Abstract
Adenovirus serotype 5 protein IX (pIX) has two domains connected by a flexible linker. Three N-terminal domains form triskelions on the capsid facets that cement hexons together, and the C-terminal domains of four monomers form complexes toward the facet periphery. Here we present a cryoelectron microscopy structure of recombinant adenovirus with a peptide tag added to the C terminus of pIX. The structure, made up by several C termini of pIX, is longer at both ends than the wild-type protein, and Fabs directed against the tag bind to both ends of the oligomer, demonstrating that the pIX C termini associate in an antiparallel manner.
Adenoviruses (Ad) are double-stranded DNA viruses which are found in all vertebrate species (6). More than 50 human serotypes provoke mild diseases and induce a robust and long-lasting immunity, which can be detrimental to the use of Ad vectors in substitutive gene therapy and anticancer treatments (1). The viral capsid is built up by 7 out of the 12 polypeptides present in the virion. The major structural component is the trimeric hexon, with 240 copies per virion that make up the 20 facets of the icosahedron (11). At each of the 12 vertices is a pentameric penton base and the externally projecting trimeric fiber. Among the four remaining minor proteins, protein IIIa and protein IX (pIX) have been tentatively positioned in the first three-dimensional (3D) electron microscopy (EM) map. It had been assumed that both protein IIIa and pIX were located on the outside of the capsid: IIIa would bridge two capsid facets, whereas pIX would stabilize the group-of-nine hexons as four pIX trimers per group-of-nine (11).
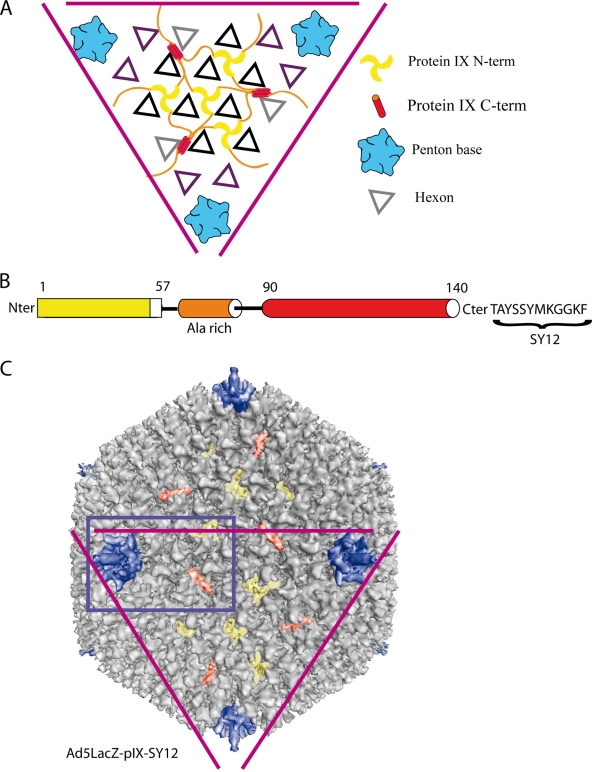
More recently, a new location for protein IIIa and pIX was proposed, based on a high-resolution cryo-EM 3D reconstruction of the Ad37 capsid, combined with secondary structure predictions. Protein IIIa is located inside the capsid and connected to the N-terminal domain of the penton base oriented inwards (9). pIX would form a connecting network of four trimers in the middle of each facet (9). The N-terminal domains of three pIX monomers form a triskelion structure that cements three hexons together (Fig. 1A and B). The C-terminal domains would be located near the edge between two facets and would form a tetramer at that position (Fig. 1A and B). The N- and C-terminal domains would be connected by the central alanine-rich domain. In this model, three out of the four C-terminal domains of pIX would associate together in a parallel structure, whereas the fourth domain would associate to the trimer in an antiparallel manner (Fig. 1A).
FIG. 1.
Structure of recombinant Ad5LacZ-pIX-SY12. (A) Schematic view of a human adenovirus facet, showing the protein organization as described by Saban et al. (9). (B) Schematic representation of the three domains of pIX, with the dodecapeptide (SY12) fused at its C-terminal end. (C) Eleven-Å-resolution 3D reconstruction of recombinant Ad5LacZ-pIX-SY12. The purple triangle highlights the same facet as in panel A. The blue rectangle corresponds to the part of the virus surface that is shown in Fig. 2A, B, and E.
