Abstract
The hepatitis C virus (HCV), which currently infects an estimated 3% of people worldwide, has been present in some human populations for several centuries, notably HCV genotypes 1 and 2 in West Africa and genotype 6 in Southeast Asia. Here we use newly developed methods of sequence analysis to conduct the first comprehensive investigation of the epidemic and evolutionary history of HCV in Asia. Our analysis includes new HCV core (n = 16) and NS5B (n = 14) gene sequences, obtained from serum samples of jaundiced patients from Laos. These exceptionally diverse isolates were analyzed in conjunction with all available reference strains using phylogenetic and Bayesian coalescent methods. We performed statistical tests of phylogeographic structure and applied a recently developed “relaxed molecular clock” approach to HCV for the first time, which indicated an unexpectedly high degree of rate variation. Our results reveal a >1,000-year-long development of genotype 6 in Asia, characterized by substantial phylogeographic structure and two distinct phases of epidemic history, before and during the 20th century. We conclude that HCV lineages representing preexisting and spatially restricted strains were involved in multiple, independent local epidemics during the 20th century. Our analysis explains the generation and maintenance of HCV diversity in Asia and could provide a template for further investigations of HCV spread in other regions.
Hepatitis C virus (HCV) is a prevalent and globally distributed human pathogen that currently infects an estimated 120 to 180 million people, or 2 to 3% of the world's population (4, 39). Chronic HCV infection significantly increases the risk of liver cirrhosis and hepatocellular carcinoma (18), causing substantial morbidity and mortality worldwide. HCV is the most common chronic blood-borne infection in the United States, causing an estimated 8,000 to 10,000 deaths per year, and this figure is expected to increase substantially over the next 20 years (67).
HCV is a positive-sense RNA virus belonging to the genus Hepacivirus in the family Flaviviridae that, at present, has not been found to naturally infect any species other than humans. The virus exhibits a very high degree of genetic diversity that is classified by phylogenetic analysis into six genotypes, denoted 1 to 6, each of which contains numerous subtypes, denoted 1a, 2c, 3d, 6f, etc. (50, 51). The recent discovery of a putative seventh genotype (33) suggests that further HCV diversity remains to be characterized.
The various genotypes and subtypes of HCV have been associated with different epidemiological and geographical patterns (31, 51, 54). For example, a high proportion of infections are caused by just a few strains that are globally distributed but genetically conserved, notably subtypes 1a, 1b, 2a, 2b, and 3a. Evolutionary molecular clock methods suggest that these so-called epidemic subtypes originated around 100 years ago (43, 45, 54, 59). These probably spread due to their association with new and efficient routes of viral transmission that arose during the 20th century, notably blood transfusion, hemodialysis, use of blood products, injection drug use, and nonsterile medical injections (e.g., see references 13, 15, and 45). Much research attention has focused on these strains, not least because they account for most HCV infections in Europe, North America, Japan, and Australasia. However, an understanding of the evolution and genetic diversity of all genotypes is necessary for the development of successful drug and vaccine treatments. For example, genotype 1 infections respond more poorly than genotypes 2 and 3 to antiviral drugs (11, 30), and the efficacies of cellular immune responses may also vary among strains.
In contrast to the epidemic scenario described above, HCV lineages in areas of endemicity are highly divergent and are typically isolated from residents or emigrants of restricted (and sometimes remote) geographic regions, suggesting a long duration of continuous infection in these areas. Endemic strains belonging to genotypes 1 and 2 are found in West Africa (2, 20, 47, 66). Similar regional patterns of endemic diversity have been found for genotype 3 in South Asia, for genotype 4 in Central Africa and the Middle East, and for genotype 6 in East Asia (28, 32, 36). However, no region has yet been found to contain high levels of HCV genotype 5 genetic diversity (64). Molecular clock analyses suggest that these strains have been present in their respective geographical regions for at least several centuries (43, 54).
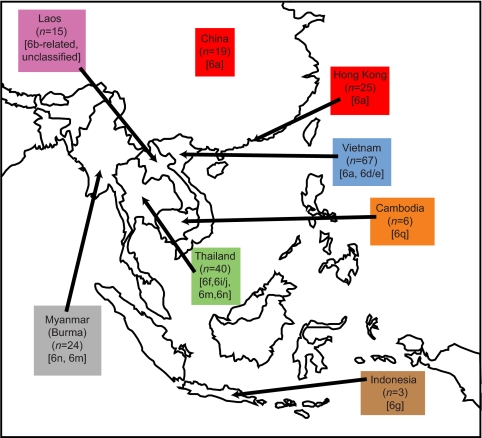
Genotype 6 provides a striking example of HCV diversity in endemic areas. Indeed, the first HCV isolates from East Asia were so divergent that they were initially classified as separate genotypes, designated 7, 8, 9, and 11 (1, 50, 61, 62, 63). These strains have since been reclassified as individual subtypes within genotype 6 (52). Genotype 6 infections are also of considerable epidemiological importance: there are an estimated 62 million HCV-infected people in the WHO-defined Western Pacific region (68), which represents approximately one-third of all infections worldwide. As illustrated in Fig. 1, genotype 6 isolates have been obtained from residents or emigrants of Thailand, India, Cambodia, Laos, Myanmar (Burma),Vietnam, China, Hong Kong, and Indonesia (25, 46). The prevalence of HCV in the general population is variable among East Asian countries, ranging from about 0.5% in Singapore and Hong Kong, to around 6% in Vietnam and Thailand (69), and exceeding 10% in Myanmar (29). The reported prevalence in China is approximately 2 to 3%, which amounts to approximately 30 million people (22, 70). Of course, not all HCV infections in East Asia are caused by genotype 6, and the genotype distribution of HCV infection is variable among and within different East Asian countries. For example, genotype 6 appears to be the most frequent genotype in Myanmar (49% of infections) (29) and Vietnam (52% of infections) (37), but not in Thailand, where the globally distributed subtype 3a, which is associated with injection drug use, is twice as common as genotype 6 (23). The most common strain in China is the global subtype 1b (5), although subtype 6 is found at higher frequencies in southern China (27) and Hong Kong (42, 71).
FIG. 1.
A map of East Asia illustrating the geographic distribution of the NS5B gene data set (see also Fig. 3). Arrows indicate the countries of origin of the sequences and values in parentheses indicate the number of sequences from each country. Common subtypes present in each country (>20% of sequences) are shown in brackets. Note that the subtype distribution of NS5B sequences does not necessarily reflect the relative prevalence of infection by each subtype. Twelve isolates were sampled outside of Asia; available information indicates that at least seven of these were obtained from individuals of Asian origin.
