Abstract
The conserved membrane-associated tegument protein pUL11 and envelope glycoprotein M (gM) are involved in secondary envelopment of herpesvirus nucleocapsids in the cytoplasm. Although deletion of either gene had only moderate effects on replication of the related alphaherpesviruses herpes simplex virus type 1 (HSV-1) and pseudorabies virus (PrV) in cell culture, simultaneous deletion of both genes resulted in a severe impairment in virion morphogenesis of PrV coinciding with the formation of huge inclusions in the cytoplasm containing nucleocapsids embedded in tegument (M. Kopp, H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 78:3024-3034, 2004). To test whether a similar phenotype occurs in HSV-1, a gM and pUL11 double deletion mutant was generated based on a newly established bacterial artificial chromosome clone of HSV-1 strain KOS. Since gM-negative HSV-1 has not been thoroughly investigated ultrastructurally and different phenotypes have been ascribed to pUL11-negative HSV-1, single gene deletion mutants were also constructed and analyzed. On monkey kidney (Vero) cells, deletion of either pUL11 or gM resulted in ca.-fivefold-reduced titers and 40- to 50%-reduced plaque diameters compared to those of wild-type HSV-1 KOS, while on rabbit kidney (RK13) cells the defects were more pronounced, resulting in ca.-50-fold titer and 70% plaque size reduction for either mutant. Electron microscopy revealed that in the absence of either pUL11 or gM virion formation in the cytoplasm was inhibited, whereas nuclear stages were not visibly affected, which is in line with the phenotypes of corresponding PrV mutants. Simultaneous deletion of pUL11 and gM led to additive growth defects and, in RK13 cells, to the formation of large intracytoplasmic inclusions of capsids and tegument material, comparable to those in PrV-ΔUL11/gM-infected RK13 cells. The defects of HSV-1ΔUL11 and HSV-1ΔUL11/gM could be partially corrected in trans by pUL11 of PrV. Thus, our data indicate that PrV and HSV-1 pUL11 and gM exhibit similar functions in cytoplasmic steps of virion assembly.
The structure of the herpesvirus virion is highly complex. The core which contains the double-stranded DNA genome is enclosed in an icosahedral capsid shell. The nucleocapsid is embedded in a proteinaceous tegument which is surrounded by a cell-derived lipid bilayer envelope containing virally encoded (glyco)proteins (51). Tegument and envelope each comprise more than 10 different virally encoded proteins (57). Although the composition of tegument and envelope has been studied in some detail (19, 45, 46, 59, 60), the molecular processes underlying their assembly are still under investigation. Final tegumentation and envelopment start after intranuclear nucleocapsids gain access to the cytoplasm by budding at the inner nuclear membrane, thereby acquiring a primary envelope, and subsequent fusion of the primary envelope with the outer nuclear membrane. Although the tegument has long been considered unstructured, it appears to be composed of several layers, and tegument proteins have tentatively been divided into capsid-proximal (inner) or envelope-proximal (outer) components (43, 44). Tegument-coated nucleocapsids bud into vesicles presumably derived from the trans-Golgi network (TGN) which contain viral glycoproteins and outer tegument proteins, resulting in a mature virion located in a transport vesicle. After fusion of the transport vesicle with the plasma membrane, virions are released into the extracellular space. Evidence from different alpha-, beta-, and gammaherpesviruses suggests that this fundamental pathway is conserved throughout the Herpesviridae (reviewed in reference 42). Although tegument and glycoprotein compositions in the envelope vary widely between the different herpesvirus subfamilies, and also even within subfamilies, several tegument and envelope proteins are conserved, indicating that these proteins might be key players in virion assembly. Among these proteins are the homologs of the UL10 (gM) and UL11 gene products of herpes simplex virus type 1 (HSV-1).
HSV-1 pUL11 is a small, myristoylated, and palmitoylated tegument protein which was shown to be located at membranes, preferentially the TGN (2, 30). As predicted by amino acid sequence analysis, myristoylation and palmitoylation are conserved in the pUL11 homologs and are required for their targeting to membranes (52, 53, 54). Also, pUL11 was reported to associate with pUL16 in HSV-1 and pseudorabies virus (PrV) (31), and it has been suggested that pUL11 could be involved in recruiting viral proteins to the site of virus assembly at the TGN. In line with this hypothesis, deletion of pUL11 homologs resulted in impairment of secondary envelopment in HSV-1, PrV, and human cytomegalovirus (HCMV) (2, 14, 23, 56, 57). However, in PrV and HSV-1 the absence of pUL11 reduced titers only approximately 10-fold, whereas the homologous ppUL28 (pUL99) in HCMV is essential for viral replication (55, 56), which may be indicative of functional differences between these proteins.
HSV-1 mutants lacking pUL11 have been analyzed ultrastructurally in two studies with different results. Baines and Roizman (1) reported that, in the absence of pUL11, capsids accumulated at the inner nuclear membrane, while in a more recent study Fulmer et al. (14) reported the presence of large amounts of nonenveloped cytoplasmic nucleocapsids with no detectable defect in nuclear egress. The latter results are similar to those observed for pUL11-deficient PrV (23). Thus, a clear picture on the role of pUL11 in the different viruses is still lacking.
Glycoprotein M (gM) is a conserved type III integral membrane protein with multiple transmembrane domains. It forms a complex with the product of the conserved UL49.5 gene, which is glycosylated in several herpesviruses, including PrV, and designated as gN (18, 25, 28, 33). Transgenic expression of gM of HSV-1 and PrV was found to alter cellular membrane trafficking, causing relocalization of membrane proteins from the cell surface to the TGN (8, 20). Thus, gM may be involved in retaining viral glycoproteins at the TGN or retrieving them from the plasma membrane and moving them to the TGN, thereby supporting virion maturation at the budding site. Moreover, PrV gM interacts via its cytoplasmic tail with the major tegument protein pUL49 (12). Interestingly, in the mammalian alphaherpesviruses HSV-1, PrV, and equine herpesvirus 1 (EHV-1) deletion of gM only slightly affected replication (6, 10, 35, 49), whereas the gM homologs of Marek's disease virus, HCMV, Epstein-Barr virus (EBV), EHV-4, and murine gammaherpesvirus 68 are required for efficient virus replication in cell culture (17, 29, 36, 58, 61), indicating differing requirements within the Herpesviridae also for this protein.
