Abstract
The nucleotide sequence of the highly divergent small-ruminant lentivirus genotype E has been determined. The full genome consists of 8,418 nucleotides and lacks two large portions corresponding nearly to the entire dUTPase subunit of the pol and vpr-like accessory genes. Moreover, the 70-bp repeat of the U3 region of the long terminal repeat was observed to be deleted. Interestingly, this lentivirus genotype is able to persist in a local breed population, and retrospective analysis revealed its presence in milk samples collected in 1999. gag sequences obtained from a flock coinfected with the B1 and E genotypes revealed that the evolutionary rates of the two viruses were quite similar. Since a reduced viral load and/or disease progression was observed for viruses with artificially deleted dUTPase and vpr-like genes, it is proposed that this viral cluster be designated a low-pathogenicity caprine lentivirus.
The small-ruminant lentiviruses (SRLVs) are a genetically and antigenically heterogeneous group of viruses infecting sheep and goats, leading to persistent infection and chronic debilitating diseases. The majority of SRLV isolates can be classified into two main phylogenetic clusters: genotype A, involving maedi-visna virus-like strains, and genotype B, including caprine arthritis-encephalitis virus (CAEV)-like isolates, originally isolated from sheep and goats, respectively. Additional genotypes include Norwegian (genotype C) (4) and Swiss and Spanish (genotype D) (15, 18) isolates and the recently described Italian caprine isolates (genotype E) identified in flocks in which the local Roccaverano goat breed was prevalent (5). Interestingly, as for other indigenous goat breeds, typical clinical signs of lentiviral infection had never been observed in Italy before the introduction of imported breeds carrying the B1 subtype in the early 1980s.
SRLVs possess a complex genome comprising the gag, pol, and env structural genes and the vif, tat, and rev accessory genes. The low-pathogenicity SRLVs characterized so far have shown that deletions or mutations in the long terminal repeat (LTR) may be associated with variations in virulence, likely due to the presence of replication enhancer elements such as AP1, AML, tumor necrosis factor-α, and gamma interferon response elements (1, 11). Additional information about virulence factors has been produced in in vitro and in vivo studies by using genetic manipulation of infectious molecular clones (7, 8, 10, 19, 23). The dUTPase subunit, encoded by the pol gene, has been found to be dispensable for viral replication (12); however, dUTPase-negative strains produce less-severe lesions, restricted to the injection site (20). The tat gene of SRLV has been recently designated vpr-like, based on its primary protein structure and some functional similarities to human immunodeficiency virus type 1 Vpr protein (21). The CAEV tat (hereafter named vpr-like) gene increases the viral load, tissue distribution, and inflammatory lesion severity over that of the vpr deletion counterpart (9).
In the last few decades, the increasing interest in the development of live attenuated viruses capable of inducing resistance to superinfection has focused on specific deletion mutants for safe and efficacious live vaccine. In this report, we describe the genetic features of genotype E, a novel goat lentivirus which, although naturally deleted for dUTPase and the vpr-like gene, can persist in the population.
Three flocks of Roccaverano breed goats (It-02, It-06, and It-09) were selected from among a population of 3,200 head where genotype E was identified by using a previously described gag PCR (5). Blood samples (n = 70) were collected and sera and buffy coats obtained. A 7-year-old goat, clinically healthy and highly reactive against the type E immunodominant epitope of capsid antigen (sequence KLNKEAETWMRQNPQPP), was selected for virus isolation. After euthanasia, tissue explants were obtained from the mammary gland, mammary lymph nodes, lung, mediastinal lymph nodes, synovial membrane, choroid plexus, and spleen. Cultures were maintained over five passages. Giemsa staining was carried out at weekly intervals with replicate 24-well microplates, while gag PCR was performed without a cytopathic effect from the third passage. Virus isolation was successfully carried out with a restricted number of tissue explantations. PCR and cytopathic effect assays were positive for mammary primary cultures from the third and fifth passages, respectively, while PCR signals were observed in spleen and mammary lymph node explants. Synovial membrane, choroid plexus, and lung and mediastinal lymph nodes remained negative until the fifth passage. This field isolate is hereafter named Roccaverano.
Supernatants from PCR-positive cultures were collected, and DNA and RNA were extracted and used for genome amplification.
