Abstract
The highly conserved LWYIK motif located immediately proximal to the membrane-spanning domain of the gp41 transmembrane protein of human immunodeficiency virus type 1 has been proposed as being important for the surface envelope (Env) glycoprotein's association with lipid rafts and gp41-mediated membrane fusion. Here we employed substitution and deletion mutagenesis to understand the role of this motif in the virus life cycle. None of the mutants examined affected the synthesis, precursor processing, CD4 binding, oligomerization, or cell surface expression of the Env, nor did they alter Env incorporation into the virus. All of the mutants, particularly the ΔYI, ΔIK, and ΔLWYIK mutants, in which the indicated residues were deleted, exhibited greatly reduced one-cycle viral replication and the Env trans-complementation ability. All of these deletion mutant proteins were still localized in the lipid rafts. With the exception of the Trp-to-Ala (WA) mutant, which exhibited reduced viral infectivity albeit with normal membrane fusion, all mutants displayed loss of some or almost all of the membrane fusion ability. Although these deletion mutants partially inhibited in trans wild-type (WT) Env-mediated fusion, they were more effective in dominantly interfering with WT Env-mediated viral entry when coexpressed with the WT Env, implying a role of this motif in postfusion events as well. Both T20 and L43L peptides derived from the two gp41 extracellular C- and N-terminal α-helical heptad repeats, respectively, inhibited WT and ΔLWYIK Env-mediated viral entry with comparable efficacies. Biotin-tagged T20 effectively captured both the fusion-active, prehairpin intermediates of WT and mutant gp41 upon CD4 activation. Env without the deletion of the LWYIK motif still effectively mediated lipid mixing but inhibited content mixing. Our study demonstrates that the immediate membrane-proximal LWYIK motif acts as a unique and distinct determinant located in the gp41 C-terminal ectodomain by promoting enlargement of fusion pores and postfusion activities.
The surface envelope (Env) glycoprotein of human immunodeficiency virus type 1 (HIV-1) is initially synthesized as a gp160 precursor, which is subsequently endoproteolytically cleaved, probably in the trans-Golgi network, by a furin- or subtilisin-like cellular convertase(s) into the noncovalent heterodimer that consists of the surface gp120 subunit and the transmembrane (TM) anchor gp41 subunit. gp120 determines viral tropism through binding to cell surface CD4 and CXCR4/CCR5 chemokine receptors, and gp41 mediates membrane fusion through the induction of a transient nonlamellar structure at the point where viral and cellular membranes merge (for reviews, see references 23 and 27).
HIV-1 gp41 has common structural features shared by other retroviral TM proteins; these include the N-terminal hydrophobic fusion domain, the N-terminal α-helical leucine zipper-like heptad repeat (HR) sequence, a cysteine loop, the C-terminal α-helix, a TM region, and a long cytoplasmic domain (Fig. 1A). Binding of the extracellular subunit gp120 to receptors triggers a series of conformational changes in gp41 that promote membrane fusion between the virus and the host plasma membrane. Multiple sequences within the ectodomain of gp41 have been implicated in the membrane fusion process. A stretch of about 15 amino acids located at the N terminus of gp41, termed the fusion peptide, is believed to insert into and destabilize cellular membranes, thus triggering viral and cellular membrane fusion. During the virus-cell membrane fusion process, the two α-helical HR segments, termed NHR and CHR, assemble into a stable, six-helix bundle (9, 65, 71). This trimer of hairpin structure likely represents a fusion-active conformation of gp41 after receptor binding by exposing both the N-terminal fusion peptide and the C-terminal TM domain at the same site as a stable protein rod and bringing the cellular and viral membranes into close contact in order to form a fusion pore.
FIG. 1.
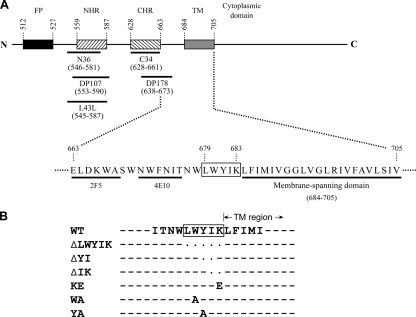
(A) Schematic representation of gp41. The rectangles marked FP, NHR, CHR, and TM indicate the locations of the fusion peptide, N-terminal HR sequence, C-terminal HR sequence, and TM domain, respectively. Residues 663 to 683 of the extracellular domain, which constitute the Trp-rich region, and the TM domain are shown in the single-amino-acid code. The immediate membrane-proximal LWYIK motif, located at residues 679 to 683, is boxed. The epitopes for the 2F5 and 4E10 MAbs and the locations of the inhibitor peptides are underlined with dark lines. (B) Construction of the LWYIK motif mutant proviruses. Site-specific, oligonucleotide-directed mutagenesis was performed based on the infectious pHXB2RU3 provirus to generate proviral variants with mutations in the LWYIK motif as indicated. Dashes and dots indicate that the residue in that position of the mutant proviruses is identical to or absent from that of the WT provirus, respectively.
The region located in the C-terminal segment of the gp41 ectodomain contains highly conserved hydrophobic residues and an unusually high content of tryptophan (Trp) residues. This conserved, predicted α-helical Trp-rich, or pre-TM, region overlaps the C-terminal part of the DP178 peptide (residues 638 to 673) (also called T20; trade name, Fuzeon or enfuvirtide), which causes potent inhibition of membrane fusion and viral entry, as well as the epitopes for two cross-clade neutralizing monoclonal antibodies (MAbs), 2F5 and 4E10 (Fig. 1A). ELDKWAS (47, 48, 66), the epitope of MAb 2F5, overlaps the C terminus of DP178, and the core epitope of 4E10, which is immediately C terminal to the 2F5 epitope, maps to residues NWFNIT (residues 671 to 676) (62, 78). A peptide representing the Trp-rich domain partitions into membrane interfaces and aggregates within them, perturbing the bilayer architecture (59, 63, 64), and mutational analyses have implicated this pre-TM region in several phases of virus replication, including membrane fusion, Env incorporation into the virus, and viral infectivity (60). Individual substitutions of Trp-667, Trp-672, Phe-673, and Ile-675 with Ala residues were also shown to greatly compromise competence for viral entry, although these mutations did not affect gp120-gp41 association or cell-cell fusion (2). Recently, it was further shown that the membrane-perturbing property of the pre-TM region is a key factor for its fusion function (70).
The sequence immediately adjacent to the TM region and C terminal to the Trp-rich region is the LWYIK motif (Fig. 1A), which is similar to those of the cholesterol recognition/interaction amino acid consensus patterns of -L/V-(X)1-5-Y-(X)1-5-R/K-, as identified by Li and Papadopoulos (36). This LWYIK motif, spanning residues 679 to 683, is highly conserved among HIV-1 isolates and is also found in the Env protein of the cpzant strain of simian immunodeficiency virus (SIV) (35, 69). Differing from those of HIV-1 isolates, the putative cholesterol-binding motifs of most SIV and HIV-2 strains consist of six amino acids, L(AS/TS)WI(K/R), which share a unique feature in that a positively charged residue (K/R) adjacent to the TM region is separated from an aromatic amino acid by an isoleucine residue.
Lipid rafts, also called detergent-resistant membranes (DRMs), are highly specialized cellular membrane microdomains characterized by being insoluble in detergents, having a light density, and being enriched in cholesterol, glycosphingolipids, and glycosylphosphatidylinositol-linked proteins that are anchored in the membrane by their attached glycosylphosphatidylinositol moiety (for reviews, see references 1 and 61). Rafts are generally believed to act as portals for HIV entry, facilitating interactions between CD4, coreceptors, and incoming virions (14, 41), although some studies suggested that HIV-1 may enter target cells via nonraft membrane domains (54, 55). Evidence for the role of rafts as platforms for the assembly of viral components and/or budding has also accumulated (38, 49, 52). Cholesterol in target cells was shown to play a crucial role in HIV-1 Env-mediated membrane fusion and viral infectivity (37, 41, 68). Also, HIV-1-associated cholesterol is important in maintaining the virion structure, integrity, and infectivity and for viral internalization (7, 30). Vincent et al. showed that the LWYIK motif, in the format of a fusion protein with the maltose-binding protein, binds to cholesteryl groups in vitro, and they thus surmised that this LWYIK motif may play a critical role in Env's association with lipid rafts and membrane fusion (69). However, this hypothesis has never been tested.
In the present study, we performed mutagenic studies to determine the role of the LWYIK motif in HIV-1 replication. We demonstrate here that the juxtamembrane LWYIK motif is not essential for Env maturation, Env association with lipid rafts, or Env incorporation into the virus and that localization of the Env in lipid rafts does not necessarily give Env its membrane fusion ability. Although the action of this motif in fusion is independent of the formation of the fusogenic six-stranded coiled-coil structure, this motif participates in enlargement of fusion pores. Moreover, as judged by the disparate characteristics of Env mutants of this motif compared to those of Trp-rich domain mutants (60), this LWYIK motif is separable from the Trp-rich domain and acts as a distinct determinant for modulating membrane fusion and postfusion activities of Env.
MATERIALS AND METHODS
Cells, hybridomas, antibodies, and fluorescent probes.