In order to verify this hypothesis, we fused the dodecapeptide TAYSSYMKGGKF (abbreviated SY12) (5) to the C terminus of pIX (Fig. 1B), and a recombinant Ad5LacZ-pIX-SY12 vector was constructed as described previously (2, 8), amplified, and purified by CsCl gradient ultracentrifugation (3). Ad5LacZ-pIX-SY12 showed the same infectivity efficiency as wild-type (WT) virions, i.e., a ratio of 1:25 to 1:50 in terms of PFU to physical particles, and was as stable as the WT virus, as observed generally for other vectors carrying ligands at the pIX C terminus (12, 13).
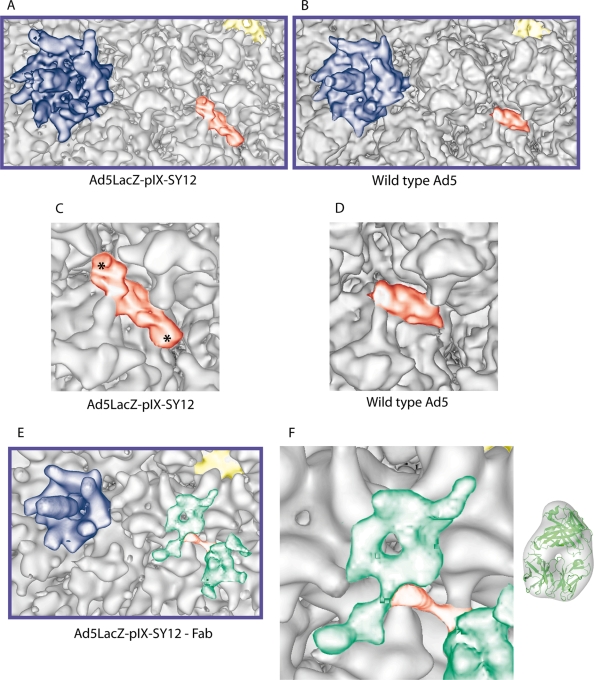
A sample of the Ad5LacZ-pIX-SY12 virion was prepared for cryo-EM observation as described previously (10). Two thousand, nine hundred forty-three single Ad particles out of a total of 4,905 coming from 18 micrographs imaged with a JEOL 2010 FEG electron microscope at a magnification of ×30,000 were used to generate the 3D structure of the pIX-modified virus (Fig. 1C). The overall 3D structure of this virus calculated at an 11-Å resolution (0.3 cutoff for the Fourier shell correlation) was similar to that of WT human Ad5 (4). However, the structure formed by the C-terminal domains of pIX had changed significantly (Fig. 2A to D). The cylindrical density in the Ad5LacZ-pIX-SY12 virus was lying more parallel to the capsid wall than the corresponding density in WT Ad5 (Fig. 2C and D) and was longer at both ends (Fig. 2C). The amino acid sequence of SY12-liganded pIX is 25% longer than that of WT pIX (62 versus 50 residues), whereas the C-terminal domain of SY12-liganded pIX is about 50% longer than the corresponding domain of WT pIX (Fig. 2C). This suggested that the extra density added by the dodecapeptide was present at both ends of the structure and that at least one of the four C-terminal domains of pIX associated with the others in an antiparallel way.
FIG. 2.
Comparison between the complexes formed by the C-terminal domains of pIX from Ad5LacZ-pIX-SY12 and WT Ad5. (A) Detailed view of the structure of SY12-tagged pIX within the capsid of recombinant Ad5LacZ-pIX-SY12 at an 11-Å resolution. The penton is in blue, and pIX is colored yellow and red (N- and C-terminal domains, respectively). (B) Detailed view of the structure of pIX in the WT human Ad5 capsid structure (EBI EM database accession number 1113) at a 10-Å resolution (4) in the same colors as used for panel A. (C) High magnification of SY12-tagged pIX C-terminal domain within the capsid of recombinant Ad5LacZ-pIX-SY12. The extra densities compared with the equivalent structure of WT Ad5 virus (D) are indicated with asterisks. (D) High magnification of WT pIX C-terminal domain in the capsid of Ad5. (E) Detailed view of SY12-tagged pIX within the complex formed between Ad5LacZ-pIX-SY12 virions and anti-SY12 Fabs, at a 22-Å resolution. The penton is in blue, and pIX is colored yellow and red (N and C-terminal domains, respectively). The Fab molecules are green. (F) Comparison between the green density in the EM map attributed to a Fab and a Fab crystal structure filtered to 22 Å.