The complete genome of the Roccaverano isolate was amplified by using standard PCR and reverse transcription-PCR (RT-PCR) with the primers listed in Table 1, resulting in six overlapping products. Sequences from two independent PCR (spleen and mammary gland) were obtained using standard dye terminator chemistry.
TABLE 1.
Nucleotide sequences of primers
| Amplicon | Primer sequence 5′→3′
|
Reference or source | |
|---|---|---|---|
| Forward | Reverse | ||
| LTR | TGACACAGCAAATGTAACCGCAAG | CCACGTTGGGCGCCAGCTGCGAGA | 22 |
| LTR-Gag | TGACACAGCAAATGTAACCGCAAG | CTTGCCTGATCCATATTTGCCTGTG | This study |
| Gag | TGGTGARKCTAGMTAGAGACATGG | CATAGGRGGHGCGGACGGCASCA | 5 |
| Gag-Pol | AAAACCCGGCCACTTAGCAAG | CTATCCAGAGAACCTGTCCTG | This study |
| Pol | GGTGCCTGGACATAAAGGGATTC | GCCACTCTCCTGRATGTCCTCT | 18 |
| Pol-LTR | CCTAGGGACAAGTCCTATGG | GCCACCTGCGAGGACCGCACC | This study |
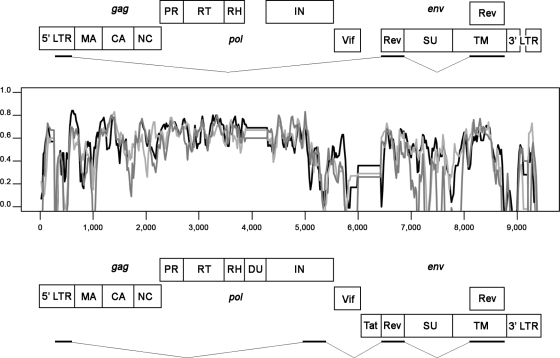
The genome consisted of 8,418 bp (accession number EU293537) including a functional LTR, three structural genes (gag, pol, and env), and two accessory genes required for the replication-competent SRLV (vif and rev). However, three deletions were found when the genome's size was compared with the canonical sizes of the SRLV genomes so far described (∼9.2 kb). The first deletion was identified in the pol gene, corresponding to nearly the whole dUTPase subunit. The second deletion, corresponding to the vpr-like accessory gene, was identified between the vif and env genes. Finally, the lack of a 70-bp repeat in the LTR U3 region was observed (Fig. 1). While traces of dUTPase in the pol gene suggest that this subunit might have been lost during evolution, it is difficult to speculate whether the vpr gene had ever been present in the genome. To assess whether these deletions may represent a unique genetic marker of this genotype, specific PCRs of the sequences flanking dUTPase (18) and vpr (this study) were carried out. Both deletions were confirmed for five animals belonging to epidemiologically unrelated flocks (accession number FJ389754).
FIG. 1.
Complete genome scheme of the genotype E Roccaverano strain (top panel) and the B1 prototype CAEV Cork strain (bottom panel), with deletions highlighted. A similarity plot (SimPlot software) of complete SRLV reference sequences (black line, accession number M33677; gray line, accession number M51543; light gray line, accession number AF322109) and the Roccaverano strain (accession number EU293537) is shown. Each plotted point represents the percentage of identity within a sliding window 100 bp wide centered on the plot position as shown, with a 20-bp step size between points.
Since the indicated viral sequence was obtained by overlapping the PCR fragments, it may not reflect the sequence of a single provirus. However a comparison of the LTR, gag, and pol fragments obtained from different tissue explants (see above-described samples) showed a divergence ranging from 0.4% (LTR) to 0.88% (pol). In addition, the rev sequence, as well as the pol-env 1.2-kb RT-PCR fragment, representing the replication-competent virus, showed a divergence of less than 1% compared with the proviral sequence.