Since it is known that herpesvirus proteins may function in a redundant fashion, deletion mutants lacking two or more proteins have been isolated. PrV triple mutants lacking gM and the likewise nonessential complex of gE and gI (gE/gI) (4) or gM and only the C-terminal part of gE (5) were strongly inhibited in secondary envelopment, producing large intracytoplasmic accumulations of nonenveloped nucleocapsids associated with tegument proteins. In contrast, the absence of gM in gE/gI-deleted HSV-1 did not result in a similar phenotype (6). Instead, comparable intracytoplasmic aggregations of tegument proteins and capsids were observed in cells infected with an HSV-1 mutant simultaneously lacking gD and gE/gI (11). We also demonstrated that the presence of either pUL11 or gM is crucial for secondary envelopment of PrV (24). Simultaneous deletion of pUL11 and gM drastically impaired virion maturation, resulting in the formation of huge intracytoplasmic inclusions containing capsids and tegument, while the single gene deletion mutants showed only moderate defects in cell culture (24). In light of the differences observed in PrV and HSV-1 mutants simultaneously lacking gM and gE/gI and considering the similarity in phenotypes between PrV lacking gM/gE/gI and PrV lacking pUL11/gM, we wanted to analyze HSV-1 mutants lacking pUL11 and/or gM for their phenotype in virion morphogenesis.
Here, we report the construction of HSV-1 pUL11 and gM single and double deletion mutants based on a newly established infectious clone of HSV-1 strain KOS. Quantitative analyses of in vitro replication properties and ultrastructural investigations of virion morphogenesis were performed in different cell lines, including cells which provided pUL11 of HSV-1 or PrV in trans. These studies continue our recently started attempt to determine the functional homology or divergence of gene products conserved between PrV and HSV-1 (26).
MATERIALS AND METHODS
Cells and viruses.
All virus mutants were derived from HSV-1 strain KOS (HSV-1 KOS; kindly provided by P. Spear, Chicago, IL). Viruses were grown on African green monkey (Vero) or rabbit kidney (RK13) cells in minimum essential medium supplemented with 5% or 10% fetal calf serum, respectively. Generation of an RK13 cell line expressing PrV pUL11 has been described previously (23). An HSV-1 pUL11-expressing cell line was isolated after transfection of Vero cells with plasmid pcDNA-HUL11 (see below) using FuGene HD transfection reagent (Roche, Mannheim, Germany). After 48 h the transfected cells were trypsinized and seeded into microtiter plates with medium containing 500 μg of Geneticin (Invitrogen, Karlsruhe, Germany) per ml. Resistant cell colonies were tested for complementation of HSV-1ΔUL11 (see below). One positive cell clone, designated Vero-UL11(HSV-1), was further used.
Construction of HSV-1 UL10 and UL11 expression plasmids.
The complete open reading frames (ORFs) UL10 and UL11 (Fig. 1A) were PCR amplified from HSV-1 KOS DNA using Pfx DNA polymerase (Invitrogen) and primers deduced from the genome sequence of HSV-1 strain 17 (37) (GenBank accession no. X14112). In primers HUL10For1 (5′-CACAGAATTCATGGGACGCCCG-3′, nucleotides [nt] 23204 to 23315), HUL10Rev (5′-CACAGCGGCCGCCGGTCGGGTTAAACA-3′, complementary to nt 24656 to 24670), HUL11For1 (5′-CACAGAATTCCCTGATCATTACCCCCG-3′, complementary to nt 25168 to 25185), and HUL11Rev (5′-CACAGCGGCCGCTATACAGAACATTGTTTTG-3′, nt 24778 to 24796), EcoRI and NotI sites (underlined) were introduced for convenient cloning. The UL10 start codon is shown in bold. After cleavage the amplification products were inserted into the EcoRI/NotI-digested eukaryotic expression vector pcDNA3 (Invitrogen), giving rise to pcDNA-HUL10 and pcDNA-HUL11 (Fig. 1B and C). For prokaryotic expression, the 3′ part of UL10 was amplified by PCR using primers HUL10For2 (5′-CACAGAATTCAAGCGCGTACGCAG-3′, nt 24312 to 24327) and HUL10Rev, and UL11 was amplified with primers HUL11For2 (5′-CACAGAATTCATGGGCCTCTCGTTCTCC-3′, complementary to nt 25074 to 25091) and HUL11Rev. Both PCR products were digested with EcoRI and NotI and cloned into the appropriately cleaved vector pGEX-4-T1 (GE Healthcare, Freiburg, Germany).
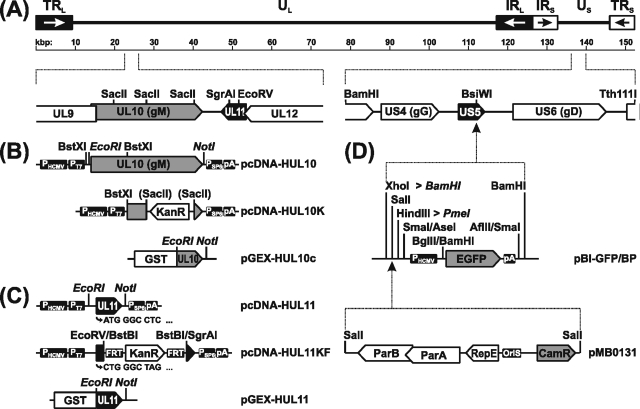
FIG. 1.
Construction of expression plasmids and virus mutants. (A) A schematic map of the HSV-1 genome shows the unique long (UL) and unique short (US) regions flanked by terminal (TRL and TRS) and internal inverted (IRL and IRS) repeat sequences. Numbers denote kilobase pairs. Relevant ORFs (pointed rectangles) and restriction sites are indicated. (B) Eukaryotic expression plasmid pcDNA-HUL10 contains the gM gene flanked by the HCMV immediate-early promoter (PHCMV) and a polyadenylation signal (pA). In pcDNA-HUL10K part of UL10 was replaced by a kanamycin resistance gene (KanR), and the PCR product obtained with PT7- and PSP6-specific primers was used for construction of gM deletion mutants. For preparation of a monospecific rabbit antiserum, the C terminus of gM was expressed as a bacterial fusion protein with GST from pGEX-HUL10. (C) Corresponding prokaryotic and eukaryotic expression plasmids were also generated for the complete UL11 gene (pGEX-HUL11 and pcDNA-HUL11). Deletion plasmid pcDNA-HUL11KF permitted removal of the KanR gene using flanking FRTs and introduction of a mutated start codon (CTG) as well as an artificial stop codon (TAG) to prevent expression of the 5′ part of UL11. (D) The genome of HSV-1 strain KOS was cloned as a BAC after insertion of a mini-F plasmid vector (pMBO131), together with an expression cassette for EGFP at the nonessential US5 gene encoding gJ. Artificial BamHI and PmeI restriction sites (shown in italic) were created to facilitate cloning and mutagenesis.
Generation of HSV-1 gM- and pUL11-monospecific antisera.
After transformation of Escherichia coli DH5α with pGEX-HUL10c and pGEX-HUL11 (Fig. 1B and C), expression of the glutathione S-transferase (GST) fusion proteins was induced by IPTG (isopropyl-β-d-thiogalactopyranoside) according to the manufacturer's protocol. The approximately 38-kDa GST-HUL10c and 37-kDa GST-HUL11 fusion proteins were eluted from sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and used for immunization of rabbits as described previously (21). Sera obtained after the third immunization were used throughout this study. A monospecific rabbit antiserum against HSV-1 pUL48 was similarly prepared and will be described elsewhere.
