Abstract
For many papillomaviruses, the viral protein E2 tethers the viral genome to the host mitotic chromosomes to ensure persistent, long-term maintenance of the genome during cell division. Our previous studies of E2 proteins from different genera of papillomaviruses have shown that they bind to different regions of the host chromosomes during mitosis. For example, bovine papillomavirus type 1 (BPV-1) E2 binds to all chromosomes as small speckles in complex with the cellular protein Brd4. In contrast, the human papillomavirus type 8 (HPV-8) E2 protein binds as large speckles at the pericentromeric regions of chromosomes. Here we show that these speckles do not contain Brd4, and unlike that of BPV-1, the N-terminal Brd4-interacting domain of HPV-8 E2 is not required for chromosome binding. In contrast to BPV-1 E2, the HPV-8 E2 protein targets the short arms of acrocentric mitotic chromosomes. Furthermore, the E2 protein interacts with the repeated ribosomal DNA genes found in this location and colocalizes with UBF, the RNA polymerase I transcription factor. Therefore, HPV-8 E2 genome tethering occurs by a Brd4-independent mechanism through a novel interaction with specific regions of mitotic chromosomes. Thus, a wide range of viruses have adopted the strategy of linking their genomes to host chromosomes, but individual viruses use different chromosomal targets. Characterization of these targets will enable the development of antiviral therapies to eliminate the viral genomes from infected cells.
Papillomaviruses infect the basal layer of stratified epithelia. The viral genomes are stably maintained in the nuclei of the dividing cells as low copy number, extrachromosomal elements for many years. Error-free replication and equal distribution of the replicated copies of the viral DNA to the dividing cells are key to the persistence of papillomavirus infection. The mechanism of viral genome partitioning is different from that of host chromosomes, as the viral DNA does not possess any sequence equivalent to the centromere to utilize the mitotic segregation machinery of the host. Instead, E2, a multifunctional viral protein, attaches the viral DNA to the host chromosomes (15, 21, 34). As the host cell chromosomes are divided equally into daughter cells during mitotic division, the viral DNA is passively segregated by being tethered to the host chromosomes. Chromosomal tethering mediated by a virus-encoded protein is a common tactic for maintaining the genomes of other persistent viruses. Gammaherpesviruses such as Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus use this strategy, which is mediated by the EBNA-1 and LANA proteins, respectively (reviewed in reference 23).
Among the papillomaviruses, bovine papillomavirus type 1 (BPV-1) is best studied for chromosomal segregation. The multifunctional viral protein E2 has been shown to play an important role in this process. Both the BPV-1 E2 protein and viral genomes are localized on mitotic chromosomes as small speckles on the arms of all chromosomes, in complex with Brd4 (3, 15, 21, 34, 40). Furthermore, BPV-1 E2 mediates the attachment and segregation of plasmids containing E2 binding sites (4, 15). There is much evidence that Brd4, a cellular bromodomain protein involved in many cellular functions, including transcription regulation, is involved in the chromosomal binding complex of BPV-1 E2. Both E2 and Brd4 colocalize on mitotic chromosomes in punctate spots (3, 25, 40), and mutations in the E2 transactivation domain compromise both the interaction of E2 with Brd4 and the association of E2 with the chromosome (3, 32). Inhibition of the Brd4-E2 interaction by the overexpression of a dominant negative C-terminal domain of Brd4 inhibits the association of E2 with the chromosome, viral genome maintenance, and BPV1-induced cellular transformation (16, 24, 41). Finally, in Saccharomyces cerevisiae, E2-mediated plasmid segregation can be reconstituted only in the presence of Brd4 (4).
For most papillomaviruses, there is no good cell-based system available to study genome segregation. However, because it has been clearly shown that BPV-1 E2 partitions genomes by tethering them to mitotic chromosomes, the localization of other papillomavirus E2 proteins to the mitotic chromosome has been analyzed to determine whether they use a similar mechanism. A systematic study was carried out in our laboratory to investigate the localization of the E2 proteins of 13 animal and human papillomaviruses (HPVs) from 7 different papillomavirus genera to the mitotic chromosome (27). Many of the E2 proteins, such as those from European elk papillomavirus and HPV type 1a (HPV-1a), show colocalization with Brd4 on mitotic chromosomes similar to that of BPV-1 E2. However, the HPV-8 E2 protein was found to associate with the pericentromeric region of chromosomes as larger, often dimeric, speckles (27). The E2 proteins of the human alphapapillomaviruses HPV-11, HPV-16, and HPV-31 also showed localization patterns similar to that of HPV-8 E2 under certain fixation conditions. These E2 proteins did not colocalize with Brd4, and mutations in the HPV-31 E2 protein which abrogate Brd4 binding did not affect mitotic chromosome binding (24). In another study, the HPV-11, HPV-16, and HPV-18 alphapapillomavirus E2 proteins were reported to be associated with the mitotic spindle (37). So, while the chromosomal target of BPV has been established, the binding partners of many human papillomaviruses remain elusive, and these viruses may interact with diverse binding partners for genome segregation.
The association of HPV-8 E2 with the host chromosome is one of the most stable and distinctive of those of the papillomavirus E2 proteins that we have studied. HPV-8 is a betapapillomavirus that subclinically infects the cutaneous epithelium in normal individuals. In individuals with the genetic disorder epidermodysplasia verruciformis, infection causes abundant lesions which have a propensity to transform into carcinomas (13). Several viral types from the genus Papillomavirus have also been implicated in nonmelanoma skin cancers (11). Considering the carcinogenic potential of HPV-8 and the distinctive localization of the E2 protein of HPV-8 on mitotic chromosomes, we further investigated the mechanism of HPV-8 genome segregation.
In the present work, we show that the HPV-8 E2 protein associates with mitotic chromosomes in the absence of the N-terminal domain, which has been shown to be a critical determinant for BPV-1 E2 chromosome targeting (2, 3, 34). On human host chromosomes, HPV-8 E2 binds to the short arms of the acrocentric chromosomes, to a region containing both β-satellite DNA and the ribosomal DNA (rDNA) loci. Further mapping demonstrated that the HPV-8 E2 protein binds to the rDNA genes and colocalizes with the ribosomal transcription factor UBF1.
MATERIALS AND METHODS
Plasmids.