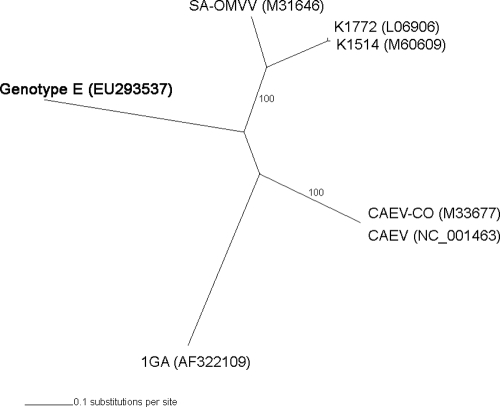
The phylogenetic relationship between genotype E and the above-referenced complete SRLV genomes was analyzed. By using evolutionary model estimation (13) and Bayesian approaches with MrBayes version 3.1.2 software (16), sequences belonging to genotypes A (accession numbers L06906, M60609, and M31646), B (accession numbers M33677 and NC001463), and C (accession number AF322109) were compared with those of genotype E. Deletions of the dUTPase and vpr gene were considered, and their sequences were eliminated from the reference samples. Nucleotide diversity was 42% compared to all SRLV complete genome sequences known so far. The overall ratio of nonsynonymous to synonymous substitutions (ω) was always low (gag ω, 0.120; pol ω, 0.065; and env ω, 0.195), showing the presence of a strong purifying selection among genotype E and reference strains (DnaSP version 4.10.9 software) (17). Bayesian trees based on the gag, env, and concatenate genes showed the same topology, confirming the high divergence of genotype E (Fig. 2). These results enabled us to consider the gag gene a good target for inferring SRLV phylogeny and evolution.
FIG. 2.
Bayesian tree based on concatenate genes of six reference strains and genotype E. Accession numbers of reference sequences are shown in parentheses. The newly described Italian genotype E is shown in boldface. The gag-, pol-, and env-based trees showed the same topology (not shown).
Since specific deletions of dUTPase and Vpr in different lentiviral models had shown a key role in RT fidelity (20), it was important to evaluate whether nucleotide diversity in the Roccaverano strain was due to an altered evolutionary rate. To address this issue, we evaluated additional gag sequences obtained from flock It-02 based on preliminary serological screening to record the B and E genotype coinfection. Sequences obtained from the DNA from 13 milk samples collected from the same flock in 1999 allowed us to determine the evolutionary rate of both genotypes, as well as the number and proportion of G-to-A substitutions. Epidemiological data exclude virus reintroduction and possible polyclonal origins in the flock. Evolutionary rates estimated using the best-fit clock and parametric demographic models based on the gag gene have been conducted and include taxa belonging to genotypes E (8 sequences from 1999, 4 from 2006, and 10 from 2007) and B1 (8 sequences from 1999, and 9 from 2007). The rates estimated were 0.781·10−3 substitutions per site per year (95% highest posterior density interval, 1.207·10−4 to 1.486·10−3) for genotype E and 0.555·10−3 (7.836·10−5 to 1.175·10−3) for genotype B1 (BEAST version 1.4.7 software) (3).
As shown in Table 2, there were no differences in G-to-A transition proportions between genotype E and subtype B1 (accession numbers EU726488 through EU726525).
TABLE 2.
Comparison of diversities and proportions of G-to-A transitions of genotypes and collection periodsa
| Sample genotype | Yr (no. of sequences) | Mean no. of substitutions | Uncorrected P value (%) | Mean no. of G-to-A substitutions | Proportion (%) of G-to-A substitutions |
|---|---|---|---|---|---|
| E | 1999 (8) | 5.86 | 1.12 | 1.5 | 27.0 |
| 2007 (10) | 11.93 | 2.27 | 3.0 | 29.9 | |
| B1 | 1999 (8) | 8.39 | 1.60 | 2.32 | 27.3 |
| 2007 (9) | 14.21 | 2.71 | 4.61 | 32.9 |
Data show pairwise nucleotide diversity and proportion of G-to-A transitions between samples belonging to different genotypes and different collection periods.
Our results bore out the fact that the Roccaverano SRLV strain, even with natural deletions of the dUTPase and vpr-like genes, showed genetic features that were very similar to those of classical SRLV strains, reflecting the expected mutation rate of a prone-to-error RT. It is debatable whether CAEV RT might have affected the fidelity of genotype E replication; however, coinfection at a cellular level is a rare event, since only 1 × 105 to 1 × 106 monocytes are estimated to be infected by SRLV (2). Furthermore, the mean nucleotide diversity between sequences in flock It-09, in which only genotype E was present, was similar to that recorded in the coinfected flock.
