Abstract
Hepatitis C virus (HCV) F protein is encoded by the +1 reading frame of the viral genome. It overlaps with the core protein coding sequence, and multiple mechanisms for its expression have been proposed. The full-length F protein that is synthesized by translational ribosomal frameshift at codons 9 to 11 of the core protein sequence is a labile protein. By using a combination of genetic, biochemical, and cell biological approaches, we demonstrate that this HCV F protein can bind to the proteasome subunit protein α3, which reduces the F-protein level in cells in a dose-dependent manner. Deletion-mapping analysis identified amino acids 40 to 60 of the F protein as the α3-binding domain. This α3-binding domain of the F protein together with its upstream sequence could significantly destabilize the green fluorescent protein, an otherwise stable protein. Further analyses using an F-protein mutant lacking lysine and a cell line that contained a temperature-sensitive E1 ubiquitin-activating enzyme indicated that the degradation of the F protein was ubiquitin independent. Based on these observations as well as the observation that the F protein could be degraded directly by the 20S proteasome in vitro, we propose that the full-length HCV F protein as well as the F protein initiating from codon 26 is degraded by an ubiquitin-independent pathway that is mediated by the proteasome subunit α3. The ability of the F protein to bind to α3 raises the possibility that the HCV F protein may regulate protein degradation in cells.
The proteasome is a large, multisubunit complex that is important for protein degradation in eukaryotic cells. It exists in cells in several forms (54). The 20S proteasome core consists of four stacked heptameric rings (20). The two outer rings, which each contain seven α subunits, serve as binding sites for regulatory particles and control the entry of target proteins into the proteasome. The two inner rings, which each contain seven β subunits (33), have trypsin-like, chymotrypsin-like, and caspase-like enzymatic activities (13). The catalytic activities of the β rings are sequestered in the central cavity by the α rings at each side to prevent nonspecific protein entry and degradation (18). Each of the two α rings of the 20S core may bind to a 19S regulatory particle to form the 26S proteasome. The 19S regulatory particle has several activities, including ATPase, polyubiquitin binding, deubiquitination, and protein denaturation (7, 51, 52). These activities are required for the translocation of target proteins into the catalytic lumen of the 20S core for degradation (16, 54). In addition to the 19S regulatory particle, the 20S core may also bind to the REG complex, which is also known as 11S or PA28. The REG complex does not contain the ATPase activity and promotes the degradation of primarily short peptides (22).
The majority of cellular proteins degraded by the proteasome are tagged by polyubiquitin, which serves as a signal for recognition by the 19S regulatory particle for entry into the 20S core (16). The ubiquitination process consists of three steps. In the first step, the single ubiquitin-activating enzyme, known as E1, adenylates ubiquitin monomers. In the second step, the E1 enzyme transfers the adenylated ubiquitin to one of the many E2 ubiquitin-conjugating enzymes. In the final step, one of a diverse class of enzymes known as E3 ubiquitin ligases transfers the ubiquitin monomer to the protein substrate (19, 45). Ubiquitin is attached to the amino group of either the N-terminal residue (2, 8, 37) or an internal lysine residue of the target protein (16). Additional ubiquitin molecules are added to the lysine-48 residue of the previous ubiquitin on the target protein, to result in polyubiquitination (9). A polyubiquitin chain that contains at least four ubiquitin molecules is required to target a protein to the proteasome (45). After binding to the 19S regulatory particle, the subsequent protein degradation step requires that the polyubiquitinated protein be denatured and deubiquitinated for entry into the channel of the 20S core (7, 27, 45). Only a limited number of proteins have been found to be degraded by proteasomes in an ubiquitin-independent manner (1, 5, 23, 32, 41).
Hepatitis C virus (HCV) is a positive-stranded RNA virus with a genome size of 9.6 kb. Its genome codes for a large polyprotein, which is cleaved into 10 protein products by cellular and viral proteases, and a small protein named F protein or alternative reading frame protein (48, 55, 59). The F protein is encoded by the +1/−2 reading frame embedded in the 5′ end of the polyprotein coding sequence and was initially reported to be expressed by translational ribosomal frameshift at codons 9 to 11 of the polyprotein coding sequence (11, 59). Recent studies suggest that other mechanisms, which include translational ribosomal frameshift at codon 42, transcriptional slippage at codons 9 to 11, and internal initiation of translation at codon 26 or codons 85 and 87 (4, 6, 36, 50), may also be involved in the expression of different forms of the F protein. The biological function of the F protein is unclear. Antibodies and T cells specifically recognizing the F protein have been identified in HCV patients, indicating its expression during natural infection (3, 12, 25, 47, 55, 59). However, abolishing the expression of several forms of the F protein does not abolish the replication of HCV in a chimpanzee or cell cultures, indicating that the F protein may not be essential for productive HCV replication (31). The F protein may have regulatory functions, as it has been shown to induce the expression of proinflammatory cytokines (15) and c-myc (57), enhance the activity of c-myc (30), and suppress the expression of p53 (57). The full-length F protein has a short half-life and is degraded by proteasomes (38, 58). To further understand the biological functions of the F protein, we have conducted yeast two-hybrid screening studies to identify cellular proteins that the F protein may bind to. We identified the proteasome α3 subunit as one such protein. To understand the biological significance of the interaction between the F protein and α3, we coexpressed these two proteins in cells. This unexpectedly led us to the findings that the full-length F protein is degraded by the proteasome via an ubiquitin-independent pathway and that different truncated forms of the F protein have different stabilities. These findings may have important implications in HCV replication and pathogenesis.
MATERIALS AND METHODS
DNA plasmids.
Unless otherwise indicated for the mutation or truncation experiments, the F-protein sequence used in our studies was full length and included 10 amino acids from the polyprotein sequence followed by the sequence encoded by the F reading frame. The F protein derived from the HCV-1 isolate (genotype 1a) was used throughout our entire studies. The construction of pCDEF-HAF, which contains the hemagglutinin (HA)-tagged, full-length F protein, had been described previously (58). For the construction of pCDNA3.1/NTGFP-F(20-60), the F-protein cDNA fragment encoding amino acids 20 to 60 was isolated by PCR using primers 5′ ACC CGA ATT CAG ACG TCA AGT TCC CGG GT 3′ and 5′ CCC CTC TAG ACT CGA GGT TGC GAC CG 3′ and inserted into EcoRI and XbaI sites of pCDNA3.1/NTGFP (Invitrogen). For the construction of pCDNA3.1/NTGFP-F(40-60), the two oligonucleotides 5′ AAT TCA GGC CCT AGA TTG GGT GTG CGC GCG ACG AGA AAG ACT TCC GAG CGG TCG CAA CCT CGA GT 3′ and 5′ CTA GAC TCG AGG TTG CGA CCG CTC GGA AGT CTT TCT CGT CGC GCG CAC ACC CAA TCT AGG GCC TG 3′ were annealed and ligated into EcoRI and XbaI sites of pCDNA3.1/NTGFP. These two oligonucleotides encode amino acids 40 to 60 of the F-protein sequence. For the construction of pCDEF-HAFΔK, the first 11 codons of the F-protein coding sequence were deleted and the lysine codon at position 159 was mutated into an arginine codon using the sense primer 5′ CCC CAA GCT TAC ATT TGC TTC TGA CAC AAC TGT GTT CAC TAG CAA CCT CAA ACA GAC ACC ATG TAC CCA TAC GAC GTC CCA GAC TAC GCT ACA AAC GTA ACA CCA ACC 3′ and the antisense primer 5′ ACC ATC TAG AAA TCA CGC CGT CCT CCA GAA C 3′. The PCR product was then inserted into HindIII and XbaI sites of the pCDEF vector. For the construction of pGST-α3, the proteasome subunit α3 coding sequence was cloned into the BamHI and EcoRI sites of pGEX-4P-1 (Amersham Biosciences) using the forward primer 5′ CAT GGA TCC TTT AGC ACG ATG AGC TCA ATC GGC ACT G 3′ and the reverse primer 5′ CAT GAA TTC CAT ATT ATC ATC ATC TGA TT 3′. The α3 cDNA was PCR amplified from the plasmid pGAD57, which was isolated by yeast two-hybrid screening. For the construction of pCMV-mtFlag-α3, the α3 coding sequence was PCR amplified and cloned into NdeI and EcoRI sites of pCMV-mtFlag (a gift of Lucio Comai).
Yeast two-hybrid screening.