The pMEP-HPV-8 E2 plasmid, which expresses FLAG-tagged E2 from the metallothionein promoter, has been described previously (24). DNA that encodes subdomains of HPV-8 E2 was amplified, using PCR primers with appropriate restriction sites, and was cloned into the pMEP4 vector. Details of the cloning procedures will be provided on request. All E2 proteins except the HPV-8 E2 C domain, were encoded by an N-terminal FLAG epitope tag. The tag on the C domain consisted of the FLAG tag, followed by CBP (calmodulin binding protein) and SBP (streptavidin binding protein) tags. The NH and HC domains of HPV-8 E2 correspond to amino acids 1 to 404 and 206 to 498, respectively. The NC domains encode the E2 protein with amino acids 1 to 205 fused to amino acids 404 to 498. The N, H, and C domains correspond to amino acids 1 to 205, 207 to 405, and 404 to 498, respectively. The R37AI73A mutation was generated in the full-length E2 gene, using the QuikChange multi site-directed mutagenesis kit (Stratagene). The pMEP-9 BPV-E2 expression vector is similar to pMEP-4 BPV-E2, except that the hygromycin resistance marker has been replaced by the neomycin resistance gene. pA and p21β7 plasmids, carrying rDNA and β-satellite DNA, respectively, have been described previously (10, 14).
Cell culture.
HPV-8- and BPV-1 E2-expressing CV-1 cells were derived, as described previously (25), in the experiment shown in Fig. 1, with double selection with hygromycin and G418. C33A cells expressing wild-type and different truncated versions of HPV-8 E2 were selected with 80 μg/ml hygromycin B (Roche) and maintained in 40 μg/ml hygromycin B.
FIG. 1.
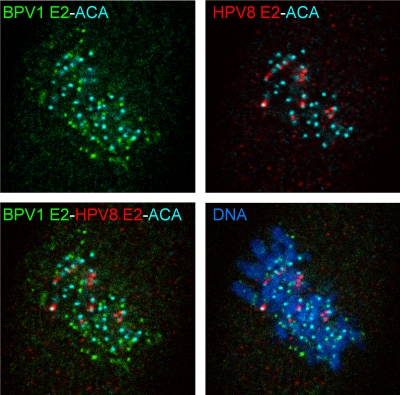
BPV-1 and HPV-8 E2 proteins bind to different chromosomal locations. BPV-1 E2 (green) and HPV-8 E2 (red) proteins were detected in CV-1 cells expressing both proteins by indirect immunofluorescence, using B201 and anti-FLAG antibodies, respectively. The kinetochore was detected with an anticentromere antibody (aqua blue). Cellular DNA was counterstained with DAPI (blue).
Brd4-E2 binding assay.
The following procedure has been described previously (3, 5). Purified Brd4 protein (0.64 μg in 5 μl) was bound to M2 anti-FLAG agarose by incubation in 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% Tween 20, 1 mM dithiothreitol, and 1× Complete EDTA-free protease inhibitor cocktail (Roche) at room temperature for 1 h. E2 proteins were synthesized and labeled with [35S]-methionine, using Promega's TNT quick-coupled reticulocyte lysate system, and the concentrations of each were normalized to one another based upon the number of methionines. Equivalent amounts of 35S-labeled E2 proteins were incubated with Brd4 agarose beads for 1 h. Bound proteins were eluted in 2% sodium dodecyl sulfate (SDS)-62.5 mM Tris-HCl (pH 6.8)-25% glycerol and separated by SDS- polyacrylamide gel electrophoresis. The amount of 35S-E2 bound to Brd4 was measured using a Typhoon imager (Molecular Dynamics).
Indirect immunofluorescence.
pMEP plasmids containing cells were grown on Superfrost Plus slides. For mitotic synchronization, cells were blocked with 2 mM thymidine and fixed with 4% paraformaldehyde 9 h after release, as described previously (24). Four hours before fixation, E2 expression was induced with 6 μM CdSO4. For metaphase chromosome spreads, cells were treated with 30 ng/ml colchicine for 3 h before fixation and swollen in hypotonic buffer 1 (10 mM Tris [pH 7.4], 10 mM NaCl, 5 mM MgCl2) and hypotonic buffer 2 (25% phosphate-buffered saline [vol/vol]) for 15 min each at room temperature, and the slides were cytocentrifuged at 1,500 rpm for 3 min and fixed as described above (42). Cells were permeabilized and stained as described previously (24). HPV-8 E2, UBF, Brd4, and centromeres were detected with monoclonal anti-FLAG M2 antibody (1:500; Sigma), rabbit polyclonal anti-UBF antibody (1:100; Santa Cruz Biotechnologies, Inc.), rabbit polyclonal antiserum against Brd4 2290 (9), and human anticentromere antiserum (1:1,000; ImmunoVision), respectively. Fluorescent secondary antibodies were from Jackson ImmunoResearch Laboratories. Slides were mounted in Vectashield containing 1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole). Images were collected with a Leica TCS-SP5 laser scanning confocal imaging system. Maximal projection images of 15 to 25 0.2 μM Z-stack images were obtained, using Imaris software (Bitplane AG).
FISH.