293T, HeLa, and HeLa-CD4 (clone 1022) cells were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum (FBS). In particular, HeLa-CD4 cells were grown in medium supplemented with G418. Hybridomas 902, Chessie 8, 183 (clone H12-5C), SIM2 (which secretes a MAb specifically recognizing human CD4), and sheep anti-gp120 were as previously described (10, 11). The OKT4 hybridoma was purchased from the Food Industry Research and Development Institute (Hsinchu, Taiwan). All hybridomas and CEM-SS, SupT1, and H938 cells were cultured in RPMI 1640 supplemented with 10% FBS. A MAb directed against glycoprotein G of vesicular stomatitis virus (VSV) was purchased from Sigma (St. Louis, MO). Rabbit anti-caveolin-1 antibody and anti-flotillin-1 MAb were obtained commercially from BD Biosciences (San Jose, CA). Fluorescent probes 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO), 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI), calcein-AM, and chloromethyl-benzoyl-amino-tetramethyl-rhodamine (CMTMR) were all purchased from Invitrogen (Carlsbad, CA).
Construction of LWYIK motif mutants.
For cloning purposes, the HindIII site located at nucleotide position 6027 of the HXB2 provirus was destroyed by the PCR overlap extension method. The EcoRI- and StuI-cut PCR product was then inserted in the corresponding sites of the pEGM/EB vector, which was first generated by insertion of the EcoRI-to-BamHI fragment (positions 5743 to 8475 in the provirus) in a pGEM7ZF vector (Promega, Madison, WI). The resultant construct was named pGEM/EB(ΔH). Next, the PCR overlap extension method was used to generate LWYIK motif mutants using paired sense and antisense primers specific for each mutation. The HindIII (nucleotide 8141)-to-BamHI (nucleotide 8475) fragments obtained from the PCR products were inserted in the corresponding sites of pGEM/EB(ΔH) to obtain each of the mutations. The SalI (position 5786)-to-BamHI (position 8475) fragments isolated from mutated pGEM/EB(ΔH) were then cloned in the corresponding sites in pHXB2RU3 to generate various mutants. The SalI-BamHI fragment isolated from pGEM/EB(ΔH) was cloned in pHXB2RU3 in parallel, and the resultant clone was used as the wild-type (WT) construct in this study. To obtain mutant env expression plasmids, the 2.7-kb KpnI-to-BamHI fragments isolated from mutant proviruses were cloned in the same sites within pSVE7puro, an HIV-1 long terminal repeat (LTR)-directed env expression plasmid. DNA autosequencing with appropriate primers was performed to confirm the mutations in each of the pGEM/EB(ΔH), pHXB2RU3, and pSVE7puro constructs.
Plasmid DNA transfection.
Subconfluent 293T cells grown in 10-cm petri dishes were mock transfected (used as the control) or transfected with 10 μg of pHXB2RU3 proviruses using a standard calcium phosphate coprecipitation method. Subconfluent HeLa cells grown in 10-cm petri dishes were transfected with 8 μg of proviruses by the Lipofectamine Plus transfection method (Invitrogen, Carlsbad, CA). For preparation of VSV glycoprotein G trans-complemented or HIV-1 Env-pseudotyped viral stocks, 293T cells were cotransfected with 7.5 μg each of proviruses and pHCMV-VSV G, a human cytomegalovirus promoter-directed VSV G protein expression plasmid, or with 7.5 μg each of the pHXB2ΔBglCAT (or pNL4-3R−E−Luc) and WT or mutant pSVE7puro plasmids, respectively. For the WT and mutant coexpression analyses, 293T cells were cotransfected with 1.5 μg of pIIIextat (for membrane fusion) or 7.5 μg of pHXB2ΔBglCAT (for viral entry) together with 5 μg of the WT plasmid or with 5 μg each of the WT and mutant plasmids. The pSVE7puro(ΔKS) plasmid, which is a defective env construct in which the KpnI (at nucleotide position 6351 of the HXB2 sequence)-to-StuI (at nucleotide position 6834) site was deleted, was added to the transfection mixtures to ensure that the total DNA amounts in all transfections were the same.
Virus infection studies.
Culture supernatants obtained from transfection were filtered through 0.45-μm membrane discs and normalized by reverse transcriptase (RT) activity. For viral replication assays, cell-free viruses containing 2 × 104 cpm of RT activity were used to challenge 106 CEM-SS cells. For VSV G trans-complemented viral infection, 106 SupT1 cells were challenged with VSV G trans-complemented viruses containing 2 × 106 cpm of RT activity. For assessment of single-cycle viral replication, viruses containing 106 cpm of RT activity were used to challenge 106 H938 cells. For the HIV-1 env trans-complementation assays, Env-pseudotyped reporter viruses containing 3 × 105 to 6 × 105 cpm of RT activity were used to challenge 106 CEM-SS cells. For assessment of virus entry into H938 cells and for VSV G trans-complemented or HIV-Env-pseudotyped viral infection studies, the previously described spinoculation method (50) was followed. For the analyses of Env expressed in acutely infected CD4+ T cells, CEM-SS cells were inoculated with WT or mutant viruses as described above. Culture supernatants and cells were retained in initial feedings to scale up the cell numbers, and culture volumes were eventually maintained at about 20 ml. At or near the peak of infection, as determined by observations of syncytium formation and confirmed by RT activity, cell lysates and culture supernatants were prepared and analyzed for Env association with lipid rafts and assembly into virions.
SDS-PAGE and Western blot analyses.
Two days after proviral DNA transfection or infection with VSV G trans-complemented recombinant viruses, virions were isolated from culture supernatants over a 20% (wt/wt) sucrose cushion prepared in phosphate-buffered saline (PBS). Cell and viral lysates were prepared and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis using mouse 902, Chessie 8, and 183 MAbs, respectively. For the analysis of Env incorporation into viral particles from infected CEM-SS cells, culture supernatants at or near the peak of infection were collected, filtered through 0.45-μm filters, concentrated by ultracentrifugation at 100,000 × g for 2 h at 4°C through a 20% sucrose cushion, and resuspended in RPMI with 1% FBS at 1/100 of the starting volume. The p24 contents of stocks of reconstituted viral concentrates were determined by enzyme-linked immunosorbent assay (ZeptoMetrix, Buffalo, NY), and equal amounts of WT or mutant viruses (0.5 μg) were analyzed by SDS-PAGE and subsequent Western blotting.
Enzymatic analyses.
Cell-free viruses produced from proviral transfection or from HIV-1 Env-pseudotyped or VSV G trans-complemented viruses were assayed for virion-associated RT activity as previously described (16). Cell extracts prepared from HXBΔBglCAT and NL4-3R−E−Luc pseudotype infection analyses were assayed for chloramphenicol acetyltransferase (CAT) activity as previously described (18) and for firefly luciferase activity using the luciferase assay system (Promega), respectively.
Env cell surface expression.
Proviral DNA-transfected HeLa cells grown in 6-cm petri dishes were metabolically labeled with [35S]methionine for 4 h and then surface biotinylated with 0.25 mg/ml of membrane-impermeable sulfo-N-hydroxysuccinimide-biotin as previously described (10, 11). Cell lysates were first immunoprecipitated with pooled anti-HIV-1 antisera and protein A-Sepharose beads. After extensive washing, the Env proteins were released from beads by boiling with Laemmli loading dye without bromophenol blue. The samples were divided into two portions. One portion was directly resolved by SDS-PAGE; the other was precipitated with neutravidin-agarose (Pierce. Rockford, IL), and the Env proteins were released from beads by boiling with loading dye prior to SDS-PAGE.
CD4-binding assay.
A previously described procedure (15) was followed to assess the CD4-binding ability of Env proteins. Briefly, HeLa cells cotransfected with the WT or mutant pSVE7puro plasmids in the presence of pIIIextat, an HIV-1 LTR-driven Tat expression plasmid, were metabolically labeled with [35S]methionine, and cell lysates were prepared. A portion of the cell lysates was immunoprecipitated with pooled AIDS patients' antisera preadsorbed onto protein A-Sepharose beads. After extensive washing with radioimmunoprecipitation assay buffer, the immune complexes were resolved by SDS-PAGE. Another portion of the lysates was incubated with SupT1 cell lysates, which were used as the source of CD4. The samples were then precipitated with OKT4 and SIM2 MAb-preadsorbed mixtures of protein A- and protein G-Sepharose beads, and the immune complexes were separated by SDS-PAGE.
Hetero-oligomerization study.
HeLa cells grown in 6-cm petri dishes were transfected with 2 μg of WT or TC pSVE7puro (17) or with 2 μg of the TC plasmid together with 2 μg of the WT or mutant pSVE7puro plasmids in the presence of 1 μg of pIIIextat. pSVE7puro(ΔKS) was added to the transfection mixtures to maintain the same amounts of total plasmids. Aliquots containing an equal volume of cell lysates from each transfection were directly subjected to Western blotting using the 902 MAb. Other aliquots were first precipitated with protein G-Sepharose beads coated with 5 μl of Chessie 8 ascitic fluid, and the precipitates were washed, eluted from the beads, and analyzed by Western blotting using goat anti-gp120.
Analysis of Env association with lipid rafts.
To examine Env expression in 293T cells, sucrose gradient equilibrium ultracentrifugation was performed as previously described (10) to assess Env localization in lipid rafts. To examine Env expression from acutely infected CEM-SS cells, a scaled-down version of the sucrose density gradient technique was performed using a Beckman SW60 rotor. Sample fractions were collected and trichloroacetic acid precipitated as described above for SDS-PAGE and Western blot analysis.
Peptide inhibition study.
