Abstract
During human immunodeficiency virus type 1 (HIV-1) infection, patients develop various levels of neutralizing antibody (NAb) responses. In some cases, patient sera can potently neutralize diverse strains of HIV-1, but the antibody specificities that mediate this broad neutralization are not known, and their elucidation remains a formidable challenge. Due to variable and nonneutralizing determinants on the exterior envelope glycoprotein (Env), nonnative Env protein released from cells, and the glycan shielding that assembles in the context of the quaternary structure of the functional spike, HIV-1 Env elicits a myriad of binding antibodies. However, few of these antibodies can neutralize circulating viruses. We present a systematic analysis of the NAb specificities of a panel of HIV-1-positive sera, using methodologies that identify both conformational and continuous neutralization determinants on the HIV-1 Env protein. Characterization of sera included selective adsorption with native gp120 and specific point mutant variants, chimeric virus analysis, and peptide inhibition of viral neutralization. The gp120 protein was the major neutralizing determinant for most sera, although not all neutralization activity against all viruses could be identified. In some broadly neutralizing sera, the gp120-directed neutralization mapped to the CD4 binding region of gp120. In addition, we found evidence that regions of the gp120 coreceptor binding site may also be a target of neutralizing activity. Sera displaying limited neutralization breadth were mapped to the immunogenic V3 region of gp120. In a subset of sera, we also identified NAbs directed against the conserved, membrane-proximal external region of gp41. These data allow a more detailed understanding of the humoral responses to the HIV-1 Env protein and provide insights regarding the most relevant targets for HIV-1 vaccine design.
Most successful antiviral vaccines elicit neutralizing antibodies as a correlate of protection (45, 46). Often these vaccines do not need to deal with viral heterogeneity. Many viruses display an acute pattern of infection and do not acquire the more complex mechanisms of evasion of the humoral immune responses that evolve during the process of chronic infection in successive hosts. A notorious example of this is the human immunodeficiency virus (HIV), which possesses both extreme variability and immune evasion mechanisms, presenting unprecedented challenges for vaccine development (21).
Mapping the neutralization specificities of broadly neutralizing sera, and for HIV in particular, is a difficult but critical process for illuminating both mechanisms of pathogenesis and pathways leading to successful vaccine development. In terms of immunogen design and improvement, binding antibody analysis is also useful to determine immunodominance and, if undesired, subsequent immunogen modification. During the course of HIV type 1 (HIV-1) infection, neutralizing antibodies appear to be an important component of the host immune response (14, 34, 41). The sole viral targets for neutralizing antibodies are the trimeric and noncovalently associated HIV-1 envelope glycoproteins, gp120 and gp41, which together mediate receptor binding and entry. The exterior envelope glycoprotein gp120 binds to the primary receptor, CD4 (7), and coreceptor, either CCR5 or CXCR4 (1, 5, 9, 11-13, 35-37). Receptor-induced conformational changes in gp120 are transmitted in a yet-to-be-defined manner to the transmembrane glycoprotein gp41, which undergoes further conformational changes driving viral to target cell membrane fusion and entry.
Two classes of neutralizing antibodies specific for the HIV-1 Env protein are eventually elicited in most infected individuals: strain-restricted and broadly cross-reactive antibodies. The strain-restricted antibodies appear early after infection and are generally directed against linear determinants within the gp120 variable regions, including the second (V2) and the third (V3) regions (or loops) (23, 33); in principle, accumulation of an array of variable region-directed responses might mediate some increases in neutralization breadth. To date, only four broadly neutralizing antibodies isolated from HIV-infected individuals have been identified. The two most broadly neutralizing antibodies, 2F5 and 4E10, are directed against the membrane-proximal external region (MPER) of the HIV gp41 transmembrane glycoprotein (31, 48) (see Fig. S1 in the supplemental material). The other two broadly neutralizing antibodies, b12 and 2G12, are directed against the gp120 envelope glycoprotein. The b12 antibody (4) recognizes an epitope that overlaps with the CD4 binding site (CD4bs), and 2G12 (40) recognizes a cluster of glycans on the outer domain of gp120 (see Fig. S1 in the supplemental material).
These antibodies identify conserved regions on the envelope glycoproteins which currently are the only known broad neutralization targets. Therefore, it is logical that the initial attempts to map broadly neutralizing sera from HIV-infected individuals focus on these regions of the HIV envelope glycoproteins as we and others recently described (10, 27). The elucidation of the specific targets for broadly neutralizing antibodies elicited during natural HIV-1 infection will be informative to guide vaccine design to elicit such antibodies. Information from such analysis will be applicable to the analysis of neutralizing antibodies elicited by current vaccine candidates in either animal models or humans.
In HIV-infected individuals, the range of antibodies that bind to the HIV Env protein is extremely complex. The array of antibodies is comprised to a large extent of antibodies that bind to subunits of the functional spike, such as monomeric gp120 (42) or the gp41 six-helix bundle postfusogenic conformation (2, 44), but these antibodies do not neutralize the virus because their epitopes are not exposed or formed on the functional viral spike. Within gp120, many antibody responses are directed against immunodominant conserved regions involved in trimeric interactions in the context of the functional spike or against immunodominant variable sequences, some of which are occluded on most circulating isolates (as is, for example, the so-called V3 loop) (17, 20, 47), or others that are highly variable (the V1V2 loops). Within gp41, many antibodies are directed against the immunodominant cluster 1 region and recognize a postfusogenic conformation of gp41 not relevant to the prefusogenic epitopes displayed on that static viral envelope glycoprotein spike (2, 44). Thus, the presence of high levels of Env-binding, but nonneutralizing antibodies, increases the difficulty in analyzing the diverse array of antibodies circulating in the serum of HIV-1-infected individuals.
Here we describe a more detailed characterization of the broad neutralizing specificity detected in selected, but representative, HIV-1 patient sera that extends our previously reported results to better define both the binding and neutralizing specificities of selected sera previously partially analyzed (27) and in newly identified broadly neutralizing antisera. The analysis is predicated first upon the identification of relatively rare HIV-1 patient serum with broad neutralizing capacity as has recently been reported by several groups (10, 14, 27). Then, we use selective inhibition of the neutralization capacity, employing selected subcomponents of Env, to map the specificity of neutralization by what we have termed the process of selective adsorption (or selective peptide inhibition; see Fig. 1A). Following the selective adsorption, the neutralization capacity of the serum before and after the adsorption process is analyzed. In addition, we have employed chimeric viruses sensitive to a defined neutralization specificity within the gp41 MPER neutralizing determinant of Env, followed by peptide inhibition of the neutralization on wild-type isolates. Some details of this analysis we described in regards to the two most broadly neutralizing sera relatively recently (27); however, here we extend the analysis of these sera and broaden the scope of the study to other representative sera. The knowledge of protein structure, mutagenic analysis, high-throughput protein production, virologic modifications, and high-throughput neutralization assays are combined efficiently to ensure a comprehensive analysis of neutralizing specificities against known broad neutralizing determinants or against those not previously defined by monoclonal antibodies (MAbs) or in HIV-1 patient sera. When applicable, and importantly, each reagent and assay is “validated” using conformationally intact proteins and HIV-1 broadly neutralizing MAbs, extending significantly our original means of analysis. Here, we present data for the first time that indicate that there is perhaps accessibility of highly conserved regions of Env not recognized in previous studies. Rational approaches to dissect the neutralizing specificity in complex sera may be generally applicable to other variable viruses to define conserved neutralizing determinants for vaccine improvement (e.g., influenza virus) or development (e.g., hepatitis C virus).
FIG. 1.
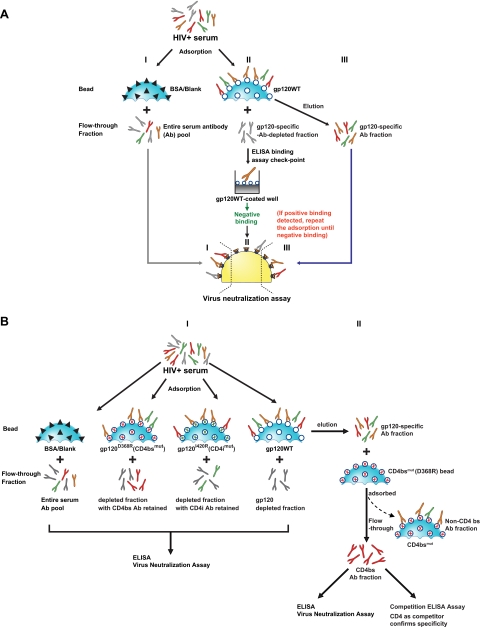
Schematic diagram of the solid-phase adsorption process. HIV+ serum, HIV-positive serum. (A) gp120-directed neutralizing antibody specificity analysis using solid-phase paramagnetic beads covalently conjugated to control protein, gp120, or gp120 point mutants. Serum antibodies are adsorbed with BSA (I) (black triangles) or wild-type gp120 protein (II) (open circles) conjugated to paramagnetic, solid-phase beads (blue sphere, cut-away segment depicted). The adsorbed serum is then analyzed by ELISA to confirm complete removal of gp120-specific antibodies, and the effect of adsorption on the serum HIV-1 neutralization titer (ID50) is then measured. A substantial decrease in ID50 following adsorption with the gp120WT beads indicates gp120-directed neutralizing activity. (III) gp120-specific antibodies are eluted and tested for binding to gp120 and HIV-1 neutralization capacity. (B) Experimental schema for the fine mapping of gp120-directed neutralizing specificity. (I) Differential adsorption using gp120WT or selected and specific gp120 mutant proteins. Following adsorption, ELISA binding or neutralization assays are performed to analyze the effects on the serum. CD4bs mutant protein D368R (red cross) and coreceptor binding site (CD4i) mutant I420R (green cross) protein are used to determinate the cognate specificity. (II) Schematic depiction of the process to further fractionate antibodies in the serum. Following adsorption, the antibodies are eluted from gp120WT-coupled beads followed by adsorption with D368R-coupled beads to enrich for CD4bs antibodies in the “flowthrough” which can be confirmed by increases in neutralization potency and more efficient cross competition by the fractioned antibodies in regards to the binding of sCD4 to gp120 target proteins.