Despite the recent completion of full genome sequences for several HCV genotype 6 subtypes (28), our understanding of the evolutionary and epidemic history of genotype 6 is still deficient, for several reasons. First, genotype 6 diversity is well characterized for some East Asian countries (e.g., Vietnam and Thailand) but is poorly sampled or absent for other countries (Fig. 1). In this paper we improve this situation by reporting the isolation and sequencing of a panel of highly divergent genotype 6 strains from Laos. This is the first time the genetic diversity of HCV in Laos has been characterized. Second, phylogenetic analyses have been typically small in scale and performed on ad hoc bases, whereas in this paper we use all previously published core and NS5B gene sequences to investigate the epidemic history of genotype 6. Third, there has been little consideration of the spatial structure of past HCV infection in Asia. The phylogenetic distribution of strains from different locations—viral phylogeography—contains information about the historical movement of viral lineages (16, 35), which in turn sheds lights on contemporary patterns of transmission. To rectify this we used newly developed statistical tests (38) to investigate the geographic structure of genotype 6 diversity. Fourth, previous estimates of the time scale of HCV infection in Asia (43, 54) used limited data sets and potentially unrealistic assumptions during the analyses. Most importantly, earlier analyses relied on the assumption of a constant rate of HCV evolution (a strict molecular clock). We show here that the strict clock hypothesis is inappropriate for HCV and we instead have employed recently developed relaxed clock evolutionary models that incorporate variation in evolutionary rates (8). In addition, previous estimates of genotype 6 history were based on single reconstructed phylogenies (43, 54) and therefore underestimated the statistical error that may arise from phylogenetic reconstruction. We address this situation here by using more advanced Bayesian inference methods that explicitly incorporate phylogenetic uncertainty (8).
We have investigated the transmission history of HCV in Asia by conducting a comprehensive evolutionary analysis of available HCV genotype 6 gene sequences. By combining sophisticated molecular clock, coalescent, and geographical analyses, we show that genotype 6 in Asia is structured by both geography and by an explosion of local epidemics that occurred during the 20th century. The genetic diversity of our new Lao isolates indicates a pattern of past HCV transmission distinct from that found elsewhere in Southeast Asia.
MATERIALS AND METHODS
Sampling, isolation, and sequencing of Lao HCV infections.
Serum samples were obtained from 31 patients admitted to Mahosot Hospital, Vientiane, Laos, and were stored at −80°C. The patients presented with jaundice or elevated liver transaminases (>3 times the upper normal limit) and were positive for HCV antibodies (Serodia HCV antibody microtiter particle agglutination kit; Fujirebio Inc). Informed consent was obtained from each patient, and ethical approval was granted by the Ethical Review Committee of the Faculty of Medical Sciences, National University of Laos, Vientiane. Viral RNA was extracted from 140 μl of serum, using the QIAmp viral RNA extraction kit (Qiagen) according to the manufacturer's protocol.
Viral RNA was reverse transcribed using the SuperScript II reverse transcriptase protocol (Invitrogen). In brief, 10 μl viral RNA, 0.5 μl of deoxynucleoside triphosphates (25 mM each), 0.5 μl random primers (500 μg/ml), and 1 μl sterile water were heated to 65°C for 5 min and then quick-chilled on ice. Four μl of 5× first-strand buffer, 2 μl dithiothreitol (0.1 M), and 1 μl RNasin were added and incubated at room temperature for 2 min, followed by addition of 1 μl SuperScript II reverse transcriptase. The mixture was incubated at 42°C for 50 min and then at 70°C for 15 min. cDNA was amplified by nested PCR, using the High-Fidelity Expand PCR system (Roche). Serum samples from healthy individuals were used as negative controls.
Table 1 provides full details of the primers used during each round of amplification for each subgenomic region. Amplification of the 5′ untranslated region (UTR; 236 bp) was used to define HCV RNA positivity; 5′UTR PCR conditions for both rounds were 94°C for 2 min, 30 cycles of 94°C for 25 s, 55°C for 25 s, and 72°C for 25 s, and then 72°C for 2 min. HCV core gene (464 bp) amplification was performed; PCR conditions for both rounds were 94°C for 2 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s, and then 72°C for 7 min. HCV NS5B gene (377 bp) amplification was performed; PCR conditions for both rounds were 94°C for 2 min, 28 rounds of 94°C for 15 s, 60°C for 30 s, and 72°C for 45 s, and then 72°C for 7 min. Amplicons were run on 1% agarose gels stained with ethidium bromide, and DNA was purified using the Qiagen gel extraction kit. Purified DNA was sequenced in both forward and reverse directions using the ABI Prism Big Dye Terminator cycle sequencing kit (Perkin-Elmer Applied Biosciences).
TABLE 1.
Details of HCV primers for each subgenomic region
| Viral region | PCR round | Primer name | Primer sequence |
|---|---|---|---|
| 5′UTR | First | 5′UTR ExF 1 | CCCTGTGAGGAACTWCTGTCTTCACGC |
| 5′UTR ExR 2 | GGTGCACGGTCTACGAGACCT | ||
| Second | 5′UTR InF 3 | TCTAGCCATGGCGTTAGTRYGAG | |
| 5′UTR InR 4 | CACTCGCAAGCACCCTATCAGGCAGT | ||
| CORE | First | GEN6_EXF | ATCACTCCCCTGTGAGGAACTACTGT |
| GEN6_EXR2 | CCCTGTTGCATA(AG)TT(AG)ATCCCGTC | ||
| Second | GEN6_INTF | ACTGCCTGATAGGGTGCTTGCG | |
| GEN6_INTR | ATGTACCCCATGAGGTCGG | ||
| NS5Ba | First | ENO2_NS5BF1 | TGGG(GC)TT(CT)(GT)C(GC)TATGA(CT)AC(CT)CG(AC)TG(CT)TTTGA |
| ENO4_NS5BR1 | A(AG)TACCT(AG)GTCATAGCCTCCGTGAA | ||
| Second | NS5S3_NS5BF2 | TATGATACCCGCTGCTTTGACTCCAC | |
| NS5A5_NS5BR2 | GTCATAGCCTCCGTGAAGGCTC |
As previously described by Lu et al. (27).
Phylogenetic analysis of the core and NS5B data sets.
All available genotype 6 sequences were downloaded from the Los Alamos HCV database (25). Database sequences were retained if they spanned either the core or NS5B subgenomic regions obtained for the Lao isolates (see above). Only one sequence from each infected individual was retained, and sequences from experimental chimpanzee infections were excluded. Database reference sequences were then cropped, collated with the newly generated Lao sequences, and aligned by hand in Se-Al (available from http://tree.bio.ed.ac.uk). The resulting core alignment was 399 nt long and contained 230 sequences, and the NS5B alignment was 378 nt long and contained 211 sequences. A third alignment was constructed by concatenating the core and NS5B sequences of all isolates that had been sequenced in both genome regions. This concatenated alignment was 777 nt long and contained 127 sequences.
Maximum likelihood phylogenies were estimated for the core and NS5B data sets using PAUP (58). The most appropriate nucleotide substitution model for phylogenetic analysis was determined using the model selection procedure implemented in the program MODELTEST; for both data sets the best-fitting model was GTR+Γ+I (41). Under this model, phylogenies were heuristically searched using the SPR (subtree pruning and regrafting) and NNI (nearest neighbor interchange) perturbation algorithms. The statistical robustness levels of phylogenetic groupings were subsequently assessed using bootstrap analyses (500 replicates). Phylogeographic structure was then identified using FigTree (available from http://tree.bio.ed.ac.uk), and clades and lineages were colored according to their location of sampling.
Estimation of evolutionary time scale.
We used an evolutionary molecular clock to estimate the time scale of HCV epidemic history in East Asia. Previous evolutionary analyses of HCV have employed the simplest strict clock model, which assumes that all phylogeny branches evolve at exactly the same rate. This potentially unrealistic assumption can be avoided by using a relaxed clock, which allows evolutionary rates to vary among lineages according to some probability distribution (8).