T20 (residues 638 to 673 in Env) and L43L (residues 545 to 587) were synthesized in an automatic mode using a solid-phase synthesizer from Applied Biosystems (Foster City, CA) as previously described (12), and their N and C termini were capped by acetyl and amide groups, respectively. The peptides were cleaved from the resin and purified by high-pressure liquid chromatography with a reverse-phase C18 column, and the primary sequences of peptides were confirmed by matrix-assisted laser desorption-ionization mass spectrometry. Peptide inhibition analyses were performed as previously described (44) with modifications. Both T20 and L43L were dissolved in dimethyl sulfoxide and quantified by measuring the absorbance at 280 nm. On the day of infection, HeLa-CD4 cells were seeded at 5 × 104 cells per well in a 96-well flat-bottom plate at least 4 h prior to infection. For each well inoculation, 50 μl of concentrated WT or ΔLWYIK Env-pseudotyped NL4-3R−E−Luc reporter viruses containing 1 × 105 cpm (for WT pseudotype) or 1 × 106 cpm (for ΔLWYIK pseudotype) of RT activity was mixed with 20 μl of T20 or L43L to arrive at the specified final concentrations of inhibitors and a constant concentration of dimethyl sulfoxide in all mixtures. The peptide-virus mixtures were incubated for 1 h at 37°C before being overlaid onto HeLa-CD4 monolayers, and the cultures were subjected to spinoculation. The cultures were then incubated at 37°C overnight, washed once with PBS, replenished with 100 μl of fresh medium per well, and allowed to recover for 24 h before luciferase activity was assayed in triplicates. Data were plotted on SigmaPlot and curve-fitted with a model of three-parameter logistic functions. Percentage inhibition was calculated with the following formula: % inhibition = (averageno inhibition − point value)/(averageno inhibition − averagemaximum inhibition) × 100%.
Capture of gp41 fusion-active conformation.
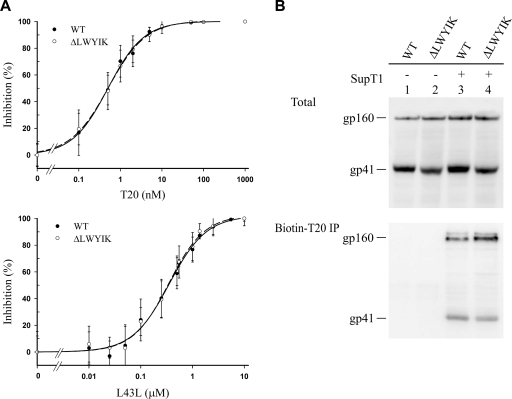
Capture of the gp41 fusion intermediate was performed as previously described (26) with an N-terminally biotinylated T20 derivative (residues 635 to 673), which was synthesized and purified by reverse-phase high-pressure liquid chromatography to >95% purity by Genemed Synthesis, Inc. (San Antonio, TX). One day after 293T cells were transfected with pCAGGS-based WT or ΔLWYIK expression plasmid, 1 × 106 Env-expressing 293T cells were incubated with 50 μg of biotinylated T20 in the presence or absence of 5 × 106 SupT1 cells at 37°C for 2 h. After being washed with PBS three times, cells were lysed with 50 mM Tris-HCl (pH 7.5) containing 0.15 M NaCl and 1% TritonX-100, and aliquots of cell lysates were incubated with 30 μl of neutravidin agarose suspension at 4°C for 12 h. The precipitated complexes were analyzed by Western blotting with Chessie 8 MAb.
Membrane fusion and fluorescent probe transfer assays.
The syncytium-forming ability of Env proteins was evaluated as previously described (11). In brief, 293T cells were transfected with 5 μg of the WT or mutant pSVE7puro plasmids together with 1.5 μg of pIIIextat. One day after transfection, 106 H938 cells were added to transfected cells. Two days after coculture, cell lysates were prepared and the CAT activity was measured. Dye transfer assays were performed as previously described (2). Briefly, HeLa effector cells were transfected with the WT or ΔLWYIK mutant plasmids. At 18 h posttransfection, cells were labeled with 2 μM of the lipophilic probe DiO (green fluorescence, excitation at 484 nm, and emission at 501 nm), whereas target CEM-SS cells were labeled with 2 μM of the lipophilic DiI (red fluorescence, excitation at 550 nm, and emission at 565 nm) for 15 min at room temperature. After extensive washing with PBS, the target cells were added to the Env-expressing effector cells at a ratio of 6:1. For content mixing experiments, Env-expressing HeLa and CEM-SS cells were labeled with 0.5 μg/ml each of the cytosolic probes calcein-AM (green fluorescence, excitation at 496 nm, and emission at 517 nm) and CMTMR (red fluorescence, excitation at 540 nm, and emission at 566 nm) for 5 min at room temperature and then extensively washed and cocultured. Following 5 h of coculture at 37°C, the suspension of CEM-SS cells was harvested and fixed with 4% paraformaldehyde in PBS. Two-color flow cytometric analyses were performed using a FACSCalibur (BD Bioscience, San Jose, CA) with Cell-Quest software (BD Biosciences).
RESULTS
Construction and replication of LWYIK motif mutant viruses.
To study the role of the LWYIK motif (Fig. 1A) (located immediately adjacent to the TM region and positioned at residues 679 to 683 of Env) in the virus life cycle, substitutions and deletions were introduced into this motif by site-directed mutagenesis based on the pHXB2RU3 molecular clone (67). For ease of cloning, the HindIII site located at nucleotide 6027 of the pHXB2RU3 infectious clone was removed by a C-to-T substitution which was outside the env open reading frame. Although this substitution gave rise to Val-to-Ala and Leu-to-Phe mutations at residues 67 and 21 of the tat and rev genes, respectively, these substitutions were conservative and did not affect HIV-1 replication (data not shown). The resultant provirus was therefore used as the WT construct in the present study and as the construct from which the mutant proviruses were derived (Fig. 1B). The provirus that encoded an Env in which this motif was deleted was referred to as the ΔLWYIK mutant. Mutants with two amino acids deleted, i.e., the ΔYI and ΔIK mutants, were also constructed. To examine the specificity of this motif in viral replication, substitutions of Glu for Lys-683 and Ala for Trp-680 or Tyr-681 were also generated, and these were termed the KE, WA, and YA mutants, respectively.
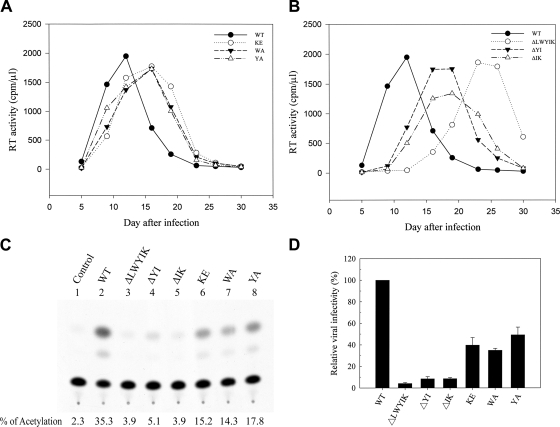
To examine the replication capacities of these mutant viruses, 293T cells were transfected with each of the WT and mutant proviruses, and the resultant cell-free viruses, containing equal amounts of RT activity, were used to challenge a human T4-lymphoblastoid CEM-SS cell line. Postinfection viral production as measured by RT activity in the culture medium was monitored. The peak of RT production for the three single-substitution mutants KE, WA, and YA shifted to day 16, from day 12 for the WT virus (Fig. 2A). Nevertheless, these three point substitution mutants replicated with kinetics similar to that of the WT virus (Fig. 2A). The RT peak for the ΔLWYIK mutant virus occurred on day 23, whereas those for the ΔYI and ΔIK mutant viruses occurred at around days 16 to 19 after infection (Fig. 2B). All three of these deletion mutants eventually replicated productively in cell culture (Fig. 2B).
FIG. 2.
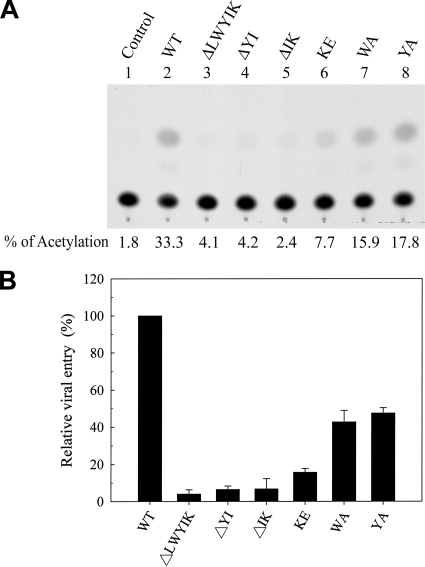
Effects of mutations in the LWYIK motif on viral infectivity. (A and B) Replication kinetics of viruses. Cell-free WT and mutant viruses containing equal amounts of RT activity were used to challenge CEM-SS cells, and virus production in the culture media as measured by RT activity was monitored after infection. (C and D) One-cycle viral replication assay. H938 cells were infected with RT-normalized WT or mutant viruses. Two days after infection, cell lysates were prepared, and CAT activity was determined. A typical CAT activity analysis is shown in panel C. The background CAT level detected in mock-infected cells (control) was subtracted from the CAT activity of the WT or mutant virus infection. The relative infectivities of mutant viruses are expressed as percentages of that of the WT virus. The results from four independent experiments (mean ± standard deviation) are shown in panel D.
To determine the effects of these mutations on viral entry, equal amounts of the WT and mutant viruses normalized by RT activity were used to infect H938 (24), a cat-harboring reporter H9 cell line, which allows assessment of nearly one-cycle viral entry based on the ability of Tat, upon viral infection, to transactivate HIV-1 LTR-linked cat gene expression in H938 cells (Fig. 2C). When the results from four individual experiments were quantitated and averaged, the viral infectivities of the KE, WA, and YA mutants were reduced to 40% to 50%, whereas those of the three deletion mutants were reduced to <10% of that of the WT virus (Fig. 2D).
Viral protein expression of the LWYIK motif mutants.