A combined immunofluorescence and fluorescence in situ hybridization (FISH) technique was developed for the studies described herein. After immunofluorescence detection as described above, slides were treated with methanol:acetic acid (3:1) for 10 min, 2% paraformaldehyde for 1 min, and a 70%, 90%, and 100% ethanol series for 3 min each. Fluorescein isothiocyanate-labeled α-satellite probes specific for the centromeres of chromosomes 1, 5, and 19; 13 and 21; or 14, 22, and 15 (Qbiogene) were used to identify specific chromosomes. Rhodamine-labeled α-satellite probes (2 μl) specific for chromosomes 13 and 21; 14 and 22; or 1, 5, 9, and 15 (Qbiogene) were hybridized in 8 μl hybridization buffer (Qbiogene), and the cellular DNA and probe were denatured by incubating the slides at 75°C for 5 min. Hybridization was carried out overnight at 37°C. The slides were washed with 1× phosphate-buffered detergent (Qbiogene) for 5 min at room temperature, 1× wash buffer (0.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]-0.1% SDS) for 5 min at 65°C, and 1× phosphate-buffered detergent for 5 min at room temperature and mounted as described above. Probes for rDNA and β-satellite DNA were prepared from pA and p21β7 plasmids, respectively. DNA (2 μg) was labeled with Alexa Fluor 594, using the Ulysis nucleic acid labeling kit according to the manufacturer's instructions. The probe DNA was precipitated with 4 mg/ml salmon sperm DNA and resuspended at 10 μg/ml in hybridization buffer (50% formamide, 10% dextran sulfate, 4× SSC). After indirect immunofluorescence staining and cross-linking, the slides were treated with 100 μg/ml RNaseA in 2× SSC. Subsequently, the slides were denatured in 70% formamide in 2× SSC at 72°C for 5 min, dehydrated in ice-cold 70%, 80%, and 100% ethanol for 2 min each, and rinsed with 100% ethanol for 1 min at room temperature. After denaturation, 10 μl of 10 μg/ml probe DNA was hybridized overnight to the denatured slide. For rDNA FISH, the slides were washed three times with washing solution I (50% formamide-2× SSC) for 5 min each at 45°C and three times with washing solution II (1× SSC) for 5 min each at 65°C and were rinsed briefly in washing solution III (4× SSC-0.1% Tween 20) at room temperature. For β-satellite FISH, the slides were washed two times with washing solution I (50% formamide-2× SSC) for 5 min each at 40°C and rinsed briefly in washing solution III (4× SSC-0.1% Tween 20) at room temperature. The slides were mounted in DAPI-containing Vectashield as described above. Images were collected and processed as described in the previous section.
Deconvolution and colocalization.
Three-dimensional reconstruction, maximal projection, and colocalization of the sequential Z-sections were performed with Imaris software (Bitplane AG). Collected data for analysis were deconvolved with Huygens essential software (Scientific Volume Imaging BV). Values for 2-D line profiles of maximal projections were obtained by using IPlab software (BD Biosciences Bioimaging).
RESULTS
HPV-8 and BPV-1 E2 proteins bind different regions of mitotic chromosomes.
In previous studies, the HPV-8 and BPV-1 E2 proteins showed distinct patterns of localization on mitotic chromosomes (27). To analyze this further, a CV-1-derived cell line that expressed both E2 proteins was established. The location of each protein was determined by using an antibody against the N-terminal FLAG tag on the HPV-8 E2 protein and a monoclonal antibody specific for the untagged BPV-1 E2 protein. The centromeres of the host chromosomes were identified with an antibody against the kinetochore. As shown in Fig. 1, each E2 protein bound to distinct regions of mitotic chromosomes, and no colocalization was observed. The HPV-8 E2 protein was observed to always be located adjacent to the centromeres of the host chromosomes, whereas BPV-1 E2 had a random distribution with respect to the centromere. Thus, HPV-8 E2 has a unique chromosomal target that is distinct from that of BPV-1 E2.
The H and C domains of HPV-8 E2 are sufficient for mitotic chromosome binding.
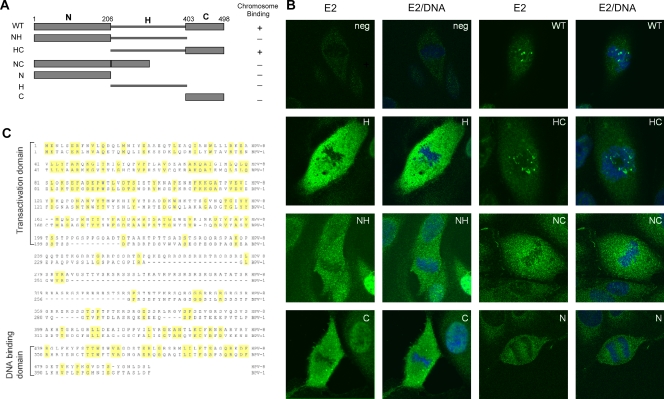
The E2 protein consists of three domains. The N-terminal transactivation domain (N) and the C-terminal DNA-binding domains (C) are conserved among different papillomaviruses. These conserved domains are connected by a flexible, nonconserved hinge region (H). The N-terminal domain of BPV-1 E2 has been shown to be important for binding to mitotic chromosomes, while the C-terminal domain binds specifically to sites in the viral genome (2). The differences between HPV-8 E2 and BPV E2 chromosome localization suggested that the mechanism of chromosome binding could be different for these two proteins. To investigate the regions of the HPV-8 E2 protein required for chromosome association, FLAG-tagged proteins consisting of different domains of HPV-8 E2 (Fig. 2A) were expressed in CV-1 cells, and their localization was studied by indirect immunofluorescence. As shown in Fig. 2B, localization of the HPV-8 E2 HC protein lacking any N-terminal domain was comparable to that of the full-length protein. However, proteins consisting of NC, NH, or the individual N, H, and C domains showed no specific chromosome binding. Therefore, the hinge region and C terminal domain of HPV-8 E2 are necessary and sufficient for chromosome targeting. Alignment of the BPV-1 and HPV-8 E2 proteins is shown in Fig. 2C.
FIG. 2.
Both full-length and HC HPV-8 E2 proteins bind mitotic chromosomes. (A) Diagram showing the structure of wild-type (WT) HPV-8 E2 and the individual domains used for chromosome localization. N, H, and C represent the N-terminal, hinge, and C-terminal domains, respectively. The amino acid positions delineating these domains are shown. (B) Immunolocalization of HPV-8 E2 proteins and derived domains expressed in CV-1 cells. HPV-8 E2 proteins were detected with anti-FLAG antibody (green), and cellular DNA was counterstained with DAPI (blue). neg, negative. (C) Alignment of the amino acid sequences of the BPV-1 and HPV-8 E2 proteins.
HPV-8 E2 mitotic chromosome binding does not require Brd4.