Because there was insufficient temporal structure in the study sequences to directly estimate rates of molecular evolution, we used the external rate calibration approach previously employed by Pybus et al. (44) and Hue et al. (19). First, we obtained an external estimate of the evolutionary rate of our 399-nt core gene fragment from an independent, previously published HCV data set (59) that does contain significant temporal structure. Posterior distributions of this rate were estimated under three clock models using the Bayesian MCMC approach implemented in the program BEAST: (i) strict clock, (ii) relaxed clock with an uncorrelated log normal rate distribution, and (iii) relaxed clock with an uncorrelated gamma rate distribution (8, 9). The exponential relaxed clock model is a special case of the gamma relaxed clock model. Second, these external rates were subsequently used to define normally distributed priors for the core gene evolutionary rate during our strict and relaxed clock analyses of the concatenated data set. Third, evolutionary rates for the NS5B gene region were estimated using a relative rate approach. Briefly, once we have specified a rate for the core region, we can use the relative diversity of the core and NS5B regions to estimate an NS5B rate, because the time scales for both regions are identical. Additionally, the core and NS5B gene regions were given independent among-site Γ rate distributions. In summary, the sequence evolution model used incorporated multiple levels of rate heterogeneity: among nucleotide sites, among genes, and among lineages.
Evolutionary analysis of the concatenated data set.
To estimate the epidemic history of HCV genotype 6, we analyzed the concatenated core and NS5B alignment using the Bayesian Skyline plot (BSP) approach, as implemented in the program BEAST (for details, see Pybus et al. [44] and Drummond et al. [8]). To test the robustness of our results to model specification, we estimated epidemic history under a number of different model combinations and then used Bayes factors to choose the statistically most appropriate model (57). Marginal posterior distributions were estimated for each model parameter using Bayesian MCMC inference. All MCMC analyses were run for at least 50 million states. MCMC chain convergence, effective sample sizes, and Bayes factors were computed and investigated using the program Tracer v1.4 (9). In addition, an independent maximum likelihood relative rates analysis was performed using HYPHYv0.99 (40), which confirmed that relative rates were sampled appropriately in the Bayesian MCMC analyses.
A Bayesian estimate of phylogeny was obtained from the posterior distribution of trees arising from the best-fitting BEAST analysis (see above). First, the program TreeAnnotator (9) was used to calculate the Bayesian posterior probabilities of each internal node. Second, these probabilities were multiplied for each phylogeny sampled during the MCMC analyses. Third, the phylogeny with the highest total was located. This phylogeny best summarizes the set of credible trees and is called the maximum clade support phylogeny. Because a relaxed clock was used in the Bayesian MCMC analysis, the branch lengths and node heights of the maximum clade support phylogeny are in units of years. See Drummond et al. (8) for further details.
Lastly, the program BaTS (38) was used to test for the presence of statistically significant phylogeographic structure. BaTS tests the null hypothesis of panmixis (i.e., no correlation between phylogeny and taxa location) by performing randomization tests on two tree-shaped statistics: the parsimony score (PS) (53) and the association index (AI) (65). The randomizations are performed across a credible set of trees; hence, BaTS correctly incorporates phylogenetic uncertainty when testing for phylogeographic structure. For our BaTS analyses we used the posterior distribution of trees arising from the best-fitting BEAST analysis of the concatenated data set (see above). Further methodological details have been provided by Parker et al. (38).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the new HCV sequences from Laos are EU420957 to EU420986.
RESULTS
Detection of HCV RNA in Laos samples.
HCV RNA was detected by amplification of the 5′UTR in 18 out of 31 patients with jaundice and detectable anti-HCV antibodies. As this assay is highly sensitive for the detection of HCV RNA across all genotypes, it is most likely that the HCV RNA-negative patients had spontaneously controlled HCV replication, either at the time of presentation (if they were acutely infected) or at some time in the past (if patients presented with jaundice due to other causes). Our spontaneous resolution rate of 42% is in line with previous reports of acute HCV infection in jaundiced patients (12). Of the 18 patients with detectable viremia, the core and NS5B regions were amplified in 16 and 14 patients, respectively. Our failure to amplify these regions in a few instances reflects the genotype 6-specific primers used and the higher variability of these regions compared to the 5′UTR.
Phylogenetic analysis of the core and NS5B data sets.
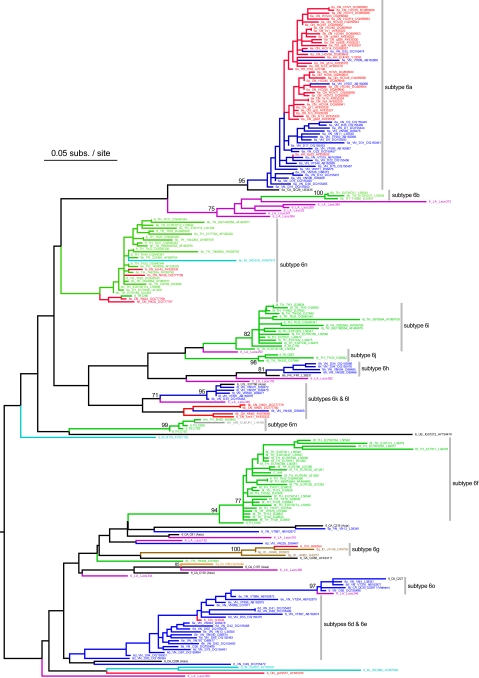
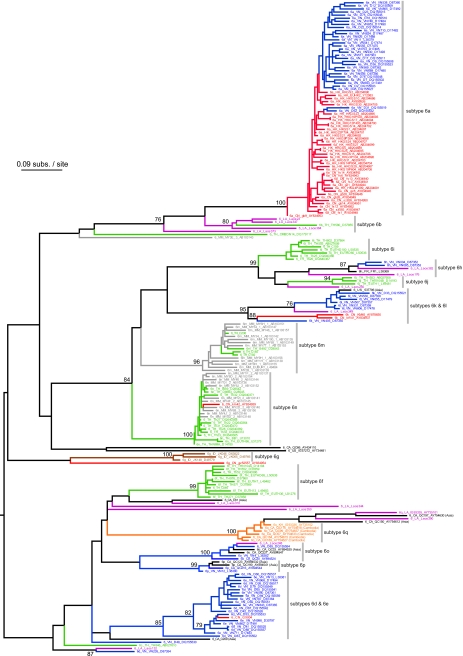
Figures 2 and 3 show the maximum likelihood phylogenies estimated from the core and NS5B alignments, respectively. As expected, the NS5B gene phylogeny is deeper and has a greater number of well-supported clades, reflecting the greater genetic variation of this region. In most cases sequences correctly group into their respective subtypes, although in both phylogenies strains belonging to subtypes 6d and 6e are mixed; we suggest this could be due to database annotation errors. In the core phylogeny, subtype 6o is incorrectly placed as a monophyletic ingroup of subtypes 6d and 6e; this is a phylogenetic estimation error most likely arising from limited sequence diversity.
FIG. 2.