MATERIALS AND METHODS
HIV-positive sera and virus neutralization assay.
Sera from chronically HIV-1-infected patients, infected prior to 1999, were acquired from a cohort of patients studied at the National Institutes of Health Bethesda campus as previously described. Based on previously described criteria of plasma RNA copy number and peripheral blood CD4 T-cell counts, the patients were classified as long-term nonprogressors (sometimes referred to as elite controllers), slow progressors (SP), or progressors (30). In addition to the previous set of sera, three other sera derived from SP individuals were added to the analysis (US12, US21, and B7B5). Sera were prepared as previously described and incubated at 55°C for 1 h to heat inactivate complements prior to the virus neutralization assay (29). HIV-1 neutralization assays were mostly conducted using a single round of infection with the HIV-1 Env pseudovirus assay described previously (26, 29). In Fig. 2 and elsewhere, the clade B molecular clones used were HXBc2, SF162, ADA, JRFL, BaL.01, YU2, SC422661.8, 6535.3, and PVO.04, molecular clone RW20.02 was used for clade A, and ZA012.29 was used for clade C.
FIG. 2.
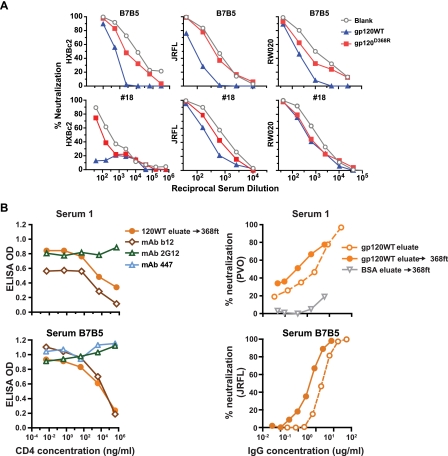
CD4bs neutralizing antibody specificity of sera analyzed by differential adsorption. The CD4bs point mutant protein harboring a D-to-R substitution at residue 368 (gp120D368R) was coupled to paramagnetic beads for differential adsorption of serum samples and compared to adsorption using gp120WT protein as depicted schematically in Fig. 1B. (A) Neutralization curves for serum B7B5 and serum 18 are shown against the neutralization-sensitive clade B virus HXBc2, the more resistant clade B virus JRFL, and the clade A virus RW020.2. Control adsorptions with blank beads are shown. Note that the less effective serum adsorption by gp120D368R protein compared to that of gp120WT protein suggests that antibodies to the CD4bs of gp120 are contributing to virus neutralization. This pattern was seen for serum B7B5 against all three viruses tested. However, the neutralization in serum 18 mapped strongly to the CD4bs against HXBc2 virus but less so for JRFL, and there was no difference against RW020.2 virus. (B) Characterization of serum B7B5 and 1 antibodies eluted from gp120WT-coupled beads followed by adsorption with D368R-coupled beads to enrich for CD4bs antibodies. The enriched CD4bs antibody fraction is verified by competition ELISA assay (bottom left) using soluble CD4 as a competitor for binding to the CD4 binding region of BaL gp120 (coated on the plate). MAbs b12 and 2G12 are controls, and the data show that increasing concentrations of sCD4 have no effect on 2G12 binding but completely block binding of the CD4bs b12 antibody to gp120. The antibodies eluted from gp120WT and then “flowed through” the gp120D368R mutant protein beads are denoted as gp120WT eluate → 368ft. As shown (left panels), this process results in the isolation of a fraction of antibodies that are highly enriched for binding to the CD4bs of gp120. The panels on the right show data from neutralization assays with either gp120WT eluate or gp120WT→368ft against the MAb b12-resistant virus PVO for serum 1 or JRFL for serum B7B5. As seen, the two-step process increased the neutralizing capacity of IgG relative to one-step elution, consistent with the enrichment of potently neutralizing CD4bs antibodies from both sera. The IgG concentration (in micrograms per millliliter) is shown on the x axes.
Neutralization assays with the Env pseudovirus variants were performed using the TZM-bl cell single-round infectivity measured by luciferase reporter gene activity by luciferase assays performed on target cell lysates (28). Diverse HIV-1 isolates including isolates from clades A, B, C, and E were included in the assays similar to a previous study (27). In addition, the chimeric HIV-2 clone containing the MPER of HIV-1 was derived from the parental HIV-2 7312A clone in which the HIV-2 Env MPER sequence QKLNSWDVFGNWFDLASWVKYIQ was replaced by the HIV-1 MPER sequence LALDKWASLWNWFDITKWLWYIK (8, 15). Heat-inactivated serum or antibody was serially diluted in the assay. Fifty percent infective dose (ID50) values of the sera or 50% inhibitory concentrations (IC50s) of antibodies or purified immunoglobulin G (IgG) were derived by determination of the serum dilution or antibody concentration that 50% of the infectious virus was neutralized. Values were calculated through a dose-response curve fit with nonlinear function (four-parameter logistic equations) using GraphPad prism software (San Diego, CA). During the initial screening, some neutralization assays were done using a single round of infection and flow cytometric assay as previously described, using a replication-competent virus cultured in peripheral blood mononuclear cells (PBMCs) and PBMC target cells (29). Viral infection of the target cells was measured by intracellular p24 antigen staining as described previously (29).
Expression and purification of HIV-1 gp120 proteins.
Monomeric gp120 envelope glycoproteins were expressed from stably transfected Drosophila S2 lines as previously described (24) or by transient transfection of FreeStyle 293 cells (Invitrogen, Carlsbad, CA) that were adapted to serum-free FreeStyle 293 expression medium (Invitrogen, Carlsbad, CA), per the supplier's instructions. Typically, 250 μg of mammalian cell codon-optimized plasmid DNA encoding gp120 combined in mixture with 1 ml of 293 Fectin (Invitrogen) transfection reagent was used to transfect FreeStyle 293 cells in a 1-liter volume at a density of 1.2 × 106 cells/ml in FreeStyle 293 expression medium. Four days posttransfection, cell culture supernatants were collected, and two protease inhibitor tablets (Roche) per liter of supernatant were added to limit proteolysis. The gp120-containing supernatants were stored at 4°C prior to purification.
The gp120 glycoproteins were purified by antibody affinity purification (16), alternatively by a two-step lentil-lectin chromatography followed by chelation chromatography. The supernatants containing the His-tagged gp120 proteins were applied to columns containing 15 ml of lentil lectin Sepharose 4B beads (GE Healthcare) that we packed (versus prepacked). The column was then washed sequentially with 10 column volumes of phosphate-buffered saline (PBS) (pH 7.4) containing 0.5 M NaCl, followed by 10 column volumes of PBS (pH 7.4). The lectin-bound glycoproteins were eluted with a total of 14 column volumes of elution buffer (PBS buffer [pH 7.4] with 1 M methyl-α-d-mannopyranoside and 10 mM imidazole). In detail, lectin elution was performed by repeatedly applying 1 column volume of the elution buffer to the column and gently rocking on a nutating mixer for 5 min followed by the collection of the eluate. The mannoside-eluted glycoproteins were pooled, and additional protease inhibitor was added. The eluate was applied to a second purification over a nickel-charged, Sepharose-based chelating affinity column (GE Healthcare) purification procedure as described previously (28).
The purified glycoproteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blue native gel, or gel filtration analysis, and the purity of the HIV-1 Env protein approached 95% homogeneity. Denatured YU2 gp120 (denoted gp120DEN) was prepared by boiling in the presence of 100 mM dithiothreitol, followed by alkylation at 37°C for 1 h with the addition of 20 mM of iodoacetamide, followed by extensive dialysis against PBS (pH 7.4). The two previously described gp120 proteins containing amino acid mutations in the CD4 binding pocket of gp120 (D368R) or coreceptor binding site (I420R) were also expressed in this manner. These mutations selectively eliminate the binding of most CD4bs antibodies (D368R) or most coreceptor site antibodies (I420R) (32, 38, 39, 43). The I420R mutant was generated in both the BaL and YU2 gp120 background (37). We also expressed the YU2 gp120 core protein, which lacks N- and C-terminal gp120 residues and the variable regions V1, V2, and V3, while preserving the highly conserved CD4bs (24, 42).
Protein-paramagnet bead coupling and antigenic characterization.
To couple gp120 glycoproteins to a solid phase, we selected the paramagnetic polystyrene, tosylactivated magnet MyOne Dynabeads (Invitrogen). The relatively small diameter and high binding capacity were preferred for treatment of the patient sera in a relatively small volume. Coupling was performed according to the manufacturer's instructions as follows. Typically, 1 mg of protein (extensively dialyzed against PBS [pH 7.4] and at a concentration greater than 5 mg/ml) was coupled to 50 mg (0.5-ml volume) of tosylactivated magnet MyOne Dynabeads. Coupling was done at 37°C in a total volume of 1.25 ml in coupling buffer (0.1 M sodium borate buffer [pH 9.5] with 1 M ammonium sulfate) with gentle rocking for 8 to 12 h. The Dynabeads and bound protein were separated from the coupling buffer with a magnet and resuspended with 5 ml of blocking buffer (PBS [pH 7.4] with 0.5% [wt/vol] bovine serum albumin [BSA] and 0.05% Tween 20) for an additional 8 to 12 h. The blocking buffer was removed by aspiration, and the protein-coupled Dynabeads were washed two times with 5 ml of buffer (PBS [pH 7.4] with 0.1% [wt/vol] BSA and 0.05% Tween 20). The beads were then resuspended in 0.5 ml of storage buffer (PBS [pH 7.4] supplemented with 0.1% [wt/vol] BSA, 0.05% Tween 20, and 0.02% sodium azide and containing protease inhibitors) and stored at 4°C.