Generation of BAC pHSV-1ΔgJ.
For cloning of the HSV-1 KOS genome as a bacterial artificial chromosome (BAC), an expression cassette encoding enhanced green fluorescent protein (EGFP) and a mini-F plasmid vector was inserted at the nonessential gJ gene locus (Fig. 1D). To this end, the multiple cloning site between the HCMV immediate-early promoter and the EGFP ORF of pEGFP-N1 (Clontech) was removed by double digestion with BamHI and BglII and religation. Then, the modified expression cassette was recloned as an AseI/AflII fragment after Klenow treatment into the SmaI-digested vector pBluescript SK(−) (Stratagene). To facilitate subsequent cloning, a second BamHI site was created by cleavage of the obtained plasmid pBl-GFP with XhoI, Klenow treatment, and insertion of the self-annealed linker sequence CGGGATCCCG. Furthermore, a unique PmeI site was introduced by insertion of the self-annealed linker AGCTGTTTAAAC into the HindIII-digested plasmid. The resulting construct, pBl-GFP/BP, was digested with SalI and ligated with the similarly digested mini-F plasmid pMBO131 (48) to obtain pBl-GFP/MBO/BP (Fig. 1D).
To remove unwanted restriction sites, pUC-BamJ, which contains a genomic BamHI fragment of HSV-1 KOS (corresponding to nt 136289 to 142746 of GenBank accession no. X14112), was cleaved with Tth111I and EcoRI and religated after Klenow treatment. The resulting plasmid, pUC-BamJET, was digested with BsiWI, and an 8,040-bp BamHI fragment of pBl-GFP/MBO/BP was inserted after blunt ending of the noncompatible 5′ overhangs (Fig. 1D). The obtained transfer plasmid, pΔgJ-GFP/MBO/Pme, and HSV-1 KOS virion DNA were used for calcium phosphate-mediated cotransfection of Vero cells (15). An EGFP-expressing HSV-1 recombinant was plaque purified from the virus progeny. Circular viral DNA was prepared from infected cells and used for electroshock transformation of E. coli strain DH10B as described previously (13). Bacteria carrying BACs were selected on agar plates containing 30 μg chloramphenicol per ml, and full-length clones of the HSV-1 genome were identified by restriction analyses of the BAC DNA and Southern blotting. One clone, named pHSV-1ΔgJ, was further propagated and used for mutagenesis of UL10 and UL11.
Construction of HSV-1ΔgM.
For generation of HSV-1ΔgM, plasmid pcDNA-HUL10 (Fig. 1B) was digested with BstXI and religated to remove an unwanted SacII site, resulting in plasmid pcDNA-HUL10B. The kanamycin resistance gene was amplified by PCR from pACYC177 (New England Biolabs, Frankfurt, Germany) using primers KAN950For (5′-TCCGGATCCCGATTTATTCAACAAAGCCACG-3′) and KAN950Rev (5′-TTCGAATTCGCCAGTGTTACAACCAATTAA-3′) and inserted into SacII-cleaved pcDNA-HUL10B, thereby deleting 625 bp corresponding to codons 234 to 442 of UL10 (Fig. 1B). The complete insert of pcDNA-HUL10K was amplified by PCR with T7 and SP6 primers (New England Biolabs) and Pfx DNA polymerase. The 1.3-kbp product was used for mutagenesis of the BAC clone pHSV1-ΔgJ in E. coli as described earlier (23). Briefly, bacteria containing pHSV1-ΔgJ were transformed with helper plasmid pKD46, which carries the Red recombinase of bacteriophage lambda under the control of an arabinose-inducible promoter (9). After induction, the cells were transformed by electroshock with the pcDNA-HUL10K PCR product, and recombinant clones were selected on agar plates containing 30 μg chloramphenicol and 50 μg kanamycin per ml. For efficient removal of the mini-F plasmid vector and the adjacent EGFP expression cassette from the gJ locus, BAC DNA was linearized by digestion at the unique, artificially introduced PmeI site and used for cotransfection of Vero cells together with plasmid pUC-BamJET, which contains the authentic gJ gene of HSV-1 KOS (Fig. 1A). Homologous recombination between this plasmid and the cleaved BAC DNA repairs the discontinuity of the viral genome, resulting in a high proportion of gJ rescuants. The desired recombinant HSV-1ΔgM was isolated from nonfluorescent progeny virus plaques.
Generation of HSV-1ΔUL11.
The UL11 ORF was mutagenized with the synthetic oligonucleotide HUL11-MUT (5′-GGGTCCCGGAGAACGActaGCCCAgAGCTCGGCGA GCGTGTC-3′), which corresponds to nt 25067 to 25108 of the HSV-1 genome but contains four base exchanges (shown in lowercase) to mutate the UL11 start codon, to generate a stop codon at amino acid position 3, and to introduce a SacI marker site (underlined; Fig. 1C). For site-directed mutagenesis (27), uracil-containing single-stranded DNA of pcDNA-HUL11 was prepared after transformation of the dUTPase and uracil-DNA-glycosylase-negative E. coli strain BW313 and infection with f1 helper phage R408 (Stratagene) according to the distributor's protocol. After hybridization with primer HUL11-MUT, the second DNA strand was completed in vitro using Klenow polymerase and T4 DNA ligase, and E. coli XL1Blue MRF′ (Stratagene) was transformed. By double digestion of the resulting plasmid pcDNA-HUL11MUT with EcoRV and SgrAI, 113 bp representing UL11 codons 38 to 75 (Fig. 1C) was removed and, after Klenow polymerase treatment, replaced by a 1,258-bp BstBI fragment of pKD13 (9), which contains a kanamycin resistance gene flanked by flp recombinase recognition target (FRT) sites, giving rise to plasmid pcDNA-HUL11KF (Fig. 1C). The 1.6-kbp insert of pcDNA-HUL11KF was amplified by PCR with vector-specific T7 and SP6 primers, and the product was used for mutagenesis of pHSV-1ΔgJ in E. coli as described above. Subsequently, the kanamycin resistance gene was excised from pHSV-1ΔUL11 after transformation with helper plasmid pCP20 (7) expressing the FRT site-specific flp recombinase. Finally, the mini-F plasmid vector and the adjacent EGFP expression cassette were also removed from the HSV-1 genome after cotransfection of Vero cells with BAC DNA and plasmid pUC-BamJET, and mutant HSV-1ΔUL11 was isolated from nonfluorescent progeny virus plaques.
Generation of HSV-1ΔUL11/gM.