Estimated maximum likelihood phylogeny for the core gene alignment. Bootstrap scores of >70% are shown next to well-supported nodes, and the phylogeny is midpoint rooted. Sequence names are given in the Los Alamos HCV Database format, as follows: subtype/country code/strain name/accession number. Where available, information is given in parentheses about the country of origin of emigrants from Southeast Asia to other countries. Gray bars indicate major subtypes of HCV genotype 6. Phylogeny branches and sequence names are colored according to the country of origin of the sampled individual. Country codes and colors for Asian strains are as follows: CN, China (red); VN, Vietnam (dark blue); HK, Hong Kong (red); TH, Thailand (green); IN, India (light blue); MM, Myanmar (gray); ID, Indonesia (brown); KH, Cambodia (orange); LA, Laos (magenta). Strains sampled outside of Asia are colored black (CA, Canada; FR, France; US, United States).
FIG. 3.
The estimated maximum likelihood phylogeny for the NS5B gene alignment. Bootstrap scores of >70% are shown next to well-supported nodes, and the phylogeny is midpoint rooted. See Fig. 2 for sequence naming details and the coloring scheme.
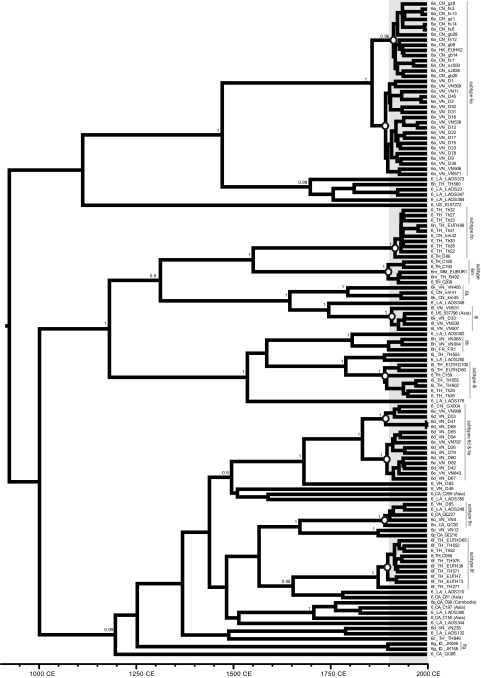
As previously suggested by Mellor et al. (32), HCV genotype 6 phylogenies show phylogeographic structure, that is, sequences tend to group together according to their location of sampling. In one instance, spatial structure can even be seen within a single subtype; subtype 6a strains from China and Vietnam are phylogenetically distinct in both trees. Subtype 6b and related strains are from Thailand and Laos. Subtype 6m and 6n strains are mostly from Thailand and Myanmar, and occasionally from China. Subtypes 6f, 6i, and 6j are dominated by Thai isolates, whereas subtypes 6d, 6e, 6o, 6p, and 6h are mostly from Vietnam. Subtype 6q and closely related sequences are from Cambodia and Laos, whereas subtype 6g strains are from Indonesia. Subtypes 6k and 6l are frequently from Vietnam but are also found in China and Laos. Several strains were sampled from patients residing outside Asia; whenever information on these individuals was available, it always indicated an Asian ethic origin or background.
Our new isolates from Laos are genetically diverse and are interspersed among many different genotype 6 lineages. There is only one well-supported cluster of Lao sequences (strains Laos373, Laos394, Laos259, Laos23, Laos347, and Laos38), which is most closely related to subtype 6b. The locations of the remaining Lao strains are as follows: Laos250 falls between subtypes 6i and 6j; Laos382 groups most closely with subtype 6 h; Laos349 clusters near subtype 6l; Laos248 belongs to subtype 6o; Laos132 groups with the divergent Vietnamese strain VN235; Laos310 clusters with strain C81 sampled in Canada from an Asian immigrant; Laos390 is similar to the Laos strain IG93335 and groups with subtype 6q sequences from Cambodia. Laos344, Laos350, and Laos176 are highly divergent and do not closely group with any other strains.
Estimation of evolutionary time scale.
We estimated a time scale for the evolution of HCV genotype 6 from an independent, previously published set of HCV sequences sampled at different times (59). Under the strict clock model, the estimated rate for our 399-nt core gene region was 1.78 × 10−4 substitutions/site/year (95% credible region, 1.11 × 10−4 to 2.6 × 10−4). A very similar estimate was obtained under the relaxed clock models, 1.72×10−4 substitutions/site/year (95% credible region, 0.91 × 10−4 to 2.7×10−4). These rate estimates were subsequently used as prior distributions in all subsequent BEAST analyses (see Materials and Methods for details).
Evolutionary analysis of the concatenated data set.
Evolutionary analysis of the concatenated core plus NS5B data set was performed in BEAST under a range of molecular clock and coalescent model combinations. Simple coalescent models (i.e., constant size, exponential growth) consistently performed very poorly in comparison to the Bayesian Skyline plot (log10 Bayes factors, >25) and are therefore not reported here (results available on request). Six remaining models were well-supported: (i) a strict clock with BSP of 5 steps, (ii) a strict clock with BSP of 10 steps, (iii) a log normal relaxed clock with BSP of 5 steps, (iv) a log normal relaxed clock with BSP of 10 steps, (v) a gamma relaxed clock with BSP of 5 steps, and (vi) a gamma relaxed clock with BSP of 10 steps.
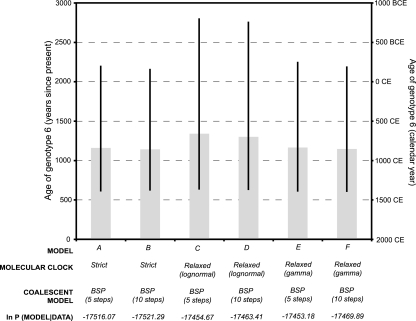
As shown in Fig. 4, all six model combinations gave similar median estimates for the age of HCV genotype 6, which was dated to ∼1,100 to 1,350 years ago. However, the 95% credible intervals for these estimates are large, ranging from ∼600 years ago to nearly 3,000 years ago, with the lower interval being less variable among models than the higher interval. However, these limits more accurately portray the true extent of statistical error than those reported in previous analyses (43, 54), which failed to use realistic models of sequence evolution or to incorporate uncertainty arising from phylogeny estimation. Our estimate of the date of the most recent common ancestor of genotype 6 is 400 years older than that reported by Pybus et al. (43), which likely reflects the much greater diversity of isolates considered here. Figure 4 also gives the estimated marginal likelihood of each model, calculated using Tracer v1.4, which represent the probability of each model combination given the data. Models C and E had substantially higher marginal likelihoods than the other models (log10 Bayes factors of >3.5). Model E has a slightly greater marginal likelihood than model C, but this difference is not considered significant (log10 Bayes factor, ∼0.5). For each clock model, the BSP with 5 steps was statistically favored over the BSP with 10 steps (Fig. 4).
FIG. 4.
Estimated dates of origin of genotype 6, as obtained under model combinations A to F (see text for details). The composition and marginal posterior log likelihood of each model combination are provided. The error bars are the 95% highest posterior density credible intervals for each estimate.