Following coupling, the antigenic integrity and specificity of each protein were verified by flow cytometry using the well-characterized MAbs b12, 2G12, 447-52D, and 17b and polyclonal HIVIG. Briefly, 5 to 10 μl of the coupled bead slurry was washed three times with 200 μl of fluorescence-activated cell sorting (FACS) buffer (PBS [pH 7.4] with 2% fetal bovine serum) in a 96-well V-bottom plate with a magnet, followed by the addition of 50 μl of FACS buffer containing MAbs at 1 to 40 μg/ml or HIVIG at 0.1 to 4 mg/ml and incubated at room temperature for 1 h. The beads were then washed three times with FACS buffer, stained in 100 μl of FACS buffer containing phycoerythrin-conjugated goat anti-human Fc secondary antibody at room temperature for 1 h, followed by three washes with FACS buffer. The antibody-stained beads were analyzed with a FACSCalibur flow cytometer (Becton Dickinson), and data analysis was performed with FlowJo software (Tree Star, Inc., San Carlos, CA).
Serum/antibody adsorptions.
For adsorptions, the protein-coupled beads were washed three times with Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and incubated in DMEM containing 10% FBS at room temperature for 30 min to block potential nonspecific binding to the beads prior to adsorption. HIV-positive sera were diluted 1:20 to 1:50 in DMEM containing 10% FBS, and 800 to 1,000 μl of the diluted sera was incubated with 400 to 500 μl of beads at room temperature for 30 min. In most cases, two or three rounds of bead adsorption would result in the nearly complete removal of gp120-specific antibodies in the diluted serum sample. The removal of binding antibodies to a given gp120 variant was evaluated by an enzyme-linked immunosorbent assay (ELISA). Typically, from initial endpoint titer values exceeding 1:100,000, “self-adsorption” reduced endpoint titer values to less than 1:100. Following adsorption, the protein-coupled beads were removed from the treated serum samples with a magnet. The protein-coupled beads, which might contain gp120-specific antibodies were stored in PBS containing 0.2% BSA and 0.02% sodium azide at 4°C before further elution analysis. The remaining adsorbed serum was then centrifuged three times at 16,100 × g for 5 min each time to remove any residual Dynabeads and used subsequently for ELISA and neutralization assays. For validation of bead adsorption for gp120 MAbs, selected MAbs were diluted at concentrations of 3 to 10 μg/ml in DMEM containing 10% FBS in a volume of 100 μl. Adsorptions with 10 to 30 μl of the gp120-coupled Dynabeads were performed in the same manner as described above, and the remaining MAbs in solution were detected by ELISA. Note that in all serum or antibody adsorptions, BSA-coated beads or blank beads were used as negative controls for the solid phase.
Elution and quantification of adsorbed IgG.
To recover and characterize the bead-bound antibodies, the beads were washed three times with PBS containing 500 mM NaCl and one time with PBS, and the antibodies were eluted by a stepwise decrease in pH. First, the beads were mixed with 100 mM glycine-HCl elution buffer (pH 2.7) for 30 s. The beads were then microcentrifuged for 30 s and held in place on the bottom of the tube with a magnet. The acid-eluted solution containing IgG was quickly removed and placed into a separate tube containing 1 M Tris (pH 9.0) buffer to reach pH 7.0 to 7.4. This process was repeated for a total of three times. The eluted IgG was diluted in DMEM and concentrated over a 30-kDa Centricon plus filter (Millipore Corp.). Subsequently, the same procedure was performed on the beads at an elution pH of 2.2 to recover any IgG resistant to elution at pH 2.7. The IgG fractions recovered by both acid elution steps were combined, and the concentration of the combined IgG fractions were measured with a radial immunodiffusion kit to quantify the human IgG (Binding Site Corp.). The recovered and quantified IgG from the protein-coupled beads was further characterized by ELISA and neutralization assays. Typically, from 50 μl of adsorbed serum, 10 to 30 μg of IgG was eluted from wild-type gp120 (gp120WT) protein-coupled beads. Virus neutralization assays with the eluted IgG were performed with IgG concentrations that started at 10 to 40 μg/ml with threefold serial dilutions.
Antibody binding assay (ELISA).
The presence of serum binding antibodies and MAbs in adsorption experiments were quantified by an ELISA using standard methods as follows. All the binding/coating reactions were performed in a 100-μl volume. Plates were coated with the specified protein at 1 μg/ml in PBS and reacted with serial dilutions of sera or MAbs. PBS containing 2% dry milk and 5% FBS was used to block the wells on an ELISA plate, and PBS with 0.2% Tween was used to wash the wells. The secondary antibody was a goat anti-human IgG Fc antibody (Jackson Labs) conjugated to horseradish peroxidase. The optical density (OD) was determined using a microplate reader (Molecular Devices) at 450 nm, and the endpoint titers of the serum antibodies were defined as the last reciprocal serum dilution at which the OD signal was greater than twofold over the signal detected with a negative-control serum. The CD4bs competition ELISA was done similarly as follows. ELISA plates were coated with sheep anti-gp120 C5 region polyclonal antibodies (Aalto, Dublin, Ireland) at 5 μg/ml, followed by blocking with blocking buffer (PBS containing 2% dry milk and 5% FBS), and incubation with gp120 at 1 μg/ml. The competitor ligands used were either two-domain soluble CD4 (sCD4) (with a concentration ranging from 500 μg/ml to 0.005 ng/ml in 100-fold serial dilutions) or the CD4bs antibody F105 Fab (with a concentration ranging from 20 μg/ml to 0.0002 ng/ml in 100-fold serial dilutions). These ligands were incubated with gp120 protein at 1 μg/ml for 1 h prior to the addition of the eluted IgG fractions at final concentrations of 100 to 300 ng/ml. The eluted IgG bound to coated gp120 protein to achieve approximately 0.7 OD unit at 450 nm in the absence of competitor.
Peptide inhibition of neutralization.
The serum neutralizing antibodies were mapped to specific HIV-1 Env regions by a method of selective peptide inhibition of neutralization as follows. Individual or selected pools of overlapping peptides derived from gp120 variable regions (see Table S1 in the supplemental material), including the BaL.01 V3-specific peptide (TRPNNNTRKSIHIGPGRAFYTTG) (see Table 3), and gp41-specific peptides (see Table S2 in the supplemental material) were added to the serum or control MAbs for 30 min at a concentration of 25 μg/ml prior to the addition of virus. The assay format was the same as described for the single round of infection with HIV-1 Env pseudovirus assay above. All the peptides were confirmed to have minimal effect on virus entry before being included in the peptide inhibition of neutralization assay. “Mock” peptide (defined as an equivalent volume of PBS) was used as negative control. In addition, a scrambled V3 region peptide (scrambled V3) (NKGTHNIPTARNIYGFPTSRRGT), was also used as a reference control for the effect on neutralization. For the gp41 overlapping peptides, a pool of peptides derived from nonhuman proteins was also included as negative control (see Table S2 in the supplemental material). The effect of the peptide on virus neutralization was reported in two ways either as the change between the ID50 or IC50 of the serum or MAb with or without the test peptides or the reference peptide (scrambled V3) or as the percent inhibition of neutralization, which was calculated as [(percent neutralization observed with no peptide − percent neutralization observed with test peptide)/percent neutralization observed with no peptide] × 100. In addition, at a single dilution of serum or of the V3-directed MAbs (22) 2182, 2191, 2219, 2442, 2456, and 1.3D and the conformational V3 antibody 447-52D, the serum, or antibody were tested against the sensitive primary isolate SF162 and the BaL.01 and 6535.3 isolates with the V3 peptides to validate the specificity of the assay (see Table 3).
TABLE 3.
V3 peptide effects on serum neutralization ID50 titers against the clade B V3 neutralization-sensitive isolate JRFL-Δ301a
| Serum | Mock peptide ID50 | Scrambled V3 peptide
|
% Neut. to scrambled peptidec | BaL V3 peptide
|
% Neut. to V3 peptidee | Net % neut. to V3f | ||
|---|---|---|---|---|---|---|---|---|
| ID50 | ID50 fold decreaseb | ID50 | ID50 fold decreased | |||||
| Strong/broadly neutralizing sera | ||||||||
| 1 | 5,704 | 10,019 | 0.6 | 0 | 3,657 | 1.6 | 38 | 38 |
| 18 | 901 | 807 | 1.1 | 9 | 1,167 | 0.8 | 0 | 0 |
| 20 | 507 | 421 | 1.2 | 17 | 291 | 1.7 | 41 | 25 |
| 30 | 786 | 833 | 0.9 | 0 | 615 | 1.3 | 23 | 23 |
| 45 | 3,291 | 3,631 | 0.9 | 0 | 2,445 | 1.3 | 23 | 23 |
| Weak/limited neutralizing sera | ||||||||
| 6 | 465 | 502 | 0.9 | 0 | 126 | 3.7 | 73 | 73 |
| 8 | 531 | 553 | 1 | 0 | 162 | 3.3 | 70 | 70 |
| 15 | 89 | 91 | 1 | 0 | 2.5 | 35.6 | 97 | 97 |
| 19 | 204 | 277 | 0.7 | 0 | 28 | 7.3 | 86 | 86 |
| 29 | 1,889 | 1,126 | 1.7 | 41 | 443 | 4.3 | 77 | 36 |
| 31 | 80 | 60 | 1.3 | 23 | 2.5 | 32 | 97 | 74 |
The assay was validated against a set of V3-directed antibodies (2182, 2191, 2219, 2442, 2456, 447-52D, and 1.3D) (22), and complete inhibition of antibody-mediated neutralization against the BaL.01, 6535, and SF162 isolates was demonstrated by the BaL.01 V3-derived peptide, while the V3 scrambled peptide had no effect on neutralization. Neither peptide demonstrated any impact on 2G12- or 2F5-mediated neutralization of the BaL.01, 6535, or SF162 pseudotyped virus. The final concentration of the V3 peptides in the presence of serum and virus was 25μg/ml.
Serum ID50 value ratio in the absence and presence of scrambled V3 peptide, calculated as (ID50 with mock peptide/ID50 with scrambled V3 peptide).
Indicator of percentage of ID50 change by the presence of scrambled V3 peptide, calculated as [1 − (ID50 with mock peptide/ID50 with scrambled V3 peptide)] × 100.
Serum ID50 value ratio in the absence and presence of V3 peptide, calculated as ID50 with mock peptide/ID50 with V3 peptide.
Indicator of percentage of ID50 change by the presence of V3 peptide, calculated as [1 − (ID50 with mock peptide/ID50 with V3 peptide)] × 100.