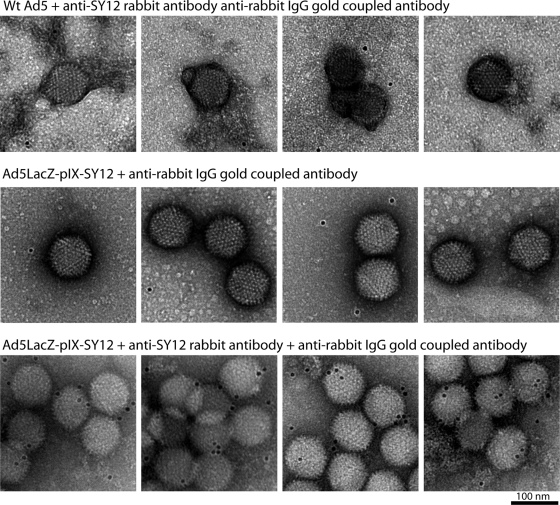
We then generated antibodies against the SY12 dodecapeptide (Eurogentec, Belgium). Synthetic SY12 was coupled to keyhole limpet hemocyanin and injected into rabbits according to a standard protocol (7). Anti-SY12 immunoglobulin G (IgG) was purified from hyperimmune serum by affinity chromatography on a Sepharose column with immobilized SY12 peptide. The absence of cross-reactivity between the anti-SY12 antibody and WT Ad5 was verified by immunogold labeling (Fig. 3). Purified antibody was then cleaved into Fab fragments by papain digestion and anti-SY12 Fabs recovered after chromatography on a protein A column. Ad5LacZ-pIX-SY12 was incubated with anti-SY12 Fabs in large excess (1:10 molar ratio), and the complex was imaged as described above. Using 949 particles out of 2,548 scanned Ad particles from 11 micrographs, we calculated a 3D reconstruction of the Ad5LacZ-pIX-SY12-Fab complex at a 22-Å resolution (0.3 cutoff for the Fourier shell correlation) (Fig. 2E). The cylindrical density attributed to the C-terminal domain of pIX mostly disappeared from the Ad5LacZ-pIX-SY12-Fab complex. Instead, two densities with the approximate sizes of Fab fragments were apparent on top of both ends of the previously present density Fig. 2E and F). The fact that there were two Fab densities located above both ends of the C-terminal density of pIX implied that certain polypeptide chains corresponding to the C-terminal domain of pIX were in a head-to-tail arrangement. These results, as well as results from our previous study of CAV-2 virions (10), confirm the model suggested by Saban et al. (9) shown in Fig. 1A; the three polypeptide chains of pIX that associate in a parallel orientation represent the C-terminal extensions of three out of the four triskelions present on one facet, whereas the fourth pIX C-terminal domain binds in an antiparallel orientation and comes from a neighboring facet.
FIG. 3.
Immunogold labeling of WT Ad5 and Ad5LacZ-pIX-SY12 viruses reacted with antirabbit gold-coupled IgG with or without primary anti-SY12 antibody. WT Ad5 incubated for 2 h with anti-SY12 antibody (top row), SY12-liganded virus incubated without primary antibody (middle row), and SY12-liganded virus incubated with anti-SY12 antibody (bottom row) were deposited on an electron microscopy grid, washed with phosphate-buffered saline, and incubated with anti-rabbit IgG antibody coupled to 6-nm gold particles for 1 min before uranyl acetate staining. WT Ad5 (top row) was not recognized by anti-SY12 antibody and showed isolated particles without gold labeling. A control sample of SY12-liganded virus reacted only with secondary antibody also showed unlabeled isolated particles. In contrast, SY12-liganded virus incubated with both primary and secondary antibodies showed aggregation due to interparticle bridging by bivalent primary antibody and gold labeling due to the secondary antibody.
pIX has been used as a platform for ligand insertion at its C terminus, with the aim of developing cell-targeted vectors for gene therapy (12, 13). One of the theoretical advantages for liganded pIX versus fiber was the high number of pIX copies per virion (240) compared to the fiber subunits (36). The model of Saban et al. (9), which is confirmed by the data presented here, implies that pIX-liganded vectors would expose two groups of ligands (in a head-to-tail orientation) on the two sides of each capsid edge. Considering that there are 30 edges per icosahedron, this would give 120 groups of ligands per adenoviral capsid that could be modified for retargeting purposes. In contrast, by increasing the distance between the ligand and the pIX C terminus via a long peptide linker, one might contribute to a better display and accessibility of each individual ligand of the cluster.