Figure 5 shows the maximum clade support phylogeny for the concatenated core plus NS5B data set, reconstructed from the phylogenies sampled under the best-supported model (i.e., combination E). Concatenation of the two genome regions leads to high posterior probabilities (P > 0.9) for many internal nodes. There is strong evidence (P = 0.96) that the Chinese subtype 6a sequences are monophyletic, but there is no such evidence for the Vietnamese subtype 6a strains (P = 0.5). Therefore, full-length sequences will be required to determine if subtype 6a originated in China/Hong Kong and then spread to Vietnam, or vice versa. Many lineages show rapid diversification during the 20th century (Fig. 5), giving rise to clusters of related sequences that are typically from the same location (Fig. 5). These clusters mostly correspond to the current subtype definitions. Subtypes such as 6q, 6 h, and 6k, which are poorly represented in the concatenated tree but well-represented in the core and NS5B trees, exhibit similar levels of genetic diversity and will therefore also have a 20th century origin. In contrast, many other genotype 6 lineages (e.g., Laos350, TH846, or QC66) show no such evidence of diversification during the 20th century.
FIG. 5.
The maximum clade support phylogeny of the concatenated (core plus NS5B) data set, obtained under the best-fitting model (combination E). Branch lengths represent time (see time scale at the bottom of the figure). Posterior probability scores (>0.9) are shown next to well-supported nodes. The shaded area corresponds to the 20th century, during which the lineages denoted by white circles showed rapid diversification. See Fig. 2 for sequence naming details.
We also obtained estimates of the molecular clock model parameters under the best-fitting model (combination E). The evolutionary rate of the core gene (1.8 × 10−4; 95% credible region, 0.9 × 10−4 to 2.9 × 10−4) is roughly two-thirds that of the NS5B gene (3.3 × 10−4; 95% credible region, 1.6 to × 10−4 to 5.1 × 10−4), in line with previous estimates (48). Rate heterogeneity among sites is slightly higher for the core region (α = 0.22) than for NS5B (α = 0.32). Rate heterogeneity among lineages is represented by the relaxed clock coefficient of variation (COV) parameter. Smaller COV values represent less rate variation among lineages and more clock-like evolution, hence the credible region of COV should abut zero if evolution follows a strict molecular clock. Our COV estimate is 0.37 (95% credible region, 0.28 to 0.45), which represents significant among-lineage rate variation and is similar to recently reported values for Dengue virus, human influenza A virus, and human immunodeficiency virus type 1 (8, 26). However, we might have expected to obtain lower COV values for HCV, because some previous studies reported that the hypothesis of a strict molecular clock is not always rejected (43). We therefore suggest that the failure of previous analyses to reject the strict clock was due to the comparatively small sample sizes used. In order to ensure accuracy, future evolutionary analyses of HCV data sets of sufficient size should incorporate rate variation among lineages. The relaxed clock covariance parameter is 0.02 (95% credible region, −0.11 to 0.14). This parameter measures the degree to which among-lineage rate variation is randomly distributed across the phylogeny, as opposed to being localized to specific clades. Our data suggest the former is true for HCV, because the estimated covariance is not significantly different from zero. Very similar COV and covariance values were obtained under the other relaxed clock models (combinations C, D, and F).
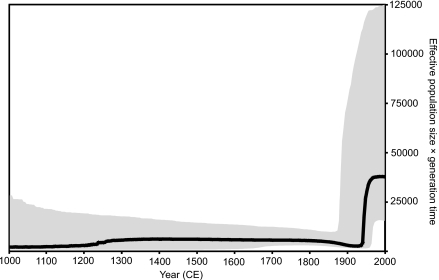
Figure 6 shows the BSP estimated from the concatenated data set. The BSP is a flexible, nonparametric estimate of past changes in effective population size (7). It is based on the coalescent process, a population genetic model that describes the relationship between the demographic history of a population and the ancestral relationships of sequences sampled from it (explained further in reference 6). The most notable feature of Fig. 6 is the change at the onset of the 20th century, from a low and relatively constant effective population size to rapid, epidemic growth. This change coincides with the onset of rapid diversification in the lineages highlighted in Fig. 3. The rate of growth appears to slow from the 1980s to the present, matching the change in HCV transmission that followed the virus' isolation in 1989, although this recent decrease is not statistically significant given the large confidence intervals. However, BSPs should be interpreted carefully when, as in this case, sequences have been sampled from a geographically structured population (3). Specifically, the 20th century growth phase shown in Fig. 5 was estimated from a heterogenous collection of lineages, some of which show diversification during the 20th century and others which do not (Fig. 3). Consequently, the exponential growth rate during this recent phase (i.e., between 1900 and 1980) (Fig. 6) is ∼0.035 year−1, considerably lower than equivalent rates previously estimated for more specific populations, such as genotype 4 in Egypt (∼0.25 year−1 [44]), subtype 1b in China (∼0.3 year−1 [34]), and subtypes 1a and 3a in the United Kingdom and United States (0.1 to 0.2 year−1 [45, 60]). The growth rate we have estimated is therefore likely to reflect an average rate for the whole of genotype 6 and should not be extrapolated to individual epidemic subtypes, which will have spread comparatively faster. For example, the exponential growth rate of subtype 6a in Hong Kong has been estimated at ∼0.17 year−1 (60), in agreement with the population-specific and subtype-specific rates listed above.
FIG. 6.
Bayesian skyline plot, showing the epidemic history of genotype 6 estimated from the concatenated (core plus NS5B) data set (see text for details). The thick black line represents the estimated effective population size through time. The gray area represents the 95% highest posterior density confidence intervals for this estimate.
Lastly, we undertook statistical tests for the presence of phylogeographic structure, using the PS and AI statistics, as implemented in the program BaTS (38). When all sequences were labeled according to their country of origin, both statistics strongly rejected the null hypothesis of panmixis (observed AI of 3.9, expected AI of 11.04 [P < 0.0001]; observed PS of 31.6, expected PS of 69.3 [P < 0.0001]). We also tested whether isolates from the same country were clustered together on the tree. Isolates from Vietnam, Thailand, and China showed very strong phylogenetic clustering (AI statistic P < 0.0001; PS statistic P < 0.0001). Isolates from Laos were also clustered but less significantly (AI, P = 0.01; PS, P = 0.04), while isolates from Canada, which represent emigrants from various Asian countries, were not significantly clustered (AI, P > 0.5; PS, P > 0.5).
DISCUSSION
Taken together, our phylogenetic, geographic, and coalescent analyses provide a coherent picture of the epidemic history of hepatitis C virus in East Asia, which can be split into two distinct phases, pre- and post-1900. It is currently thought that HCV spread rapidly worldwide during the 20th century via multiple transmission routes, including blood transfusion, blood products, injection drug use, and unsafe medical injections (15). Our results demonstrate that the effect on genotype 6 of this explosion in transmission was to rapidly increase the prevalence of some, but not all, preexisting lineages in areas of endemicity, giving rise to many distinct clusters of sequences (Fig. 5) that largely equate to the current HCV subtype definitions. As Fig. 2 and 3 show, these clusters are typically dominated by sequences from a single country (e.g., subtype 6d from Vietnam or subtype 6q from Cambodia) or less often, from two neighboring countries (e.g., subtype 6a from China/Vietnam or subtype 6n from Thailand/Myanmar). Thus, both the distinctive shape and the spatial structuring of the genotype 6 tree are products of the same underlying epidemiological process.
