deleted. wt, wild type.
FIG. 5.
Autoradiogram showing that gene H in trans (LM3671) is able to complement a deletion of H (Δ329 to 1152) in the phage genome with respect to the effects of J. However, H in trans does not restore the downregulation of the L transcript. wt, wild type.
The loss of H can be complemented in trans by gene Hb in a plasmid (pLM3102) so as to restore the levels of the S and M transcripts, but in this case, the transcript of L is not completely downregulated (Fig. 5 and 6). It appears that the targeting of the L transcript requires a cis element. If H is deleted from segment L and moved to segment M, the transcript level of segment M is found to be diminished and that of L is elevated (not shown).
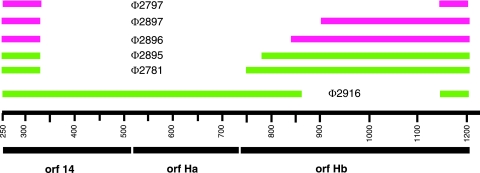
FIG. 6.
Map showing deletion constructs involving the fusion of the N terminus of the ORF 14 product with constructs with deletions of Hb. The designations of viruses with corresponding deletions are shown. Pink lines indicate constructs that do not downregulate the L transcript even when Hb is supplied in trans. Green lines indicate deletions that are able to partially downregulate the L transcript when Hb is supplied in trans.
Fine structure of region H.
There is an ORF starting at position 507 in segment L that continues to nucleotide 1211. We designate this ORF H. It contains 234 amino acids. There is a smaller ORF that starts at position 514 and continues to position 744 that we designate ORF Ha (Fig. 1) and whose product is 76 amino acids, and there is an ORF in the same reading frame as ORF H that starts at position 741 and continues to position 1211 that we designate ORF Hb and whose product is 156 amino acids. Although both ORF H and ORF Ha have reasonable Shine-Dalgarno sequences preceding an initiating methionine codon, we have not seen gene products for ORF H or ORF Ha in infected cells, but we do see a product of the size of ORF Hb that correlates with the presence of that gene. Phage mutants that are missing ORF Hb behave as if they are missing the entire H region; however, mutants missing gene 14 and part of ORF Ha can plate on normal cells and show downregulation of the L transcript. An amber mutation in ORF Ha does not show any abnormalities in terms of RNA levels or reproduction. A fusion of the N terminus of P14 with Hb that is missing the first five amino acids (Φ2781) behaves as if the Hb protein is intact but that the target of H is partially missing. The resulting fusion protein can be seen on labeled protein gels. Fusions that lack the first 34 or 54 amino acids behave as if both the protein and the target are missing (Fig. 6). This suggests that the target(s) for the action of Hb on the turnover of the L transcript is between gene 14 and part of gene Hb. We believe that ORF Hb makes an active protein and that several parts of the region involving genes Ha and Hb are the target for the action of protein Hb and other cellular components. Deletions of regions within segment L result in partial loss of downregulation of L transcript abundance when Hb is in trans.
Transposon knockouts of rnr.