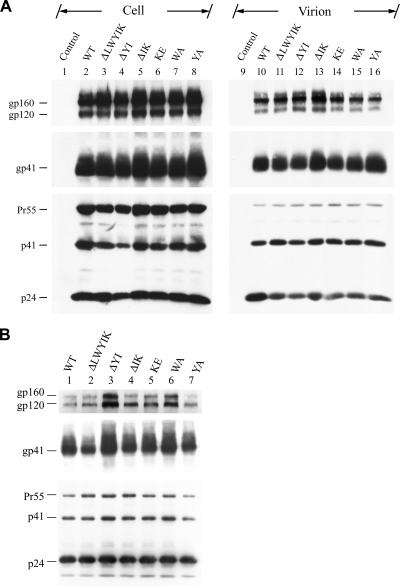
To study the protein expression profiles of these mutants, HeLa cells were mock transfected or transfected with each of the WT and mutant proviruses, and cell and virus lysates were analyzed by SDS-PAGE followed by Western blotting using the 902, Chessie 8, and 183 MAbs. The 902 MAb specifically detects gp160 and gp120, the Chessie 8 MAb reacts with gp41, and the 183 MAb recognizes the p24 capsid protein. All of the WT and mutant proviruses produced comparable amounts of intracellular and virion-associated Gag Pr55 precursor and its cleavage products (p41 and p24) upon transfection (Fig. 3A). The WT and all mutant proviruses also produced comparable amounts of intracellular gp160, gp120, and gp41 (Fig. 3A, lanes 2 to 8) as well as virion-associated gp120 and gp41 (Fig. 3A, lanes 10 to 16).
FIG. 3.
Assessment of viral protein expression. (A) HeLa cells were mock transfected (control) or transfected with the WT or mutant proviruses, and equal volumes of cell and virion lysates were subjected to SDS-PAGE followed by Western blotting analysis using the 902, Chessie 8, and 183 MAbs. (B) Cell-free culture supernatants obtained from acutely infected CEM-SS cells at the peak of viral replication were concentrated by ultracentrifugation and reconstituted with RPMI containing 1% FBS, and p24 levels were determined. WT and mutant virions containing equal amounts of p24 were resolved by SDS-PAGE followed by Western blotting using the 902, Chessie 8, and 183 MAbs.
We then determined the viral protein expression of the mutants in CD4+ T cells, which are natural target cells for HIV-1 infection, by a high-level transient HIV-1 expression system based on trans-complementation with the VSV G protein. All of the mutants produced levels of intracellular Gag and Env proteins comparable to those produced by the WT virus upon infection (see Fig. S1, lanes 2 to 8, in the supplemental material). Also, Gag assembly/budding and incorporation of gp120 and gp41 into the mutant viruses were normal compared to those observed in the WT virus (see Fig. S1, lanes 10 to 16, in the supplemental material).
To further address mutant Env incorporation into viruses, the amounts of p24 in cell-free WT and mutant virions recovered from supernatants of acutely infected CEM-SS cells collected at or near the peak of virus replication were normalized prior to Western blotting. Comparable amounts of Gag products were detected for WT and mutant viruses, and mutations in this motif did not greatly affect the assembly of gp120 and gp41 into the mutant virions (Fig. 3B).
In these studies, considerable amounts of WT and mutant gp160 proteins were detected in the viral particles produced, which was in agreement with previous observations (33, 42). Since Env is heavily and differentially N glycosylated in the Golgi apparatus, resulting in a mixture of complex, hybrid, and high-mannose carbohydrate structures (28, 43), this may explain why a gp160 doublet was detected on virions when expressed in HeLa and SupT1 cells (Fig. 3; see Fig. S1 in the supplemental material).
Cell surface expression of Env mutants.
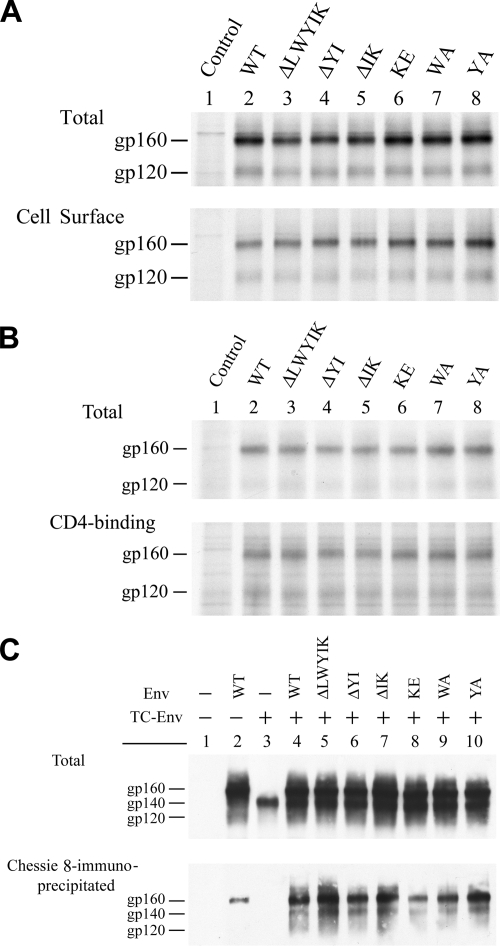
To assess Env cell surface expression, HeLa cells transfected with each of the WT and mutant proviruses were labeled with [35S]methionine and then surface biotinylated using the membrane-impermeative sulfo-N-hydroxysuccinimide-biotin. This method was shown to detect bona fide expression of viral Env glycoproteins on the cell surface (10, 11). All of the mutant Env proteins were expressed on the cell surface as effectively as was the WT Env (Fig. 4A), implying that this motif is not directly involved in the proper folding of Env or its intracellular trafficking to the cell surface.
FIG. 4.
Characterization of LWYIK motif mutant Env proteins. (A) Cell surface expression. HeLa cells mock transfected (control) or transfected with the WT or mutant proviruses were metabolically labeled with [35S]methionine and surface biotinylated with sulfo-N-hydroxysuccinimide-biotin. Levels of Env proteins expressed in cells (top panel) or on the cell surface (bottom panel) were assessed as described in Materials and Methods. (B) CD4-binding ability. HeLa cells transfected with pSVE7puro(ΔKS) (control), WT, or mutant pSVE7puro plasmids in the presence of pIIIextat were metabolically labeled with [35S]methionine. Levels of Env proteins expressed in cells (top panel) and the CD4-binding ability of Env proteins (bottom panel) were determined. (C) Oligomerization ability. HeLa cells were transfected with pSVE7puro(ΔKS) (control), WT, or TC mutant pSVE7puro plasmid or cotransfected with the TC plasmid along with the WT or LWYIK motif plasmids in the presence of pIIIextat. Levels of Env proteins expressed in cells (top panel) and the self-assembly abilities of the Env proteins (bottom panel) were determined.
CD4-binding ability of Env mutants.
To determine whether mutations in the LWYIK motif affect the ability of Env to bind to the cellular receptor CD4, a previously described procedure (15) was followed. No difference in the CD4-binding ability was noted among the WT and mutant proteins (Fig. 4B), indicating that this LWYIK motif is not required for the efficient binding of Env to its primary CD4 receptor.
Oligomerization ability of Env mutants.
We then determined the oligomerization abilities of these Env mutants by assessing their interactions with the TC mutant Env, which contains the normal LWYIK sequence but lacks the entire gp41 cytoplasmic domain (17), as previously reported (19). This TC Env was not precipitated by the Chessie 8 MAb, which maps to residues 727 to 732 in the cytoplasmic domain, unless it forms a hetero-oligomer with an Env containing the normal cytoplasmic domain. Transfection with the env-defective ΔKS plasmid did not produce Env proteins (Fig. 4C, top panel, lane 1). A truncated gp160 precursor, labeled like gp140, was produced by the TC construct (Fig. 4C, top panel, lane 3). gp160 as well as gp140 and gp120 were produced by cotransfection with the TC and each of the WT or mutant plasmids (Fig. 4C, top panel, lanes 4 to 10). When expressed alone, WT gp160, but not TC gp140, was precipitated by Chessie 8 (Fig. 4C, bottom panel, lanes 2 and 3, respectively). Nevertheless, similar amounts of TC gp140 precursor were coprecipitated with this MAb when it was coexpressed with the WT or each of the mutant proteins (Fig. 4C, bottom panel, lanes 4 to 10). This demonstrates that all of these LWYIK motif mutants are able to self-assemble into an oligomeric structure.
Env trans-complementation ability of the mutants.
Since these mutant proteins were effectively expressed on the cell surface and incorporated into the virus, we determined whether these mutant proteins might confer one-cycle viral entry into CEM-SS cells by generating env-defective HIV-1 cat-encoding HXBΔBglCAT reporter viruses from cotransfection with pSVE7puro(ΔKS) (which was used as the control), WT, or mutant Env plasmids. A representative CAT assay is shown in Fig. 5A. When the effects of these mutations on the ability of Env to mediate one-cycle viral infectivity was quantitated, the WA and YA mutants showed reduced trans-complementation ability to 40% to 50% of that of the WT Env, and the KE mutant showed reduced trans-complementation ability to 16% of that of the WT Env (Fig. 5B). The three deletion mutants also showed greatly reduced trans-complementation abilities to <10% of that of the WT Env (Fig. 5B). These results suggest that mutations in the LWYIK motif may inhibit membrane fusion and/or postfusion events.
FIG. 5.
Env trans-complementation assay of mutant Env proteins. Env pseudotypes produced from 293T cells cotransfected with pHXB2ΔBglCAT and each of the ΔKS (control), WT, or mutant Env plasmids were normalized for RT activity before being used to challenge CEM-SS cells. CAT activity was measured 3 days after virus infection. A representative result is shown in panel A. The background CAT level detected in the defective virus produced from ΔKS pSVE7puro cotransfection was subtracted from the CAT activity of the WT or mutant pseudotypes. The relative viral entry ability mediated by mutant proteins is expressed as a percentage of that mediated by the WT Env. The results from three independent experiments are shown as the mean ± standard deviation in panel B.