The genotype 6 clusters highlighted in Fig. 2 can be described as “local epidemic” strains, which transmitted rapidly during the 20th century in specific locations but did not spread internationally in the manner of the “global epidemic” subtypes 1a, 1b, and 3a (56). It is therefore probable that local epidemic lineages are characterized by transmission routes that are different and more varied than the routes associated with global epidemic subtypes. Local epidemic strains have previously been noted in Africa, particularly subtype 4a in Egypt, which has been associated with large-scale injectable antischistosomiasis treatment campaigns (10, 44). Our results indicate that local epidemic subtypes are also common to Asia. Since it is reasonable to assume that similar epidemiological factors have affected other HCV genotypes, we further argue that all common HCV subtypes are the result of selective amplification of endemic lineages, either locally or globally, during the 20th century. This hypothesis can explain why HCV subtypes contain unusually similar levels of genetic diversity and why they are highly phylogenetically distinct.
In the context of the scenario described above, the pattern of HCV genetic diversity in Laos is unusual, since there are no discernible “20th century” clusters of Lao sequences (Fig. 4). This is unlikely to be a sample size artifact, since smaller or equivalent-sized samples from other countries are sufficient to identify tight sequence clusters. Our phylogeographic analysis indicates that the Lao strains are clustered, but less strongly than strains from Vietnam, Thailand, or China. Five Lao isolates group together with the subtype 6b isolate TH580, but the branching events in this cluster substantially predate the 20th century (Fig. 4). Therefore, HCV in Laos appears to have been less involved with whatever events amplified endemic genotype 6 lineages elsewhere in Asia; the low prevalence of HCV in Laos (∼1%) compared to nearby countries also supports this notion (21, 69). Although we can only speculate on the reasons for this, differences in health care infrastructure among countries may be important, particularly if iatrogenic and nosocomial transmission has contributed to Asian HCV spread. We have been unable to find formal comparisons of investment in public health campaigns (such as vaccinations and mass treatment with injectable drugs) during the first half of the 20th century. However, the information available suggests that the French colonial authorities in Laos invested less in such public health campaigns than other mainland Southeast Asian countries (24, 55). In the second half of the 20th century Laos suffered civil war, extraordinarily destructive aerial bombing by the United States, and economic hardship in the wake of the war and the 1975 revolution, resulting in relatively low investment in public health intervention until the mid-1990s. Furthermore, before the 1990s the country had few reliable transport links (49). Hence, it is possible that the Lao population has experienced comparatively lower levels of mass exposure to blood-borne viruses such as HCV.
Our analyses show that genotype 6 infections worldwide are descended from a common ancestor that existed around 1,100 to 1,350 years earlier (95% credible region, 600 to >2,500 years ago) (Fig. 4). This long time scale is based on extrapolation of HCV evolution observed over a much shorter time span of about 25 years. Although there is no obvious reason why HCV rates should significantly vary through time—and our relaxed clock results suggest that they do not—we note that such estimates should be interpreted cautiously and are more likely to underestimate clade age than to overestimate it (17).
Prior to the 20th century, HCV transmission in Asia appears to have been characterized by long-term low-level infection in areas of endemicity (Fig. 6). We currently have almost no understanding of how stable, endemic transmission of HCV can be maintained for many centuries (46) and no idea how the virus spread across Asia from a common ancestor. Our results do indicate that endemic genotype 6 lineages were historically associated with different locations (e.g., subtype 6f in Thailand, 6g in Indonesia, 6d in Vietnam, and 6q in Cambodia), with multiple genotype 6 lineages being present in modern-day Vietnam, Thailand, and Laos (Fig. 5). In addition, the presence of old phylogenetic nodes that connect different country-specific lineages (Fig. 2, 3, and 5) suggests that at least some historical gene flow occurred. However, the significant spatial structure we observed demonstrates that genotype 6 gene flow is restricted. Furthermore, it appears to be limited by distance, as sequences from pairs of nearby nonadjacent countries (Thailand-Vietnam, Myanmar-Vietnam, Myanmar-Cambodia, China-Cambodia, or China-Thailand) do not tend to cluster with each other (Fig. 2 and 3). In contrast, isolates from Laos group with strains from several neighboring countries, as expected given Laos' geographically central position in mainland Southeast Asia. Similarly, strains from Myanmar are often found intermingled with those from neighboring Thailand. Of course, historical patterns of movement may not be well represented by classifications based on current political borders.
The evolutionary models employed in our analyses included a relaxed molecular clock that estimated the degree to which the rate of molecular evolution varies across a phylogenetic tree (8). The analyses indicated a larger-than-expected amount of rate variation for HCV. Incorporating this rate heterogeneity increases the accuracy of our estimates (8) and the realism of our analysis, and therefore we recommend that future phylogenetic analyses of HCV gene sequences use this, or a similar, approach. However, such methods cannot be reliably applied to small data sets or short sequences. It is common for HCV molecular epidemiological surveys to produce short subgenomic fragments around 300 nt long, which by themselves are insufficient to reliably estimate phylogenetic groupings (34, 52). The compromise solution used here was to increase statistical power by concatenating multiple subgenomic sequences, but at the expense of a reduction in the number of available reference strains for comparison. Since viral sequences in databases can prove useful for many years after their initial investigation, we encourage the standardization of the subgenomic regions used and the production of longer gene fragments per isolate.
Acknowledgments
This research was directly supported by a 2006 Royal Society Research Grant to O.G.P. and P.K. E.B. is supported by the Medical Research Council (United Kingdom), P.K., R.P., and P.N.N. are supported by The Wellcome Trust, and P.L. is supported by a Marie Curie IEF fellowship. P.K. was funded by the NIHR Biomedical Research Centre Programme and the James Martin School for the 21st Century. The research in the Lao PDR was part of the Wellcome Trust-Mahosot Hospital-Oxford Tropical Medicine Research Collaboration, funded by The Wellcome Trust.
We are very grateful to the participating patients, and to the doctors, nurses, and technical staff of Mahosot Hospital: Vimone Soukkhaserm, Mayfong Mayxay, Nicholas J. White, Amphay Phyaluanglath, Somphone Phannouvong, Pathila Inthepphavong, and Martin Stuart-Fox.
Footnotes
Published ahead of print on 29 October 2008.
REFERENCES
- 1.Bukh, J., R. H. Purcell, and R. H. Miller. 1993. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc. Natl. Acad. Sci. USA 908234-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Candotti, D., J. Temple, F. Sarkodie, and J. Allain. 2003. Frequent recovery and broad genotype 2 diversity characterize hepatitis C virus infection in Ghana, West Africa. J. Virol. 777914-7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrington, C. V., J. E. Foster, O. G. Pybus, S. N. Bennett, and E. C. Holmes. 2005. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J. Virol. 7914680-14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1998. Recommendations for prevention and control of hepatitic C virus (HCV) infection and HCV-related chronic disease. MMWR Recomm. Rep. 47(RR-19)1-39. [PubMed] [Google Scholar]
- 5.Chen, Y. D., M. Y. Liu, W. L. Yu, J. Q. Li, M. Peng, Q. Dai, X. Liu, and Z. Q. Zhou. 2002. Hepatitis C virus infections and genotypes in China. Hepatobiliary Pancreat. Dis. Int. 1194-201. [PubMed] [Google Scholar]
- 6.Drummond, A. J., O. G. Pybus, A. Rambaut, R. Forsberg, and A. G. Rodrigo. 2003. Measurably evolving populations. Trends Ecol. Evol. 18481-488. [Google Scholar]
- 7.Drummond, A. J., A. Rambaut, B. Shapiro, and O. G. Pybus. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 221185-1192. [DOI] [PubMed] [Google Scholar]
- 8.Drummond, A. J., S. Y. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, C., M. K. Mohamed, G. T. Strickland, D. Lavanchy, R. R. Arthur, L. S. Magder, T. El Khoby, Y. Abdel-Wahab, E. S. Aly Ohn, W. Anwar, and I. Sallam. 2000. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet 355887-891. [DOI] [PubMed] [Google Scholar]
- 11.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Lu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347975-982. [DOI] [PubMed] [Google Scholar]
- 12.Gerlach, J. T., H. M. Diepolder, R. Zachoval, N. H. Gruener, M. C. Jung, A. Ulsenheimer, W. W. Schraut, C. A. Schirren, M. Waechtler, M. Backmund, and G. R. Pape. 2003. Acute hepatitis C: high rate of both spontaneous and treatment-induced viral clearance. Gastroenterology 12580-88. [DOI] [PubMed] [Google Scholar]
- 13.Goedert, J. J., B. E. Chen, L. Preiss, L. M. Aledort, and P. S. Rosenberg. 2007. Reconstruction of the hepatitis C virus epidemic in the US hemophilia population, 1940-1990. Am. J. Epidemiol. 1651443-1453. [DOI] [PubMed] [Google Scholar]
- 14.Reference deleted.