The Matchmaker yeast two-hybrid screening kit and the human liver cDNA library were purchased from Clontech. The HCV-1 F-protein coding sequence, which was generated by deleting one adenosine at codon 10 of the HCV core protein coding sequence to mimic a +1 ribosomal frameshift, was used as the bait for the screening following the manufacturer's protocols. The inserts of the positive cDNA clones isolated in the screening procedures were sequenced, and their sequences were searched against the National Center for Biotechnology Information database for identification.
In vitro transcription and translation.
For C-terminal truncations, the HCV-1 F-protein coding sequence was PCR amplified using the forward primer 5′ AAT ACG ACT CAC TAT AGG GAG AGC CAC CAT GGC CCT AGA TTG GGT GTG CG 3′, which contains the T7 promoter at the 5′ end, and various reverse primers. For N-terminal truncations, the reverse primer 5′ AAC AGA ATT CTC ACG CCG TCT TCC AGA AC 3′ and various forward primers that contain the T7 promoter sequence were used for PCR. The PCR products or the plasmid pCDNA3.1-HAF that had been linearized with XbaI was used as the template for RNA synthesis using Mega Script (Ambion) following the manufacturer's instruction. The protein synthesis was carried out at 30°C for 1 h using the following conditions: 0.5 to 1 μg RNA, 10 μl of rabbit reticulocyte lysates (Promega), 0.5 μl of 1 mM amino acid mixture without methionine, and 50 μCi [35S]methionine (>1,000 Ci/mmol; ICN). For the in vitro pulse-chase labeling experiment, the F protein was pulse-labeled for 15 min using the same translation condition. The protein synthesis was then stopped by the addition of cycloheximide to a final concentration of 1 μg/μl, and chase was performed for different lengths of time.
GST pull-down assay.
The fusion protein of glutathione S-transferase and α3 (GST-α3) was purified by using the GST purification kit (Amersham Biosciences) following the manufacturer's instructions. Briefly, the expression of the GST-α3 fusion protein or the control GST protein in Escherichia coli BL21 was induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside. Cells were then lysed by sonication, and GST-α3 or GST was purified by using glutathione-Sepharose CL-4B beads (Pharmacia Biotech). For the pull-down assay, 5 μl of the translational mixture containing the [35S]methionine-labeled F protein was mixed with GST-α3 or GST and then diluted in 1 ml radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate). After incubation at 4°C for 1 h, glutathione beads were pelleted and washed with RIPA buffer four times. Protein samples were then boiled in Laemmli buffer for gel electrophoresis.
Cell lines and DNA transfection.
Huh7 and Huh7.5 human hepatoma cell lines and the mouse ts85 mouse mammary carcinoma cell line were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). The ts85 cell line expresses a temperature-sensitive E1 ubiquitin-activating enzyme and thus was maintained at 33°C. Huh7 cells growing in a 10-cm dish were transfected with DNA plasmids using the Lipofectamine Plus reagent (Invitrogen). For the transfection of ts85 cells, cells growing in a 10-cm dish were trypsinized and washed with Opti-MEM (Invitrogen) three times. Five million cells in 500 μl OPTI-MEM were mixed with 50 μg DNA plasmids and electroporated at 300 V using a Bio-Rad Gene Pulser. Cells were then plated in DMEM supplied with 10% FBS and harvested 48 hours after transfection in Laemmli buffer for Western blot analysis. Alternatively, cells were used for the pulse-chase labeling experiments.
Pulse-chase labeling and small interfering RNA (siRNA) knockdown experiment.
Cells were incubated in methionine-free DMEM supplied with 10% dialyzed FBS for 30 min, pulse-labeled with [trans-35S]methionine (ICN) for 15 min, washed with phosphate-buffered saline, and chased in complete DMEM containing 10% FBS for different lengths of time. Cells were then lysed in RIPA buffer containing a protease inhibitor cocktail (Roche Diagnostics), and the cell lysates were incubated with the mouse anti-HA antibody (USC Norris Cancer Center Cell Culture Core), mouse anti-green fluorescent protein (anti-GFP) antibody (Santa Cruz Biotechnology) or anti-hypoxia-inducible factor 1α (anti-HIF-1α) antibody (Santa Cruz Biotechnology). After incubation at 4°C overnight, the immune complex was precipitated with 30 μl Gammabind Plus (Amersham). The immunoprecipitates were washed four times with RIPA buffer, followed by electrophoresis in an 8% or 15% gel. The protein bands were visualized by autoradiography.
For the pulse-chase labeling experiment involving siRNA knockdown of α3, Huh7.5 cells were electroporated with HCV JFH-1 genomic RNA, and 8 days after electroporation cells were transfected with siRNA specific for α3 subunit (Dharmacon) or a nonspecific negative control siRNA (Invitrogen) using Lipofectamine 2000 following the manufacturer's protocol. Twenty-four hours after siRNA transfection, cells were transfected with pCDEF-9Acore, an expression plasmid for the full-length F protein without the HA tag (11). Cells were harvested 48 hours after siRNA transfection for Western blot analysis or pulse-chase labeling experiment. For the pulse-chase-labeling experiment, the F protein was immunoprecipitated using the mouse monoclonal anti-F antibody.
Coimmunoprecipitation analysis.
Huh7 cells in a 10-cm petri dish were transfected with pCDEF-HAF and pCMV-mtFlag-α3 or their control expression vectors in different combinations using Lipofectamine Plus. Forty-eight hours after transfection, cells were rinsed twice with TBS (10 mM Tris-HCl [pH 7.0], 150 mM NaCl) and lysed in 1 ml TBS containing 0.5% Nonidet P-40 (NP-40). The cell lysates were briefly centrifuged to remove cell debris. The supernatant was incubated with 3 μg mouse anti-HA antibody or rabbit anti-Flag (Sigma) antibody. After incubation at 4°C overnight, the immune complex was precipitated with 30 μl Gammabind Plus (Amersham). The immunoprecipitates were washed four times with TBS containing 0.5% NP-40, followed by electrophoresis in a 15% gel. The protein gel was Western blotted to a nitrocellulose membrane, which was then hybridized with the rabbit anti-Flag or mouse anti-HA antibody and then with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse secondary antibody.
Confocal microscopy.
Huh7 cells grown on coverslips in a six-well dish were transfected with pCDEF-HAF, pCMV-mtFlag-α3, or their control expression vectors in different combinations. Forty-eight hours after transfection, cells were fixed with acetone and double stained with rabbit anti-Flag, for Flag-tagged α3, and mouse anti-HA, for the HA-tagged F protein, primary antibodies and rhodamine-conjugated goat anti-rabbit and fluorescein-conjugated goat anti-mouse secondary antibodies. Cells were then analyzed by confocal microscopy.
Expression and purification of the F protein.
The F-protein coding sequence of the HCV-1 isolate was generated by deleting an adenosine residue from codon 10 of the core protein sequence to mimic a +1 ribosomal frameshift. This sequence was inserted into the yeast expression vector to create the plasmid pBS24UB.F9a.his-14912bp. A His tag coding sequence was fused to the 3′ end of the F-protein sequence to facilitate purification. After fermentation, yeast cells were broken by glass beads in Dynomill and centrifuged. The insoluble pellet, which contained the F protein, was successively washed in 2 M, 4 M, and 8 M urea in 20 mM Tris-HCl (pH 7.5) and 150 mM NaCl. The F protein, which was soluble in both 4 M and 8 M urea washes, was purified with a nickel affinity column and then with an SP Fractogel ion-exchange column. The eluted F protein was finally concentrated by using the nickel column again. The F protein was loaded on the nickel column in 10 mM imidazole and eluted with 0.5 M imidazole. The F protein was finally dialyzed against 20 mM Tris-HCl (pH 7.5) containing 0.5% Tween 80. The F protein thus purified did not react with any monoclonal antibodies that recognize epitopes in amino acids 10 to 160 of the HCV core protein sequence (data not shown).
Proteasome degradation assay.
The proteasome degradation assay was performed as previously described with slight modifications (26). Briefly, 125 nM purified F protein, HBV core protein (29), or mouse immunoglobulin was incubated with 30 nM purified 20S proteasome (Boston Biochem) in the presence or absence of the proteasome inhibitor MG132 (2 μM final concentration) in reaction buffer (20 mM Tris-HCl [pH 7.0], 70 mM NaCl, 0.25 mM ATP, and 1 mM dithiothreitol) at 37°C. An aliquot was then removed at 0, 15, 30, and 60 min after the initiation of the reaction for Western blot analysis for the determination of the protein degradation rate.