Lipid raft association of the mutants.
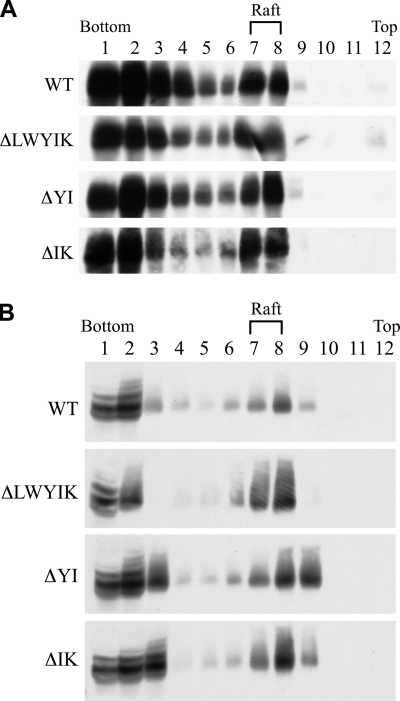
The HIV-1 Env has been shown to be located in lipid raft membranes (4, 10, 49, 57). To determine whether exclusion from lipid rafts may be responsible for the inhibited one-cycle viral replication and Env trans-complementation ability of the mutants, 293T cells were transfected with the WT or each of the three deletion mutant proviruses, since these deletion mutants exhibited greater reduction in virus infectivity and Env trans-complementation ability than substitution mutants (Fig. 2 and 5). Cells were extracted with cold Triton X-100, and the lysates were analyzed using sucrose gradient equilibrium ultracentrifugation. To determine the reliability of this lipid raft flotation method that we employed in separating raft-associated proteins from detergent-soluble proteins, localization of CD4 and VSV G protein in lipid rafts was assessed. VSV G was distributed predominantly to the detergent-soluble bottom fractions (see Fig. S2, second panel, in the supplemental material), and CD4 could be detected in lipid raft fractions 7 and 8 (see Fig. S2, first panel, in the supplemental material), which was consistent with their membrane localization properties (5, 53). Two other raft-associated cellular proteins, caveolin-1 and flotillin-1, were also analyzed. A significant fraction of caveolin-1 was associated with DRMs, while flotillin-1 was located predominantly to lipid rafts (see Fig. S2, third and fourth panels, respectively, in the supplemental material). When the WT and mutant Env proteins were assessed, a significant fraction of these proteins was detected in DRMs (Fig. 6A). To determine whether Env localization to lipid rafts is Tat dependent or not, 293T cells were cotransfected with pHP, which carries gag-pol genes (13), and with HIV-1 Tat-independent, pCAGGS-based rev/env expression plasmids encoding each of WT and mutant proteins (data not shown). The pCAGGS vector contained the cytomegalovirus early enhancer/chicken β-actin promoter. The WT and three deletion mutant proteins could be detected in DRM fractions (see Fig. S3 in the supplemental material).
FIG. 6.
Effects of deletions in the LWYIK motif on Env localization in lipid rafts. (A) 293T cells were transfected with 10 μg each of the WT and mutant proviruses, and transfected cells were extracted with 1% Triton X-100 at 4°C. The lysates were subjected to lipid raft membrane flotation assay followed by Western blotting using the Chessie 8 MAb. The distribution profile of gp41 is shown. (B) CEM-SS cells acutely infected with WT or mutant viruses were collected at or near the peak of virus infection, extracted with 1% cold Triton X-100, and then subjected to a lipid raft fractionation analysis as described for panel A.
To exclude the possibility that Env fractionation into lipid rafts is due to overexpression of Env, which may result in Env distribution into raft fractions, cold Triton X-100-extracted cell lysates containing WT and ΔLWYIK mutant proteins were either left untreated or treated with 1% SDS at 4°C for 30 min prior to membrane flotation. Treatment with SDS, which drastically disrupts the Env-raft association, abolished localization of WT and mutant proteins as well as caveolin-1 and flotillin-1 in lipid rafts (see Fig. S4 [compare left and right panels] in the supplemental material).
To understand whether WT and mutant Env proteins are associated with DRMs in the context of HIV infection, cold detergent-extracted cell lysates from acutely infected CEM-SS cells were analyzed by lipid raft membrane flotation. The three mutants as well as WT Env were still localized in the lipid raft membranes (Fig. 6B). These results together indicate that flotation into DRMs is an intrinsic property of WT and mutant Env proteins and that the LWYIK motif does not play a critical role in Env's association with DRMs.
In these lipid raft flotation analyses, only a fraction of WT and mutant proteins was localized in DRMs. Since the vast majority of the Env after synthesis is retained in the endoplasmic reticulum or a cis-Golgi compartment in an endoglycosidase H-sensitive state (3, 11, 20) and only a small fraction of Env is cleaved to produce the mature gp120-gp41 complex and then transported to the cell surface (75), it is likely that in the early stage of the Env exocytic transport pathway, Env is not associated with lipid rafts until it reaches a later stage of its maturation process. This may explain why only a fraction of Env is associated with lipid rafts.
Membrane fusion ability of Env mutants.
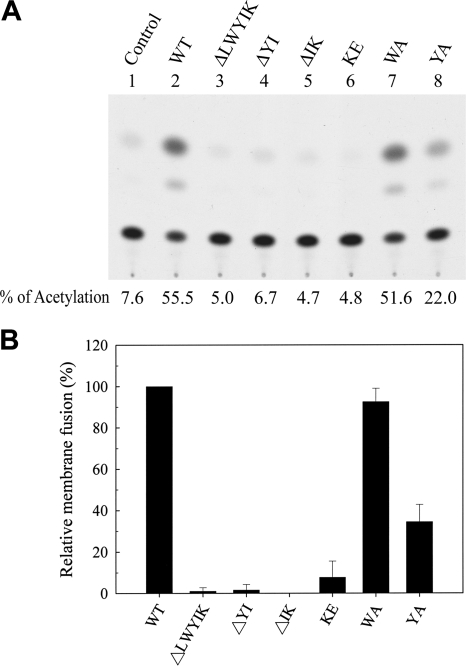
To determine whether the inhibited viral infectivity of these mutants can be accounted for by their reduced membrane fusion, 293T cells coexpressing Tat and Env were cocultured with H938 cells, and the CAT activity was measured (Fig. 7A). This assay was based on the ability of Tat expressed in effector cells to transactivate LTR-driven cat reporter gene expression in H938 cells upon Env-mediated membrane fusion. When the results from three individual studies were quantitated, the membrane fusion abilities of the YA and KE mutants were reduced to about 35% and <10% of that of the WT Env, respectively, but that of the WA mutant was unaffected (Fig. 7B). Also, the three deletion mutants showed significantly reduced membrane fusion, to almost background levels (Fig. 7B).
FIG. 7.
Effects of mutations in the LWYIK motif on the Env membrane fusion ability. 293T cells were cotransfected with pIIIextat along with the ΔKS (control), WT, or mutant pSVE7puro plasmids as indicated and cocultured with 106 H938 cells, and CAT activity was assessed. A representative result is shown in panel A. The background CAT level detected in the absence of Env (control) was subtracted from the CAT activity detected in the presence of the WT or mutant proteins. The relative membrane fusion activity of the mutant proteins is expressed as a percentage of that of the WT Env. The results from three independent experiments (mean ± standard deviation) are shown in panel B.
Dominant negative inhibition in WT Env-mediated membrane fusion and viral entry by deletion mutants.
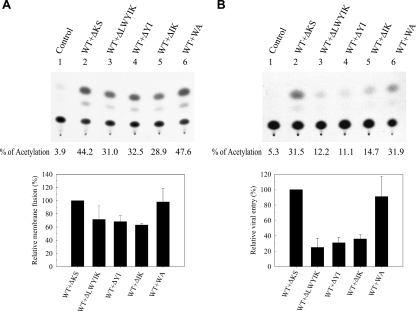
The observation that the WA mutant showed reduced viral infectivity and trans-complementation ability, albeit with normal fusogenicity (Fig. 2, 5, and 7), implied that the LWYIK motif may also play a role in postfusion events in addition to a role in cell-cell fusion. To further define the role of this motif in virus infection, the ability of these deletion mutants and the WA mutant, which did not display a significant effect on membrane fusion (Fig. 7), to trans-dominantly interfere with WT Env-mediated membrane fusion and viral infectivity was determined. As a control, the WA did not show a significant dominant interference effect on WT Env-mediated fusion and viral entry when coexpressed with the WT Env (Fig. 8). Although the deletion mutants partially inhibited in trans WT Env-mediated membrane fusion when coexpressed with the WT Env (Fig. 8A), they were more effective at dominantly interfering with WT Env-mediated viral infectivity (Fig. 8B), implying that this motif is critical not only for membrane fusion but also for postfusion events.
FIG. 8.
Ability of deletion mutants to dominantly interfere with WT Env-mediated membrane fusion and viral entry. (A) 293T cells were cotransfected with 1.5 μg of pIIIextat together with 10 μg of pSVE7puro(ΔKS) (control) (lane 1), 5 μg each of the WT and ΔKS plasmids (lane 2), or 5 μg each of the WT and mutant plasmids (lanes 3 to 6). Transfected cells were cocultured with H938 cells, and CAT activity was assayed. (B) 293T cells were cotransfected with 7.5 μg of pHXBΔBglCAT and 10 μg of pSVE7puro(ΔKS) (control) (lane 1), 5 μg each of the WT and ΔKS plasmids (lane 2), or 5 μg each of the WT and mutant pSVE7puro plasmids (lanes 3 to 6). Cell-free viruses normalized by RT activity from each transfection were used to challenge 106 CEM-SS cells, and CAT activity was assayed. In each case, a representative CAT assay is shown in the top panel, whereas the results from four (for membrane fusion) and three (for viral entry) independent experiments (mean ± standard deviation) are shown in the bottom panel.