To obtain HSV1-ΔUL11/gM, pHSV1-ΔUL11 was mutagenized with the 1.3-kbp PCR product of pcDNA-HUL10K as described above.
Virus purification and immunoblotting.
For virus purification, Vero cells were infected with HSV-1 KOS, HSV-1ΔUL11, HSV-1ΔgM, or HSV-1ΔUL11/gM and incubated until complete cytopathic effect developed. Remaining intact cells were lysed by freezing (−70°C) and thawing (37°C), cellular debris was removed by low-speed centrifugation, and virions were sedimented by centrifugation for 1 h at 20,000 rpm (Beckman SW32 rotor) through a 40% sucrose cushion in phosphate-buffered saline. Lysates of purified virions (3 μg per lane) were separated in SDS-10% or 15% polyacrylamide gels and electrotransferred onto nitrocellulose membranes (Mini-Protean II and Transblot SD cell; Bio-Rad). Blots were blocked with 5% low-fat milk in Tris-buffered saline (150 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.25% Tween 20) and incubated for 1 h with monospecific antisera against pUL11 and gM at a dilution of 1:100,000 in Tris-buffered saline-Tween. Bound antibody was detected with peroxidase-conjugated anti-rabbit antibodies (Dianova, Hamburg, Germany) and visualized by chemiluminescence (SuperSignal; Pierce, Bonn, Germany) recorded on X-ray films.
One-step growth analysis and plaque assays.
To monitor one-step growth, Vero, Vero-UL11(HSV-1), RK13, and RK13-UL11(PrV) cells were infected with the indicated viruses at a multiplicity of infection (MOI) of 5 and incubated on ice for 1 h. Thereafter, the inoculum was replaced with prewarmed medium, and virus was allowed to penetrate for 60 min at 37°C. The remaining extracellular virus was then inactivated by low-pH treatment (40), and incubation was continued at 37°C. After 0, 4, 12, 24, 36, 48, and 72 h cells were harvested by being scraped into the medium and lysed by freezing (−70°C) and thawing (37°C). The virus progeny was titrated on Vero cells. Average values and standard deviations were determined from three independent experiments.
For plaque assays, infected Vero, Vero-UL11(HSV-1), RK13, and RK13-UL11(PrV) cells were incubated in semisolid minimum essential medium containing 5% fetal calf serum and 6 g of methylcellulose per liter. After 3 days of incubation at 37°C, cells were fixed with 2% formaldehyde and stained with 1% crystal violet in 50% ethanol. For each combination of virus and cells, 30 plaques were measured microscopically, and the average plaque sizes and standard deviations were determined and compared to the mean size of plaques induced by HSV-1 KOS, which was set as 100%.
Electron microscopy.
Vero and RK13 cells were infected with HSV-1 KOS, HSV-1ΔUL11, HSV-1ΔgM, or HSV-1ΔUL11/gM at an MOI of 1, incubated for 14 h (for HSV-1 KOS) or 24 h (for mutant viruses) at 37°C, and processed for electron microscopy as described previously (16). Counterstained ultrathin sections were analyzed with an electron microscope (Tecnai 12; Philips, Eindhoven, The Netherlands).
RESULTS
Generation of pUL11 and gM deletion mutants of HSV-1.
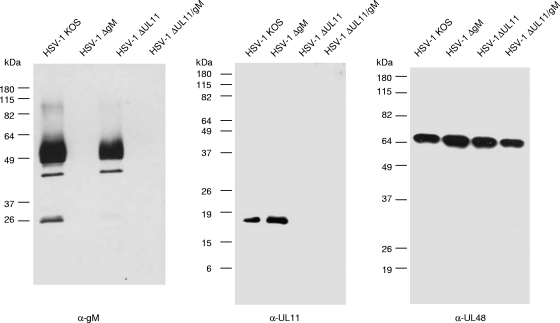
All mutants analyzed in this study were generated via Red-mediated mutagenesis (9) of the BAC clone pHSV-1ΔgJ in E. coli. This infectious clone contains a mini-F plasmid vector and an expression cassette encoding EGFP at the nonessential gJ gene locus of HSV-1 strain KOS (Fig. 1D). Since the 5′ region of UL11 encodes known functional domains (30) but overlaps with UL12 (Fig. 1A), construction of a complete deletion without impairing UL12 expression is difficult. Therefore, only UL11 codons 38 to 75 (of 97) were removed and replaced by an FRT site consisting of 36 bp of foreign DNA. Expression of the remaining N-terminal part of pUL11 was prevented by mutation of the UL11 initiation codon from ATG to CTG and introduction of a stop codon at the third codon position (Fig. 1C). None of these alterations affected the deduced amino acid sequence of the overlapping UL12 gene product. The UL10 gene, which encodes gM, was mutated by deletion of codons 234 to 442 (of 474) and concomitant insertion of a kanamycin resistance gene (Fig. 1B). A double mutant containing both deletions was also constructed. Subsequent removal of the mini-F plasmid and the EGFP expression cassette from the virus genome was facilitated by an introduced unique PmeI restriction site (Fig. 1D). After cotransfection of cells with PmeI-digested BAC DNA and a plasmid containing the authentic gJ gene, the majority of progeny viruses exhibited rescue of the gJ gene locus (results not shown). Correct mutations in the obtained virus recombinants HSV-1ΔUL11, HSV-1ΔgM, and HSV-1ΔUL11/gM were verified by restriction enzyme analysis and Southern blot hybridization of virion DNA, as well as by PCR amplification and DNA sequencing of the mutated genome regions (data not shown). Furthermore, purified virions were characterized by Western blotting with monospecific antisera (Fig. 2). As expected, the 50- to 60-kDa gM was detectable only in virions of HSV-1 KOS and HSV-1ΔUL11 but not in deletion mutants HSV-1ΔgM and HSV-1ΔUL11/gM (Fig. 2, left panel). In contrast, the 18-kDa pUL11 was found in HSV-1 KOS and HSV-1ΔgM particles but was absent from HSV-1ΔUL11 and HSV-1ΔUL11/gM (Fig. 2, middle panel). As loading control, a parallel blot was probed with a serum specific for the tegument protein pUL48 (VP16) of HSV-1 (Fig. 2, right panel).
FIG. 2.
Western blot analyses. Purified virions of HSV-1 KOS or deletion mutants HSV-1ΔgM, HSV-1ΔUL11, and HSV-1ΔgM/UL11 were separated by SDS-polyacrylamide gel electrophoresis. After transfer to nitrocellulose filters, blots were probed with monospecific antisera against gM, pUL11, and pUL48. Locations of molecular mass markers are indicated on the left.
In vitro growth properties of HSV-1 gM and UL11 mutants.