RESULTS
Binding of HCV F protein to proteasome subunit α3.
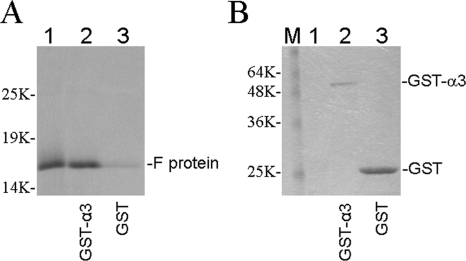
To identify the cellular proteins that the HCV F protein may bind to, we screened 2.5 × 106 human liver cDNA clones using the full-length F protein as the bait. Thirty-three positive cDNA clones were identified, and 12 of them were found to encode the proteasome subunit α3. To confirm the interaction between the F protein and α3, we performed GST pull-down experiments. The GST-α3 fusion protein and the GST control protein were expressed in E. coli, separately incubated with [35S]methionine-labeled F protein that had been synthesized in vitro using rabbit reticulocyte lysates, and then precipitated using glutathione beads. As shown in Fig. 1A, a significantly higher level of the F protein could be pulled down by GST-α3 than by the control GST protein, even though the amount of the latter used for the pull-down assay was much higher (Fig. 1B).
FIG. 1.
GST pull-down analysis of the interaction between the HCV F protein and the proteasome subunit α3. GST-α3 and GST proteins expressed in E. coli were purified and incubated with [35S]methionine-labeled F protein for the pull-down assay. The F protein was synthesized from HCV-1 RNA with the deletion of an adenosine residue at codon 10 of the polyprotein coding sequence to mimic a +1 ribosomal frameshift. (A) Autoradiography of the F protein pulled down by the glutathione beads. (B) The same protein gel shown in panel A was stained with Coomassie blue to reveal GST-α3 and GST protein bands before autoradiography. The locations of the F protein in panel A and of GST-α3 and GST in panel B are indicated. Lanes 1, F protein translational mixture prior to the pull-down assay; lanes 2, GST-α3 pull-down assay; lanes 3, control GST pull-down assay; lane M, molecular weight markers.
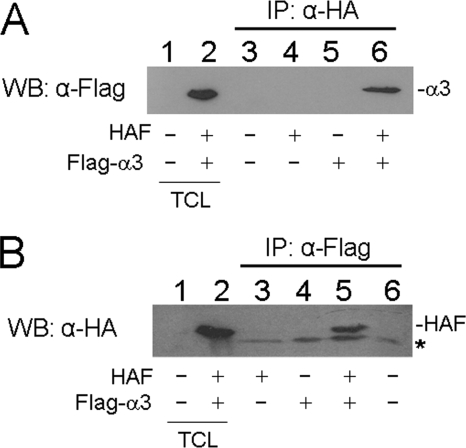
To confirm that the F protein and α3 could indeed bind to each other, we also performed coimmunoprecipitation experiments. Huh7 hepatoma cells were cotransfected with the HA-tagged F-protein expression plasmid, the Flag-tagged α3 expression plasmid, and their control expression vectors in different combinations. Cells were then lysed and immunoprecipitated with the mouse anti-HA antibody, followed by Western blot analysis of the immunoprecipitates using the rabbit anti-Flag antibody. As shown in Fig. 2A, the Flag-tagged α3 protein could be precipitated by the anti-HA antibody only in the presence of the HA-tagged F protein and not in the absence of it. In a reciprocal experiment, cell lysates were immunoprecipitated with the anti-Flag antibody, followed by Western blot analysis of the immunoprecipitates with the anti-HA antibody. As shown in Fig. 2B, the HA-tagged F protein could be precipitated by the anti-Flag antibody only in the presence of Flag-tagged α3 and not in the absence of it. Thus, the results shown in Fig. 2 confirmed that the F protein and α3 could indeed bind to each other.
FIG. 2.
Coimmunoprecipitation analysis of the F protein and the α3 protein. Huh7 cells were cotransfected with expression plasmids for HA-tagged F protein and Flag-tagged α3 protein or their control vectors in different combinations as indicated below the gel. Cell lysates were then immunoprecipitated (IP) with anti-HA (A) or anti-Flag (B) antibody, followed by Western blot (WB) analysis using anti-Flag (A) or anti-HA (B) antibody. The asterisk in panel B marks the location of a nonspecific protein band. TCL, total cell lysates.
Colocalization of the F protein and α3 in Huh7 cells.
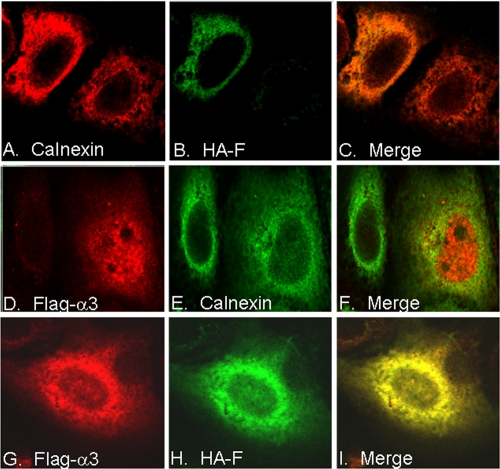
To further confirm the interaction between the F protein and α3 in cells, we performed confocal microscopy. As shown in Fig. 3, the F protein expressed in Huh7 cells by itself was present in the cytoplasm and colocalized with calnexin, a resident protein of the endoplasmic reticulum (top panels). This result is consistent with previous reports that the F protein is associated with the endoplasmic reticulum membranes (49, 58). When α3 was expressed in Huh7 cells, it displayed both nuclear and cytoplasmic staining patterns with little colocalization with calnexin (middle panels). This result is also consistent with previous observations that proteasomes are diffusely distributed in both the nucleus and the cytoplasm (34, 35). Importantly, when the F protein and α3 were coexpressed in the cell, most of the F protein and α3 colocalized to the cytoplasm (bottom panels). Thus, the results shown in Fig. 3 together with the results shown in Fig. 2 indicated that the F protein could bind to α3 in Huh7 cells.
FIG. 3.
Colocalization of the F protein and the α3 protein in cells. Huh7 cells were transfected with the HA-tagged F protein expression plasmid (A to C), the Flag-tagged α3 expression plasmid (D to F), or both (G to I); stained for calnexin (A and E), HA-tagged F (B and H), or Flag-tagged α3 (D and G); and then analyzed by confocal microscopy. Panels C, F, and I are merged pictures of panels A and B, D and E, and G and H, respectively.
Reduction of the F-protein level by α3 in a dose-dependent manner.
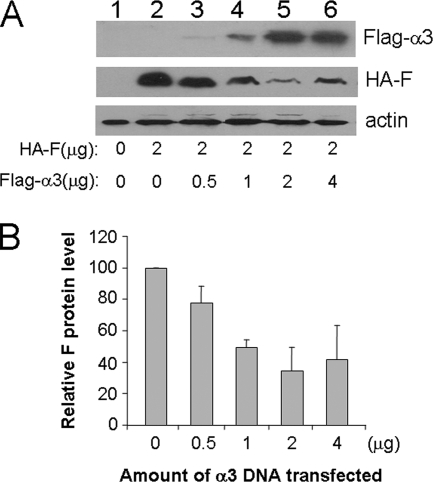
In our studies, we noticed that the coexpression of α3 with the F protein frequently led to a reduced expression level of the F protein. To determine whether α3 could indeed reduce the expression level of the F protein, we coexpressed the F protein with an increasing amount of α3 in Huh7 cells. As shown in Fig. 4, the increase of the α3 expression level led to the decrease of the F-protein level. This dose-dependent reduction of the F protein by α3 reached a peak when the ratio of the expression plasmids of these two proteins reached 1. Further increase of the α3-to-F-protein plasmid ratio did not further decrease, but rather slightly increased, the F-protein expression level. The reason for this slight increase is unclear, but it may be due to the inability of excessive amounts of α3 to assemble into proteasomes, which led to the stabilization of the F protein that bound to it (see below). The ability of α3 to reduce the F-protein expression level in a dose-dependent manner and the fact that α3 is an important component of the proteasome raised the possibility that the F protein might be degraded by the proteasome via its direct interaction with α3.