There is no occurrence of revertants or second-site mutations in ΔLWYIK mutant virus infection.
We noted in Fig. 2B that the ΔLWYIK mutant virus, which had the most drastic mutation among the mutants analyzed, eventually replicated productively in CEM-SS cells, albeit with slower replication kinetics than the WT virus. To determine whether revertants or second-site mutations may arise during this culture period, cell-free culture supernatants were collected from CEM-SS cells infected with the WT or ΔLWYIK mutant virus at their replication peaks (i.e., days 12 and 24 postinfection, respectively, in this repetitive experiment). Viral RNAs were isolated using the QIAamp viral RNA minikit (Qiagen, Valencia, CA), and cDNAs were synthesized with an oligo(dT)20 primer using the Superscript III RT-PCR kit (Invitrogen, Carlsbad, CA). The sequence between nucleotides 5971 to 9153, which encompassed the entire env gene, was amplified by PCR using PfuUltra II HS high-fidelity DNA polymerase mix (Stratagene, Cedar Creek, CA), phosphorylated with T4 polynucleotide kinase, and subcloned in the pcDNA3 vector (Invitrogen). Eleven WT and mutant clones were picked and sequenced with a set of primers. We found that the WT and mutant env sequences from 11 independent clones were identical to those of the input viruses and that no additional mutations other than deletion of the LWYIK motif were noted in these individual mutant clones. These results together indicated that the delayed viral replication of the ΔLWYIK mutant virus arises not from outgrowth of revertants or compensatory, second-site mutations in the env gene but rather from multiple infections by the residual infectivity of the mutant virus after a long period of culture. This notion was further evidenced by the observations that the one-cycle viral infectivity, env-mediated trans-complementation ability, and membrane fusion ability of the ΔLWYIK mutant virus were not completely blocked (Fig. 2C and D, 5, and 7).
Independence of the action of the LWYIK motif from that of the NHR and CHR during fusion.
Synthetic peptides derived from the CHR, e.g., T20 and C34, inhibit HIV infectivity and cell-cell fusion at nanomolar concentrations (9, 34, 74), and that a CHR peptide binds gp41 following receptor activation (26). Peptides corresponding to the NHR, e.g., DP107 (also called T21) and N36 (Fig. 1A), also block virus infection and cell-cell fusion (12, 73). These CHR and NHR peptides are believed to act in a trans-dominant negative manner to bind to the gp41 fusion intermediates prior to formation of the six-helix bundle, thus preventing membrane fusion and virus entry (9, 72).
To address whether the action of the LWYIK motif in fusion is independent of that of the NHR and CHR, the sensitivities of the WT and ΔLWYIK mutant Env-pseudotyped NL4-3R−E−Luc reporter viruses to T20 and L43L were assessed. L43L (12) is a 43-mer peptide (residues 545 to 587) that encompasses the NHR of gp41, and its sequence partially overlaps that of DP107 (residues 553 to 590) (Fig. 1A). Both the WT and ΔLWYIK mutant pseudotypes were equivalently inhibited by the T20 and L43L peptides, respectively, as similar responsiveness to each peptide concentration was obtained for these two pseudotypes (Fig. 9A, top and bottom panels, respectively).
FIG. 9.
Sensitivities of WT and ΔLWYIK mutant viruses to CHR and NHR peptides. (A) Inhibition of WT and ΔLWYIK mutant Env pseudotypes to L43L and T20 peptides. Replication-incompetent NL4-3R−E−Luc viruses pseudotyped with WT or ΔLWYIK mutant proteins were incubated with T20 or L43L peptide at the concentrations indicated, and cell lysates were assayed for luciferase activity. Ten times more ΔLWYIK mutant pseudotyped virus, in terms of RT activity, than the WT pseudotype was examined. The percent inhibition of WT and mutant pseudotypes at each peptide concentration was calculated as described in Materials and Methods from at least five independent analyses, and the averages with standard deviations are shown. (B) 293T cells transiently expressing WT or ΔLWYIK mutant Env were incubated with biotinylated T20 in the presence or absence of SupT1 cells. The cell lysates were directly analyzed by Western blotting (top panel) or subjected to coprecipitation with neutravidin beads prior to Western blotting with Chessie 8 MAb (bottom panel).
To confirm that receptor activation triggers formation of a fusion-active, prehairpin intermediate that is accessible to a peptide inhibitor in mutant Env, binding of biotinylated T20 to WT or mutant Env-expressing cells cocultured with or without SupT1 cells was assessed, followed by precipitation of the gp41-tagged T20 complex with neutravidin beads. Similar amounts of the gp160 precursor and gp41 of WT and mutant Env proteins were captured by tagged T20 in the presence, but not in the absence, of SupT1 cells (Fig. 9B). These results together indicate that upon receptor activation the ΔLWYIK mutant Env expressed on the cell surface, though impaired in virus infectivity and membrane fusion, is still able to undergo conformational changes to a fusogenic intermediate that is accessible to and neutralized by NHR and CHR peptides.
Deletion of the LWYIK motif inhibits fusion pore enlargement.
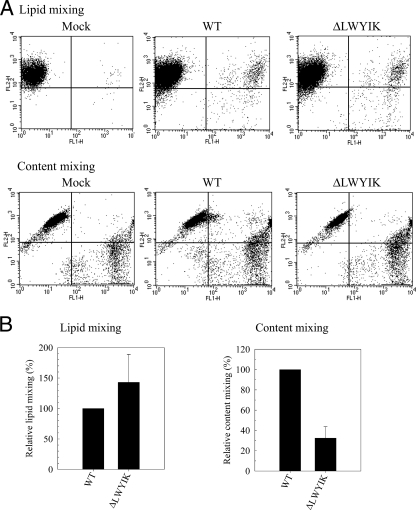
To further provide insights into the impaired membrane fusion of the LWYIK motif deletion mutant, dye transfer assays based on the redistribution of fluorescent probes between Env-expressing effector and CD4+ CEM-SS target cells following coculture (45, 46) were performed. Although Env with a deletion of the LWYIK motif still mediated lipophilic probe transfer as effectively as the WT Env (Fig. 10A, top panel, and B, left panel), this deletion mutant showed reduced cytosolic probe transfer to 32% of that of the WT Env (Fig. 10A, bottom panel, and B, right panel). These results collectively indicate that the LWYIK motif plays a crucial role in membrane fusion by promoting the enlargement of fusion pores.
FIG. 10.
Fluorescent probe exchange assays. (A) HeLa cells transfected with pCAGGS-based WT or ΔLWYIK mutant plasmids and labeled with DiO (green) were cocultured with CEM-SS cells labeled with DiI (red) as described in Materials and Methods. CEM-SS cells stained with both lipophilic fluorescent dyes were quantified by flow cytometry, and a representative two-color flow cytometic analysis is shown in the top panel. In addition, WT or ΔLWYIK mutant Env-expressing HeLa cells labeled with calcein-AM (green) and CEM-SS cells labeled with CMTMR (red) were cocultured, and CEM-SS cells stained with both cytosolic probes were quantified by flow cytometry; a representative result is shown in the bottom panel. (B) The background percentage of lipophilic dye transfer detected in pCAGGS vector transfection (control) was subtracted from that detected in WT or ΔLWYIK Env plasmid transfection, and the degree of lipid mixing of the ΔLWYIK mutant is expressed as a percentage of that of the WT Env. Results from three independent experiments (mean ± standard deviation) are shown (left panel). Also, the degree of content mixing of the ΔLWYIK mutant was quantified from four separate analyses and is expressed as a percentage of that of the WT Env (right panel).
DISCUSSION
Salzwedel et al. previously performed a detailed mutational study of the Trp-rich domain and found that substitutions of all five or the first three Trp residues in this domain with Ala residues, but not substitutions of other combinations of Trp residues or of single Trp residues, and deletion of the entire Trp-rich sequence, i.e., the Δ665-682 mutant, abrogated the membrane fusion of Env (60). Intriguingly, all mutations in this region greatly inhibited Env incorporation into the virus, which may result in a tendency for greater sensitivity of viral infectivity than membrane fusion with mutations in this sequence. Compared to the Δ678-682 mutant observed by Salzwedel et al., which still retained approximately a fusogenicity of 20% of that of the WT Env, deletions in the LWYIK motif, which is merely an amino acid shift toward the TM region, greatly reduced the Env fusion ability to almost the background level (Fig. 7). Unlike the Trp-rich domain mutants examined by Salzwedel et al., all of the mutants examined here still retained the WT phenotype of Env incorporation into the virus (Fig. 3; see Fig. S1 in the supplemental material). The observation that the KE mutant significantly inhibited membrane fusion (Fig. 7) was in accordance with the previous findings that the Lys-683 residue, which forms the N-terminal border of the TM domain, is critical for membrane fusion (31, 60). As shown by Salzwedel et al., we also found that substitution of Ala for Trp-680, i.e., the WA mutant, did not affect membrane fusion (Fig. 7). Our results revealed that except for the WA mutant, the reduced fusion abilities of the mutant proteins were in accordance with their reduced viral replication capacities (compare Fig. 7 to Fig. 2 and 5).