HSV-1 pUL11 and gM have previously been shown to be nonessential for viral replication (1, 35). Successful isolation of the novel mutants HSV-1ΔUL11, HSV-1ΔgM, and HSV-1ΔUL11/gM on noncomplementing Vero cells confirmed that none of the introduced mutations was fatal for virus replication in cell culture. To investigate the in vitro growth properties of the three mutants in more detail, replication kinetics and plaque sizes were analyzed on Vero and RK13 cells, as well as on trans-complementing cell lines.
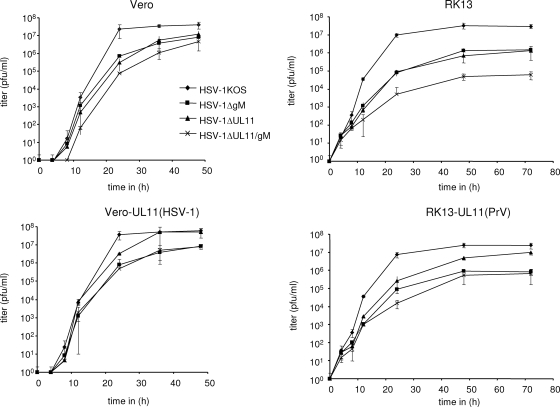
One-step growth studies revealed that in Vero cells HSV-1ΔUL11 and HSV-1ΔgM replicated to approximately fivefold-lower final titers than did the parental strain HSV-1 KOS (Fig. 3). Simultaneous deletion of pUL11 and gM led to a 10-fold reduction, suggesting additive effects of the mutations. On trans-complementing Vero-UL11(HSV-1) cells, growth properties of HSV-1ΔUL11 were similar to those of HSV-1 KOS and maximum titers of HSV-1ΔUL11/gM were comparable to those of HSV-1ΔgM (Fig. 3). Several attempts to isolate a Vero cell line constitutively expressing HSV-1 gM were unsuccessful, indicating that the protein might be deleterious for the cells. On RK13 cells the growth defects of HSV-1ΔUL11 and HSV-1ΔgM were more pronounced, since the single mutants reached only approximately 105 PFU per ml compared to 5 × 107 PFU/ml for HSV-1 KOS. The double mutant again showed an additive defect resulting in maximum titers of ca. 5 × 104 PFU/ml (Fig. 3). The UL11-related growth defects of HSV-1ΔUL11 and HSV-1ΔUL11/gM were partially compensated on RK13 cells expressing pUL11 of PrV (Fig. 3), indicating similar functions of these homologous proteins.
FIG. 3.
One-step growth analyses. Vero, Vero-UL11(HSV-1), RK13, and RK13-UL11(PrV) cells were infected with HSV-1 KOS, HSV-1ΔgM, HSV-1ΔUL11, and HSV-1ΔUL11/gM at an MOI of 5, harvested at the indicated times after infection, and titrated on Vero cells. Average titers and standard deviations from three independent experiments are shown.
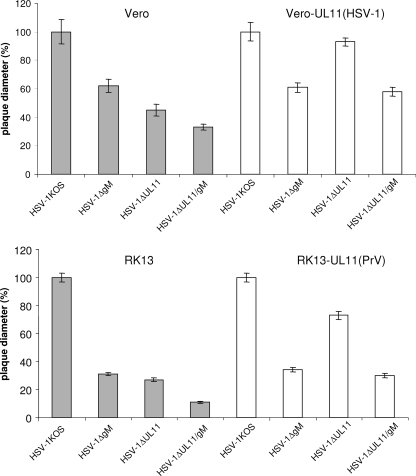
To analyze direct cell-to-cell spread of the virus mutants, Vero, Vero-UL11(HSV-1), RK13, and RK13-UL11(PrV) cells were infected under plaque assay conditions and plaque diameters were measured microscopically 3 days postinfection (p.i.). On Vero cells the plaque sizes of HSV-1ΔgM, HSV-1ΔUL11, and HSV-1ΔUL11/gM were reduced by ca. 40%, 55%, and 70%, respectively, compared to those of HSV-1 KOS (Fig. 4). As expected, the defect caused by deletion of UL11 could be corrected on complementing Vero-UL11(HSV-1) cells, on which the plaques of HSV-1ΔUL11 were comparable to those of HSV-1 KOS, and plaque sizes of HSV-1ΔUL11/gM were restored to those of HSV-1ΔgM (Fig. 4). These findings confirmed that pUL11 as expressed by Vero-UL11(HSV-1) is functional and that no unwanted mutations affect the phenotype of the UL11-deleted HSV-1 mutants. Paralleling the results of one-step growth analyses, the reduction in plaque size of all virus mutants was more pronounced on RK13 than on Vero cells. On RK13 cells plaques of HSV-1ΔUL11 and HSV-1ΔgM reached only 30% of the size of wild-type virus plaques (Fig. 4). Plaque diameters of HSV-1ΔUL11/gM were reduced by 90%, again indicating an additive effect of the double deletion (Fig. 4). On RK13-UL11(PrV) cells, plaque sizes of HSV-1ΔUL11 were increased to approximately 75% of the wild-type size and plaque diameters of HSV-1ΔUL11/gM were comparable to those of HSV-1ΔgM (Fig. 4).
FIG. 4.
Determination of plaque sizes. Plaque diameters of HSV-1 KOS, HSV-1ΔgM, HSV-1ΔUL11, and HSV-1ΔgM/UL11 on Vero, Vero-UL11 (HSV-1), RK13, and RK13-UL11 (PrV) cells were measured microscopically 3 days p.i. For each cell line, relative plaque sizes of all mutants were calculated by comparison to those of HSV-1 KOS, which were set as 100%. Average values and standard deviations from three independent experiments are shown.
Taken together, these data demonstrate that simultaneous deletion of UL11 and gM causes additive effects on replication and direct viral cell-to-cell spread of HSV-1 which are more pronounced on RK13 than on Vero cells. Furthermore, PrV pUL11 functionally complemented the replication defect of UL11-negative HSV-1 to a significant extent. However, a reverse complementation of UL11-negative PrV by HSV-1 pUL11 was not observed (data not shown), indicating that the two proteins are functionally related but not identical.
HSV-1 gM and pUL11 are involved in secondary envelopment.