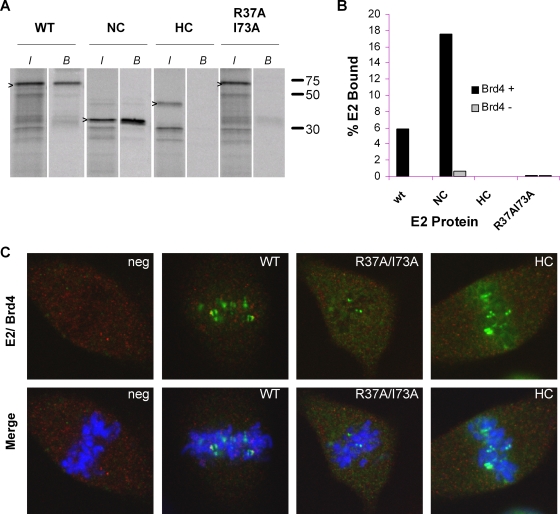
The N-terminal domain of BPV-1 E2 is critical for Brd4 binding and hence for chromosome association. The amino acid substitutions R37A and I73A in the N-terminal domain of BPV-1 result in the loss of Brd4 interaction and chromosome binding in all E2 proteins tested (3, 24, 32). Our previous observations that BPV-1 and HPV-8 E2 proteins bind to different regions of mitotic chromosomes and that the N-terminal domain of HPV8-E2 is not required indicated that HPV-8 E2 probably associates with mitotic chromosomes independently of its interaction with Brd4. To confirm this, we investigated the localization of Brd4 in CV-1 cells expressing the HPV-8 E2, HPV-8 E2 R37AI73A, and HPV8-E2 HC proteins. All three proteins bound to mitotic chromosomes without any significant colocalization with Brd4 (Fig. 3C). In vitro binding assays confirmed that as for BPV-1 E2, the N-terminal domain of HPV-8 E2 is required for Brd4 binding, that the R37AI73A mutation disrupts this binding, and that HPV8-E2 HC is unable to bind Brd4 (Fig. 3A and B). Therefore, the association of HPV-8 E2 with mitotic chromosomes does not require Brd4 and thus is different from that of BPV-1 E2.
FIG. 3.
Interaction of HPV-8 E2 protein in the absence of Brd4 binding. (A) In vitro-translated E2 proteins were tested for their abilities to bind Brd4. Aliquots (10 μl) of E2-containing reticulocyte lysate (adjusted for the concentration of E2) were assayed for binding to 5 μl of Brd4 protein extract prebound to anti-FLAG immunobeads. Bound E2 proteins (B) were eluted and analyzed by SDS-polyacrylamide gel electrophoresis. The input lanes (I) contain a 1/5 volume of lysate added to the binding reactions. Molecular size markers are at the right. (B) Percentage of input protein bound for each E2 protein in the presence or absence of Brd4 protein. wt, wild type. (C) Immunolocalization of HPV-8 E2 proteins and Brd4 on mitotic chromosomes in CV-1 cells. E2 was detected with anti-FLAG antibody (green) and Brd4 with anti-Brd4 antiserum (red). DNA was counterstained with DAPI (blue). WT, wild type; neg, negative.
HPV-8 E2 binds to the pericentromeric region of a subset of mitotic chromosomes.
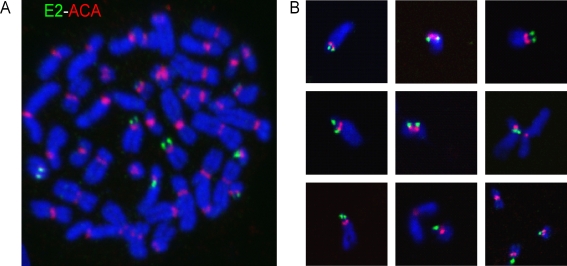
To further characterize the mitotic binding site of HPV-8 E2, E2-expressing cell lines were derived from human cervical carcinoma-derived C33A cells. Stable cell lines express detectable E2 protein in about 20 to 30% of interphase cells after induction of the metallothionein promoter. The E2 protein can be detected in about 10% of mitotic cells and was localized on mitotic chromosomes in a pattern similar to that observed previously in CV-1 cells. To analyze this localization in further detail, chromosome spreads were prepared, and HPV-8 E2 and centromeres were detected by indirect immunofluorescence. E2 staining was observed as paired or single foci of variable size, located adjacent to the centromeres (Fig. 4A). The number of E2 signals observed per cell ranged from three to eight signals, with an average of six signals. The intensity of the E2 staining on each chromosome was variable; however, each focus within a paired signal had a similar intensity. Figure 4B shows a panel of individual, well-isolated chromosomes with E2 and ACA staining. These chromosomes were randomly selected from different metaphase spreads. For the majority of the E2-bound chromosomes, both E2 and ACA staining was located very close to the end of the chromosomes, suggesting that E2 is associated with centromeres on the short arms of the chromosomes.
FIG. 4.
Distribution of HPV-8 E2 protein on mitotic chromosomes. (A) Immunolocalization of E2 protein and centromeric ACA antigen on a metaphase chromosome spread of C33A cells expressing HPV-8 E2. E2 was detected with an anti-FLAG antibody (green), and centromeres were detected with an anti-ACA centromeric antibody (red). DNA was counterstained with DAPI (blue). (B) Images of isolated, individual chromosomes showing HPV-8 E2 and ACA staining.
HPV-8 E2 associates with acrocentric chromosomes.
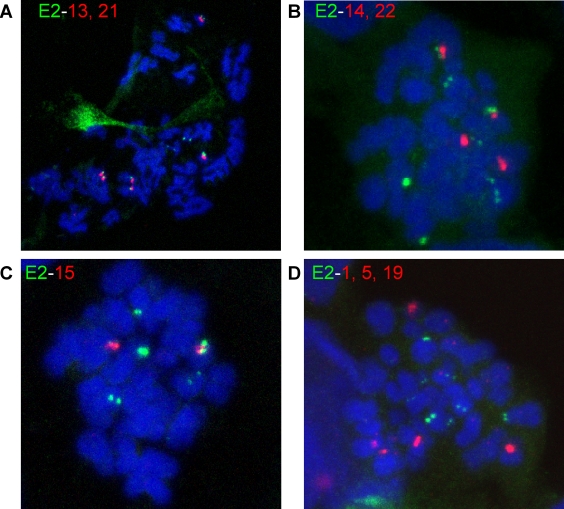
By definition, chromosomes with the centromere close to one end are known as acrocentric chromosomes. To further prove that E2 was binding specifically to such chromosomes, a combined immunofluorescence/FISH technique was developed. HPV-8 E2 was detected on metaphase chromosomes by indirect immunofluorescence with an anti-FLAG antibody. The antibodies were cross-linked, and FISH was performed with α-satellite probes specific for the acrocentric chromosomes 13 and 21, 14 and 22, or 15. In mitotic cells expressing E2, the immunofluorescence signal for E2 was observed adjacent to the FISH signal specific for each of the acrocentric chromosomes (Fig. 5). However, the distributions of E2 on different acrocentric chromosomes were not identical. E2 was localized next to the centromere of chromosomes 13 and 21 in 100% of cases (Table 1). In contrast, only 43% of chromosomes 14 and 22 and 73% of chromosomes 15 showed E2 binding. To further prove the specificity of E2 binding to acrocentric chromosomes, similar immuno-FISH experiments were carried out with probes specific for the nonacrocentric chromosomes 1, 5, and 19. E2 staining was never observed adjacent to the FISH signal of these chromosomes, confirming the specificity for acrocentric chromosomes (Fig. 5D).