FIG. 4.
Dose-dependent reduction of the F-protein level by α3. Huh7 cells were transfected with 2 μg of the HAF expression plasmid and different amounts of the Flag-tagged α3 expression plasmid as indicated. Control vectors were also included in the transfection to ensure that the total amount of DNA plasmids used for the transfection for each experiment was the same. Cells were lysed 48 hours after transfection for Western blot analysis. A representative result is shown in panel A. Lane 1 is Huh7 cells transfected with only the control vectors. The relative F-protein levels were quantified by Sigma Scan, and the averages and standard deviations from three independent experiments are shown in panel B. The F-protein level in the absence of α3 was arbitrarily defined as 100%.
Identification of the α3-binding domain in the F protein.
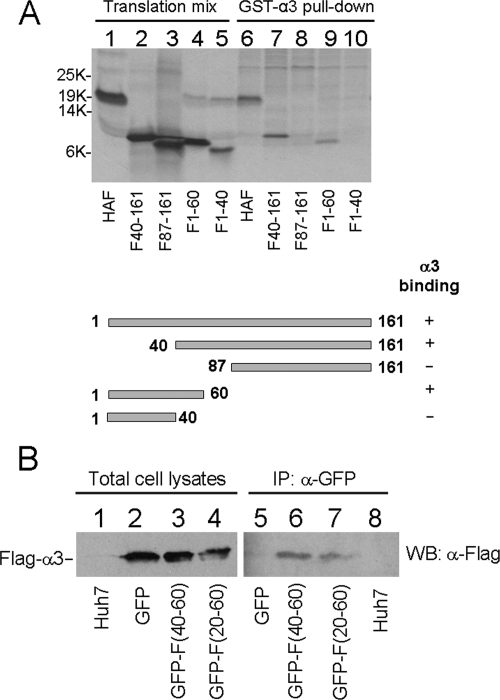
To understand how α3 reduced the expression level of the F protein, we performed deletion-mapping experiments to identify the F-protein domain that the α3 protein may bind to. The F protein with different N-terminal or C-terminal deletions was synthesized and labeled with [35S]methionine in vitro using rabbit reticulocyte lysates. The ability of these truncated F proteins to bind to α3 was then analyzed by the GST pull-down assay. As shown in Fig. 5A, the F protein with the N-terminal truncation at amino acid 40, but not the F protein with the N-terminal truncation at amino acid 87, was able to bind to α3. This result indicated the importance of amino acids 40 to 87 for the binding of F protein to α3. In the C-terminal truncation studies, the F protein with the truncation at amino acid 60 was able to bind to α3. However, further truncation at amino acid 40 abolished the binding of the F protein to α3. Thus, these deletion-mapping results together indicated that amino acids 40 to 60 of the F protein were required for the F protein to bind to α3.
FIG. 5.
Identification of the α3-binding domain in the F protein. (A) Deletion-mapping analysis. The F protein was truncated at different locations as indicated and synthesized using rabbit reticulocyte lysates. The deletion mutants were then incubated with purified GST-α3, precipitated using glutathione beads, and analyzed by gel electrophoresis and autoradiography. Lanes 1 to 5, translational mixtures; lanes 6 to 10, GST-α3 pull-down experiments. The deletion-mapping results are summarized below the gel. (B) Coimmunoprecipitation analysis of α3 with GFP, GFP-F(40-60), or GFP-F(20-60). Naïve Huh7 cells (lanes 1 and 8) or Huh7 cells cotransfected with the expression plasmids for Flag-tagged α3 and GFP (lanes 2 and 5), Flag-tagged α3 and GFP-F(40-60) (lanes 3 and 6), or Flag-tagged α3 and GFP-F(20-60) (lanes 4 and 7) were lysed 48 hours after transfection and immunoprecipitated (IP) with the anti-GFP antibody, followed by Western blot (WB) analysis using the anti-Flag antibody.
To determine whether amino acids 40 to 60 of the F protein serve as the domain for binding to α3, we fused amino acids 40 to 60 and 20 to 60 to the C terminus of GFP to create the fusion proteins GFP-F(40-60) and GFP-F(20-60). These two GFP-F fusion proteins were then separately coexpressed with Flag-tagged α3 in Huh7 cells for coimmunoprecipitation experiments. As shown in Fig. 5B, the anti-GFP antibody could immunoprecipitate α3 in the presence of either GFP-F(40-60) or GFP-F(20-60) but not the control GFP protein. These results confirmed that amino acids 40 to 60 of the F protein indeed served as the binding domain for α3.
Destabilization of a GFP reporter by the F-protein sequence.
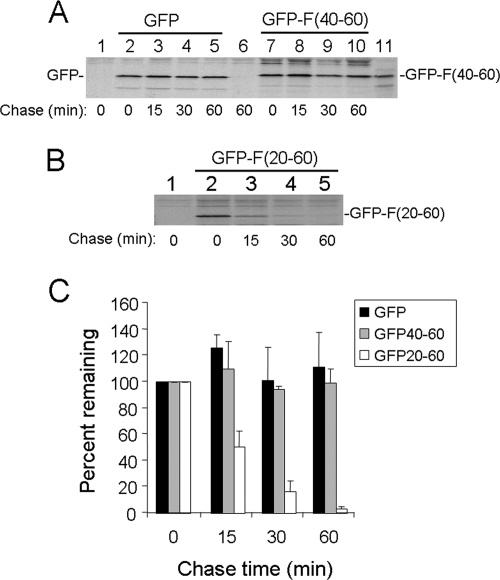
If α3, as a subunit of the proteasome, reduced the expression level of the F protein by facilitating its degradation by the proteasome through their physical interaction, then fusion of the α3-binding domain of the F protein to a stable protein may also destabilize that protein. To test this possibility, we determined the half-life of the GFP-F(40-60) fusion protein and that of the control GFP in Huh7 cells by performing pulse-chase labeling experiments. As shown in Fig. 6A, neither the GFP nor the GFP-F(40-60) fusion protein level was significantly reduced during the 1-hour chase period, indicating that the α3-binding domain of the F protein was insufficient to destabilize the GFP reporter. Interestingly, when the same pulse-chase labeling experiment was repeated using GFP-F(20-60), this protein was found to have a short half-life of approximately 15 minutes, similar to that of the wild-type F protein (Fig. 6B and C). These results indicated that the α3-binding domain (i.e., amino acids 40 to 60) of the F protein and its upstream sequence (i.e., amino acids 20 to 40) were required to destabilize GFP.
FIG. 6.
Stability analysis of GFP and F fusion proteins. (A) Pulse-chase labeling of GFP and GFP-F(40-60). Huh7 cells transfected with the GFP (lanes 2 to 5) or GFP-F(40-60) (lanes 7 to 10) expression plasmid were pulse-labeled with [35S]methionine for 15 min and chased for 0, 15, 30, and 60 min. Cell lysates were then immunoprecipitated with the anti-GFP antibody. Lanes 1 and 6, naïve Huh7 cells pulse-labeled for 15 min and chased for 0 and 60 min, respectively. Lane 11, [35S]methionine-labeled GFP-F(40-60) marker, which was synthesized in vitro using rabbit reticulocyte lysates. (B) Pulse-chase labeling of GFP-F(20-60). Lane 1, control Huh7 cells. (C) Quantification of the relative protein levels at different chase time points. The protein levels of GFP and GFP-F fusion proteins shown in panels A and B were quantified using Sigma Scan. The results represent the averages and standard deviations from three independent experiments.
Ubiquitin-independent degradation of the F protein.
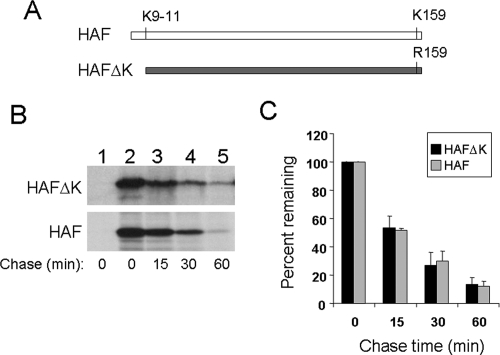
With a few exceptions, ubiquitination of proteins is required for their degradation by the proteasome. The F-protein sequence between amino acids 20 and 60 does not contain any lysine residue, which is the ubiquitination site. This observation prompted us to investigate whether the F protein is degraded by an ubiquitin-independent mechanism. The F protein that we used in our studies for Fig. 1 to 4 was full length and contained four lysine residues at amino acids 9 to 11 and 159 (Fig. 7A). To remove these putative ubiquitination sites, we created an HA-tagged, F protein lacking lysine (HAFΔK) by deleting its first 11 amino acids and mutating lysine-159 to arginine (56). This F protein without lysine was expressed in Huh7 cells, and its half-life was determined by pulse-chase labeling experiments. As shown in Fig. 7B and C, this F protein without lysine has a half-life of approximately 15 minutes, similar to that of the wild-type F protein (38, 58). This result indicates that the degradation of the F protein is lysine independent and supports the possibility that the F protein is degraded by an ubiquitin-independent pathway.