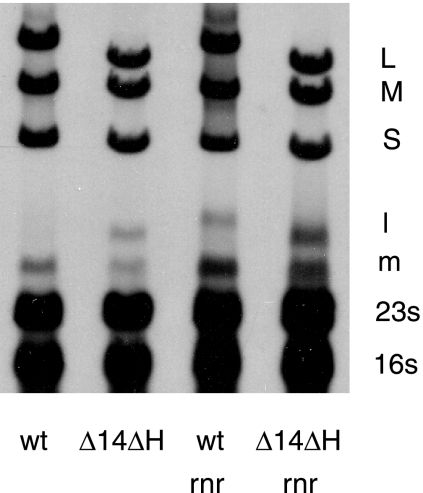
In parallel experiments, we used transposon mutagenesis in a search for host genes involved in the propagation of bacteriophage Φ8. The EZ::TN system of Epicentre (23) was used to create insertions in the genome of P. syringae strain LM2489. Kanamycin-resistant colonies were produced after electroporation of the transposon-transposase complex into cells. The colonies were cross-streaked against Φ8, Φ13, and Φ6. Strains that were resistant to Φ8 were further tested by plating phage dilutions on lawns. Several promising cultures that produced very small plaques were isolated. Chromosomal DNA was prepared from these, restricted with EcoRI, ligated, and PCR amplified with primers supplied by Epicentre. The PCR products were then sequenced with the forward primer supplied by Epicentre. Several of the inserts were in genes involved with surface proteins and were not pursued further. Among the isolates were insertions into mdoH and mgoG, which are glycosyltransferase genes in P. syringae pv. phaseolicola listed under accession number AAZ34542.1 in GenBank. Another set of inserts was in a DedA family protein which is probably an integral membrane protein, and another was in a glucosyl UTP transferase. However, several of the inserts were in a gene with high identity to P. syringae pv. phaseolicola rnr, which codes for RNase R (locus tag AAZ33054.1). This enzyme plays a role in the modification of the structure and regulation of RNA production in E. coli (2, 3). The gene was cloned from the chromosomal DNA of LM2489 and was found to complement the knockout strain with respect to plaque size when it was expressed on an expression plasmid, pLM254. We found that the rnr knockouts showed a less dramatic downregulation of the L transcript late in Φ8 infection than the wild type, indicating that RNase R acts with the gene H product to downregulate the L transcript (Fig. 7). The loss of RNase R does not entail as strong an effect as the loss of gene H, suggesting that other elements may also participate or substitute in the decay of the L transcript. It was of interest that the rnr knockouts did not suppress the downregulation shown by the gene J product, suggesting that the J product acts either alone or with an element apart from RNase R.
FIG. 7.
Autoradiogram showing that wild-type Φ8 downregulates the L transcript (l) but that the Δ14 ΔH deletion mutant (Φ2797) does not downregulate and that the rnr knockout strain shows poor downregulation even in infections with wild-type (wt) virus.
DISCUSSION
Temporal control of gene expression is a common feature of virus infection. In the case of the Cystoviridae, there is a dramatic difference in the relative quantities of segment L transcripts at early versus late times of infection. The genes of segment L code for the proteins of the procapsid or internal core of the virions, and these proteins are expressed early in infection. In Φ6, transcription of segment L is dependent upon a host protein. Φ6 nucleocapsids do not transcribe segment L in vitro unless host protein or manganese ions are present; however, changing the second 5′ base in segment L from U to G to make GG facilitates the transcription of segment L (Fig. 2). In Φ8, nucleocapsids transcribe all three genomic segments in vitro without special assistance (Fig. 2). Yet there is little L transcript present in the late period of Φ8 infection. Whereas most members of the Cystoviridae regulate transcription, Φ8 regulates the amount of the L transcript by its degradation during the late period of infection.
The H region apparently plays two roles in Φ8 infection. (i) The Hb product acts with RNase R, and probably with other host elements, to degrade the transcript of L late in infection. This works effectively, since the amount of protein Hb must reach a particular level before it has a significant effect, and therefore it does not cut off the translation of L at early times in infection. (ii) At late times in infection, it appears to be necessary to bolster the amounts of the M and S transcripts, since these are under attack from the product of gene J (Fig. 4).
The involvement of RNase R in the regulation of the L transcript is of special interest because the role of RNase R in bacteria is not very clear. There is evidence that it is involved in the degradation of defective rRNA and that it enhances the expression of genes on pathogenicity plasmids in E. coli and Shigella spp. (2, 4). The enzyme appears to act in both negative and positive manners. In the latter case, it seems that it must inactivate or interfere with a repressor system of some sort. Since RNase R is involved in large complexes, it is possible that the regulation of L is also mediated by a large complex. RNase R has also been found to be associated with the SsrA RNA complex, which is for peptide degradation (13). We do not know how the combination of Hb and RNase R affects the degradation of the L transcript, but our working model is that Hb guides the RNase, probably in a complex to a site within the H region.