- 15.Hauri, A. M., G. L. Armstrong, and Y. J. Hutin. 2004. The global burden of disease attributable to contaminated injections given in health care settings. Int. J. STD AIDS 157-16. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, E. C. 2004. The phylogeography of human viruses. Mol. Ecol. 4745-756. [DOI] [PubMed] [Google Scholar]
- 17.Holmes, E. C. 2003. Molecular clocks and the puzzle of RNA virus origins. J. Virol. 773893-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36S21-S29. [DOI] [PubMed] [Google Scholar]
- 19.Hué, S., D. Pillay, J. P. Clewley, and O. G. Pybus. 2005. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc. Natl. Acad. Sci. USA 1024425-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeannel, D., C. Fretz, Y. Traore, N. Kohdjo, A. Bigot, E. P. Gamy, G. Jourdan, K. Kourouma, G. Maertens, F. Fumoux, J. J. Fournel, and L. Stuyver. 1998. Evidence for high genetic diversity and long-term endemicity of hepatitis C virus genotypes 1 and 2 in West Africa. J. Med. Virol. 5592−97. [PubMed] [Google Scholar]
- 21.Jutavijittum, P., A. Yousukh, B. Samountry, K. Samountry, A. Ounavong, T. Thammavong, J. Keokhamphue, and K. Toriyama. 2007. Seroprevalence of hepatitis B and hepatitis C virus infections among Lao blood donors. Southeast Asian J. Trop. Med. Public Health 38674-679. [PubMed] [Google Scholar]
- 22.Kang, L. Y., Y. D. Sun, L. J. Hao, X. Y. Cao, and Q. C. Pan. 1997. Studies on epidemiology of the population with HCV and HEV infections and their epidemic factors in china. Chin. J. Infect. Dis. 1571−75. [In Chinese.] [Google Scholar]
- 23.Kanistanon, D., M. Neelamek, T. Dharakul, and S. Songsivilai. 1997. Genotypic distribution of hepatitis C virus in different regions of Thailand. J. Clin. Microbiol. 351772-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klauss, R. 1997. Laos: the case of a transitioning civil service system in a transitional economy. Civil Service Systems in Comparative Perspective, School of Public and Environmental Affairs, Indiana University, 5 to 8 April 1997. http://www.indiana.edu/∼csrc/klauss1.html.
- 25.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21379-384. [DOI] [PubMed] [Google Scholar]
- 26.Lemey, P., A. Rambaut, and O. G. Pybus. 2006. HIV evolutionary dynamics within and among hosts. AIDS Rev. 8125-140. [PubMed] [Google Scholar]
- 27.Lu, L., T. Nakano, Y. He, Y. Fu, C. H. Hagedorn, and B. H. Robertson. 2005. Hepatitis C virus genotype distribution in China: predominance of closely related subtype 1b isolates and existence of new genotype 6 variants. J. Med. Virol. 75538-549. [DOI] [PubMed] [Google Scholar]
- 28.Lu, L., C. Li, Y. Fu, F. Gao, O. G. Pybus, K. Abe, H. Okamoto, C. H. Hagedorn, and D. Murphy. 2007. Complete genomes of hepatitis C virus (HCV) subtypes 6c, 6l, 6o, 6p and 6q: completion of a full panel of genomes for HCV genotype 6. J. Gen. Virol. 881519-1525. [DOI] [PubMed] [Google Scholar]
- 29.Lwin, A. A., T. Shinji, M. Khin, N. Win, M. Obika, S. Okada, and N. Koide. 2007. Hepatitis C virus genotype distribution in Myanmar: predominance of genotype 6 and existence of new genotype 6 subtype. Hepatol. Res. 37337-345. [DOI] [PubMed] [Google Scholar]
- 30.Manns, M. P., J. G. McHutchison, S. C. Gordon, et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358958-965. [DOI] [PubMed] [Google Scholar]
- 31.McOmish, F., P. Yap, B. Dow, E. Follett, C. Seed, A. Keller, T. Cobain, T. Krusius, E. Kolho, and R. Naukkarinen. 1994. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J. Clin. Microbiol. 32884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellor, J., E. C. Holmes, L. M. Jarvis, P. L. Yap, and P. Simmonds. 1995. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. J. Gen. Virol. 762493-2507. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, D. G., B. Willems, M. Deschenes, N. Hilzenrat, R. Mousseau, and S. Sabbah. 2007. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5′ untranslated region sequences. J. Clin. Microbiol. 451102-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano, T., L. Lu, Y. He, Y. Fu, B. H. Robertson, and O. G. Pybus. 2006. Population genetic history of hepatitis C virus 1b infection in China. J. Gen. Virol. 8773-82. [DOI] [PubMed] [Google Scholar]
- 35.Nakano, T., L. Lu, P. Liu, and O. G. Pybus. 2004. Viral gene sequences reveal the variable history of hepatitis C virus infection among countries. J. Infect. Dis. 1901098-1108. [DOI] [PubMed] [Google Scholar]
- 36.Ndjomou, J., O. G. Pybus, and B. Matz. 2003. Phylogenetic analysis of hepatitis C virus isolates indicates a unique pattern of endemic infection in Cameroon. J. Gen. Virol. 842333-2341. [DOI] [PubMed] [Google Scholar]
- 37.Noppornpanth, S., T. X. Lien, Y. Poovorawan, S. L. Smits, A. D. Osterhaus, and B. L. Haagmans. 2006. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J. Virol. 807569-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker, J., A. Rambaut, and O. G. Pybus. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8239-246. [DOI] [PubMed] [Google Scholar]
- 39.Perz, J. F., L. A. Farrington, C. Pecoraro, Y. J. F. Hutin, and G. L. Armstrong. 2004. Estimated global prevalence of hepatitis C virus infection. 42nd Annu. Meet. Infect. Dis. Soc. Am., Boston, MA, 30 September to 3 October 2004. Infectious Diseases Society of America, Arlington, VA.