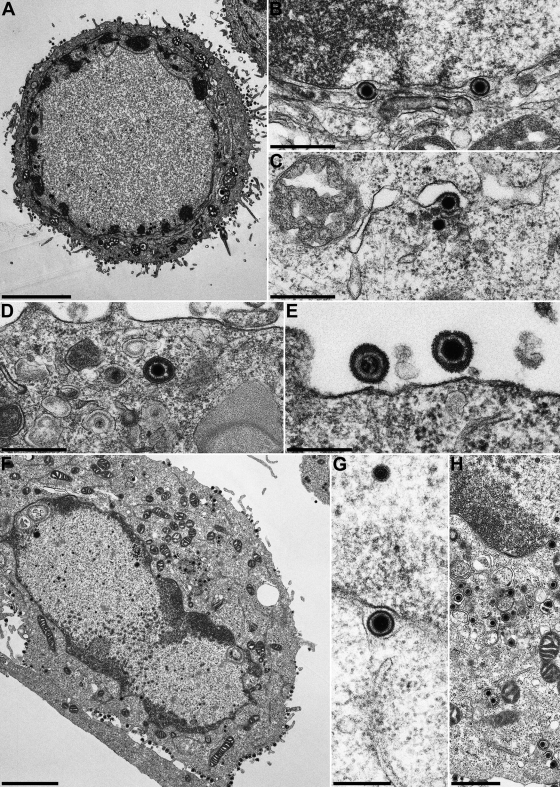
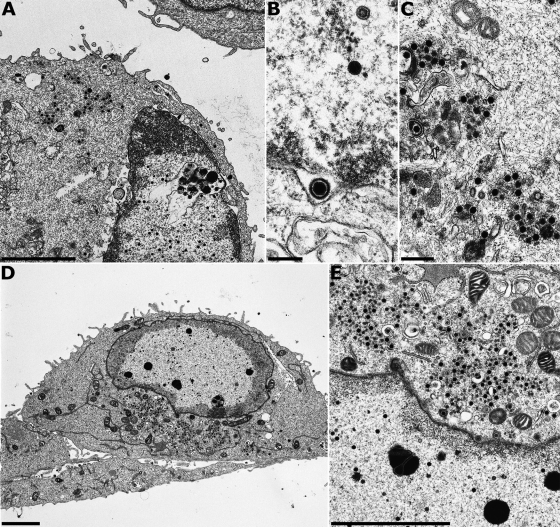
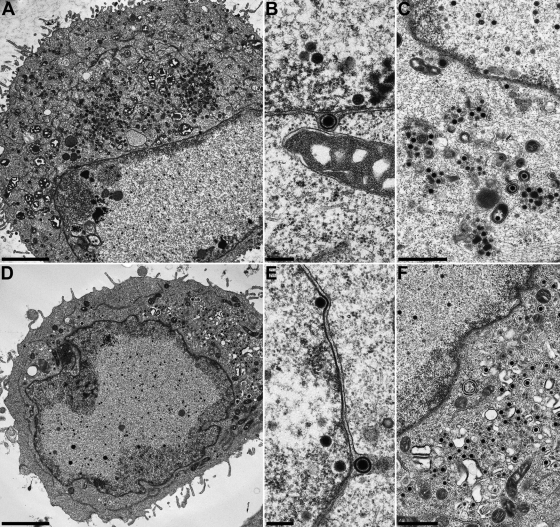
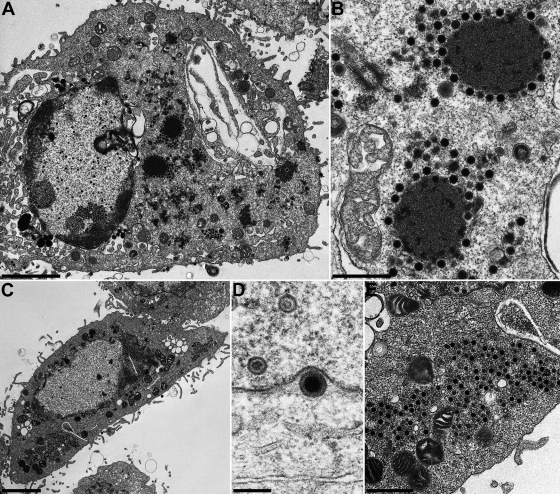
The relevance of HSV-1 pUL11 and gM for virion morphogenesis was analyzed by electron microscopy of Vero and RK13 cells 14 h after infection with HSV-1 KOS and 24 h after infection with HSV-1ΔUL11, HSV-1ΔgM, or HSV-1ΔUL11/gM. Different incubation times were used since the virus mutants exhibited delayed replication compared to wild-type HSV-1 KOS (Fig. 3). In RK13 (Fig. 5A to E) and Vero (Fig. 5F to H) cells infected with HSV-1 KOS, all stages of virion morphogenesis including intranuclear capsid assembly (Fig. 5A and F), transit of primary enveloped nucleocapsids through the nuclear membrane (Fig. 5B and G), and secondary envelopment of nucleocapsids in the cytoplasm (Fig. 5C, D, and H) as well as numerous mature virions in the extracellular space (Fig. 5A, E, and F) were observed. Infection with HSV-1ΔgM (Fig. 6) and HSV-1ΔUL11 (Fig. 7) revealed that nucleocapsid assembly and nuclear egress were not detectably affected either in RK13 (Fig. 6A and B and 7A and B) or in Vero (Fig. 6D and E and 7D and E) cells. However, compared to cells infected with HSV-1 KOS, the number of extracellular virus particles was significantly reduced (Fig. 6A and D and 7A and D). In HSV-1ΔgM-infected cells, viral particles present in the cytosol were mostly unenveloped nucleocapsids which formed loose aggregates (Fig. 6C and E), sometimes in association with electron-dense material (Fig. 6C). After infection of cells with HSV-1ΔUL11, also mostly naked nucleocapsids were present in the cytoplasm, indicating a defect in secondary envelopment (Fig. 7C and F). In RK13 cells infected with the double deletion mutant HSV-1ΔUL11/gM, large intracytoplasmic inclusions were formed which consisted of nucleocapsids associated with electron-dense material, presumably tegument (Fig. 8A and B). These structures exhibit a striking similarity to those found in pUL11 and gM-deleted (24) and gE/I- and gM-deleted (4, 5) PrV. In Vero cells, an aggregation of nucleocapsids was also observed but without the pronounced association of electron-dense material (Fig. 8C and E). Nuclear egress apparently still occurred in the simultaneous absence of pUL11 and gM (Fig. 8A and D).
FIG. 5.
Virion morphogenesis of HSV-1 KOS. RK13 (A to E) and Vero (F to H) cells were infected with HSV-1 KOS at an MOI of 1 and fixed and processed for electron microscopy 14 h after infection. Bars, 5 μm (A), 3 μm (F), 1 μm (H), 500 nm (B to E), and 300 nm (G).
FIG. 6.
Virion morphogenesis of HSV-1ΔgM. RK13 (A to C) and Vero cells (D and E) were infected at an MOI of 1 and fixed 24 h p.i. Bars, 3 μm (A and D), 500 nm (C), 300 nm (E), and 250 nm (B).
FIG. 7.
Virion morphogenesis of HSV-1ΔUL11. RK13 (A to C) and Vero cells (D to F) were infected at an MOI of 1 and fixed 24 h p.i. Bars, 3 μm (A and D), 1 μm (C and F), and 250 nm (B and E).
FIG. 8.
Virion morphogenesis of HSV-1ΔUL11/gM. RK13 (A and B) and Vero cells (C to E) were infected at an MOI of 1 and fixed 24 h p.i. Bars, 3 μm (A and C), 1 μm (E), 700 nm (B), and 200 nm (D).