Indicator of percentage of neutralization directed against the V3 epitope, calculated as the percentage of ID50 change by the presence of V3 peptide − percentage of ID50 change by the presence of scrambled V3 peptide. Values greater than 50% are shown in boldface type.
RESULTS
Virologic screening.
We previously reported the neutralization activities of 32 HIV-1-positive sera against a large panel of virus isolates from clade A (n = 11), clade B (n = 24), clade C (n = 11), and clade E (n = 1). Five sera, designated as sera 1, 18, 20, 30, and 45 neutralized most viruses tested and were designated as broadly neutralizing. Two sera (sera 1 and 45) were particularly potent and were previously studied (27). Six sera with weak neutralization and limited breadth were also identified. The potency of the broadly neutralizing sera was confirmed by determining the serum ID50 titer against a selected panel of virus isolates (see Fig. S2A and B in the supplemental material). We subsequently identified three additional broadly neutralizing sera (B7B5, US12, and US21), all derived from SP individuals. These three sera, along with sera 18 and 20, were analyzed with an expanded panel of adsorption and peptide competition reagents. This included single amino acid mutant gp120 reagents to identify antibodies directed against the CD4bs and the coreceptor binding region, as well as peptides designed to detect antibodies with specificities similar to MAbs 2F5 and 4E10. In these analyses, we occasionally utilized the previously characterized sera 1 and 45 as positive controls. Where appropriate, selected sera that were relatively limited in neutralization breadth were used to compare and contrast the antibody specificities that mediated virus neutralization relative to the more broadly neutralizing sera.
Cross-clade neutralization is gp120 directed.
We initially analyzed whether the neutralizing activity of the broadly neutralizing sera was directed against known determinants present on native gp120 (see Fig. S1 in the supplemental material). This analysis involves adsorption of the sera with native, solid-phase-coupled gp120, followed by a comparison of the binding and neutralizing properties of the adsorbed sera, as well as analysis of the gp120-eluted antibodies (27) (Fig. 1A). The coupling efficiency of gp120 to the beads was determined to be between 70 and 100% by SDS-PAGE analysis (see Fig. S3A in the supplemental material). We next confirmed the integrity of the gp120WT coupled to the beads by a FACS-based binding analysis using a panel of ligands. The panel of ligands included the conformational, anti-gp120 monoclonal antibodies b12, 2G12, and 447-52D, the CD4-induced antibody 17b, and the conformational ligand CD4 (see Fig. S3B in the supplemental material) (not shown). We further validated the specificity of the gp120WT beads by demonstrating their ability to adsorb the binding activity of the 2G12 and 447-52D antibodies and to adsorb the neutralizing capacity of the CD4bs antibody (b12) (see Fig. S3C in the supplemental material).
We performed adsorptions of sera using the gp120-coupled beads and verified that the flowthrough contained nearly undetectable levels of gp120-specific antibodies by ELISA, indicating that the absorbed sera were fully depleted of gp120-binding antibodies (see Fig. S4B in the supplemental material). For specificity controls, adsorption of BSA-conjugated beads and blank beads was performed in parallel, and the beads showed only minor effects on serum neutralization capacity compared to untreated serum (Table 1) (see Fig. S3D in the supplemental material). In contrast, we observed a significant decrease in the neutralizing activity of the gp120 flowthrough (gp120ft) fraction for several of the broadly neutralizing sera. Table 1 shows serum neutralization ID50 titers after adsorption with gp120WT-coupled beads or negative-control beads, including BSA-coupled beads or blank beads. The proportion of gp120-specific neutralization against a particular virus isolate was determined by the percent reduction in ID50 by gp120WT relative to blank beads (when available) or BSA-coupled beads. For instance, the flowthrough antibody fraction for serum B7B5 had reciprocal ID50 titers against JRFL of 2,281 (blank beads) and 240 (gp120WT beads), respectively. Thus, 89% [(2,281 − 240)/2,281] of JRFL neutralization was reduced by gp120WT bead adsorption and can be termed gp120-directed neutralization. For all seven broadly neutralizing sera, the majority of neutralization of most viruses tested was gp120-directed neutralization. Furthermore, since a single clade B monomeric gp120 protein (derived from YU2) removed the neutralizing activity against representative clade A and C isolates (RW020 and ZA12), the data suggested that the gp120-specific activity was directed against well-conserved surfaces of the clade B HIV-1 gp120 protein. Accordingly, we then turned to mapping of the neutralizing specificity to the most conserved and exposed regions on gp120, either the CD4 binding site or the coreceptor binding region.
TABLE 1.
Pre- and postadsorption neutralization ID50 values for 10 sera with various neutralization breadths and potencies with wild-type or CD4 binding site mutant D368R gp120-coupled beads
| Serum | Virus | ID50
|
% Neutralization activity
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Untreated serum | Blanka | BSAb | gp120WTc | gp120D368Rd | gp120 directede | gp120D368R directedf | gp120 neut. to CD4bsg | Total neut. to CD4bsh | ||
| 1 | HXBc2 | 5,598 | 50 | 1,885 | 99 | 66 | 33 | 33 | ||
| SF162 | 493 | 20 | 96 | |||||||
| BaL.01 | 4,092 | 5,174 | 554 | 1,315 | 86 | 68 | 22 | 19 | ||
| JRFL | 1,051 | 1,088 | 216 | 1,210 | 79 | 0 | 100 | 79 | ||
| ADA | 584 | 895 | 50 | 651 | 91 | 0 | 100 | 91 | ||
| PVO.04 | 1,932 | 286 | 166 | 362 | 91 | 81 | 11 | 10 | ||
| RW20.2 | 922 | 872 | 139 | 360 | 85 | 61 | 28 | 24 | ||
| ZA012.29 | 175 | 172 | 30 | 486 | 83 | 0 | 100 | 83 | ||
| 6 | HXBc2 | 1,356 | 20 | 99 | ||||||
| SF162 | 1,575 | 65 | 96 | |||||||
| 18 | HXBc2 | 309 | 422 | 288 | 20 | 127 | 95 | 70 | 27 | 25 |
| SF162 | 1,421 | 1,093 | 1,617 | 740 | 447 | 32 | 59 | 0 | 0 | |
| BaL.01 | 379 | 396 | 377 | 107 | 144 | 73 | 64 | 13 | 9 | |
| JRFL | 968 | 913 | 828 | 239 | 515 | 74 | 44 | 41 | 30 | |
| PVO.04 | 330 | 396 | 453 | 156 | 132 | 61 | 67 | 0 | 0 | |
| RW020.2 | 1,818 | 1,240 | 1,453 | 534 | 638 | 57 | 49 | 15 | 8 | |
| 19 | HXBc2 | 2,597 | 45 | 511 | 98 | 80 | 18 | 18 | ||
| SF162 | 114 | 83 | 330 | 27 | 0 | 100 | 27 | |||
| SC422661.8 | 227 | 35 | 10 | 84 | 96 | 0 | 0 | |||
| 6535.3 | 358 | 199 | 181 | 44 | 49 | 0 | 0 | |||
| 20 | HXBc2 | 2,034 | 292 | 154 | 86 | 92 | 0 | 0 | ||
| SF162 | 2,095 | 499 | 203 | 76 | 90 | 0 | 0 | |||
| BaL.01 | 2,865 | 23 | 56 | 99 | 98 | 1 | 1 | |||
| JRFL | 81 | 44 | 53 | 45 | 34 | 24 | 11 | |||
| 30 | HXBc2 | 413 | 20 | 95 | ||||||
| SF162 | 4,395 | 781 | 82 | |||||||
| BaL.01 | 10,398 | 677 | 100 | 99 | ||||||
| JRFL | 432 | 136 | 113 | 74 | ||||||
| YU2 | 171 | 90 | 67 | 61 | ||||||
| 45 | HXBc2 | 1,578 | 1,792 | 260 | 518 | 84 | 67 | 20 | 16 | |
| SF162 | 3,905 | 398 | ||||||||
| BaL.01 | 1,420 | 857 | 453 | 492 | 68 | 65 | 4 | 3 | ||
| JRFL | 464 | 559 | 87 | 297 | 81 | 36 | 56 | 45 | ||
| ADA | 478 | 470 | 84 | 193 | 82 | 60 | 28 | 23 | ||
| RW20.2 | 263 | 211 | 115 | 313 | 56 | 0 | 100 | 56 | ||
| ZA012.29 | 67 | 257 | 30 | 30 | 56 | 56 | 0 | 0 | ||
| B7B5 | HXBc2 | 13,575 | 544 | 3,629 | 96 | 73 | 24 | 23 | ||
| BaL.01 | 6,417 | 374 | 2,266 | 94 | 65 | 31 | 29 | |||
| JRFL | 2,281 | 240 | 1,719 | 89 | 25 | 72 | 65 | |||
| PVO.04 | 1,628 | 267 | 1,566 | 84 | 4 | 95 | 80 | |||
| RW20.2 | 1,387 | 179 | 525 | 87 | 62 | 29 | 25 | |||
| ZA012.29 | 244 | 114 | 213 | 53 | 13 | 76 | 41 | |||
| US12 | HXBc2 | 360 | 249 | 244 | 20 | 119 | 92 | 52 | 43 | 40 |
| SF162 | 1,911 | 1,488 | 2,319 | 874 | 727 | 41 | 51 | 0 | 0 | |
| BaL.01 | 2,931 | 2,349 | 2,690 | 1,282 | 1,387 | 45 | 41 | 10 | 4 | |
| JRFL | 1,936 | 1,257 | 1,699 | 831 | 570 | 34 | 55 | 0 | 0 | |
| PVO.04 | 8,550 | 10,525 | 7,199 | 5,828 | 4,138 | 45 | 61 | 0 | 0 | |
| RW20.2 | 7,855 | 6,603 | 6,648 | 4,785 | 2,709 | 28 | 59 | 0 | 0 | |
| US21 | HXBc2 | 678 | 487 | 532 | 42 | 135 | 91 | 72 | 21 | 19 |
| SF162 | 4,424 | 2,680 | 3,927 | 900 | 895 | 66 | 67 | 0 | 0 | |
| BaL.01 | 889 | 771 | 877 | 151 | 221 | 80 | 71 | 11 | 9 | |
| JRFL | 1,168 | 1,158 | 1,207 | 672 | 647 | 42 | 44 | 0 | 0 | |
| PVO.04 | 3,896 | 4,247 | 2,906 | 2,365 | 1,476 | 44 | 65 | 0 | 0 | |
| RW20.2 | 4936 | 3674 | 3920 | 2201 | 1952 | 40 | 47 | 0 | 0 | |
Samples adsorbed on blank Dynabeads.