FIG. 5.
HPV-8 E2 associates with the short arms of acrocentric chromosomes. (A, B, and C) HPV 8 E2 was mapped to acrocentric chromosomes by combined immunostaining of E2 (green) and FISH, using α-satellite probes (red). Cellular DNA is counterstained with DAPI and shown in blue. (D) FISH, with α-satellite probes specific for nonacrocentric chromosomes 1, 5, and 19, demonstrates the specificity of the association of HPV-8 E2 with acrocentric chromosomes.
TABLE 1.
Correlation of E2 localization on acrocentric chromosomesa
 |
The green circles represent the E2 fluorescence signal observed on each chromosome, and the red circles represent the fluorescence probe that identifies the centromere of each acrocentric chromosome. The numbers of chromosomes with both signals adjacent are shown as combined red and green circles.
HPV-8 E2 binds adjacent to, but does not overlap with, β-satellite DNA on the short arms of acrocentric chromosomes.
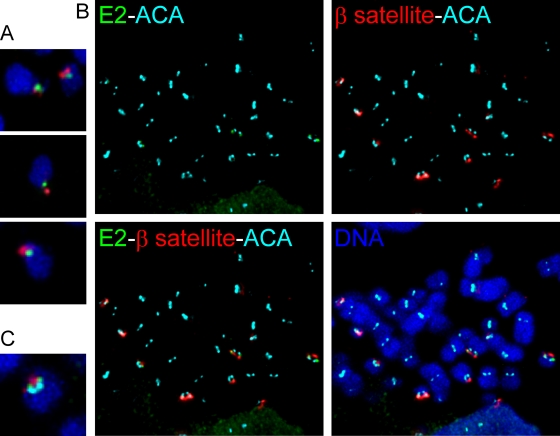
The short arms of the acrocentric chromosomes contain different families of repetitive DNA sequences arranged in tandem. The tight foci of the HPV-8 E2 immunofluorescence signals at equivalent positions on each sister chromatid prompted us to investigate whether E2 binds to these sequences on the mitotic chromosomes. β-Satellite DNA, one of the best characterized repetitive DNAs, is located on the short arms of acrocentric chromosomes (14). Approximately 250 to 500 copies of higher-order repeats of β-satellite DNA are estimated to be present per haploid genome. Two subfamilies of β-satellite DNA are organized on the short arms of acrocentric chromosomes. The p21ß7 subfamily is localized to the distal portion of the short arms, and the p21ß2 subfamily is located both at the distal end of the chromosomes and proximal to the centromeric DNA. Since the E2 signal was observed at a single locus on each chromosome, we used a FISH probe corresponding to the p21ß7 subfamily to detect β-satellite sequences. Combined immunofluorescence/FISH was performed as described above to detect HPV-8 E2 and β-satellite DNA, respectively. On chromosome spreads from C33A cells, the E2 signal was observed adjacent to the β-satellite signal (Fig. 6). When individual chromosomes were examined, the β-satellite signal was located at the distal end of the chromosome, while HPV-8 E2 was located toward the long chromosome arm (Fig. 6A). To map the location of E2 with respect to other chromosomal sequences, double immunofluorescence staining was done with anti-FLAG and ACA antibodies together with FISH by using β-satellite probes. E2 was found to be located between the centromere DNA and the β-satellite DNA on acrocentric chromosomes (Fig. 6).
FIG. 6.
β-satellite DNA and HPV-8 E2 occupy adjacent, but distinct, regions on the short arms of acrocentric chromosomes. (A) Images of individual chromosomes showing localization of E2 and β-satellite DNA. Combined immunofluorescence with anti-FLAG antibody was used to detect E2 protein (green), and FISH was used to detect β-satellite DNA (red) on metaphase spreads. DNA is counterstained with DAPI (blue). (B) An E2 binding site is located between the centromere and β-satellite DNA. Metaphase chromosome spreads with E2 (green), centromere (cyan blue), and β-satellite DNA (red) are shown. (C) Image of an individual chromosome with E2 (green), β-satellite DNA (red), and centromere (cyan blue) fluorescence signals.
HPV-8 E2 binds to the rDNA loci on mitotic chromosomes.