Protein folding and oligomerization play essential roles in the transport of viral Env glycoproteins to the plasma membrane (for reviews, see references 32 and 56). All mutant Env proteins were efficiently processed to yield gp120 and gp41 (Fig. 3; see Fig. S1 in the supplemental material), indicating that they are normally transported to the trans-Golgi network where the Env is proteolytically cleaved. The observation that comparable amounts of WT and mutant proteins were expressed on the cell surface also indicates that these LWYIK motif mutant proteins are oligomeric (Fig. 4A and C). Mutations in this region do not seem to affect the global structure of gp120, as measured by the CD4-binding ability of these mutants (Fig. 4B). Although the predicted α-helical pre-TM sequence is situated immediately adjacent to the TM α-helix of gp41, mutations in the LWYIK motif neither affect the effectiveness of the TM region in anchoring the mutant Env into membranes nor alter the gp120-gp41 interaction, as evidenced by the finding that comparable amounts of WT and mutant gp41 and gp120 proteins were associated with the virions (Fig. 3; see Fig. S1 in the supplemental material). Therefore, the inhibited membrane fusion and postfusion phenotypes (see below) of the LWYIK motif mutants can be attributed to the direct effects of these mutations on local sites, rather than to alterations of distal domains in Env.
The structural and functional roles of the pre-TM region in oligomerization and immersion into the viral membrane interface during fusion have received great attention. The dye transfer assay reinforced the notion that the pre-TM region participates in formation and expansion of fusion pores (46). Biophysical studies showed that the abilities of peptides derived from residues 664 to 683 to adopt a helical structure and form homo-oligomers in solution and membranes correlate with their abilities to induce vesicle fusion and inhibit gp41-induced cell-cell fusion (58, 64). This sequence is thought to specifically interact with cholesterol-containing membranes, since cholesterol and sphingomyelin in liposomes promote pre-TM peptide surface self-aggregation and destabilization of the membrane architecture (59). Nevertheless, in the context of full-length Env expression we found no requirement of the LWYIK motif for lipid raft association (Fig. 6). Although the ΔLWYIK mutant did not affect formation of the prehairpin structure or the step of lipid mixing (Fig. 9 and 10A), this deletion inhibited cytosolic dye exchange and severely impaired Tat redistribution (Fig. 7 and 10B). These results together indicate that deletion of the LWYIK motif arrests viral entry after the hemifusion stage by abrogating expansion of fusion pores.
Currently, the topology between the pre-TM region and the viral envelope remains controversial, with published tomographic data for SIV virions depicting conflicting models of either a “three-legged” globular stalk (77) or a “mushroom-shaped” bulb with one “single-legged” stalk (76). Nevertheless, the pre-TM sequence undergoes structural alterations during the fusion process. The epitopes of the 2F5 and 4E10 MAbs within this region are exposed in the gp120-gp41 complex prior to CD4 binding; however, they become occluded upon six-helical bundle formation (21, 22, 25, 29, 51). This antigenic change is also accompanied by a structural transition of the N-terminal segment of the pre-TM region from an extended conformation to an α-helix (6, 8, 65, 71). On the other hand, this pre-TM region may act in conjunction with other sequences in the gp41 ectodomain to maintain the native and fusion states of the Env. Lorizate et al. showed that the fusion peptide and pre-TM sequence can coassemble into a defined complex that acts as a prefusion clasp to restrict fusion peptide-mediated fusion (39, 40). The Trp-rich membrane-proximal region and the N-terminal fusion peptide-proximal polar segment were recently shown to act synergistically to form a fusion-competent prefusion gp120-gp41 complex and stabilize the membrane-interactive end of the trimeric hairpin fusion core (2). Although the pre-TM region is presumed to be inserted or immersed into the viral envelope after fusion, whether this sequence plays a role in a postfusion step remains elusive.
Among the mutants examined, only the WA mutant, albeit with decreased viral entry ability (Fig. 2C and D and 5), still possessed WT-like fusion ability (Fig. 7), suggesting that this motif may also play a role in a postfusion step. Remarkably, the three deletion mutants exhibited more significant dominant negative inhibitory effects on WT Env-mediated viral entry than membrane fusion when coexpressed with the WT Env (Fig. 8). As a control, the WA mutant did not greatly affect WT Env-mediated fusion and infectivity in these dominant interference analyses (Fig. 8). These observations indicate that the fusion ability of Env is less sensitive to a structural change in the trimeric Env complex formed between the WT and deletion Env subunits. However, this structural alteration is more effective in reducing the ability of the WT Env to mediate viral infectivity, implying that this LWYIK motif also plays a critical postfusion role as well. It is likely that this LWYIK motif may participate in a step during viral genome uncoating after gp41-mediated fusion pore formation and expansion. This motif in the membrane may modulate dissociation of the gp41 cytoplasmic domain and Gag and promote the dismantlement of the Gag core, resulting in release of the viral genome into the cytoplasm of infected cells.
Based on the results obtained, we conclude that the mutants analyzed here phenotypically differ from Trp-rich region mutants, by differing in the patterns of Env incorporation into the virus and in the sensitivities of membrane fusion and virus infectivity to mutations within the Trp-rich sequence and LWYIK motif, respectively. Moreover, the action of this motif is independent of that of the NHR and CHR during fusion. In short, this motif acts as a unique and distinct structural determinant located in the C terminus of the gp41 ectodomain in modulating membrane fusion and postfusion events.
Supplementary Material
Acknowledgments
The following cells and plasmids were obtained from the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health: SupT1 (James Hoxie), CEM-SS (Peter L. Nara), H938 (Barbara K. Felber and George N. Pavlakis), HeLa-CD4 and hybridomas 902 and 183 (Bruce Chesebro), hybridoma Chessie 8 (George K. Lewis), CD4 hybridoma SIM2 (James E. K. Hildreth), and pNL4-3R−E−Luc (Nathaniel Landau). We also thank Tun-Hou Lee and Lung-Ji Chang for providing pHXB2RU3 and pHP, respectively, and Chia-Hung Chang for technical assistance.
This work was supported by grants (NHRI-EX95-9431SII and NHRI-EX96-9431SI) from the National Health Research Institute (Miaoli) and Theme Program Project grants (92-04-23-4m and 92-04-23-4b) from Academia Sinica (Taipei), Taiwan, Republic of China.
Footnotes
Published ahead of print on 5 November 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anderson, R. G. 1998. The caveolae membrane system. Annu. Rev. Biochem. 67199-225. [DOI] [PubMed] [Google Scholar]
- 2.Bellamy-McIntyre, A. K., C. S. Lay, S. Baar, A. L. Maerz, G. H. Talbo, H. E. Drummer, and P. Poumbourios. 2007. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J. Biol. Chem. 28223104-23116. [DOI] [PubMed] [Google Scholar]
- 3.Berman, P. W., W. M. Nunes, and O. K. Haffar. 1988. Expression of membrane-associated and secreted variants of gp160 of human immunodeficiency virus type 1 in vitro and in continuous cell lines. J. Virol. 623135-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya, J., P. J. Peters, and P. R. Clapham. 2004. Human immunodeficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles. J. Virol. 785500-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68533-544. [DOI] [PubMed] [Google Scholar]
- 6.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 174572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S. M., S. M. Crowe, and J. Mak. 2002. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS 162253-2261. [DOI] [PubMed] [Google Scholar]
- 8.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89263-273. [DOI] [PubMed] [Google Scholar]
- 9.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93681-684. [DOI] [PubMed] [Google Scholar]
- 10.Chan, W. E., H. H. Lin, and S. S. Chen. 2005. Wild-type-like viral replication potential of human immunodeficiency virus type 1 envelope mutants lacking palmitoylation signals. J. Virol. 798374-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, W. E., Y. L. Wang, H. H. Lin, and S. S. Chen. 2004. Effect of extension of the cytoplasmic domain of human immunodeficiency type 1 virus transmembrane protein gp41 on virus replication. J. Virol. 785157-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, D. K., V. D. Trivedi, S. F. Cheng, and S. Francis. 2001. The leucine zipper motif of the envelope glycoprotein ectodomain of human immunodeficiency virus type 1 contains conformationally flexible regions as revealed by NMR and circular dichroism studies in different media. J. Pept. Res. 57234-239. [DOI] [PubMed] [Google Scholar]
- 13.Chang, L. J., V. Urlacher, T. Iwakuma, Y. Cui, and J. Zucali. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 6715-728. [DOI] [PubMed] [Google Scholar]
- 14.Chazal, N., and D. Gerlier. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, S. S. 1994. Functional role of the zipper motif region of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 682002-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, S. S., A. A. Ferrante, and E. F. Terwilliger. 1996. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology 226260-268. [DOI] [PubMed] [Google Scholar]
- 17.Chen, S. S., S. F. Lee, C. K. Chuang, and V. S. Raj. 1999. trans-dominant interference with human immunodeficiency virus type 1 replication and transmission in CD4+ cells by an envelope double mutant. J. Virol. 738290-8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, S. S., S. F. Lee, H. J. Hao, and C. K. Chuang. 1998. Mutations in the leucine zipper-like heptad repeat sequence of human immunodeficiency virus type 1 gp41 dominantly interfere with wild-type virus infectivity. J. Virol. 724765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang, C. K., S. F. Lee, and S. S. Chen. 1999. Conferal of an antiviral state to CD4+ cells by a zipper motif envelope mutant of the human immunodeficiency virus type 1 transmembrane protein gp41. Hum. Gene Ther. 102381-2395. [DOI] [PubMed] [Google Scholar]
- 20.Crise, B., L. Buonocore, and J. K. Rose. 1990. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J. Virol. 645585-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Rosny, E., R. Vassell, S. Jiang, R. Kunert, and C. D. Weiss. 2004. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: implications for fusion-inducing conformational changes. J. Virol. 782627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimitrov, A. S., A. Jacobs, C. M. Finnegan, G. Stiegler, H. Katinger, and R. Blumenthal. 2007. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry 461398-1401. [DOI] [PubMed] [Google Scholar]
- 23.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70777-810. [DOI] [PubMed] [Google Scholar]
- 24.Felber, B. K., and G. N. Pavlakis. 1988. A quantitative bioassay for HIV-1 based on trans-activation. Science 239184-187. [DOI] [PubMed] [Google Scholar]
- 25.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2002. Antigenic properties of the human immunodeficiency virus transmembrane glycoprotein during cell-cell fusion. J. Virol. 7612123-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5276-279. [DOI] [PubMed] [Google Scholar]
- 27.Gallo, S. A., C. M. Finnegan, M. Viard, Y. Raviv, A. Dimitrov, S. S. Rawat, A. Puri, S. Durell, and R. Blumenthal. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 161436-50. [DOI] [PubMed] [Google Scholar]
- 28.Geyer, H., C. Holschbach, G. Hunsmann, and J. Schneider. 1988. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J. Biol. Chem. 26311760-11767. [PubMed] [Google Scholar]
- 29.Gorny, M. K., and S. Zolla-Pazner. 2000. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 746186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham, D. R., E. Chertova, J. M. Hilburn, L. O. Arthur, and J. E. Hildreth. 2003. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with β-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J. Virol. 778237-8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helseth, E., U. Olshevsky, D. Gabuzda, B. Ardman, W. Haseltine, and J. Sodroski. 1990. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J. Virol. 646314-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurtley, S. M., and A. Helenius. 1989. Protein oligomerization in the endoplasmic reticulum. Annu. Rev. Cell Biol. 5277-307. [DOI] [PubMed] [Google Scholar]
- 33.Iwatani, Y., K. Kawano, T. Ueno, M. Tanaka, A. Ishimoto, M. Ito, and H. Sakai. 2001. Analysis of dominant-negative effects of mutant Env proteins of human immunodeficiency virus type 1. Virology 28645-53. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365113. [DOI] [PubMed] [Google Scholar]
- 35.Korber, B., C. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. Mellors, and J. Sodroski. 1998. Amino acid alignment, p. iiA, 1-77. In B. Korber, C. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS 1998: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, NM.