FIG. 7.
Lysine-independent degradation of the F protein. (A) Illustration of HAF and HAFΔK. HAFΔK is identical to HAF, except that it does not have the N-terminal 11 amino acids and its lysine-159 has been mutated to arginine. (B) Pulse-chase labeling for the HAFΔK mutant lacking lysine and the wild-type HAF. The pulse-chase labeling experiments for HAFΔK (upper panel) and HAF (lower panel) were conducted as described in the Fig. 6 legend. Lane 1, Huh7 cells transfected with the control vector. (C) Quantification of the relative F-protein levels during the chase. The protein levels were quantified using Sigma Scan. The results represent the averages and standard deviations from three independent experiments.
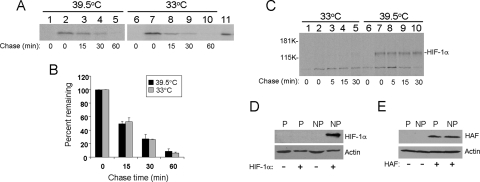
To further confirm that the F protein is indeed degraded by an ubiquitin-independent pathway, we expressed the HA-tagged F protein in ts85 cells and performed pulse-chase labeling experiments. The ts85 cell line is a mouse mammary carcinoma cell line that expresses a temperature-sensitive E1 ubiquitin-activating enzyme. The E1 enzyme is inactivated and the ubiquitin-dependent protein degradation is abolished when ts85 cells are incubated at the nonpermissive temperature (14). As shown in Fig. 8A and B, the F protein at both the permissive temperature of 33°C and the nonpermissive temperature of 39.5°C was similarly unstable, with a half-life of about 15 min. To ensure that the E1 activity was indeed inactivated at the nonpermissive temperature, we also analyzed HIF-1α at permissive and nonpermissive temperatures. HIF-1α is a labile protein with a half-life of less than 5 minutes under normoxia and is degraded by the ubiquitin-proteasome pathway (39). As shown in Fig. 8C, HIF-1α could not be detected in the pulse-chase-labeling experiment at the permissive temperature due to its instability. However, this 120-kDa protein was easily detectable at the nonpermissive temperature without a significant change of its level even after 30 min of chase. Similar results were obtained by Western blot analysis. HIF-1α was detectable at the nonpermissive temperature but not at the permissive temperature (Fig. 8D). In contrast, the F-protein expression level was not affected by the incubation temperature (Fig. 8E). Taken together, the results shown in Fig. 7 and 8 indicate that the degradation of F protein is ubiquitin independent.
FIG. 8.
E1 ubiquitin-activating enzyme-independent degradation of the F protein. (A) Pulse-chase labeling of the F protein in ts85 cells. Cells electroporated with the control vector (lanes 1 and 6) or the HAF expression plasmid were maintained at 33°C for 24 h, followed by further incubation at 39.5°C (nonpermissive temperature, lanes 1 to 5) or 33°C (permissive temperature, lanes 6 to 10) for 24 h prior to the pulse-chase labeling experiment using the procedures described in the Fig. 6 legend. Lane 11, [35S]methionine-labeled, in vitro-synthesized F protein, which served as a marker. (B) Quantification of the F-protein levels over the chase time. The F-protein bands shown in panel A were quantified using Sigma Scan. The results represent the averages and standard deviations from three independent experiments. (C) Pulse-chase labeling of HIF-1α. ts85 cells were electroporated with the control vector (lanes 1 and 6) or the HIF-1α expression plasmid (lanes 2 to 5 and 7 to 10). (D) Western blot analysis of HIF-1α expressed in ts85 cells. ts85 cells were electroporated with a control vector (−) or the HIF-1α expression plasmid (+). Cells were maintained at the permissive temperature (P) for 48 hours or maintained at the permissive temperature for the first 24 hours and then shifted to the nonpermissive temperature (NP) for another 24 h. Cells were then lysed for Western blot analysis. (E) Western blot analysis of HAF expressed in ts85 cells. The experiment was conducted as described for panel D, except the HAF expression plasmid and its control vector were used for the transfection.
Direct degradation of the F protein by the 20S proteasome.
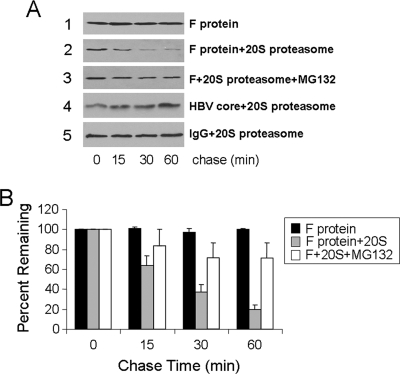
Previous studies indicate that the F protein is degraded by the proteasome (38, 58). To further investigate the molecular mechanism of the F-protein degradation by the proteasome, we expressed the F protein in yeast and subsequently purified this protein to near homogeneity as evidenced by protein gel and high-performance liquid chromatography analyses (data not shown). As shown in Fig. 9A, this F protein was stable in the absence of the 20S proteasome (panel 1). However, in the presence of the 20S proteasome, it was rapidly degraded (Fig. 9A, panel 2, and B). This degradation of the F protein by the 20S proteasome was specific, as it was inhibited by the proteasome inhibitor MG132 (Fig. 9A, panel 3), and the control HBV core protein (panel 4) and mouse immunoglobulin (panel 5) could not be degraded by the 20S proteasome. These results indicated that the F protein could be directly degraded by the 20S proteasome without the need for ubiquitination.
FIG. 9.
Degradation of the F protein by the 20S proteasome. (A) Protein degradation assay. The F protein, which was expressed in yeast cells and subsequently purified, was incubated without the 20S proteasome (panel 1), with the 20S proteasome (panel 2), or with the 20S proteasome and the proteasome inhibitor MG132 (panel 3). The control HBV core protein (panel 4) and mouse immunoglobulin G (IgG) (panel 5) were also incubated with the 20S proteasome. A protein aliquot was removed at 0, 15, 30, and 60 min after incubation and analyzed by Western blotting using the mouse anti-F antibody (panels 1 to 3), the rabbit anti-HBV core antibody (panel 4), and the goat anti-mouse antibody (panel 5). For the IgG control, both the heavy chain and the light chain were not affected by the proteasome, and hence only the result for the heavy chain is shown. (B) Quantification of the F-protein levels at various time points. The results represent the averages and standard deviations from three independent experiments.
Stabilization of the F protein in HCV cells by suppression of expression of the proteasome subunit α3.
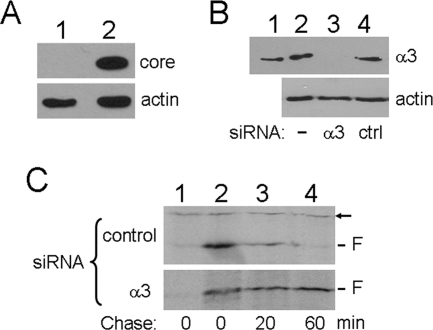
To further investigate the role of α3 and the possible effect of a replicating HCV genome on the stability of the F protein, we transfected Huh7.5 cells with HCV JFH-1 genomic RNA. This transfection resulted in the replication of the HCV JFH-1 RNA and the production of infectious viral particles (data not shown) (42). As shown in Fig. 10A, the HCV core protein was easily detectable in the HCV JFH-1 RNA-transfected cells at 8 days after transfection. These HCV JFH-1 cells were then transfected with either the α3 siRNA or a control siRNA, followed by transfection with the F-protein expression plasmid. As shown in Fig. 10B, the α3 siRNA reduced the α3 protein level to an undetectable level, whereas the control siRNA had no effect on α3. When the stability of the F protein was analyzed by pulse-chase labeling experiments, the F protein was labile, with a half-life of less than 20 minutes in cells treated with the control siRNA, indicating that the half-life of the F protein was not affected by the presence of other HCV proteins and a replicating HCV genome. In contrast, in cells treated with the α3 siRNA, the F protein was stable, with no significant reduction of its level even after 60 minutes of chase (Fig. 10C). These results indicated that the α3 protein played an important role in the degradation of the F protein.