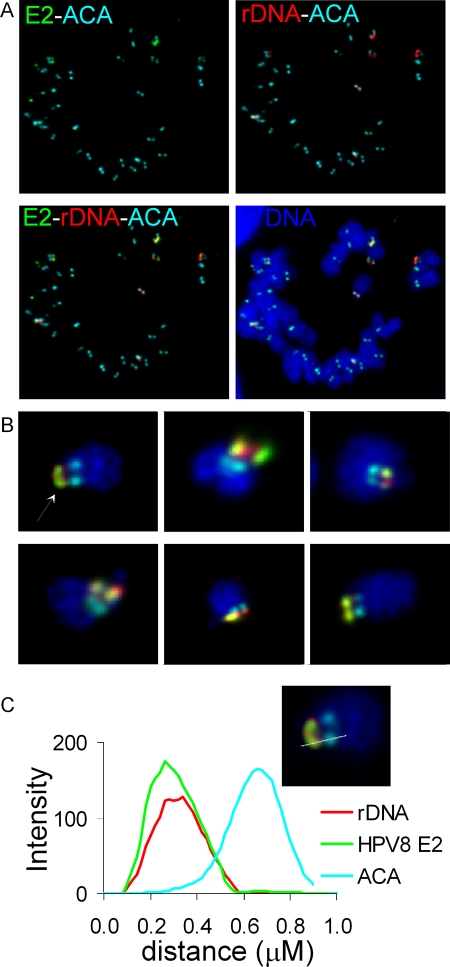
The genes carrying 5.8S, 18S, and 28S rRNA are organized as tandemly repeating units on the short arms of acrocentric chromosomes (14, 22). There are approximately 400 copies of these repeat units present per diploid genome. The p21ß7 subfamily is known to be localized to the distal end of the rDNA gene cluster (14). Localization of HPV-8 E2 relative to β-satellite DNA indicated that rDNA could be the potential chromosomal target of HPV-8 E2. To confirm this, HPV-8 E2, kinetochores, and rDNA were located on chromosome spreads by indirect immunofluorescence with anticentromeric ACA and anti-FLAG antibodies and by FISH with a probe specific for rDNA. The probe was generated from the pA plasmid that contains a 7.3-kb EcoRI fragment spanning ∼200 nucleotides of the 18S rRNA gene, the internal transcribed spacer 1 region, sequences carrying the 5.8S rRNA gene, the internal transcribed spacer 2, and ∼4,500 nucleotides of the upstream end of the 28S rRNA gene. We could detect between six and nine FISH signals per cell, representing the rDNA loci. The majority of the E2 signal was found to be colocalized with these rDNA signals (Fig. 7A). Analysis of randomly selected, well-isolated chromosomes from different mitotic chromosome spreads showed that both HPV-8 E2 and rDNA colocalize on the mitotic chromosomes and are located adjacent to the centromere DNA, as detected by ACA staining (Fig. 7B). The relative position of each locus was determined by plotting the relative intensities of each fluorescence signal along a line drawn through the signals on the acrocentric chromosome (Fig. 7C). This further demonstrated that the peak of the intensity of the E2 signal coincided with that of rDNA. The fluorescence intensity peak for the centromere (ACA) was distinct and did not overlap with either E2 or rDNA signals.
FIG. 7.
HPV-8 E2 colocalizes with rDNA on mitotic chromosomes. (A) Combined immunofluorescence and FISH were used to detect E2 (green), rDNA (red), and ACA centromere antigen (cyan blue) on metaphase chromosome spreads of HPV-8 E2-expressing C33A cells. DNA is shown in blue. (B) E2, rDNA, and ACA staining on randomly chosen isolated chromosomes from different metaphase chromosome spreads. (C) A fluorescence intensity light scan was obtained by drawing a line through the three fluorescence signals of E2, rDNA, and ACA. The chromosome used for drawing the line scan is shown in the inset.
HPV-8 E2 colocalizes with the rDNA transcription factor UBF on mitotic chromosomes.
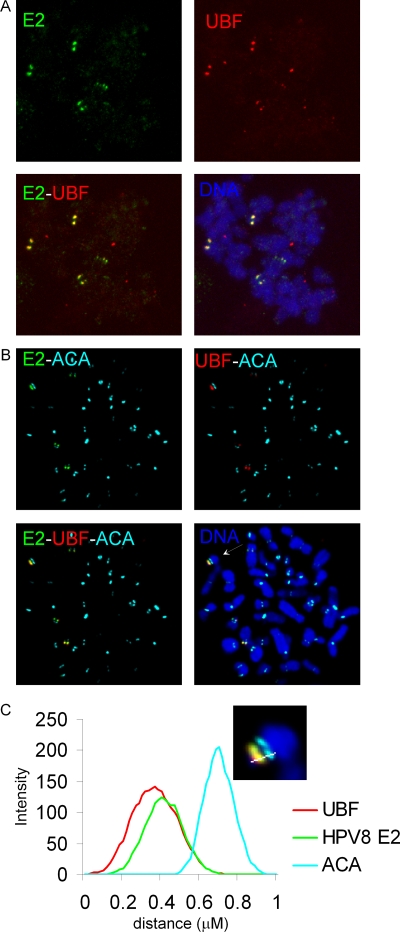
UBF is a transcription factor required for transcription of rDNA by RNA polymerase I (17). UBF has been shown to associate with the chromosomal region corresponding to rDNA and remains bound in all stages of mitosis, although transcription is silenced in mitosis (28, 29). Colocalization of E2 with UBF with reference to the centromere was investigated, using anti-FLAG, anti-UBF, and anti-ACA antibodies. HPV-8 E2 was found to associate with the chromosome at the same position as UBF (Fig. 8A). Colocalization was observed as yellow foci, equivalently positioned on the sister chromatids of the mitotic chromosomes. The numbers of UBF foci per cell ranged from four to eight, with an average of seven. Like that for E2, UBF staining could be seen as either single foci or paired dots, and the intensities of different UBF signals were variable. Immunostaining with ACA, anti-FLAG, and anti-UBF showed that E2 and UBF colocalize adjacent to the centromere (Fig. 8B). This data was confirmed by the fluorescence line scans of the E2, UBF, and ACA signals (Fig. 8C).
FIG. 8.
HPV-8 E2 and UBF1 colocalize on mitotic chromosomes. (A) E2 (green) and UBF1 (red) were detected in metaphase chromosome spreads by indirect immunofluorescence, using anti-FLAG and anti-UBF antibodies, respectively. (B) Localization of E2 and UBF relative to the centromere was determined by immunostaining with ACA (aqua blue), E2 (green), and UBF (red) antibodies. DNA was stained with DAPI (blue). (C) Fluorescence intensity light scan of E2, UBF1, and ACA signals. The chromosome used for drawing the line scan is shown in the inset.