- 36.Li, H., and V. Papadopoulos. 1998. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 1394991-4997. [DOI] [PubMed] [Google Scholar]
- 37.Liao, Z., L. M. Cimakasky, R. Hampton, D. H. Nguyen, and J. E. Hildreth. 2001. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum. Retroviruses 171009-1019. [DOI] [PubMed] [Google Scholar]
- 38.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 757913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorizate, M., I. de la Arada, N. Huarte, S. Sanchez-Martinez, B. G. de la Torre, D. Andreu, J. L. Arrondo, and J. L. Nieva. 2006. Structural analysis and assembly of the HIV-1 Gp41 amino-terminal fusion peptide and the pretransmembrane amphipathic-at-interface sequence. Biochemistry 4514337-14346. [DOI] [PubMed] [Google Scholar]
- 40.Lorizate, M., M. J. Gomara, B. G. de la Torre, D. Andreu, and J. L. Nieva. 2006. Membrane-transferring sequences of the HIV-1 Gp41 ectodomain assemble into an immunogenic complex. J. Mol. Biol. 36045-55. [DOI] [PubMed] [Google Scholar]
- 41.Manes, S., G. del Real, R. A. Lacalle, P. Lucas, C. Gomez-Mouton, S. Sanchez-Palomino, R. Delgado, J. Alcami, E. Mira, and A. C. Martinez. 2000. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep. 1190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCune, J. M., L. B. Rabin, M. B. Feinberg, M. Lieberman, J. C. Kosek, G. R. Reyes, and I. L. Weissman. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 5355-67. [DOI] [PubMed] [Google Scholar]
- 43.Mizuochi, T., T. J. Matthews, M. Kato, J. Hamako, K. Titani, J. Solomon, and T. Feizi. 1990. Diversity of oligosaccharide structures on the envelope glycoprotein gp120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. Presence of complex-type oligosaccharides with bisecting N-cetylglucosamine residues. J. Biol. Chem. 2658519-8524. [PubMed] [Google Scholar]
- 44.Munch, J., L. Standker, K. Adermann, A. Schulz, M. Schindler, R. Chinnadurai, S. Pohlmann, C. Chaipan, T. Biet, T. Peters, B. Meyer, D. Wilhelm, H. Lu, W. Jing, S. Jiang, W. G. Forssmann, and F. Kirchhoff. 2007. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell 129263-275. [DOI] [PubMed] [Google Scholar]
- 45.Munoz-Barroso, I., S. Durell, K. Sakaguchi, E. Appella, and R. Blumenthal. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munoz-Barroso, I., K. Salzwedel, E. Hunter, and R. Blumenthal. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 736089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muster, T., R. Guinea, A. Trkola, M. Purtscher, A. Klima, F. Steindl, P. Palese, and H. Katinger. 1994. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 684031-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 676642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 743264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 7410074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ofek, G., M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 7810724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 9813925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parolini, I., S. Topa, M. Sorice, A. Pace, P. Ceddia, E. Montesoro, A. Pavan, M. P. Lisanti, C. Peschle, and M. Sargiacomo. 1999. Phorbol ester-induced disruption of the CD4-Lck complex occurs within a detergent-resistant microdomain of the plasma membrane. Involvement of the translocation of activated protein kinase C isoforms. J. Biol. Chem. 27414176-14187. [DOI] [PubMed] [Google Scholar]
- 54.Percherancier, Y., B. Lagane, T. Planchenault, I. Staropoli, R. Altmeyer, J. L. Virelizier, F. Arenzana-Seisdedos, D. C. Hoessli, and F. Bachelerie. 2003. HIV-1 entry into T-cells is not dependent on CD4 and CCR5 localization to sphingolipid-enriched, detergent-resistant, raft membrane domains. J. Biol. Chem. 2783153-3161. [DOI] [PubMed] [Google Scholar]
- 55.Popik, W., and T. M. Alce. 2004. CD4 receptor localized to non-raft membrane microdomains supports HIV-1 entry. Identification of a novel raft localization marker in CD4. J. Biol. Chem. 279704-712. [DOI] [PubMed] [Google Scholar]
- 56.Rose, J. K., and R. W. Doms. 1988. Regulation of protein export from the endoplasmic reticulum. Annu. Rev. Cell Biol. 4257-288. [DOI] [PubMed] [Google Scholar]
- 57.Rousso, I., M. B. Mixon, B. K. Chen, and P. S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA 9713523-13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saez-Cirion, A., J. L. Arrondo, M. J. Gomara, M. Lorizate, I. Iloro, G. Melikyan, and J. L. Nieva. 2003. Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 853769-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saez-Cirion, A., S. Nir, M. Lorizate, A. Agirre, A. Cruz, J. Perez-Gil, and J. L. Nieva. 2002. Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J. Biol. Chem. 27721776-21785. [DOI] [PubMed] [Google Scholar]
- 60.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 732469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387569-572. [DOI] [PubMed] [Google Scholar]
- 62.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 171757-1765. [DOI] [PubMed] [Google Scholar]
- 63.Suarez, T., W. R. Gallaher, A. Agirre, F. M. Goni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 748038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suarez, T., S. Nir, F. M. Goni, A. Saez-Cirion, and J. L. Nieva. 2000. The pre-transmembrane region of the human immunodeficiency virus type-1 glycoprotein: a novel fusogenic sequence. FEBS Lett. 477145-149. [DOI] [PubMed] [Google Scholar]
- 65.Tan, K., J.-H. Liu, J.-H. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 9412303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, et al. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-immunoglobulin G. J. Virol. 696609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trujillo, J. R., W. K. Wang, T.-H. Lee, and M. Essex. 1996. Identification of the envelope V3 loop as a determinant of a CD4-negative neuronal cell tropism for HIV-1. Virology 217613-617. [DOI] [PubMed] [Google Scholar]
- 68.Viard, M., I. Parolini, M. Sargiacomo, K. Fecchi, C. Ramoni, S. Ablan, F. W. Ruscetti, J. M. Wang, and R. Blumenthal. 2002. Role of cholesterol in human immunodeficiency virus type 1 envelope protein-mediated fusion with host cells. J. Virol. 7611584-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vincent, N., C. Genin, and E. Malvoisin. 2002. Identification of a conserved domain of the HIV-1 transmembrane protein gp41 which interacts with cholesteryl groups. Biochim. Biophys. Acta 1567157-164. [DOI] [PubMed] [Google Scholar]
- 70.Vishwanathan, S. A., and E. Hunter. 2008. The Membrane-proximal external region (MPER) of HIV-1 gp41: importance of its membrane-perturbing properties to viral fusion. J. Virol. 825118-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Willey. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387426-430. [DOI] [PubMed] [Google Scholar]
- 72.Wild, C., J. W. Dubay, T. Grreenwell, T. Baird, Jr., T. G. Oas, C. McDanal, E. Hunter, and T. Matthews. 1994. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA 9112676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. J. Virol. 8910537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wild, C., D. C. Shugars, T. K. Greenwell, C. B. McDonal, and T. J. Matthews. 1994. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 919770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willey, R. L., J. S. Bonifacino, B. J. Potts, M. A. Martin, and R. D. Klausner. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA 859580-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zanetti, G., J. A. Briggs, K. Grunewald, Q. J. Sattentau, and S. D. Fuller. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu, P., J. Liu, J. Bess, Jr., E. Chertova, J. D. Lifson, H. Grise, G. A. Ofek, K. A. Taylor, and K. H. Roux. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441847-852. [DOI] [PubMed] [Google Scholar]
- 78.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 7510892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.