It is not clear why Φ8 has adopted this unique set of controls of RNA turnover. In the other members of the Cystoviridae, it appears that temporal control is mediated by the difference in sequence at positions 2 of the plus strands, which affects the relative rates of transcription (initiation) and a difference in half-life of the plus strands where the L transcript is longer lived than the other two. The Φ8 arrangement seems so much more complex. The role of J is even more obscure in that the phage does quite well without gene J. The effect of J seems to be specific for Φ8. A plasmid expressing protein J inhibits the plaque formation of Φ8 but not that of Φ6 or Φ13 (20). It appears that J and H might be involved in a control circuit that modulates the temporal control of phage protein synthesis. H seems to be responsible for the high turnover of the L transcript late in infection and the prevention of the high turnover of M and S transcripts at the same time.
The temporal control of gene expression during bacteriophage infection is usually determined by differential transcription, with early genes being transcribed by normal host RNA polymerase and late genes being transcribed by host polymerase with new or modified sigma factors or by virally encoded RNA polymerase (27). In the case of T4, two proteins that play a role in the temporal control of message degradation have been identified. One is dmd, a small protein involved in middle and late message stability but that also seems to facilitate the degradation of some middle messages (12). The other is regB, an endoribonuclease that selectively cleaves early messages (22). regB is of particular interest because it attacks message in a subset of Shine-Dalgarno sequences with the collaboration of ribosomal protein S1. Both proteins have affinity for Shine-Dalgarno sequences, and it is thought that S1 modifies the structure of the stem-loops in a manner that converts them to substrates for regB endoribonuclease activity. We have found no RNase activity in Hb so far, and we have not found any interaction in vitro with RNase E, RNase R, or protein S1 (data not shown). However, there is a strong expectation that the nuclease activity might involve a large complex that is difficult to demonstrate in vitro. The role of the rnr product would be in the further degradation of molecules cut by Hb and its cofactor.
One possible explanation for the unique transcriptional control of Φ8 is that it facilitates the ability of the phage to infect unrelated bacteria. Φ8 is the only member of the Cystoviridae that can form plaques on strains of Salmonella enterica serovar Typhimurium (17). The other members of the Cystoviridae require a host protein for transcriptional control. Although mutations to independence have been isolated, they do not confer completely normal production.
Acknowledgments
This work was supported by grant GM34352 from the National Institutes of Health.
Footnotes
Published ahead of print on 29 October 2008.
REFERENCES
- 1.Caldentey, J., and D. H. Bamford. 1992. The lytic enzyme of the Pseudomonas phage Φ6. Purification and biochemical characterization. Biochim. Biophys. Acta 115944-50. [DOI] [PubMed] [Google Scholar]
- 2.Cheng, Z. F., and M. P. Deutscher. 2002. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 27721624-21629. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, Z. F., and M. P. Deutscher. 2003. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl. Acad. Sci. USA 1006388-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, Z. F., Y. Zuo, Z. Li, K. E. Rudd, and M. P. Deutscher. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 27314077-14080. [DOI] [PubMed] [Google Scholar]
- 5.Coplin, D. L., J. L. Van Etten, R. K. Koski, and A. K. Vidaver. 1975. Intermediates in the biosynthesis of double-stranded ribonucleic acids of bacteriophage Φ6. Proc. Natl. Acad. Sci. USA 72849-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davanloo, P., A. H. Rosenberg, J. J. Dunn, and F. W. Studier. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 812035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria; construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 777347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frilander, M., M. Poranen, and D. H. Bamford. 1995. The large genome segment of dsRNA bacteriophage Φ6 is the key regulator in the in vitro minus and plus strand synthesis. RNA 1510-518. [PMC free article] [PubMed] [Google Scholar]
