Abstract
Photosynthetic organisms need defense systems against photooxidative stress caused by the generation of highly reactive singlet oxygen (1O2). Here we show that the alternative sigma factor RpoHII is required for the expression of important defense factors and that deletion of rpoHII leads to increased sensitivity against exposure to 1O2 and methylglyoxal in Rhodobacter sphaeroides. The gene encoding RpoHII is controlled by RpoE, and thereby a sigma factor cascade is constituted. We provide the first in vivo study that identifies genes controlled by an RpoHII-type sigma factor, which is widely distributed in the Alphaproteobacteria. RpoHII-dependent genes encode oxidative-stress defense systems, including proteins for the degradation of methylglyoxal, detoxification of peroxides, 1O2 scavenging, and redox and iron homeostasis. Our experiments indicate that glutathione (GSH)-dependent mechanisms are involved in the defense against photooxidative stress in photosynthetic bacteria. Therefore, we conclude that systems pivotal for the organism's defense against photooxidative stress are strongly dependent on GSH and are specifically recognized by RpoHII in R. sphaeroides.
Anoxygenic photosynthetic bacteria are widespread in aquatic habitats and show remarkable metabolic versatility that allows adaptation to changing environmental conditions, e.g., light intensity and oxygen concentration. Rhodobacter spp. have been studied extensively in regard to their adaptation to different environmental conditions (20, 31, 50). Rhodobacter sphaeroides can perform aerobic respiration. If the oxygen tension in the environment drops, photosynthetic complexes are formed, which are organized in an intracytoplasmic membrane system. This differentiation process is independent of light. In the presence of light and the absence of oxygen, anoxygenic photosynthesis enables R. sphaeroides to grow, while in the dark, anaerobic respiration or fermentation can be carried out. The redox-dependent formation of photosynthetic complexes is controlled by several regulatory systems in R. sphaeroides (31, 49). When R. sphaeroides grows at intermediate oxygen concentrations, blue-light illumination leads to a repression of photosynthesis genes (5, 32, 44). Most likely, this response reduces the risk of 1O2 formation. Under anaerobic conditions, the expression of photosynthesis genes is stimulated by light (5, 21).
Even when pigmented, R. sphaeroides grows well in the presence of oxygen and high light intensities, although bacteriochlorophylls and their precursors represent potent photosensitizers. Only recently, BChl a in the photosynthetic reaction center was shown to generate 1O2 (4, 46), which has also been measured in intact R. sphaeroides cultures harboring or lacking carotenoids (16). Upon addition of the photosensitizer methylene blue, growth transiently slows down in the presence of light but then continues at rates almost as high as those of controls in the dark (16). This implies an adaptive response to 1O2. Although, as in other organisms, carotenoids play a major role in the defense against photooxidative stress in R. sphaeroides (8), the carotenoid content shows little change in the presence of 1O2 (16). However, the mRNA levels for a glutathione (GSH) peroxidase and a putative Zn-dependent hydrolase were strongly increased by 1O2 (16). The upregulation of the genes encoding RpoE and RpoHII (previously called σ38) and of some genes with putative RpoE and RpoHII target sequences upon blue-light illumination suggested a role for these sigma factors in the organism's response to photooxidative stress (6). While RpoE belongs to the ECF (extracytoplasmic function) family of sigma 70 factors (36), RpoHI and RpoHII are members of the sigma 32 family of proteins (25). Dissociation of RpoE from the anti-sigma factor ChrR activates expression of genes involved in the response to 1O2 (2, 3). A putative RpoE target sequence upstream of the rpoHII gene suggested that its expression is under RpoE control (3, 6).
Recently, we have identified cytoplasmic proteins of R. sphaeroides which show increased synthesis rates upon 1O2 exposure (17). While this response was RpoE dependent for some of the proteins, it was independent of RpoE for others. There was only a small overlap of RpoE-dependent genes, as detected by transcriptome analysis (3), and of genes encoding proteins whose expression is dependent on RpoE (17). We wanted to test the hypothesis that RpoHII directly activates some of the RpoE-dependent genes and that RpoHII plays an important role in the 1O2 response. To this end, we constructed and characterized an rpoHII deletion mutant. Proteins exhibiting altered synthesis in the rpoHII mutant were identified by proteome analysis, and changes in expression of rpoHII and RpoE-dependent genes were monitored in response to 1O2 exposure. It was previously shown that RpoHII and RpoHI are able to recognize similar promoter regions (19); however, RpoHI does not complement RpoHII under conditions of 1O2. Our data reveal that RpoHII has a major role in 1O2 stress defense and acts downstream of RpoE in a sigma factor cascade.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
R. sphaeroides strains were grown at 32°C in minimal salt medium containing malate as the carbon source (14). Aerobic growth conditions with a concentration of 160 to 180 μM of dissolved oxygen were established by gassing cultures with air in flat glass bottles or by continuous shaking of Erlenmeyer flasks at 140 rpm with a culture volume of 20%. In semiaerobic cultures, a volume of 80% in Erlenmeyer flasks and shaking at 140 rpm led to a dissolved-oxygen concentration of approximately 25 μM. For heat shock experiments, aerobic cultures were shifted to 42°C in a water bath. When necessary, trimethoprim (50 μg ml−1) or spectinomycin (10 μg ml−1) was added to liquid and solid growth media, which contained 1.6% agar. Antibiotics were omitted from precultures, cultures, and agar plates used for R. sphaeroides strains during stress experiments and inhibition zone assays (see below) to ensure identical culture conditions. Escherichia coli strains were grown at 37°C in LB medium with continuous shaking at 180 rpm or on solid growth medium.