FIG. 10.
Effect of suppression of α3 expression on the stability of the F protein in HCV JFH-1 cells. (A) Western blot analysis of the HCV core protein in cells transfected with the replication-defective HCV JFH-1 GND mutant RNA (lane 1) or the wild-type HCV JFH-1 RNA (lane 2) (42). Cells were lysed 8 days after transfection for Western blot analysis. Actin was also analyzed to serve as a loading control. (B) Suppression of α3 expression with siRNA. HCV JFH-1 cells without siRNA transfection (lane 2), with α3 siRNA transfection (lane 3), or with control (ctrl) siRNA transfection (lane 4) were lysed for Western blot analysis. Lane 1 contained the purified 20S proteasomes, which provided the α3 protein to serve as the marker. Upper panel, Western blot using the anti-α3 antibody; lower panel, Western blot using the antiactin antibody. (C) Pulse-chase labeling of the F protein. HCV JFH-1 cells treated with the control siRNA (upper panel) or α3 siRNA (lower panel) were transfected with the empty expression vector (lane 1) or the F-protein expression plasmid (lanes 2 to 4), pulse-labeled with [35S]methionine for 15 minutes, and chased for 0 (lanes 1 and 2), 20 (lane 3), or 60 (lane 4) minutes. The F protein was immunoprecipitated with the mouse monoclonal anti-F antibody. The arrow denotes a nonspecific protein band, which served as an internal control. This nonspecific protein was relatively stable during the chase period.
Stability analysis of different forms of the F protein.
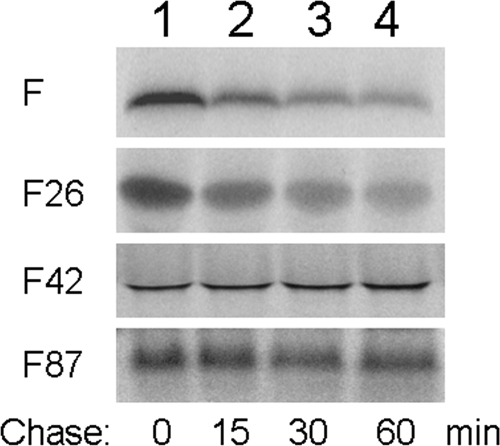
As different mechanisms, which include translational ribosomal frameshift at codon 42 and internal initiation of translation at codon 26 or codons 85 and 87 (4, 6, 50), had been proposed to explain the expression of the F protein, we also investigated the stabilities of these different truncated forms of the F protein by conducting pulse-chase labeling experiments in vitro. As shown in Fig. 11, the full-length F protein was unstable with a short half-life, in agreement with our previous report (58) and with observations in cell cultures (Fig. 7, 8, and 10). The F protein truncated at codon 26 was also unstable, with a stability similar to that of the full-length F protein. In contrast, the F protein truncated at codon 42 or codon 87 was stable, with no significant reduction of its protein level even after 60 min of chase. These results, in combination with the observations shown in Fig. 6, indicated that amino acids 26 to 60 are sufficient to mediate the instability of the F protein.
FIG. 11.
Pulse-chase labeling of different truncated forms of the F protein in vitro. The pulse-chase labeling experiment was conducted as previously described (58). Briefly, the full-length F protein and the F protein with an N-terminal truncation at codon 26 (F26), codon 42 (F42), or codon 87 (F87) were synthesized using rabbit reticulocyte lysates and pulse-labeled with [35S]methionine for 15 minutes before the translation reaction was stopped with cycloheximide. The F protein was then chased for 0 (lane 1), 15 (lane 2), 30 (lane 3), or 60 (lane 4) minutes. For F26 and F42, a methionine codon was inserted at the 5′ end for translation initiation. For F87, codon 87, which is a methionine codon, was used for translation initiation.
DISCUSSION
The HCV F protein is encoded by the +1 reading frame that overlaps with the core protein coding sequence. This protein was initially reported to be expressed by translational ribosomal frameshift at codons 9 to 11 of the core protein sequence (11, 59). However, other mechanisms have subsequently also been proposed to explain its expression (4, 6, 50). The full-length HCV F protein synthesized by translational ribosomal frameshift at codons 9 to 11 is a labile protein that is degraded by proteasomes (38, 58). We have further analyzed the molecular pathway that mediates the degradation of the F protein. By using a combination of yeast two-hybrid assays, GST pull-down experiments (Fig. 1), coimmunoprecipitation experiments (Fig. 2), and confocal microscopy (Fig. 3), we demonstrated that the HCV F protein could bind to the proteasome subunit α3, which facilitates the degradation of the F protein in a dose-dependent manner (Fig. 4). The role of α3 in the degradation of the F protein was also supported by our siRNA knockdown experiment, in which we found that the suppression of α3 expression resulted in the stabilization of the F protein, even in the presence of a replicating HCV genome (Fig. 10). Our deletion-mapping studies identified the α3-binding domain in the F-protein sequence between amino acids 40 and 60 (Fig. 5). Interestingly, this α3-binding domain was insufficient to destabilize the GFP reporter (Fig. 5 and 6), but it, together with its upstream sequence located between amino acids 20 and 40, could reduce the half-life of GFP to that of the F protein (Fig. 6). This observation is reminiscent of a recent finding by Takeuchi et al., who reported that tethering GFP to Rpn10, a 19S proteasome regulatory subunit protein, was insufficient to destabilize GFP unless the fusion protein was also linked to a loosely structured sequence to initiate insertion into the proteasome (43). Thus, it is conceivable that the role of amino acids 20 to 40, which lack apparent structures based on the structural prediction using computer programs (data not shown), is to serve as the signal to initiate translocation of the protein into the catalytic cavity after the binding of the F protein to α3.
A limited number of proteins, including ornithine decarboxylase (ODC) and p21Cip1, are degraded by proteasomes via the ubiquitin-independent pathway. ODC contains a degron, which is a 34-amino-acid sequence located at its C terminus. This degron can bind to the 19S regulatory particle, and appending this degron to various proteins can target them to the 19S regulatory particle to enhance their degradation by the 26S proteasome via the ubiquitin-independent pathway (44). In the case of p21Cip1, which is a cyclin-dependent kinase inhibitor, its degradation by the proteasome is mediated by its interaction with REGγ (10), although an earlier report also indicated that that it could bind to the proteasome subunit α7 (46). For both ODC and p21Cip1, the ability of these two proteins to bind directly to a proteasome subunit is essential for their degradation by the proteasome. If the F protein follows the same rule and is targeted to the proteasome by α3, then its degradation by the proteasome will also be ubiquitin independent. This apparently is the case, based on three lines of evidence: first, the F protein mutant without lysine, which lacks the ubiquitination site, is as unstable as the wild-type F protein (Fig. 7); second, the F protein expressed in ts85 cells, which have a temperature-sensitive E1 ubiquitin-activating enzyme, is not stabilized when the cell is incubated at the nonpermissive temperature (Fig. 8); and third, the F protein can be degraded directly by the 20S proteasome in vitro in the absence of ubiquitination machineries (Fig. 9).
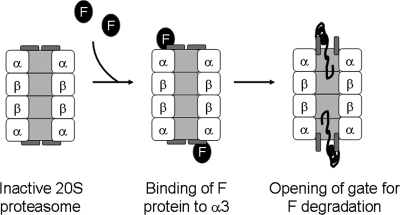
The α3 protein is an essential component of the 20S proteasome. Its N-terminal tail plays a critical role in the formation of a gate that blocks unregulated access of proteins to the catalytic cavity of the proteasome. The deletion of this tail results in the constitutive activation of the 20S proteasome (18). Based on the biological functions of α3, we propose that the binding of the F protein to α3 results in the conformational change of α3 and the opening of the 20S proteasome gate. The poorly structured region of the F protein (i.e., amino acids 20 to 40) then initiates the translocation of the F protein into the catalytic cavity of the 20S proteasome for degradation. In this regard, amino acids 20 to 60 of the F protein are functionally similar to the degron of ODC, which consists of a proteasome association element and a loosely structured region for initiating the translocation of the protein into the catalytic cavity of the 20S core (28, 60). The model for F-protein degradation by the 20S proteasome is shown in Fig. 12.