The finding that genotype E persists in the population offers new insights for understanding the pathogenesis of SRLV infection and the role of dispensable viral proteins as virulence factors under natural conditions. Circulating monocytes latently infected by the dUTPase-deficient virus may be the result of an infection occurring in the myeloid precursors, providing a cellular environment typical of actively dividing cells, known to be necessary for compensating the lack of viral dUTPase (20). Therefore, it is important to establish the role of bone marrow as a virus reservoir in genotype E infection, which is still controversial in dUTPase-positive strains (6, 14). In addition, in all previous studies in which dUTPase, vpr-like, and U3 70-bp repeat sequences were independently deleted from the CAEV genome, a reduction in viral load and/or disease progression was recorded, thus providing indirect evidence that genotype E exhibits a low-pathogenicity potential in vivo (9, 11, 20).
Preliminary observations for the coinfected flock suggest that the presence of genotype E could hinder arthritis induced by CAEV strains, since animals showed no symptoms. Animals from the same area and the goat breed infected with only the B1 genotype showed an increased arthritic clinical index (personal observation), excluding possible breed resistance. Previous reports suggested that strains deleted of both dUTPase and vpr might induce superinfection resistance or decrease cell-associated viremia in different animal models (9, 23). Further studies are needed to elucidate the in vitro properties of the Roccaverano isolate as well as to evaluate its potential as a live attenuated vaccine strain.
Acknowledgments
This work was supported by Regione Piemonte, Progetto di Ricerca Sanitaria Finalizzata 2007.
We thank A. Quasso, veterinarian, Local Health Unit ASL AT-19, and the goat breeders for invaluable collaboration.
Footnotes
Published ahead of print on 5 November 2008.
REFERENCES
- 1.Barros, S. C., V. Andresdottir, and M. Fevereiro. 2005. Cellular specificity and replication rate of Maedi Visna virus in vitro can be controlled by LTR sequences. Arch. Virol. 150201-213. [DOI] [PubMed] [Google Scholar]
- 2.de Andres, D., N. J. Klein, E. Watt, S. Berriatua, B. Torsteinsdottir, B. A. Blacklaws, and G. D. Harkiss. 2005. Diagnostic tests for small ruminant lentiviruses. Vet. Microbiol. 10749-62. [DOI] [PubMed] [Google Scholar]
- 3.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gjerset, B., A. K. Storset, and E. Rimstad. 2006. Genetic diversity of small-ruminant lentiviruses: characterization of Norwegian isolates of caprine arthritis encephalitis virus. J. Gen. Virol. 87573-580. [DOI] [PubMed] [Google Scholar]
- 5.Grego, E., L. Bertolotti, A. Quasso, M. Profiti, D. Lacerenza, D. Muz, and S. Rosati. 2007. Genetic characterization of small ruminant lentivirus in Italian mixed flocks: evidence for a novel genotype circulating in a local goat population. J. Gen. Virol. 883423-3427. [DOI] [PubMed] [Google Scholar]
- 6.Grossi, P., C. Giudice, I. Bertoletti, G. Cioccarelli, E. Brocchi, G. Cammarata, and D. Gelmetti. 2005. Immunohistochemical detection of the p27 capsid protein of caprine arthritis-encephalitis virus (CAEV) in bone-marrow cells of seropositive goats. J. Comp. Pathol. 133197-200. [DOI] [PubMed] [Google Scholar]
- 7.Harmache, A., P. Russo, C. Vitu, F. Guiguen, J. F. Mornex, M. Pepin, R. Vigne, and M. Suzan. 1996. Replication in goats in vivo of caprine arthritis-encephalitis virus deleted in vif or tat genes: possible use of these deletion mutants as live vaccines. AIDS Res. Hum. Retrovir. 12409-411. [DOI] [PubMed] [Google Scholar]