Samples adsorbed on Dynabeads coated with BSA.
Samples adsorbed on Dynabeads coated with YU2 p120WT protein.
Samples adsorbed on Dynabeads coated with YU2 p120D368R protein (CD4bs knockout mutation).
Percent reduction in ID50 by gp120WT relative to blank beads (when available) or BSA-coupled beads as an indication of the proportion of gp120-specific neutralization against a particular virus isolate.
Percent reduction in ID50 by gp120D368R relative to blank beads (when available) or BSA-coupled beads as an indication of the proportion of non-CD4bs-directed neutralization.
Percentage total gp120-directed neutralization (neut.) that is via the CD4bs, i.e., [(percentage of gp120WT-directed neutralization − percentage of gp120D368R-directed neutralization)/percentage of gp120-directed neutralization] (values of 20% or greater are shown in boldface type).
Percentage total serum neutralization (neut.) that is via the CD4bs; i.e., (percentage of gp120 neutralization that is directed against CD4bs × percentage of gp120-directed neutralization).
Mapping neutralizing specificity to the CD4 binding site.
We screened for the presence of CD4bs-directed neutralization activity using the well-defined gp120 point mutation D368R (32, 38, 39). This mutation eliminates the binding of both CD4 and the CD4bs antibodies, such as b12 and F105 (27, 38, 39). We previously confirmed that the critical gp120D368R point mutant protein coupled to the Dynabeads was well recognized by the gp120-specific antibodies 2G12 and 447-52D, but not by the CD4bs neutralizing antibody, b12 (27). To further confirm the specificity of this analysis, we demonstrated that the gp120D368R bead could adsorb all binding activity of the broadly neutralizing antibody (2G12), the CD4-induced (CD4i) antibody (17b) (see Fig. S4A in the supplemental material), and the V3 antibody (447-52D) (data not shown), but not the b12 antibody (see Fig. S4A in the supplemental material).
We extended the differential solid-phase adsorption to the five broadly neutralizing sera 18, 20, US12, US21, and B7B5 (Fig. 1B). We also included one weakly neutralizing serum (serum sample 19) in the analysis. After adsorption with gp120WT and gp120D368R beads in parallel, serum antibody binding was tested both for binding to gp120WT and gp120D368R and for virus neutralization capacity. As expected, an ELISA confirmed that binding to the homologous protein, either gp120WT or gp120D368R, was undetectable after adsorption of each of the serum samples. However, for some sera, a significant level of gp120WT binding remained after the gp120D368R bead adsorption (see Fig. S4B in the supplemental material), suggesting that a fraction of the binding antibodies are CD4bs directed. This is consistent with our previous data regarding sera 1 and 45. In fact, such a differential binding assay can be used to detect the presence of CD4bs antibodies in polyclonal sera.
To test whether this subset of antibodies mediates virus neutralization, neutralization assays against clade B and non-clade B viruses were performed. We observed a range of CD4bs-directed neutralization potency profiles (Fig. 2A and summarized in Table 1). CD4bs-directed neutralizing activity was assessed by comparing the serum ID50 titer after gp120WT and gp120D368R mutant bead adsorption relative to blank beads or BSA beads, as summarized in Table 1. Similar to the calculation of the proportion of gp120-directed neutralizing activity, the gp120D368R-directed (non-CD4bs-directed) neutralizing proportion was calculated as percent reduction in ID50 by gp120D368R relative to blank beads (when available) or BSA-coupled beads. Then, the portion of CD4bs-directed neutralizing activity in the total gp120-directed neutralization was calculated as [(percentage of gp120WT-directed neutralizing activity − percentage of gp120D368R-directed neutralizing activity)/percentage of gp120-directed neutralizing activity]. For example, when tested against virus JRFL, serum B7B5 has 89% gp120WT-directed neutralizing activity [(2,281 − 240)/2,281] and 25% gp120D368R-directed neutralizing activity [(2,281 − 1,719)/2,281]. Therefore, approximately 72% of the B7B5 gp120-directed neutralizing activity is focused on CD4bs [89% − 25%)/89%].
The B7B5 serum was the most broadly neutralizing of the newly assessed sera and also displayed the most predominantly CD4bs-focused neutralization. The large majority of B7B5 neutralizing activity was gp120-directed activity, and depending on the virus, most of the gp120 neutralization was specific for the CD4bs (Fig. 2A and Table 1). The lower fraction of B7B5 CD4bs-directed neutralization for HXBc2 and SF162 may be related to the fact that these two viruses are sensitive to many other gp120 antibody specificities in sera. Serum 19, US12, and US21 contained CD4bs-directed neutralizing antibodies, but this fraction of antibodies neutralized only the very sensitive HXBc2 virus, suggesting a limited potency for the CD4bs antibody subset against more resistant primary viruses (Table 1). Neutralization by serum 20 was largely gp120 directed (except against JRFL), but little of this gp120-directed neutralization was CD4bs directed. Of note, these types of serum fractionation experiments involve several steps, and we consider the values in Table 1 to be general estimates of the proportion of antibody specificities rather than hard and fast numbers. Also, when the proportion of gp120-directed neutralization was less than 50% (such as for serum 20 against JRFL), further calculations of the proportion of gp120 activity are likely to be of limited accuracy. Hence, in this example, serum sample 20 had 34% gp120-directed activity against JRFL; we would not suggest that the value of 24% CD4bs-directed activity is highly accurate. In contrast, serum B7B5 demonstrated 89% gp120-directed neutralization of JRFL, and of this fraction, 72% was CD4bs directed. Here, we can be more confident that 65% (i.e., 72% of 89%) of the total neutralizing activity of this serum is CD4bs directed.
CD4bs-directed neutralization is mediated by immunoglobulin.
To further characterize the CD4bs-directed neutralizing specificity in sera, we eluted the absorbed gp120-specific binding activity from the gp120WT protein. This gp120-directed IgG was characterized as pure immunoglobulin by SDS-PAGE (data not shown), and the IgG concentration was determined by radial immunodiffusion. The binding and neutralization activity was then studied as depicted schematically in Fig. 1B. We used serum 1 as a positive control and focused on the most potent and broadly neutralizing of the five newly analyzed sera, B7B5, for this analysis.
To further enrich for the CD4bs-directed antibodies, the gp120-eluted IgG from serum B7B5 was subjected to a second adsorption with the gp120D368R-conjugated beads. This removed gp120-directed antibodies, except those directed against the CD4bs region of gp120 which would remain in the column flowthrough (Fig. 1B). The CD4bs-enriched IgG fraction was tested for its binding specificity by a competition ELISA using gp120 as the capture antigen in the absence and presence of soluble CD4 as a competitor. The CD4bs-enriched IgG fraction of B7B5 serum specifically competed with sCD4 in a manner similar to that of the CD4bs antibody, b12 (Fig. 2B). For negative controls, the MAbs 2G12 and 447-52D, which are not directed against the CD4 binding region, did not compete with sCD4 for binding to gp120. The CD4bs antibodies fractionated from the B7B5 serum were then tested for neutralization activity against selected HIV-1 primary isolates to confirm that the elute/enriched IgG can recapitulate the potent and broad neutralization of the serum sample. A severalfold increase in neutralizing potency could be observed in stepwise progression from the initial gp120 eluate IgG fraction to the CD4bs-enriched fraction of antibodies (Fig. 2B, bottom right).
The conserved gp120 coreceptor binding site and neutralization specificity.
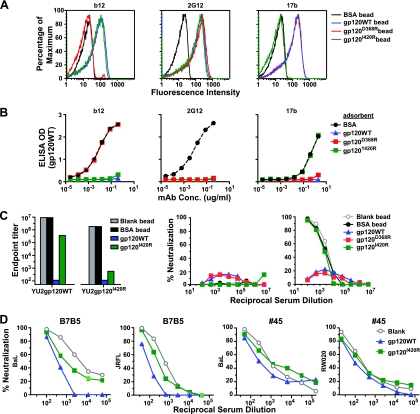
The chemokine receptor binding site of HIV-1 is highly conserved, but most known antibodies directed against this region are unable to access their epitopes on the HIV-1 functional spike due to steric or conformational constraints (6, 19, 25). Consequently, the known coreceptor site MAbs do not generally neutralize primary HIV-1 isolates (8, 25). However, the possibility exists that broadly neutralizing sera contain antibodies against this conserved region that are capable of neutralization. Therefore, we applied the selective adsorption approach using an I-to-R mutation at position 420. This residue is located in the bridging sheet region of the coreceptor binding site (see Fig. S1 in the supplemental material), and the I-to-R nonconservative mutation eliminates the binding of most known antibodies directed at the CD4i coreceptor binding sites without affecting most other binding specificities (37, 43). We confirmed these previous observations by performing binding analysis of the gp120I420R by both ELISA and FACS. By ELISA, the I420R mutation in the BaL gp120 context was recognized at levels equivalent to the levels of gp120WT recognized by MAbs b12, 2G12, and 447-52D, whereas coreceptor binding region MAbs, such as 17b, 48d, E51, and 2.8B, did not recognize the gp120I420R mutant protein (see Fig. S5 in the supplemental material). We observed a similar selective elimination of MAb 17b binding by FACS analysis of the solid-phase paramagnetic beads coupled to either gp120WT or the gp120I420R mutant proteins. For controls, recognition by the conformational antibodies b12 and 2G12 to both proteins after coupling was unaffected (Fig. 3A). We confirmed further the selective specificity of the gp120I420R beads by demonstrating that this solid-phase mutant could adsorb all the gp120-specific binding activity of the broadly neutralizing antibodies, b12 and 2G12, but not the coreceptor binding site antibody, 17b (Fig. 3B).
FIG. 3.