DISCUSSION
In the present study a novel BAC clone of HSV-1 strain KOS was used to create single and double deletion mutants lacking the conserved herpesvirus genes UL10 encoding gM and/or UL11. In vitro growth properties of the obtained virus mutants HSV-1ΔgM, HSV-1ΔUL11, and HSV-1ΔUL11/gM were analyzed in monkey (Vero) and rabbit kidney (RK13) cells, as well as in cells expressing pUL11 of HSV-1 or PrV. Furthermore, virion morphogenesis was investigated by electron microscopy. The salient findings were as follows. (i) Simultaneous deletion of gM and pUL11 did not abrogate HSV-1 replication but significantly affected cell-to-cell spread and the virus yield additive of the single mutations. (ii) The severity of the growth defects depended on the cell line used. It was more severe on RK13 than on Vero cells. (iii) Simultaneous deletion of gM and pUL11 of HSV-1 resulted in cytoplasmic accumulations of nucleocapsids, which, in RK13 cells, were associated with electron-dense material, similar to those observed in gM/pUL11-deleted PrV. (iv) Both proteins are involved in virion maturation in the cytoplasm but are not required for nucleocapsid formation or nuclear egress. (v) The function of HSV-1 pUL11 could be provided in trans also by cells expressing pUL11 of PrV.
Genome sequence analysis has revealed a core set of ca. 40 genes conserved among all members of the Herpesviridae, which are most probably inherited from a common ancestor. Most of these core gene products are involved in fundamental steps of viral replication, such as virus entry, DNA replication and packaging, or virion morphogenesis (38, 51). However, not all of these proteins are essential for viral replication in cell culture, and especially in alphaherpesviruses, several can be deleted without drastic effects on virus growth. This led to the hypothesis that important functions might be supported by functionally redundant viral proteins (42). In PrV simultaneous deletion either of gM and the alphaherpesvirus-specific envelope protein complex gE/gI or of gM and pUL11 resulted in a drastic impairment of secondary envelopment, while the single deletions showed only moderate effects (4, 10, 23). In the absence either of gM and pUL11 or of gM and gE/gI, large intracytoplasmic inclusions containing nucleocapsids embedded in tegument proteins were observed (4, 24), indicating inhibition of budding of the tegumented nucleocapsids into TGN-derived vesicles. A similar egress defect has been described for a double mutant of HSV-1 lacking gE and gD (11), whereas, unlike in PrV, an HSV-1 deletion mutant lacking gE/gI and gM did not exhibit a defect beyond those of the single mutants (6), suggesting that in HSV-1 gD might provide the redundancy in the egress function which gM provides in PrV. However, we demonstrate here that infection of RK13 cells by HSV-1 lacking pUL11 and gM leads to aggregations of nonenveloped nucleocapsids and electron-dense material, presumably tegument, in the cytoplasm similar to those that have been observed after infection of these cells with the corresponding pUL11- and gM-deleted (24) as well as gM and gE/I-deleted (4, 5) PrV. Nucleocapsids also accumulated in Vero cells after infection with the pUL11/gM-deleted HSV-1, although the effect was more modest in these cells than in RK13 cells. Thus, simultaneous deletion of pUL11 and gM from HSV-1 and from PrV resulted in similar ultrastructural phenotypes.
The defects observed in the double mutant in one-step replication and plaque size were largely additive of the deficiencies found in the single mutants. Thus, pUL11 and gM of HSV-1 apparently participate in different steps of virion morphogenesis. The membrane-associated tegument protein pUL11 of HSV-1 has been demonstrated to possess intrinsic properties of targeting to Golgi membranes, since a chimera of human immunodeficiency virus type 1 Gag protein fused to the complete HSV-1 UL11 ORF was relocated to the Golgi region (3). Therefore, it seems reasonable that pUL11 might play a role in targeting tegument proteins or tegumented capsids to the TGN. This is supported by interactions of pUL11 with tegument proteins pUL16 and pUL21 of PrV and HSV-1 (22, 31) and with gD and gE of HSV-1 (11). Taken together, the available results indicate a conserved function of pUL11 in secondary envelopment of herpesvirus particles. This is supported by the observed functional complementation of the pUL11 deficiency of the respective HSV-1 mutants by the PrV homolog, both in one-step growth and in direct cell-to-cell spread. However, reciprocal complementation of PrV pUL11 mutants by the HSV-1 homolog was not observed (data not shown), indicating that the PrV protein might possess additional functions, which cannot be executed by the homologous gene product of HSV-1. One-way complementation between homologous alphaherpesvirus proteins has also been observed with gB (41, 50) and pUL25 (26).
Our ultrastructural analyses of the HSV-1 mutants yielded no evidence for an impairment of nuclear egress, as had been described for a previous UL11 deletion mutant of HSV-1 (1). Thus, they are in line with more recent investigations of another HSV-1 UL11 deletion mutant (14), as well as of UL11-negative PrV (23), which revealed inhibition of virion formation in the cytoplasm but not of nuclear egress. These differences may be explained by the fact that the HSV-1 UL11 deletion mutant used in the first study was still able to express the N-terminal part including a dileucine motif, which has been reported to contribute to binding of pUL11 to pUL16 (32). pUL16 of HSV-1 has been shown to accumulate in the nucleus (39, 47), and binding to the truncated UL11 gene product might inhibit nuclear egress of virions. In contrast, the HSV-1 mutants described here, as well as the corresponding PrV mutants (24), are unable to express any protein products of UL11 due to mutated start codons.
Like pUL11, gM is conserved throughout the Herpesviridae, which also suggests a conserved function. gM has been shown to reduce cell fusion presumably by retention of fusogenic glycoproteins in the Golgi apparatus or efficient retrieval of these proteins from the plasma membrane for PrV, HSV-1, infectious laryngotracheitis virus, EHV-1, and human herpesvirus 8 (8, 20, 25). However, whereas gM deletion mutants of the mammalian alphaherpesviruses PrV, HSV-1, and EHV-1 are significantly attenuated in animals but exhibit only moderate replication defects in vitro (10, 35, 49), gM has been reported to be essential for viral replication of the betaherpesvirus HCMV and the avian alphaherpesvirus Marek's disease virus (17, 58), indicating that the functional significance of gM may differ among herpesviruses.
The replication defect of HSV-1ΔgM described here was comparable to that of PrV-ΔgM, which also showed moderately reduced virus titers (10) and detectable accumulations of nonenveloped virions in the cytoplasm of infected cells (5). In contrast to the present study, Browne et al. failed to observe increased amounts of naked capsids in the cytoplasm of cells infected with their gM-deleted HSV-1 mutant (6). The apparent discrepancy between the in vitro phenotypes of the two gM-negative HSV-1 mutants might be caused by the different virus strains and cell types used for the experiments. The influence of the cell type on the severity of the defects caused by gM and/or pUL11 deletion is also demonstrated by our studies.