- 40.Pond, S. L., S. D. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21676-679. [DOI] [PubMed] [Google Scholar]
- 41.Posado, D., and K. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14817-819. [DOI] [PubMed] [Google Scholar]
- 42.Prescott, L. E., P. Simmonds, C. L. Lai, N. K. Chan, I. Pike, P. L. Yap, and C. K. Lin. 1996. Detection and clinical features of hepatitis C virus type 6 infections in blood donors from Hong Kong. J. Med. Virol. 50168-175. [DOI] [PubMed] [Google Scholar]
- 43.Pybus, O. G., M. A. Charleston, S. Gupta, A. Rambaut, E. C. Holmes, and P. H. Harvey. 2001. The epidemic behavior of the hepatitis C virus. Science 2922323-2325. [DOI] [PubMed] [Google Scholar]
- 44.Pybus, O. G., A. J. Drummond, T. Nakano, B. H. Robertson, and A. Rambaut. 2003. The epidemiology and iatrogenic transmission of hepatitis C virus in Egypt: a Bayesian coalescent approach. Mol. Biol. Evol. 20381-387. [DOI] [PubMed] [Google Scholar]
- 45.Pybus, O. G., A. Cochrane, E. C. Holmes, and P. Simmonds. 2005. The hepatitis C virus epidemic among injecting drug users. Infect. Genet. Evol. 5131-139. [DOI] [PubMed] [Google Scholar]
- 46.Pybus, O. G., P. V. Markov, A. Wu, and A. Tatem. 2007. Investigating the endemic transmission of the hepatitis C virus. Int. J. Parasitol. 37839-849. [DOI] [PubMed] [Google Scholar]
- 47.Ruggieri, A., C. Argentini, F. Kouruma, P. Chionne, E. D'Ugo, E. Spada, S. Dettori, S. Sabbatani, and M. Rapicetta. 1996. Heterogeneity of hepatitis C virus genotype 2 variants in West Central Africa (Guinea Conakry). J. Gen. Virol. 772073-2076. [DOI] [PubMed] [Google Scholar]
- 48.Salemi, M., and A. M. Vandamme. 2002. Hepatitis C virus evolutionary patterns studied through analysis of full-genome sequences. J. Mol. Evol. 5462−70. [DOI] [PubMed] [Google Scholar]
- 49.Savada, A. M. (ed.). 1995. Laos: a country study, 3rd ed. Federal Research Division, Library of Congress, U.S. Government Printing Office, Washington, DC.
- 50.Simmonds, P., E. C. Holmes, T. A. Cha, S. W. Chan, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 742391−2399. [DOI] [PubMed] [Google Scholar]
- 51.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 853173-3188. [DOI] [PubMed] [Google Scholar]
- 52.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, et al. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42962-973. [DOI] [PubMed] [Google Scholar]
- 53.Slatkin, M., and W. P. Maddison. 1989. A cladistic measure of gene flow measured from the phylogenies of alleles. Genetics 123603−613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, D. B., S. Pathirana, F. Davidson, E. Lawlor, J. Power, P. L. Yap, and P. Simmonds. 1997. The origin of hepatitis C virus genotypes. J. Gen. Virol. 78321-328. [DOI] [PubMed] [Google Scholar]
- 55.Stuart-Fox, M. 1997. A history of Laos. Cambridge University Press, Cambridge, United Kingdom
- 56.Stumpf, M. P. H., and O. G. Pybus. 2002. Genetic diversity and models of viral evolution for the hepatitis C virus. FEMS Microbiol. Lett. 214143-152. [DOI] [PubMed] [Google Scholar]
- 57.Suchard, M. A., R. E. Weiss, and J. S. Sinsheimer. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 181001-1013. [DOI] [PubMed] [Google Scholar]
- 58.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 59.Tanaka, Y., K. Hanada, M. Mizokami, A. E. T. Yeo, J. Shih, T. Gojobori, and H. J. Alter. 2002. A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades. Proc. Natl. Acad. Sci. USA 9915584-15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka, Y., F. Kurbanov, S. Mano, E. Orito, V. Vargas, J. Esteban, M. Yuen, C. Lai, A. Kramvis, M. Kew, H. Smuts, S. Netesov, H. Alter, and M. Mizokami. 2006. Molecular tracing of the global hepatitis C virus epidemic predicts regional patterns of hepatocellular carcinoma mortality. Gastroenterology 13703-714. [DOI] [PubMed] [Google Scholar]
- 61.Tokita, H., H. Okamoto, F. Tsuda, P. Song, S. Nakata, T. Chosa, H. Iizuka, S. Mishiro, Y. Miyakawa, and M. Mayumi. 1994. Hepatitis C virus variants from Vietnam are classifiable into the seventh, eighth, and ninth major genetic groups. Proc. Natl. Acad. Sci. USA 9111022-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tokita, H., H. Okamoto, P. Luengrojanakul, K. Vareesangthip, T. Chainuvati, H. Iizuka, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1995. Hepatitis C virus variants from Thailand classifiable into five novel genotypes in the sixth (6b), seventh (7c, 7d) and ninth (9b, 9c) major genetic groups. J. Gen. Virol. 762329-2335. [DOI] [PubMed] [Google Scholar]
- 63.Tokita, H., H. Okamoto, H. Iizuka, J. Kishimoto, F. Tsuda, L. A. Lesmana, Y. Miyakawa, and M. Mayumi. 1996. Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J. Gen. Virol. 77293-301. [DOI] [PubMed] [Google Scholar]
- 64.Verbeeck, J., P. Maes, P. Lemey, O. G. Pybus, E. Wollants, E. Song, F. Nevens, J. Fevery, W. Delport, S. Van der Merwe, and M. Van Ranst. 2006. Investigating the origin and spread of hepatitis C virus genotype 5a. J. Virol. 804220-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, T. H., Y. K. Donaldson, R. P. Brettle, J. E. Bell, and P. Simmonds. 2001. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J. Virol. 7511686−11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wansbrough-Jones, M., E. Frimpong, B. Cant, K. Harris, M. Evans, and C. Teo. 1998. Prevalence and genotype of hepatitis C virus infection in pregnant women and blood donors in Ghana. Trans. R. Soc. Trop. Med. Hyg. 92496-499. [DOI] [PubMed] [Google Scholar]
- 67.Williams, I. 1999. Epidemiology of hepatitis C in the United States. Am. J. Med. 107(6B)2S-9S. [DOI] [PubMed] [Google Scholar]
- 68.World Health Organization. 1997. Hepatitis C. Wkly. Epidemiol. Rec. 7265-72.9115857 [Google Scholar]
- 69.World Health Organization. 1999. Hepatitis C: global prevalence (update). Wkly. Epidemiol. Rec. 74425-427. [PubMed] [Google Scholar]
- 70.Xia, G. L., C. B. Liu, H. L. Cao, et al. 1996. Prevalence of hepatitis B and C virus infections in the general Chinese population: results from a nationwide cross-sectional seroepidemiologic study of hepatitis A, B, C, D and E virus infections in China, 1992. Int. Hepatol. Commun. 562-73. [Google Scholar]
- 71.Zhou, D. X., J. W. Tang, I. M. Chu, J. L. Cheung, N. L. Tang, J. S. Tam, and P. K. Chan. 2006. Hepatitis C virus genotype distribution among intravenous drug user and the general population in Hong Kong. J. Med. Virol. 78574-581. [DOI] [PubMed] [Google Scholar]