- 8.Harmache, A., C. Vitu, P. Russo, M. Bouyac, C. Hieblot, P. Peveri, R. Vigne, and M. Suzan. 1995. The caprine arthritis encephalitis virus tat gene is dispensable for efficient viral replication in vitro and in vivo. J. Virol. 695445-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmache, A., C. Vitu, F. Guiguen, P. Russo, G. Bertoni, M. Pepin, R. Vigne, and M. Suzan. 1998. Priming with tat-deleted caprine arthritis encephalitis virus (CAEV) proviral DNA or live virus protects goats from challenge with pathogenic CAEV. J. Virol. 726796-6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy, B., D. P. Jasmer, S. N. White, and D. Knowles. 2007. Localization of a TNF-activated transcription site and interactions with the gamma activated site within the CAEV U3 70 base pair repeat. Virology 364196-207. [DOI] [PubMed] [Google Scholar]
- 11.Óskarsson, T., H. S. Hreggvidsdóttir, G. Agnarsdóttir, S. Matthíasdóttir, M. H. Ogmundsdóttir, S. R. Jónsson, G. Georgsson, S. Ingvarsson, O. S. Andrésson, and V. Andrésdóttir. 2007. Duplicated sequence motif in the long terminal repeat of maedi-visna virus extends cell tropism and is associated with neurovirulence. J. Virol. 814052-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pétursson, G., P. Turelli, S. Matthiasdottir, G. Georgsson, O. S. Andresson, S. Torsteinsdottir, R. Vigne, V. Andresdottir, E. Gunnarsson, G. Agnarsdottir, and G. Querat. 1998. Visna virus dUTPase is dispensable for neuropathogenicity. J. Virol. 721657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14817-818. [DOI] [PubMed] [Google Scholar]
- 14.Ravazzolo, A. P., C. Nenci, H. R. Vogt, A. Waldvogel, G. Obexer-Ruff, E. Peterhans, and G. Bertoni. 2006. Viral load, organ distribution, histopathological lesions, and cytokine mRNA expression in goats infected with a molecular clone of the caprine arthritis encephalitis virus. Virology 350116-127. [DOI] [PubMed] [Google Scholar]
- 15.Reina, R., M. I. Mora, I. Glaria, I. Garcia, C. Solano, L. Lujan, J. J. Badiola, A. Contreras, E. Berriatua, R. Juste, et al. 2006. Molecular characterization and phylogenetic study of maedi visna and caprine arthritis encephalitis viral sequences in sheep and goats from Spain. Virus Res. 121189-198. [DOI] [PubMed] [Google Scholar]
- 16.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 191572-1574. [DOI] [PubMed] [Google Scholar]
- 17.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP: DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 192496-2497. [DOI] [PubMed] [Google Scholar]
- 18.Shah, C., J. Boni, J. B. Huder, H. R. Vogt, J. Muhlherr, R. Zanoni, R. Miserez, H. Lutz, and J. Schupbach. 2004. Phylogenetic analysis and reclassification of caprine and ovine lentiviruses based on 104 new isolates: evidence for regular sheep-to-goat transmission and worldwide propagation through livestock trade. Virology 31912-26. [DOI] [PubMed] [Google Scholar]
- 19.Turelli, P., G. Pétursson, F. Guiguen, J. F. Mornex, R. Vigne, and G. Quérat. 1996. Replication properties of dUTPase-deficient mutants of caprine and ovine lentiviruses. J. Virol. 701213-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turelli, P., F. Guiguen, J. F. Mornex, R. Vigne, and G. Quérat. 1997. dUTPase-minus caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-A substitutions. J. Virol. 714522-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villet, S., B. A. Bouzar, T. Morin, G. Verdier, C. Legras, and Y. Chebloune. 2003. Maedi-visna virus and caprine arthritis encephalitis virus genomes encode a Vpr-like but no Tat protein. J. Virol. 779632-9638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanoni, R. G., I. M. Nauta, P. Kuhnert, U. Pauli, B. Pohl, and E. Peterhans. 1992. Genomic heterogeneity of small ruminant lentiviruses detected by PCR. Vet. Microbiol. 33341-351. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, Z., J. Guo, Y. Ni, F. W. Bazer, L. Giavedoni, and A. de la Concha-Bermejillo. 2003. Construction and characterization of a recombinant ovine lentivirus carrying the optimized green fluorescent protein gene at the dUTPase locus. Arch. Virol. 1481485-1506. [DOI] [PubMed] [Google Scholar]