The coreceptor binding region neutralizing antibody specificity of serum analyzed by differential adsorption. (A) Flow cytometric analysis of gp120WT, coreceptor binding region mutant gp120I420R, and CD4bs mutant gp120D368R proteins following the coupling to beads to confirm conformational integrity (see Materials and Methods). Three conformation-dependent antibodies, MAbs b12, 2G12, and 17b were tested for binding to the protein-coupled beads. The number of binding events as a percentage of the maximum is plotted against fluorescence intensity. All three antibodies bind gp120WT-coupled beads, while b12 and 2G12 bind the gp120I420R-coupled beads, but the coreceptor-directed antibody, 17b, binds poorly. BSA-coupled beads were used as a negative control. (B) Control MAbs b12, 2G12, and 17b were adsorbed with gp120WT protein, gp120D368R, or gp120I420R, and the residual MAb binding activity was measured by gp120 ELISA. Note that the I420R gp120 effectively adsorbed MAbs 2G12 and b12, but not the coreceptor binding region MAb, 17b. The MAb concentration (in micrograms per milliliter) is shown on the x axes. gp120 is from isolate BaL. (C) Differential adsorption of the CD4i antibody fraction in HIV-positive sera. (Left) Endpoint ELISA data for serum B7B5 following adsorption with blank beads or with beads covalently linked to BSA, YU2 gp120WT, or YU2 gp120I420R. The protein used to coat ELISA plates is indicated underneath each set of bars. Of note, adsorption with the beads conjugated with the gp120I420R mutant protein left behind a fraction of serum antibodies, presumably to the coreceptor binding region, that were detected in the gp120WT protein ELISA. (Right) Serum B7B5 neutralization against an HIV-2 isolate after adsorption with beads conjugated with various proteins or with blank beads as a negative control. Most HIV-positive sera have high titers of CD4i antibodies as detected by neutralization of the HIV-2 7312A_V434M virus with sCD4 present. Neutralization curves of HIV-2 in the absence (left) and presence of sCD4 (right) are shown. The gp120WT and gp120D368R-coupled beads removed all CD4i neutralizing activity from the serum, whereas the I420R mutant protein did not. (D) Neutralization curves of sera B7B5 and 45 are shown after adsorption with blank beads, gp120WT beads, or gp120I420R beads containing a mutation in the coreceptor binding site (CD4i region).
We then performed differential adsorption of the B7B5 serum with either gp120WT or gp120I420R and observed that the gp120WT adsorption eliminated virtually all binding to both the gp120WT and I420R mutant protein (Fig. 3C, left). However, gp120I420R adsorption eliminated binding to the I420R mutant protein, but substantial binding to the gp120WT protein was retained. This indicated that a fraction of the gp120-specific antibodies in this serum was directed against the coreceptor binding region (Fig. 3C, left). To confirm this finding of antibodies directed against the coreceptor binding site, we performed neutralization assays with an HIV-2 strain (7312A_V434M), which is an assay specific for the detection of HIV-1 coreceptor binding site-directed antibodies (8). Serum B7B5 was absorbed with gp120WT and gp120I420R, and the flowthrough fractions were tested for neutralization of HIV-2. As expected, gp120WT, but not gp120I420R, could effectively adsorb the HIV-2 neutralization (Fig. 3C, right).
By this selective adsorption analysis, we found two sera that appear to contain coreceptor binding site-directed neutralization activity (sera 45 and B7B5). In both cases, the gp120WT protein more efficiently adsorbed serum neutralization than the gp120I420R mutant did (Fig. 3D). The relative fraction of neutralizing antibodies potentially directed against the coreceptor site was calculated as described above for the CD4bs-directed activity, and values for sera 45 and B7B5 against seven isolates are shown (Table 2). The potential existence of coreceptor-directed antibodies is an intriguing result that has implications for immunogen design and awaits confirmation by the isolation of neutralizing monoclonal antibodies displaying such specificity. A caveat to this analysis is that a novel neutralizing antibody that is sensitive to both of the D368R and I420R mutations in gp120 could exist. Such an antibody would be neither a classical CD4bs or coreceptor region-specific neutralizing antibody as currently defined.
TABLE 2.
Pre- and postadsorption neutralization ID50 values for two broadly neutralizing sera with wild-type (WT) or coreceptor binding region (CD4i) mutant gp120 I420R-coupled beads
| Serum | Virus | ID50
|
% Neutralization activity
|
|||||
|---|---|---|---|---|---|---|---|---|
| Blanka | gp120WTb | gp120I420Rc | gp120 directedd | gp120I420R directede | gp120 neut. to CD4if | Total neut. to CD4ig | ||
| B7B5 | HXBc2 | 13,575 | 544 | 1,579 | 96 | 88 | 8 | 8 |
| BaL.01 | 6,417 | 374 | 1,481 | 94 | 77 | 18 | 17 | |
| JRFL | 2,281 | 240 | 941 | 89 | 59 | 34 | 31 | |
| PVO.04 | 1,628 | 267 | 947 | 84 | 42 | 50 | 42 | |
| RW20.2 | 1,387 | 179 | 368 | 87 | 73 | 16 | 14 | |
| ZA012.29 | 244 | 144 | 232 | 53 | 5 | 91 | 48 | |
| 45 | HXBc2 | 1,578 | 260 | 1,097 | 84 | 30 | 64 | 53 |
| BaL.01 | 1,420 | 453 | 2,455 | 68 | 0 | 100 | 68 | |
| RW20.2 | 263 | 115 | 229 | 56 | 13 | 77 | 43 | |
Samples adsorbed on blank Dynabeads.
Samples adsorbed on Dynabeads coated with YU2 gp120WT protein.
Samples adsorbed on Dynabeads coated with YU2 gp120I420R protein (CD4i knockout mutation).
Percent reduction in ID50 by gp120WT relative to blank beads as an indication of the proportion of gp120-specific neutralization against a particular virus isolate.
Percent reduction in ID50 by gp120I420R relative to blank beads as an indication of the proportion of non-CD4i-directed neutralization.
Percentage of total gp120-directed neutralization (neut.) that is via the CD4i; i.e., [(percentage of gp120-directed neutralization − percentage of gp120I420R-directed neutralization)/percentage of gp120-directed neutralization] (values 20% or greater are shown in boldface type).
Percentage of total serum neutralization (neut.) that is via the CD4i; i.e., (percentage of gp120 neutralization that is CD4i directed × percentage of gp120-directed neutralization).
This possibility is highlighted by inspection of the percentage of gp120-directed neutralization for both the CD4bs and the coreceptor region from serum 45 for the RW020 isolate and for serum B7B5 for the PVO and ZA012 isolates. In both cases, the percentage of gp120-directed neutralization greatly exceeds 100%, which could be explained by a single neutralization specificity sensitive to both mutations (see Table 1 compared to Table 2 for these isolates). Nevertheless, our results highlight the possibility that novel neutralizing specificities directed toward HIV-1 Env may exist in some sera.
gp120 peptide inhibition of neutralization assays specific for the major variable loops.
We further sought to determine whether the neutralizing specificity in the sera might be directed toward the major variable regions of gp120. Due to the immunodominant nature of the V3 region and because of the large variety of available V3-specific antibodies to use as controls, we initially validated and optimized our ability to detect V3-directed neutralization. Although V3-specific antibodies generally do not neutralize the majority of primary isolates, some viruses that are sensitive to V3-directed antibodies exist. Accordingly, peptides derived from the V3 region of BaL.01 were synthesized, and their ability to inhibit neutralization mediated by a panel of seven V3-directed monoclonal antibodies was analyzed. The neutralization of the V3-directed MAbs 2182, 2191, 2219, 2442, 2456, and 1.3D and the conformational V3 antibody 447-52D against primary isolate BaL.01 was efficiently and specifically inhibited by incubation with the V3 peptide prior to incubation of each antibody with virus (data not shown). V3 peptide inhibition of the V3-directed neutralizing monoclonal antibody panel was confirmed using two other primary isolates, SF162 and 6535.3, in similar experiments (data not shown).
We next tested whether there was V3-directed neutralizing activity in selected patient sera. We found no significant effect of V3 peptides for any of the potent and broadly neutralizing sera tested (Table 3). However, the neutralization activity of several weakly/narrowly neutralizing sera was appreciably inhibited by preincubation with the V3 peptide, demonstrating V3-directed neutralization in this type of sera (Table 3).
We also performed a similar analysis using peptides derived from the V1 and V2 regions of the YU2 isolate (28). When V1 and V2 peptides were tested in the peptide inhibition of neutralization assay, none of the patient sera were affected by any of the V1- or V2-derived peptides, despite the fact that selected sera have positive binding to pools of the V1 and V2 peptides (data not shown). The caveat here is that we lacked positive-control V1 or V2 neutralizing antibodies to YU2, and it is possible that linear peptides may not inhibit neutralization by an antibody to a nonlinear epitope on V1 or V2.
Analysis of potential gp41-directed neutralization.
The gp41 MPER contains two well-characterized linear, conserved epitope clusters recognized by the broadly neutralizing MAbs 2F5 and 4E10 (see Fig. S1 in the supplemental material). Because the MPER is the only major neutralizing epitope defined on gp41 and because the structure of the linear epitopes and their cognate antibody for MAbs 2F5 and 4E10 are well defined, we focused on analyzing the MPER to determine whether antibodies to this region were mediating broad neutralization. Many of the sera tested had detectable levels of antibodies binding to MPER-containing peptides (see Fig. S6 in the supplemental material) or to the MPER arrayed on hepatitis B virus surface antigen particles (data not shown).