DISCUSSION
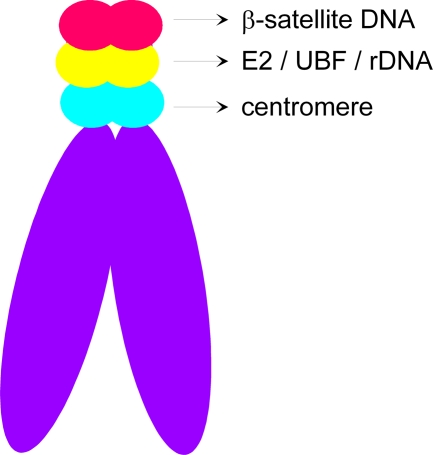
In this study, we demonstrated that the chromosomal target of the HPV-8 E2 tethering protein is quite different from that of the BPV-1 E2 protein, which binds to the chromosome arms in complex with the cellular Brd4 protein (3, 40). In contrast to BPV-1 E2, the N-terminal domain of HPV-8 E2, which is required for association with Brd4, is not required for chromosomal association. Rather, both the C-terminal DNA binding and dimerization domain and the hinge region are required. Furthermore, Brd4 does not colocalize on mitotic chromosomes with the HPV-8 E2 R37AI73A and HPV-8 E2 HC proteins that are unable to interact with Brd4. Thus, as reported previously, Brd4 is not required for genome partitioning of all papillomaviruses, although it does seem to be required for transcriptional activation and repression (16, 24, 31, 32, 39). Further characterization of the binding site of HPV-8 E2 on human chromosomes revealed that this protein binds to the rDNA genes located between β-satellite DNA and centromeric DNA on the short arms of acrocentric chromosomes (Fig. 9).
FIG. 9.
Schematic representation of E2 localization. Localization of E2 (green), rDNA (red), UBF (red), β-satellite DNA (red), and centromere DNA (aqua blue) on acrocentric chromosome. Colocalization of the E2 signal (green) and rDNA/UBF (red) is represented as yellow.
There are several reasons why rDNA might be an attractive target as a chromosomal tethering region for some papillomaviruses. The tandemly arranged repeating rDNA units (400 per diploid cell) increase the local concentration of the binding target, thereby increasing the likelihood of interaction with the E2 protein complex. Because of its repetitive nature, the rDNA locus has specialized mechanisms for replication, cohesion, condensation, and segregation (8, 12, 19, 35, 36). The rDNA transcription units are also poised for immediate RNA synthesis as cells exit mitosis, and transcription factors such as UBF remain bound throughout mitosis (26). The resulting unique chromatin structure could provide unique targets distributed among the host chromosomes. In interphase cells, all papillomavirus E2 proteins are detected in a nuclear, but nucleolar-excluded, pattern. In addition, the HPV-8 E2 protein can be observed in SC35-splicing speckles by virtue of the serine-arginine-rich hinge sequences (A. A. McBride, unpublished data), as has been shown for the related HPV-5 E2 protein (20). This separation in localization at different stages of the cell cycle could be advantageous to the virus, as the E2 protein would be free during interphase to participate in viral transcription, replication, and splicing; the high-affinity nucleolar-organizing region (NOR) targets would be sequestered in the nucleoli until the nucleolar envelope broke down in early mitosis. It is not yet known if E2 plays any functional roles at the rDNA locus other than tethering the viral genomes to the chromosome, but several viruses are known to modify rDNA expression or factors (1, 6, 30).
Notably, only 8 or 9 acrocentric chromosomes were detected by rDNA FISH instead of the expected 10 per chromosome spread. This is due to aneuploidy of the C33A cell line (Table 2). Transcription of each rDNA locus results in the formation of a nucleolus, and thus each region is termed a NOR. Within each cell, some NORs are silent, and only active NORs are associated with polymerase I transcription factors (26). Correspondingly, on average only seven UBF foci and six E2 signals were detected per metaphase spread, and each protein bound acrocentric chromosomes in the preferential order of 13, 21, >15, >14, 22 (Table 3). This implies that the E2 protein binds only to active mitotic NORs and that the E2 target may be a transcription factor or chromatin component associated with these sites. Although E2 and UBF colocalize at the NORs, no direct interaction has yet been detected by coimmunoprecipitation (data not shown), and it remains to be determined whether they have a functional interaction or simply bind to the same chromosomal locus.
TABLE 2.
Distribution of chromosomes in five karyotypes from C33A cells
| Karyotype | No. of copies of normal and marker (M) chromosomes
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | M1 | M2 | |
| 1 | 0 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 0 | 1 | 1 |
| 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 0 | 1 | 1 |
| 3 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | 1 | 0 |
| 4 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 2 | 2 | 0 | 1 | 1 |
| 5 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 0 | 1 | 1 |
TABLE 3.
Correlation of E2 and UBF signals on acrocentric chromosomesa
The green circles represent the UBF fluorescence signal observed on each chromosome, and the red circles represent the fluorescence probe that identifies the centromere of each acrocentric chromosome. The numbers of chromosomes with both signals adjacent are shown as combined red and green circles.
There are also connections between other viral tethering proteins and nucleoli or NORs. The cellular target of the Epstein-Barr virus tethering protein, EBNA, is a protein termed p40 nucleolar proliferation antigen, or hEBP2, and is involved in pre-RNA processing (33). hEBP2 is nucleolar in interphase (7) but is distributed along condensed chromosomes in mitosis (38). Recently, the C-terminal domain of the LANA tethering protein of KSHV has been shown to bind as paired dots to pericentromeric and telomeric regions of certain metaphase chromosomes (18), some of which we speculate might be acrocentric chromosomes. We have previously demonstrated that E2 proteins from the Alphapapillomavirus genus can also be observed binding to pericentromeric regions of chromosomes, although only in certain fixation conditions (27). Thus, the rDNA loci may be an attractive tethering target for several viruses, and further characterization of this interaction could result in the development of specific antiviral therapies.
Acknowledgments
This research was supported by the Intramural Research Program of the NIAID, NIH.
We thank Juraj Kabat for help with image processing.
Footnotes
Published ahead of print on 12 November 2008.