In summary, we show that pUL11 and gM play a common role in secondary envelopment but not in nuclear egress of HSV-1. Both proteins contribute to productive replication and cell-to-cell spread of HSV-1. In fact, similar functions of pUL11 are exhibited in PrV (23), EHV-1 (54), varicella-zoster virus (52), and HCMV (53). A contribution of gM to secondary envelopment has been observed for PrV (4), EHV-4 (61), EBV (29), and HCMV (34). Both gM and pUL11 represent proteins that are conserved in all herpesvirus families. Our present data demonstrate that these proteins also exhibit conserved functions within the assembly processes of HSV-1 and PrV.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm DFG Me 854/8).
We thank Charlotte Ehrlich, Petra Meyer, and Diana Werner for technical assistance and Mandy Jörn for help with the electron micrographs.
Footnotes
Published ahead of print on 12 November 2008.
REFERENCES
- 1.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 665168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowzard, J. B., R. J. Visalli, C. B. Wilson, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 748692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 735364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 744004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne, H., S. Bell, and T. Minson. 2004. Analysis of the requirement for glycoprotein M in herpes simplex virus type 1 morphogenesis. J. Virol. 781039-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1589-14. [DOI] [PubMed] [Google Scholar]
- 8.Crump, C. M., B. Bruun, S. Bell, L. E. Pomeranz, T. Minson, and H. M. Browne. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 853517-3527. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkstra, J. M., T. C. Mettenleiter, and B. G. Klupp. 1997. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycosylation. Virology 237113-122. [DOI] [PubMed] [Google Scholar]
- 11.Farnsworth, A., K. Goldsmith, and D. C. Johnson. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 778481-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 768208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulmer, P. A., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2007. UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J. Virol. 813097-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52456-467. [DOI] [PubMed] [Google Scholar]
- 16.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: a comparative ultrastructural study. J. Virol. 753675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 747720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jöns, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kattenhorn, L. M., R. Mills, M. Wagner, A. Lomsadze, V. Makeev, M. Borodovsky, H. L. Ploegh, and B. M. Kessler. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 7811187-11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 746760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 7410063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp, B. G., S. Böttcher, H. Granzow, M. Kopp, and T. C. Mettenleiter. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J. Virol. 791510-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 775339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp, M., H. Granzow, W. Fuchs, B. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 783024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koyano, S., E. C. Mar, F. R. Stamey, and N. Inoue. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 841485-1491. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn, J., T. Leege, B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2008. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J. Virol. 825725-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lake, C. M., S. J. Molesworth, and L. M. Hutt-Fletcher. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 725559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lake, C. M., and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 7411162-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 7512209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 7711417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2006. Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 8010534-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 7411881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mach, M., B. Kropff, M. Kryzaniak, and W. Britt. 2005. Complex formation glycoprotein and N of human cytomegalovirus: structural and functional aspects. J. Virol. 792160-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacLean, C. A., L. M. Robertson, and F. E. Jamieson. 1993. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J. Gen. Virol. 74975-983. [DOI] [PubMed] [Google Scholar]
- 36.May, J. S., S. Colaco, and P. G. Stevenson. 2005. Glycoprotein M is an essential lytic replication protein of the murine gammaherpesvirus 68. J. Virol. 793459-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 691531-1574. [DOI] [PubMed] [Google Scholar]
- 38.McGeoch, D. J., F. J. Rixon, and A. J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 11790-104. [DOI] [PubMed] [Google Scholar]
- 39.Meckes, D. G., and J. W. Wills. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 8113028-13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171623-625. [DOI] [PubMed] [Google Scholar]
- 41.Mettenleiter, T. C., and P. G. Spear. 1994. Glycoprotein gB (gII) of pseudorabies virus can functionally substitute for glycoprotein gB in herpes simplex virus type 1. J. Virol. 68500-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9423-429. [DOI] [PubMed] [Google Scholar]
- 44.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113163-169. [DOI] [PubMed] [Google Scholar]
- 45.Michael, K., S. Böttcher, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2006. Pseudorabies virus particles lacking tegument proteins pUL11 or pUL16 incorporate less full-length pUL36 than wild-type virus, but specifically accumulate a pUL36 N-terminal fragment. J. Gen. Virol. 873503-3507. [DOI] [PubMed] [Google Scholar]
- 46.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 801332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nalwanga, D., S. Rempel, B. Roizman, and J. D. Baines. 1996. The UL16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226236-242. [DOI] [PubMed] [Google Scholar]
- 48.O′Connor, M., M. Peifer, and W. Bender. 1989. Construction of large DNA segments in Escherichia coli. Science 2441307-1312. [DOI] [PubMed] [Google Scholar]
- 49.Osterrieder, N., A. Neubauer, C. Brandmüller, B. Braun, O. R. Kaaden, and J. D. Baines. 1996. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J. Virol. 704110-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rauh, I., F. Weiland, F. Fehler, G. M. Keil, and T. C. Mettenleiter. 1991. Pseudorabies virus mutants lacking the essential glycoprotein gII can be complemented by glycoprotein gI of bovine herpesvirus 1. J. Virol. 65621-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roizman, B., and P. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, PA.
- 52.Sadaoka, T., H. Yoshii, T. Imazawa, K. Yamanishi, and Y. Mori. 2007. Deletion in open reading frame 49 of varicella-zoster virus reduces virus growth in human malignant melanoma cells but not in human embryonic fibroblasts. J. Virol. 8112654-12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 743842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schimmer, C., and A. Neubauer. 2003. The equine herpesvirus 1 UL11 gene product localizes to the trans-Golgi network and is involved in cell-to-cell spread. Virology 30823-36. [DOI] [PubMed] [Google Scholar]
- 55.Seo, J. Y., and W. J. Britt. 2007. Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J. Virol. 816536-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 7710594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spear, P. G., R. I. Montgomery, and M. S. Warner. 1997. Virus entry: two receptors are better than one. Trends Microbiol. 5258-259. [DOI] [PubMed] [Google Scholar]
- 58.Tischer, B. K., D. Schumacher, M. Messerle, M. Wagner, and N. Osterrieder. 2002. The products of the UL10 (gM) and the UL49.5 genes of Marek's disease virus serotype 1 are essential for virus growth in cultured cells. J. Gen. Virol. 83997-1003. [DOI] [PubMed] [Google Scholar]
- 59.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 7810960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 799566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ziegler, C., F. T. Just, A. Lischewski, K. Elbers, and A. Neubauer. 2005. A glycoprotein M deleted equid herpesvirus 4 is severely impaired in virus egress and cell-to-cell spread. J. Gen. Virol. 8611-21. [DOI] [PubMed] [Google Scholar]