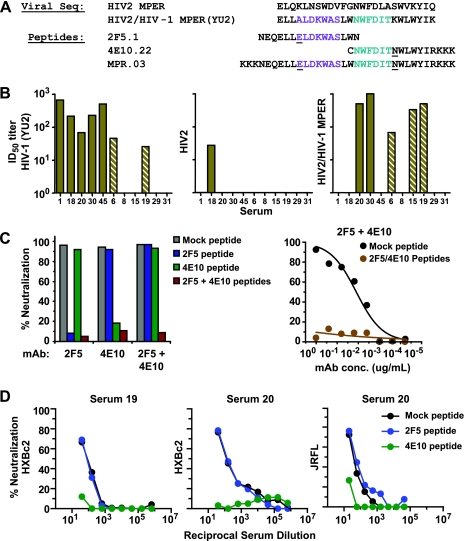
To assess whether the gp41 and MPER-specific binding antibodies were neutralizing, we used a chimeric HIV-2 virus that contains the complete 25-amino-acid sequence of the HIV-1 YU2 gp41 MPER (15) (Fig. 4A). We verified that this chimeric HIV-2/MPER virus is highly sensitive to neutralization by both MAbs 2F5 and 4E10 (IC50 of <0.2 μg/ml for each). No neutralization of the HIV-2 chimeric virus was accomplished by the two most broadly neutralizing control sera, serum 1 or 45 (Fig. 4B), consistent with the relatively low levels of MPER-specific binding activity detected in these sera (see Fig. S6 in the supplemental material). Interestingly, sera derived from patients 6, 15, 19, 20, 30, and B7B5 demonstrated a range of neutralizing titers against the HIV-2/MPER chimeric virus (Fig. 4B and Table 4). We then sought to confirm whether antibodies to the MPER were mediating neutralization of HIV-1.
FIG. 4.
Neutralization assays to detect antibodies specific for the membrane-proximal region. (A) The MPER sequence of HIV-2 and HIV-2/HIV-1 MPER chimeric virus are shown as well as peptides derived from the HIV-1 MPER that overlap with the 2F5 (purple) and 4E10 (cyan) core binding sites, respectively. Underlined residues indicate sequence polymorphisms that do not affect MAb 2F5 or 4E10 recognition. Viral Seq, viral sequences. (B) Neutralization by selected HIV-positive sera of the relatively resistant HIV-1 isolate, YU2 (left), the parental HIV-2 isolate, 7312A (center), and the HIV-2-7312A chimera which contains the HIV-1 MPER graft (HIV-2/HIV-1 MPER). The absence of a bar visible above the serum identification numbers indicates that the neutralizing titer was not detectable. (C) Control assays were performed to demonstrate the specificity of neutralization inhibition by the 2F5 and 4E10 epitope-derived peptides used to map the serum neutralization specificity toward the MPER. (Left) Neutralization curves of the HXBc2 virus by 25 μg/ml of MAb 2F5 or 4E10 in the presence or absence of the MPER-derived peptides. The assay was performed with control mock peptide, 2F5 epitope peptide only, 4E10 epitope peptide only, or with the 2F5 and 4E10 cognate peptides combined. Note the specificity of blocking, as the 2F5 epitope peptide inhibits only neutralization by MAb 2F5 and the 4E10 epitope peptide inhibits only neutralization by MAb 4E10. (Right) The 2F5 and 4E10 peptides together can completely inhibit neutralization of MAbs 2F5 and 4E10 following their addition to normal human sera. The MAb concentration (in micrograms per milliliter) is shown on the x axis. (D) The same peptides are used to inhibit neutralization by the sera from patients 19 and 20 of the HXBc2 isolate following adsorption on gp120WT beads. Similar data for neutralization of the more resistant virus, JRFL, with gp120WT-adsorbed serum 20, is shown in the rightmost panel. In each case, the majority of the non-gp120-directed neutralization of the virus was blocked by the 4E10 peptide, while the 2F5 peptide has no effect on neutralization. These data demonstrate that both sera contain 4E10 epitope-directed neutralizing antibodies that target the highly conserved gp41 MPER region of the HIV-1 envelope glycoproteins.
TABLE 4.
Serum ID50 titers in the absence and presence of MPER-derived peptides
| Serum | Sample fraction | Virus | ID50 for peptide:
|
% Neutralization inhibited by test peptide:b
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Mocka | 2F5.01 | 4E10.22 | MPR.03 | 2F5.01 | 4E10.22 | MPR.03 | |||
| 1 | Untreated plasma | HIV-2/C1 | <20 | <20 | <20 | <20 | |||
| Untreated plasma | HIV-2/C1 | 22 | 21 | 10 | 10 | 0 | 55 | 55 | |
| 6 | Untreated plasma | HIV-2/C1 | 40 | 51 | 20 | 10 | 0 | 50 | 75 |
| Untreated plasma | HIV-2/C1 | 55 | 49 | 21 | 10 | 11 | 62 | 82 | |
| gp120 Ab-depleted fractionc | SF162 | 59 | 63 | 77 | 0 | 0 | |||
| 19 | Untreated plasma | HIV-2/C1 | 281 | 276 | 42 | 72 | 2 | 85 | 74 |
| Untreated plasma | HIV-2/C1 | 243 | 183 | 69 | 110 | 25 | 72 | 55 | |
| Untreated serum | HXBc2 | 1,005 | 720 | 628 | 28 | 38 | |||
| gp120 Ab-depleted fraction | HXBc2 | 82 | 79 | 20 | 4 | 76 | |||
| gp120 Ab-depleted fraction | SF162 | <20 | <20 | <20 | |||||
| 20 | Untreated plasma | HIV-2/C1 | 563 | 512 | 34.8 | 24.9 | 9 | 94 | 96 |
| Untreated plasma | HXBc2 | 4,401 | 5,447 | 1,528 | 0 | 65 | |||
| gp120 Ab-depleted fraction | HXBc2 | 169 | 177 | 20 | 0 | 88 | |||
| gp120 Ab-depleted fraction | SF162 | 449 | 429 | 40 | 4 | 91 | |||
| gp120 Ab-depleted fraction | BaL.01 | 25 | 20 | 27 | 20 | 0 | |||
| Untreated plasma | JRFL | 1,286 | 1,583 | 977 | 0 | 24 | |||
| gp120 Ab-depleted fraction | JRFL | 40 | 56 | 10 | 0 | 75 | |||
| 30 | Untreated plasma | HIV-2/C1 | 651 | 600 | 250 | 486 | 8 | 62 | 25 |
| Untreated plasma | HIV-2/C1 | 928 | 426 | 372 | 737 | 54 | 60 | 21 | |
| 45 | Untreated plasma | HIV-2/C1 | 20.1 | 20 | 10 | 10 | 0 | 50 | 50 |
| Untreated plasma | HIV-2/C1 | 58.3 | 71.7 | 40 | 40 | 0 | 31 | 31 | |
| gp120 Ab-depleted fraction | HXBc2 | 56 | 53 | 44 | 6 | 21 | |||
| gp120 Ab-depleted fraction | SF162 | 179 | 106 | 266 | 41 | 0 | |||
| B7B5 | Untreated plasma | HIV-2/C1 | 3,204 | 1,659 | 487 | 867 | 48 | 85 | 73 |
| gp120 Ab-depleted fraction | HXBc2 | 3,953 | 3,766 | 5,413 | 4,479 | 5 | 0 | 0 | |
Mock peptide (defined as an equivalent volume of PBS) was used as a negative control.
Indicator of peptide-specific neutralizing antibody response; calculated as [1 − (ID50 with test peptide)/ID50 with mock peptide)] × 100. Values higher than 50% are shown in boldface type.
The fraction of serum that was adsorbed with gp120-coupled Dynabeads prior to the neutralization assay.
We generated soluble MPER-derived peptides overlapping the core epitopes of MAbs 2F5 and 4E10 and that specifically inhibited the virus neutralization (Fig. 4C). Of note, the 4E10 peptide is hydrophobic and can affect entry of some viruses. Hence, the peptide alone was tested in all assays and used to control for any effect on viral entry. Sera were tested against HXBc2, SF162, and JRFL, using the 2F5 and 4E10 peptides, along with mock and irrelevant peptide controls. For serum sample 20 neutralization of HXBc2, we observed an approximately twofold decrease of the serum ID50 titer in the presence of the 4E10 peptide. Since gp120-directed neutralization may mask the contribution of MPER neutralizing antibodies, selected serum samples were adsorbed with gp120WT protein to remove gp120-directed neutralizing antibodies. We first confirmed that the gp120ft fraction from two sera, 19 and 20, could still neutralize the sensitive HIV-1 isolate HXBc2, whereas the gp120ft fraction from two other sera, 6 and 30, completely lost the capacity to neutralize HXBc2 (Table 1).
We then used the MPER-containing peptides to test whether the remaining non-gp120 was targeting the gp41 MPER region. Neutralization of HXBc2 by the gp120ft fractions of sera 19 and 20 was completely eliminated in the presence of 4E10 epitope peptide but was not affected by 2F5 epitope peptide (Fig. 4D). Similar results were observed for these two serum samples when tested with virus isolate SF162 (Table 4). Furthermore, we demonstrated that the 4E10 epitope-directed antibodies in the potent serum 20 were contributing to neutralization of the relatively resistant primary virus isolate JRFL (Fig. 4D). Since we estimated that about half (45%) of the serum sample 20 neutralization of JRFL was gp120 directed (Table 1), the data here suggest that the remaining 50% of JRFL neutralization is directed against the MPER and specifically to the 4E10 epitope.
DISCUSSION
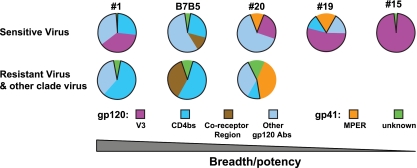
Here we have presented a stepwise analysis of complex, neutralizing HIV-1 patient sera that extends upon and validates the preliminary findings of our previous study. We demonstrate that in most sera, broad neutralizing activity was predominantly directed against gp120 (Fig. 2 and 3 and the summary pie charts in Fig. 5). In several sera, a substantial portion of this activity was directed against the CD4 binding region. We also detect, for the first time, neutralization of primary isolates that appears to map, at least in part, to the coreceptor binding region as determined by selective adsorption with the gp120I420R protein (Fig. 3D and 5). Important and as provocative as these latter data are, at this juncture, we cannot rule out the possibility that this could be a novel antibody that was not previously described that is sensitive to alterations at both gp120 residues 368 and 420. Furthermore, for the first time, with the exceptions of the MPER-directed monoclonal antibodies themselves, 2F5 and 4E10, neutralization activity targeting the highly conserved gp41 MPER region and overlapping with the MAb 4E10 epitope was detected in several sera. No activity was detected against the epitope of the gp41 MAb 2F5 in our serum samples (Table 4). Our data imply that the HIV-1 MPER is immunogenic and that it is possible to elicit neutralizing antibodies targeting this region by immunogen design, despite recent studies suggesting that homology of the MPER to self antigens, such as cardiolipin, could tolerize B cells and limit the immunogenicity of this region (18). Although the potency of these MPER-directed neutralization was modest, it is unclear whether this is due to relatively low titers present in the serum or whether the individual clones producing the MPER-directed neutralization are not as potent as 2F5 or 4E10 themselves. In Fig. 5, we show summary pie charts that illustrate that in selected sera we can map the specificity of most neutralizing activity.