3D EM map accession numbers.
The 3D EM map of recombinant Ad5LacZ-pIX-SY12 has been deposited in the European Bioinformatics Institute (EBI) EM database under accession number 1574. The 3D EM map of the complex of Fab and virus has been deposited in the EBI EM database under accession number 1575.
Acknowledgments
G.S. was supported in part by a Jeunes Chercheurs grant from the Agence Nationale pour la Recherche (ANR-06-JCJC-0126). C.M.S.F. was financed through a MENRT fellowship from the French government. P.B. was supported by a grant from the French Cystic Fibrosis Foundation Vaincre la Mucoviscidose (VLM; TG-0702). C.M. was supported by a DGE grant. M.R.-C. was supported by a Contrat d'Interface grant from the Hospices Civils de Lyon and by a grant from La Ligue Contre le Cancer. We thank the Vector Core Facility of the University Hospital of Nantes, supported by the Association Française contre les Myopathies (AFM), for the production of the Ad5LacZ-pIX-SY12 vector.
Footnotes
Published ahead of print on 12 November 2008.
REFERENCES
- 1.Campos, S. K., and M. A. Barry. 2007. Current advances and future challenges in adenoviral vector biology and targeting. Curr. Gene Ther. 7189-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 704805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Defer, C., M. T. Belin, M. L. Caillet-Boudin, and P. Boulanger. 1990. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J. Virol. 643661-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabry, C. M., M. Rosa-Calatrava, J. F. Conway, C. Zubieta, S. Cusack, R. W. Ruigrok, and G. Schoehn. 2005. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 241645-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaden, F., L. Franqueville, M. K. Magnusson, S. S. Hong, M. D. Merten, L. Lindholm, and P. Boulanger. 2004. Gene transduction and cell entry pathway of fiber-modified adenovirus type 5 vectors carrying novel endocytic peptide ligands selected on human tracheal glandular cells. J. Virol. 787227-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwitz, M. 2001. Adenoviruses, p. 2149-2171. In B. Fields and D. Knipe (ed.), Fields virology, vol. 2. Raven Press, Philadelphia, PA. [Google Scholar]
- 7.Karayan, L., B. Gay, J. Gerfaux, and P. A. Boulanger. 1994. Oligomerization of recombinant penton base of adenovirus type 2 and its assembly with fiber in baculovirus-infected cells. Virology. 202782-795. [DOI] [PubMed] [Google Scholar]
- 8.Rosa-Calatrava, M., L. Grave, F. Puvion-Dutilleul, B. Chatton, and C. Kedinger. 2001. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 757131-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saban, S. D., M. Silvestry, G. R. Nemerow, and P. L. Stewart. 2006. Visualization of alpha-helices in a 6-ångstrom resolution cryoelectron microscopy structure of adenovirus allows refinement of capsid protein assignments. J. Virol. 8012049-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoehn, G., M. El Bakkouri, C. M. S. Fabry, O. Billet, L. F. Estrozi, L. Le, D. T. Curiel, A. V. Kajava, R. W. Ruigrok, and E. J. Kremer. 2008. Three-dimensional structure of canine adenovirus serotype 2 capsid. J. Virol. 823192-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart, P. L., R. M. Burnett, M. Cyrklaff, and S. D. Fuller. 1991. Image reconstruction reveals the complex molecular organization of adenovirus. Cell 67145-154. [DOI] [PubMed] [Google Scholar]
- 12.Vellinga, J., J. de Vrij, S. Myhre, T. Uil, P. Martineau, L. Lindholm, and R. C. Hoeben. 2007. Efficient incorporation of a functional hyper-stable single-chain antibody fragment protein-IX fusion in the adenovirus capsid. Gene Ther. 14664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vellinga, J., S. Van der Heijdt, and R. C. Hoeben. 2005. The adenovirus capsid: major progress in minor proteins. J. Gen. Virol. 861581-1588. [DOI] [PubMed] [Google Scholar]