- 9.Goryshin, I. Y., J. Jendrisak, L. M. Hoffman, R. Meis, and W. S. Reznikoff. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 1897-100. [DOI] [PubMed] [Google Scholar]
- 10.Hoogstraten, D., X. Qioa, Y. Sun, A. Hu, S. Onodera, and L. Mindich. 2000. Characterization of Φ8, a bacteriophage containing three double-stranded RNA genomic segments and distantly related to Φ6. Virology 272218-224. [DOI] [PubMed] [Google Scholar]
- 11.Jaalinoja, H. T., J. T. Huiskonen, and S. J. Butcher. 2007. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages phi6 and phi8. Structure 15157-167. [DOI] [PubMed] [Google Scholar]
- 12.Kai, T., and T. Yonesaki. 2002. Multiple mechanisms for degradation of bacteriophage T4 soc mRNA. Genetics 1605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karzai, A. W., M. M. Susskind, and R. T. Sauer. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 183793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mindich, L. 2004. Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives. Virus Res. 10183-92. [DOI] [PubMed] [Google Scholar]
- 15.Mindich, L., G. MacKenzie, J. Strassman, T. McGraw, S. Metzger, M. Romantschuk, and D. Bamford. 1985. cDNA cloning of portions of the bacteriophage Φ6 genome. J. Bacteriol. 162992-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mindich, L., I. Nemhauser, P. Gottlieb, M. Romantschuk, J. Carton, S. Frucht, J. Strassman, D. H. Bamford, and N. Kalkkinen. 1988. Nucleotide sequence of the large double-stranded RNA segment of bacteriophage φ6: the genes specifying the viral replicase and transcriptase. J. Virol. 621180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mindich, L., X. Qiao, J. Qiao, S. Onodera, M. Romantschuk, and D. Hoogstraten. 1999. Isolation of additional bacteriophages with genomes of segmented double-stranded RNA. J. Bacteriol. 1814505-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagratis, N., and H. R. Revel. 1990. Detection of bacteriophage Φ6 minus-strand RNA and novel mRNA isoconformers synthesized in vivo and in vitro, by strand-separating agarose gels. Virology 177273-280. [DOI] [PubMed] [Google Scholar]
- 19.Partridge, J. E., J. L. Van Etten, D. E. Burbank, and A. K. Vidaver. 1979. RNA polymerase activity associated with bacteriophage Φ6 nucleocapsid. J. Gen. Virol. 43299-307. [Google Scholar]
- 20.Qiao, X., J. Qiao, S. Onodera, and L. Mindich. 2000. Characterization of Φ13, a bacteriophage related to Φ6 and containing three dsRNA genomic segments. Virology 275218-224. [DOI] [PubMed] [Google Scholar]
- 21.Romantschuk, M., V. M. Olkkonen, and D. H. Bamford. 1988. The nucleocapsid of bacteriophage Φ6 penetrates the host cytoplasmic membrane. EMBO J. 71821-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanson, B., R. M. Hu, E. Troitskayadagger, N. Mathy, and M. Uzan. 2000. Endoribonuclease RegB from bacteriophage T4 is necessary for the degradation of early but not middle or late mRNAs. J. Mol. Biol. 2971063-1074. [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko, Y., G. G. Bouffard, Y. S. Butterfield, R. W. Blakesley, J. L. Hartley, A. C. Young, M. A. Marra, S. J. Jones, J. W. Touchman, and E. D. Green. 2002. Systematic sequencing of cDNA clones using the transposon Tn5. Nucleic Acids Res. 302469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinclair, J. F., J. Cohen, and L. Mindich. 1976. The isolation of suppressible nonsense mutants of bacteriophage Φ6. Virology 75198-208. [PubMed] [Google Scholar]
- 25.Sun, Y., X. Qiao, J. Qiao, S. Onodera, and L. Mindich. 2003. Unique properties of the inner core of bacteriophage phi8, a virus with a segmented dsRNA genome. Virology 308354-361. [DOI] [PubMed] [Google Scholar]
- 26.Vidaver, A. K., R. K. Koski, and J. L. Van Etten. 1973. Bacteriophage Φ6: a lipid-containing virus of Pseudomonas phaseolicola. J. Virol. 11799-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong, K., G. A. Kassavetis, J. P. Leonetti, and E. P. Geiduschek. 2003. Mutational and functional analysis of a segment of the sigma family bacteriophage T4 late promoter recognition protein gp55. J. Biol. Chem. 2787073-7080. [DOI] [PubMed] [Google Scholar]
- 28.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]