FIG. 5.
Schematic summary of serum neutralization specificities. The data are shown as pie charts that summarize the fraction of the neutralization activity defined in selected sera. Both the gp120 and gp41 components and subspecificities within these two elements of Env are shown as averages for the two virus types (see below), and the values are derived from Tables 1, 2, 3, and 4. Significant neutralizing specificity pattern differences of the HIV-positive sera against sensitive viruses (top pie charts, HXBc2, SF162, BaL.01, and JRFL-Δ301) and resistant viruses (JRFL, PVO.04 and other clade viruses) are shown. For sensitive viruses, the narrowly neutralizing sera show a predominant V3-directed neutralizing specificity, while the broadly neutralizing sera display lower levels of V3-directed neutralizing specificity. Resistant viruses are neutralized only by the broadly neutralizing and potent sera. Note that most neutralizing activity maps to gp120, and several samples, including B7B5, exhibit CD4bs-directed activity. Serum B7B5 also contains neutralizing activity that is affected by the coreceptor binding site mutation. Sera 19 and 20 contain detectable MPER-directed neutralization, but of limited potency. Abs, antibodies.
In our study, the clade B gp120 proteins used for adsorption analysis were based on clade B strains BaL and YU2. The gp120 protein from each of the strains could sufficiently adsorb significant neutralization activity (Table 1) against heterologous clade B isolates and also against representatives from both clades A and C. These data were consistent with the observation that gp120 is generally the major neutralizing antibody target of envelope glycoproteins (3) and also suggested that the adsorbed antibodies were directed against conserved regions of the gp120 molecule, such as the CD4bs, or perhaps for selected sera, the coreceptor region. In this study, we analyzed the additional sera as described and found that the neutralizing breadth of CD4bs-directed antibodies in these sera is complex, varying in potency from the capacity to neutralize only sensitive isolates to CD4bs-directed antibodies capable of neutralizing relatively resistant clade B isolates. One serum, B7B5, appeared to mediate cross-clade neutralization that was CD4bs directed, similar to the two broadly neutralizing sera previously characterized. What factors determine the CD4bs-directed neutralization breadth present in several of these sera is not clear, but the data suggest that this subset of the neutralizing antibody response differs considerably in individuals. This is consistent with the fact that of the many CD4bs-directed MAbs that have been isolated, only b12 can efficiently neutralize primary isolates. The isolation of novel MAbs from B cells derived from selected patients described here is an important undertaking and is now under way to better define several of these potency, breadth, and specificity issues. In the future, as more neutralizing determinants are defined by the isolation and characterization of novel MAbs from HIV-1-infected individuals or vaccinated subjects, unequivocal mutations that eliminate a single neutralizing specificity can be incorporated into the selective adsorption process as described here. When antibodies, recombinant envelope glycoproteins, and neutralizing determinants of other chronic and variable viruses become available (e.g., hepatitis C virus), similar schema may be applicable to map patient serum neutralizing responses to aid in vaccine development for other pathogens.
Supplementary Material
Acknowledgments
We thank Brenda Hartman for help with the figures. We thank D. Burton (Scripps Research Institute) for providing MAb b12; H. Katinger (Polymun Scientific Inc.) for MAbs 2G12, 2F5, and 4E10; S. Zolla-Pazner (New York University School of Medicine) for MAb 447-52D and other V3 antibodies; and J. Robinson (Tulane Medical School) for 17b, 48d, E51, 2.8B, and 1.3D. We thank J. Sodroski and D. Gabuzda (Dana Farber Cancer Institute), J. Overbaugh (Fred Hutchinson Cancer Research Center), L. Morris (National Institute of Communicable Diseases), D. Montefiori (Duke University), J. Binley (Torrey Pines Institute for Molecular Sciences), L. Stamatatos (Seattle Biomedical Research Institute), and V. Polonis and F. McCutchan (U.S. Military Research Program) for providing virus isolates or functional Env plasmids.
This work was supported in part by the NIH intramural research program. Funding was also provided by NIH, the International AIDS Vaccine Initiative, and the Bill and Melinda Gates Foundation.
Footnotes
Published ahead of print on 12 November 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 2721955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Binley, J. M., H. J. Ditzel, C. F. Barbas III, N. Sullivan, J. Sodroski, P. W. Parren, and D. R. Burton. 1996. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res. Hum. Retrovir. 12911-924. [DOI] [PubMed] [Google Scholar]
- 3.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 7813232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 2661024-1027. [DOI] [PubMed] [Google Scholar]
- 5.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 851135-1148. [DOI] [PubMed] [Google Scholar]
- 6.Choe, H., W. Li, P. L. Wright, N. Vasilieva, M. Venturi, C. C. Huang, C. Grundner, T. Dorfman, M. B. Zwick, L. Wang, E. S. Rosenberg, P. D. Kwong, D. R. Burton, J. E. Robinson, J. G. Sodroski, and M. Farzan. 2003. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114161-170. [DOI] [PubMed] [Google Scholar]
- 7.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312763-767. [DOI] [PubMed] [Google Scholar]
- 8.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2011407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381661-666. [DOI] [PubMed] [Google Scholar]
- 10.Dhillon, A. K., H. Donners, R. Pantophlet, W. E. Johnson, J. M. Decker, G. M. Shaw, F. H. Lee, D. D. Richman, R. W. Doms, G. Vanham, and D. R. Burton. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 816548-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 851149-1158. [DOI] [PubMed] [Google Scholar]
- 12.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381667-673. [DOI] [PubMed] [Google Scholar]
- 13.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272872-877. [DOI] [PubMed] [Google Scholar]
- 14.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. USA 10218514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 816187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundner, C., Y. Li, M. Louder, J. Mascola, X. Yang, J. Sodroski, and R. Wyatt. 2005. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology 33133-46. [DOI] [PubMed] [Google Scholar]
- 17.Hartley, O., P. J. Klasse, Q. J. Sattentau, and J. P. Moore. 2005. V3: HIV's switch-hitter. AIDS Res. Hum. Retrovir. 21171-189. [DOI] [PubMed] [Google Scholar]
- 18.Haynes, B. F., J. Fleming, E. W. St. Clair, H. Katinger, G. Stiegler, R. Kunert, J. Robinson, R. M. Scearce, K. Plonk, H. F. Staats, T. L. Ortel, H. X. Liao, and S. M. Alam. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 3081906-1908. [DOI] [PubMed] [Google Scholar]
- 19.Huang, C. C., M. Venturi, S. Majeed, M. J. Moore, S. Phogat, M. Y. Zhang, D. S. Dimitrov, W. A. Hendrickson, J. Robinson, J. Sodroski, R. Wyatt, H. Choe, M. Farzan, and P. D. Kwong. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. USA 1012706-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javaherian, K., A. J. Langlois, G. J. LaRosa, A. T. Profy, D. P. Bolognesi, W. C. Herlihy, S. D. Putney, and T. J. Matthews. 1990. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science 2501590-1593. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson Hedestam, G. B., R. A. Fouchier, S. Phogat, D. R. Burton, J. Sodroski, and R. T. Wyatt. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6143-155. [DOI] [PubMed] [Google Scholar]
- 22.Krachmarov, C. P., W. J. Honnen, S. C. Kayman, M. K. Gorny, S. Zolla-Pazner, and A. Pinter. 2006. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J. Virol. 807127-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krachmarov, C. P., S. C. Kayman, W. J. Honnen, O. Trochev, and A. Pinter. 2001. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 171737-1748. [DOI] [PubMed] [Google Scholar]
- 24.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 7710557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. Hahn, and D. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 7910108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 131032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y., K. Svehla, N. L. Mathy, G. Voss, J. R. Mascola, and R. Wyatt. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 801414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascola, J. R., M. K. Louder, C. Winter, R. Prabhakara, S. C. De Rosa, D. C. Douek, B. J. Hill, D. Gabuzda, and M. Roederer. 2002. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J. Virol. 764810-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 31061-1068. [DOI] [PubMed] [Google Scholar]
- 31.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 676642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olshevsky, U., E. Helseth, C. Furman, J. Li, W. Haseltine, and J. Sodroski. 1990. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 645701-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 785205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 1004144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sattentau, Q. J., J. P. Moore, F. Vignaux, F. Traincard, and P. Poignard. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 677383-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 673978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, J. Li, and J. Sodroski. 1991. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J. Virol. 655007-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 656188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 701100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 42.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393705-711. [DOI] [PubMed] [Google Scholar]
- 43.Xiang, S. H., N. Doka, R. K. Choudhary, J. Sodroski, and J. E. Robinson. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retrovir. 181207-1217. [DOI] [PubMed] [Google Scholar]
- 44.Xu, J. Y., M. K. Gorny, T. Palker, S. Karwowska, and S. Zolla-Pazner. 1991. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 654832-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zinkernagel, R. M. 2001. Maternal antibodies, childhood infections, and autoimmune diseases. N. Engl. J. Med. 3451331-1335. [DOI] [PubMed] [Google Scholar]
- 46.Zinkernagel, R. M., A. LaMarre, A. Ciurea, L. Hunziker, A. F. Ochsenbein, K. D. McCoy, T. Fehr, M. F. Bachmann, U. Kalinke, and H. Hengartner. 2001. Neutralizing antiviral antibody responses. Adv. Immunol. 791-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zwart, G., H. Langedijk, L. van der Hoek, J. J. de Jong, T. F. Wolfs, C. Ramautarsing, M. Bakker, A. de Ronde, and J. Goudsmit. 1991. Immunodominance and antigenic variation of the principal neutralization domain of HIV-1. Virology 181481-489. [DOI] [PubMed] [Google Scholar]
- 48.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 7510892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.