REFERENCES
- 1.Banerjee, R., M. K. Weidman, S. Navarro, L. Comai, and A. Dasgupta. 2005. Modifications of both selectivity factor and upstream binding factor contribute to poliovirus-mediated inhibition of RNA polymerase I transcription. J. Gen. Virol. 862315-2322. [DOI] [PubMed] [Google Scholar]
- 2.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 with mitotic chromosomes. Virology 270124-134. [DOI] [PubMed] [Google Scholar]
- 3.Baxter, M. K., M. G. McPhillips, K. Ozato, and A. A. McBride. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 794806-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brannon, A. R., J. A. Maresca, J. D. Boeke, M. A. Basrai, and A. A. McBride. 2005. Reconstitution of papillomavirus E2-mediated plasmid maintenance in Saccharomyces cerevisiae by the Brd4 bromodomain protein. Proc. Natl. Acad. Sci. USA 1022998-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardenas-Mora, J., J. E. Spindler, M. K. Jang, and A. A. McBride. 2008. Dimerization of the papillomavirus E2 protein is required for efficient mitotic chromosome association and Brd4 binding. J. Virol. 827298-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavender, J. F., C. Mummert, and M. J. Tevethia. 1999. Transactivation of a ribosomal gene by simian virus 40 large-T antigen requires at least three activities of the protein. J. Virol. 73214-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, A., J. W. Freeman, and H. Busch. 1987. Identification and partial characterization of a Mr 40,000 nucleolar antigen associated with cell proliferation. Cancer Res. 471123-1129. [PubMed] [Google Scholar]
- 8.D'Amours, D., F. Stegmeier, and A. Amon. 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117455-469. [DOI] [PubMed] [Google Scholar]
- 9.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 206537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson, J. M., C. L. Rushford, D. J. Dorney, G. N. Wilson, and R. D. Schmickel. 1981. Structure and variation of human ribosomal DNA: molecular analysis of cloned fragments. Gene 161-9. [DOI] [PubMed] [Google Scholar]
- 11.Feltkamp, M. C., R. Broer, F. M. di Summa, L. Struijk, E. van der Meijden, B. P. Verlaan, R. G. Westendorp, J. ter Schegget, W. J. Spaan, and J. N. Bouwes Bavinck. 2003. Seroreactivity to epidermodysplasia verruciformis-related human papillomavirus types is associated with nonmelanoma skin cancer. Cancer Res. 632695-2700. [PubMed] [Google Scholar]
- 12.Freeman, L., L. Aragon-Alcaide, and A. Strunnikov. 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149811-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, P. G., T. Iftner, J. Weninger, and H. Pfister. 1986. Epidermodysplasia verruciformis-associated human papillomavirus 8: genomic sequence and comparative analysis. J. Virol. 58626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greig, G. M., and H. F. Willard. 1992. β satellite DNA: characterization and localization of two subfamilies from the distal and proximal short arms of the human acrocentric chromosomes. Genomics 12573-580. [DOI] [PubMed] [Google Scholar]
- 15.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 734404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilves, I., K. Maemets, T. Silla, K. Janikson, and M. Ustav. 2006. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 803660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jantzen, H. M., A. Admon, S. P. Bell, and R. Tjian. 1990. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 344830-836. [DOI] [PubMed] [Google Scholar]
- 18.Kelley-Clarke, B., M. E. Ballestas, T. Komatsu, and K. M. Kaye. 2007. Kaposi's sarcoma herpesvirus C-terminal LANA concentrates at pericentromeric and peri-telomeric regions of a subset of mitotic chromosomes. Virology 357149-157. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, T., D. J. Heck, M. Nomura, and T. Horiuchi. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 123821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, M. C., B. H. Teh, and W. Y. Tarn. 1999. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem. 27411832-11841. [DOI] [PubMed] [Google Scholar]
- 21.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 954338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long, E. O., and I. B. Dawid. 1980. Repeated genes in eukaryotes. Annu. Rev. Biochem. 49727-764. [DOI] [PubMed] [Google Scholar]
- 23.McBride, A. A., J. G. Oliveira, and M. G. McPhillips. 2006. Partitioning viral genomes in mitosis: same idea, different targets. Cell Cycle 51499-1502. [DOI] [PubMed] [Google Scholar]
- 24.McPhillips, M. G., J. G. Oliveira, J. E. Spindler, R. Mitra, and A. A. McBride. 2006. Brd4 is required for E2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 809530-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McPhillips, M. G., K. Ozato, and A. A. McBride. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 798920-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McStay, B. 2006. Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 201207-1214. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira, J. G., L. A. Colf, and A. A. McBride. 2006. Variations in the association of papillomavirus E2 proteins with mitotic chromosomes. Proc. Natl. Acad. Sci. USA 1031047-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Sullivan, A. C., G. J. Sullivan, and B. McStay. 2002. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roussel, P., C. Andre, C. Masson, G. Geraud, and D. Hernandez-Verdun. 1993. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci. 104327-337. [DOI] [PubMed] [Google Scholar]
- 30.Samad, M. A., M. Okuwaki, H. Haruki, and K. Nagata. 2007. Physical and functional interaction between a nucleolar protein nucleophosmin/B23 and adenovirus basic core proteins. FEBS Lett. 5813283-3288. [DOI] [PubMed] [Google Scholar]
- 31.Schweiger, M. R., J. You, and P. M. Howley. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 804276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sénéchal, H., G. G. Poirier, B. Coulombe, L. A. Laimins, and J. Archambault. 2007. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology 35810-17. [DOI] [PubMed] [Google Scholar]
- 33.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 732587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 722079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan, M., T. Higuchi, V. L. Katis, and F. Uhlmann. 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 117471-482. [DOI] [PubMed] [Google Scholar]
- 36.Torres-Rosell, J., F. Machin, S. Farmer, A. Jarmuz, T. Eydmann, J. Z. Dalgaard, and L. Aragon. 2005. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 7412-419. [DOI] [PubMed] [Google Scholar]
- 37.Van Tine, B. A., L. D. Dao, S. Y. Wu, T. M. Sonbuchner, B. Y. Lin, N. Zou, C. M. Chiang, T. R. Broker, and L. T. Chow. 2004. Human papillomavirus (HPV) origin-binding protein associates with mitotic spindles to enable viral DNA partitioning. Proc. Natl. Acad. Sci. USA 1014030-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, H., D. F. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, S. Y., A. Y. Lee, S. Y. Hou, J. K. Kemper, H. Erdjument-Bromage, P. Tempst, and C. M. Chiang. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 202383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117349-360. [DOI] [PubMed] [Google Scholar]
- 41.You, J., M. R. Schweiger, and P. M. Howley. 2005. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. J. Virol. 7914956-14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng, P. S., J. Brokaw, and A. A. McBride. 2005. Conditional mutations in the mitotic chromosome binding function of the bovine papillomavirus type 1 E2 protein. J. Virol. 791500-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]