FIG. 12.
Model for F-protein degradation by the 20S proteasome. Binding of the F-protein to α3 results in the conformational change of α3 and the opening of the proteasomal gate. The poorly structured region of the F protein (i.e., amino acids 20 to 40) then initiates the translocation of the F protein into the catalytic cavity of the 20S proteasome for degradation.
The biological functions of the F protein are unclear, although recent studies indicated that it might have regulatory functions (15, 30, 57). The instability of the F protein may be critical for limiting its regulatory activities. Alternatively, our finding that the F protein can bind to α3 also raises the possibility that the F protein may regulate cellular protein degradation. The human immunodeficiency virus type 1 (HIV-1) Tat protein has been shown to bind to the 20S proteasome and inhibits its association with the 11S REG complex (53). Since the binding of REG to the 20S proteasome is important for the generation of antigenic peptides for presentation by major histocompatibility complex class I, it has been suggested that this activity of HIV-1 Tat could suppress antigen presentation and prevent HIV-infected cells from being removed by cytotoxic T lymphocytes (17, 40). It is likely that the binding of the F protein to α3 may also inhibit the association of the REG activator to the 20S proteasome and contribute to HCV immune evasion by decreasing viral antigen presentation by host cells. Since it requires only two molecules of the F protein to suppress the association of the REG activator with both ends of the 20S proteasome, a low expression level of the F protein may be sufficient to achieve its suppressive effect. This possibility is supported by the observation that the HBV X protein, which is also a labile protein with a low expression level during viral replication (24), is capable of suppressing the degradation of cellular proteins by binding to the proteasome subunit α7 (21, 61). Further research will be required to confirm whether the F protein can indeed regulate the proteasome activities.
Besides translational ribosomal frameshift (or transcriptional slippage) at codons 9 to 11 of the polyprotein sequence, other mechanisms, which include translational ribosomal frameshift at codon 42 and internal initiation of translation at codon 26 or codons 85 and 87, have also been proposed for the expression of different forms of the F protein (4, 6, 50). As we demonstrated in Fig. 11, the F protein synthesized by internal initiation of translation from codon 26 is also unstable and has a half-life similar to that of the full-length F protein. This result indicates that the “degron” domain of the F protein can be further narrowed down to a region encompassing amino acids 26 to 60. Our finding that the F protein synthesized from codon 87 is a stable protein is also consistent with this notion, as this truncated F protein lacks this degron sequence (Fig. 11). Of particular interest is the F protein synthesized by translational ribosomal frameshift at codon 42. This protein contains an almost-complete α3-binding sequence (i.e., amino acids 40 to 60). As we showed in Fig. 11, the F protein truncated at amino acid 42 had increased stability. Thus, if this frameshifted protein is indeed expressed during natural HCV infection, it may have an increased stability and a more profound effect on the proteasome than the full-length F protein or the F protein initiated from codon 26.
In conclusion, our results demonstrated that the HCV F protein contains a binding domain for the proteasome subunit α3. This binding as well as its upstream sequence mediates the degradation of the F protein and may regulate the proteasome activities to affect HCV replication and pathogenesis. Further research in this area will generate interesting results for better understanding the biological functions of the F protein.
Acknowledgments
We thank Ben Yen for critical reading of the manuscript, David Ann for helpful discussions during the course of this work, Charlie Rice for the Huh7.5 cell line, Michelle MacVeigh and the Confocal Microscopy Core of USC Research Center for Liver Diseases for help with the confocal microscopy, Susan Ou and the Caltech Monoclonal Antibody Core for the preparation of the mouse monoclonal anti-F antibody, Zea Borok for plasmids pCEP4 and pCEP-HIF1α, Lucio Comai for pCMV-mtFlag, and Kyle Nakamura for help with the construction of the pGST-α3 plasmid.
This work was supported by research grants (T32AI007078 and R01AI060559) from the National Institutes of Health.
Footnotes
Published ahead of print on 29 October 2008.
REFERENCES
- 1.Asher, G., J. Lotem, L. Sachs, C. Kahana, and Y. Shaul. 2002. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc. Natl. Acad. Sci. USA 9913125-13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aviel, S., G. Winberg, M. Massucci, and A. Ciechanover. 2000. Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 27523491-23499. [DOI] [PubMed] [Google Scholar]
- 3.Bain, C., P. Parroche, J. P. Lavergne, B. Duverger, C. Vieux, V. Dubois, F. Komurian-Pradel, C. Trepo, L. Gebuhrer, G. Paranhos-Baccala, F. Penin, and G. Inchauspe. 2004. Memory T-cell-mediated immune responses specific to an alternative core protein in hepatitis C virus infection. J. Virol. 7810460-10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baril, M., and L. Brakier-Gingras. 2005. Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein. Nucleic Acids Res. 331474-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bercovich, Z., Y. Rosenberg-Hasson, A. Ciechanover, and C. Kahana. 1989. Degradation of ornithine decarboxylase in reticulocyte lysate is ATP-dependent but ubiquitin-independent. J. Biol. Chem. 26415949-15952. [PubMed] [Google Scholar]
- 6.Boulant, S., M. Becchi, F. Penin, and J. P. Lavergne. 2003. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J. Biol. Chem. 27845785-45792. [DOI] [PubMed] [Google Scholar]
- 7.Braun, B. C., M. Glickman, R. Kraft, B. Dahlmann, P. M. Kloetzel, D. Finley, and M. Schmidt. 1999. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1221-226. [DOI] [PubMed] [Google Scholar]
- 8.Breitschopf, K., E. Bengal, T. Ziv, A. Admon, and A. Ciechanover. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 175964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 2431576-1583. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X., L. F. Barton, Y. Chi, B. E. Clurman, and J. M. Roberts. 2007. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell 26843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, J., Z. Xu, and J. H. Ou. 2003. Triple decoding of hepatitis C virus RNA by programmed translational frameshifting. Mol. Cell. Biol. 231489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, M., L. Bachmatov, Z. Ben-Ari, Y. Rotman, R. Tur-Kaspa, and R. Zemel. 2007. Development of specific antibodies to an ARF protein in treated patients with chronic HCV infection. Dig. Dis. Sci. 522427-2432. [DOI] [PubMed] [Google Scholar]
- 13.Craiu, A., M. Gaczynska, T. Akopian, C. F. Gramm, G. Fenteany, A. L. Goldberg, and K. L. Rock. 1997. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J. Biol. Chem. 27213437-13445. [DOI] [PubMed] [Google Scholar]
- 14.Finley, D., A. Ciechanover, and A. Varshavsky. 1984. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell 3743-55. [DOI] [PubMed] [Google Scholar]
- 15.Fiorucci, M., S. Boulant, A. Fournillier, J. D. Abraham, J. P. Lavergne, G. Paranhos-Baccala, G. Inchauspe, and C. Bain. 2007. Expression of the alternative reading frame protein of hepatitis C virus induces cytokines involved in hepatic injuries. J. Gen. Virol. 881149-1162. [DOI] [PubMed] [Google Scholar]
- 16.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82373-428. [DOI] [PubMed] [Google Scholar]
- 17.Groettrup, M., A. Soza, M. Eggers, L. Kuehn, T. P. Dick, H. Schild, H. G. Rammensee, U. H. Koszinowski, and P. M. Kloetzel. 1996. A role for the proteasome regulator PA28alpha in antigen presentation. Nature 381166-168. [DOI] [PubMed] [Google Scholar]
- 18.Groll, M., M. Bajorek, A. Kohler, L. Moroder, D. M. Rubin, R. Huber, M. H. Glickman, and D. Finley. 2000. A gated channel into the proteasome core particle. Nat. Struct. Biol. 71062-1067. [DOI] [PubMed] [Google Scholar]
- 19.Haas, A. L., J. V. Warms, A. Hershko, and I. A. Rose. 1982. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 2572543-2548. [PubMed] [Google Scholar]
- 20.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67425-479. [DOI] [PubMed] [Google Scholar]
- 21.Hu, Z., Z. Zhang, E. Doo, O. Coux, A. L. Goldberg, and T. J. Liang. 1999. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 737231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, S. C., F. G. Whitby, C. Realini, M. Rechsteiner, and C. P. Hill. 1997. The proteasome 11S regulator subunit REG alpha (PA28 alpha) is a heptamer. Protein Sci. 62469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalejta, R. F., and T. Shenk. 2003. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl. Acad. Sci. USA 1003263-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. H., S. Kang, J. Kim, and B. Y. Ahn. 2003. Hepatitis B virus core protein stimulates the proteasome-mediated degradation of viral X protein. J. Virol. 777166-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komurian-Pradel, F., A. Rajoharison, J. L. Berland, V. Khouri, M. Perret, M. Van Roosmalen, S. Pol, F. Negro, and G. Paranhos-Baccala. 2004. Antigenic relevance of F protein in chronic hepatitis C virus infection. Hepatology 40900-909. [DOI] [PubMed] [Google Scholar]
- 26.Liu, C. W., M. J. Corboy, G. N. DeMartino, and P. J. Thomas. 2003. Endoproteolytic activity of the proteasome. Science 299408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, C. W., L. Millen, T. B. Roman, H. Xiong, H. F. Gilbert, R. Noiva, G. N. DeMartino, and P. J. Thomas. 2002. Conformational remodeling of proteasomal substrates by PA700, the 19 S regulatory complex of the 26 S proteasome. J. Biol. Chem. 27726815-26820. [DOI] [PubMed] [Google Scholar]
- 28.Loetscher, P., G. Pratt, and M. Rechsteiner. 1991. The C terminus of mouse ornithine decarboxylase confers rapid degradation on dihydrofolate reductase. Support for the pest hypothesis. J. Biol. Chem. 26611213-11220. [PubMed] [Google Scholar]
- 29.Lu, W., A. Strohecker, and J. H. Ou Jh. 2001. Post-translational modification of the hepatitis C virus core protein by tissue transglutaminase. J. Biol. Chem. 27647993-47999. [DOI] [PubMed] [Google Scholar]
- 30.Ma, H. C., T. W. Lin, H. Li, S. M. Iguchi-Ariga, H. Ariga, Y. L. Chuang, J. H. Ou, and S. Y. Lo. 2008. Hepatitis C virus ARFP/F protein interacts with cellular MM-1 protein and enhances the gene trans-activation activity of c-Myc. J. Biomed Sci. 15417-425. [DOI] [PubMed] [Google Scholar]
- 31.McMullan, L. K., A. Grakoui, M. J. Evans, K. Mihalik, M. Puig, A. D. Branch, S. M. Feinstone, and C. M. Rice. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl. Acad. Sci. USA 1042879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, C. L., and D. J. Pintel. 2001. The NS2 protein generated by the parvovirus minute virus of mice is degraded by the proteasome in a manner independent of ubiquitin chain elongation or activation. Virology 285346-355. [DOI] [PubMed] [Google Scholar]
- 33.Nandi, D., P. Tahiliani, A. Kumar, and D. Chandu. 2006. The ubiquitin-proteasome system. J. Biosci. 31137-155. [DOI] [PubMed] [Google Scholar]
- 34.Palmer, A., G. G. Mason, J. M. Paramio, E. Knecht, and A. J. Rivett. 1994. Changes in proteasome localization during the cell cycle. Eur. J. Cell Biol. 64163-175. [PubMed] [Google Scholar]
- 35.Palmer, A., A. J. Rivett, S. Thomson, K. B. Hendil, G. W. Butcher, G. Fuertes, and E. Knecht. 1996. Subpopulations of proteasomes in rat liver nuclei, microsomes and cytosol. Biochem. J. 316401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ratinier, M., S. Boulant, C. Combet, P. Targett-Adams, J. McLauchlan, and J. P. Lavergne. 2008. Transcriptional slippage prompts recoding in alternate reading frames in the hepatitis C virus (HCV) core sequence from strain HCV-1. J. Gen. Virol. 891569-1578. [DOI] [PubMed] [Google Scholar]
- 37.Reinstein, E., M. Scheffner, M. Oren, A. Ciechanover, and A. Schwartz. 2000. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene 195944-5950. [DOI] [PubMed] [Google Scholar]
- 38.Roussel, J., A. Pillez, C. Montpellier, G. Duverlie, A. Cahour, J. Dubuisson, and C. Wychowski. 2003. Characterization of the expression of the hepatitis C virus F protein. J. Gen. Virol. 841751-1759. [DOI] [PubMed] [Google Scholar]
- 39.Salceda, S., and J. Caro. 1997. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 27222642-22647. [DOI] [PubMed] [Google Scholar]
- 40.Seeger, M., K. Ferrell, R. Frank, and W. Dubiel. 1997. HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J. Biol. Chem. 2728145-8148. [DOI] [PubMed] [Google Scholar]
- 41.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5403-410. [DOI] [PubMed] [Google Scholar]
- 42.Sir, D., W. L. Chen, J. Choi, T. Wakita, T. S. Yen, and J. H. Ou. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 481054-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi, J., H. Chen, and P. Coffino. 2007. Proteasome substrate degradation requires association plus extended peptide. EMBO J. 26123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi, J., H. Chen, M. A. Hoyt, and P. Coffino. 2008. Structural elements of the ubiquitin-independent proteasome degron of ornithine decarboxylase. Biochem. J. 410401-407. [DOI] [PubMed] [Google Scholar]
- 45.Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 1994-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touitou, R., J. Richardson, S. Bose, M. Nakanishi, J. Rivett, and M. J. Allday. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 202367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Troesch, M., E. Jalbert, S. Canobio, M. R. Boulassel, J. P. Routy, N. F. Bernard, J. Bruneau, N. Lapointe, M. Boucher, and H. Soudeyns. 2005. Characterization of humoral and cell-mediated immune responses directed against hepatitis C virus F protein in subjects co-infected with hepatitis C virus and HIV-1. AIDS 19775-784. [DOI] [PubMed] [Google Scholar]
- 48.Varaklioti, A., N. Vassilaki, U. Georgopoulou, and P. Mavromara. 2002. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 27717713-17721. [DOI] [PubMed] [Google Scholar]
- 49.Vassilaki, N., H. Boleti, and P. Mavromara. 2007. Expression studies of the core+1 protein of the hepatitis C virus 1a in mammalian cells. The influence of the core protein and proteasomes on the intracellular levels of core+1. FEBS J. 2744057-4074. [DOI] [PubMed] [Google Scholar]
- 50.Vassilaki, N., and P. Mavromara. 2003. Two alternative translation mechanisms are responsible for the expression of the HCV ARFP/F/core+1 coding open reading frame. J. Biol. Chem. 27840503-40513. [DOI] [PubMed] [Google Scholar]
- 51.Verma, R., L. Aravind, R. Oania, W. H. McDonald, J. R. Yates III, E. V. Koonin, and R. J. Deshaies. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298611-615. [DOI] [PubMed] [Google Scholar]
- 52.Verma, R., R. Oania, J. Graumann, and R. J. Deshaies. 2004. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 11899-110. [DOI] [PubMed] [Google Scholar]
- 53.Viscidi, R. P., K. Mayur, H. M. Lederman, and A. D. Frankel. 1989. Inhibition of antigen-induced lymphocyte proliferation by Tat protein from HIV-1. Science 2461606-1608. [DOI] [PubMed] [Google Scholar]
- 54.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 681015-1068. [DOI] [PubMed] [Google Scholar]
- 55.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worthington, Z. E., and N. H. Carbonetti. 2007. Evading the proteasome: absence of lysine residues contributes to pertussis toxin activity by evasion of proteasome degradation. Infect. Immun. 752946-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, W. B., S. W. Shao, L. J. Zhao, J. Luan, J. Cao, J. Gao, S. Y. Zhu, and Z. T. Qi. 2007. Hepatitis C virus F protein up-regulates c-myc and down-regulates p53 in human hepatoma HepG2 cells. Intervirology 50341-346. [DOI] [PubMed] [Google Scholar]
- 58.Xu, Z., J. Choi, W. Lu, and J. H. Ou. 2003. Hepatitis C virus F protein is a short-lived protein associated with the endoplasmic reticulum. J. Virol. 771578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 203840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, M., C. M. Pickart, and P. Coffino. 2003. Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. EMBO J. 221488-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, Z., U. Protzer, Z. Hu, J. Jacob, and T. J. Liang. 2004. Inhibition of cellular proteasome activities enhances hepadnavirus replication in an HBX-dependent manner. J. Virol. 784566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]