Construction of an R. sphaeroides rpoHII deletion mutant.
R. sphaeroides strain 2.4.1ΔrpoHII was generated by transferring the suicide plasmid pPHU2.4.1rpoHII::ΩSpr/Str (Table 1) into R. sphaeroides 2.4.1 and screening for the insertion of the spectinomycin resistance/streptomycin resistance (ΩSpr/Str) cassette into the chromosome by homologous recombination. Parts of the rpoHII gene of R. sphaeroides 2.4.1, together with upstream and downstream sequences, were amplified by PCR using the oligonucleotides 2.4.1rpoHII_knockout-up_EcoRI, 2.4.1rpoHII_knockout-up_BamHI, 2.4.1rpoHII_knockout-down_BamHI, and 2.4.1rpoHII_knockout-down_SphI (see Table S1 in the supplemental material). The PCR fragments obtained, including the promoter region of rpoHII, were cloned into pPHU281 (23) by using the appropriate restriction endonucleases. Then, the ΩSpr/Str cassette obtained from plasmid pHP45Ω (41) was inserted into the BamHI restriction site to obtain the plasmid pPHU2.4.1rpoHII::ΩSpr/Str. The plasmid pPHU2.4.1rpoHII::ΩSpr/Str was transferred into E. coli strain S17-1 (45) and mobilized into R. sphaeroides wild-type strain 2.4.1 by biparental conjugation. Conjugants were selected on malate minimal medium agar plates containing 10 μg spectinomycin ml−1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1 | recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 tra+ Kmr Spr | 45 |
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi(lac-proAB) | New England Biolabs |
| R. sphaeroides | ||
| 2.4.1 | Wild type | 47 |
| 2.4.1(pRK415) | Wild type harboring pRK415, Tcr | This study |
| ΔchrR | chrR mutation in 2.4.1, Tpr | 36 |
| TF18 | rpoE chrR mutation in 2.4.1, Tpr | 43 |
| 2.4.1ΔrpoHII | 2.4.1rpoHII::ΩSpr/Str | This study |
| 2.4.1ΔrpoHII(pRK415) | 2.4.1ΔrpoHII harboring pRK415, Tcr | This study |
| 2.4.1ΔrpoHII(pRK2.4.1rpoHII) | 2.4.1ΔrpoHII harboring pRK2.4.1rpoHII | This study |
| 2.4.1ΔrpoHII(pRK2.4.1rpoHIIRSP0600) | 2.4.1ΔrpoHII harboring pRK2.4.1rpoHIIRSP0600 | This study |
| Plasmids | ||
| pPHU281 | Suicide plasmid for R. sphaeroides, Tcr | 23 |
| pHP45Ω | Apr Spr Smr; source of ΩSpr/Str cassette | 41 |
| pRK415 | Tcr | 26 |
| pPHU2.4.1rpoHII::ΩSp/St | pPHU281 with Spr/Str cassette, flanked by the upstream and downstream region of rpoHII | This study |
| pRK2.4.1rpoHII | pRK415 harboring a 1.0- kb fragment containing rpoHII flanked by the 104-bp upstream and 11-bp downstream regions | This study |
| pRK2.4.1rpoHIIRSP0600 | pRK415 harboring a 1.8-kb fragment of rpoHII and RSP0600 flanked by the 104-bp upstream region of rpoHII and the 14-bp downstream region of RSP0600 | This study |
| pDrive cloning vector | Apr Kmr | Qiagen |
Southern blot analysis of chromosomal DNA isolated from spectinomycin-resistant and tetracycline-sensitive conjugants were carried out to confirm that the double crossover of the ΩSpr/Str cassette into the R. sphaeroides chromosome had occurred. For this purpose, the linearized plasmid pPHU281, the rpoHII upstream region also used for construction of the deletion mutant, and the ΩSpr/Str cassette were radioactively labeled by nick translation and used as probes. By insertion of the ΩSpr/Str cassette, 473 bp of the 879-bp R. sphaeroides rpoHII gene was deleted. DNA of the rpoHII mutant did not hybridize with linearized plasmid DNA of pPHU281, and positive signals of the appropriate sizes were obtained at the same positions by hybridization with the rpoHII upstream region and the ΩSpr/Str cassette.
Complementation of the R. sphaeroides rpoHII deletion mutant 2.4.1ΔrpoHII.
For complementation of 2.4.1ΔrpoHII, a 1.0-kb PCR fragment containing the entire rpoHII gene, along with 104 bp of the upstream sequence and 11 bp of the downstream sequence, was amplified by using the oligonucleotides RSP0601_UP_EcoRI and RSP0601_DWN_EcoRI. The PCR fragment obtained was cloned into the pDrive vector (Qiagen, Hilden, Germany). Digestion of the pDrive vector containing the insert with PstI and XbaI, followed by cloning with the same restriction sites into plasmid pRK415 resulted in plasmid pRK2.4.1rpoHII. This plasmid was subsequently transformed in E. coli S17-1 and conjugated with strain 2.4.1ΔrpoHII to obtain the complemented strain 2.4.1ΔrpoHII(pRK2.4.1rpoHII). In addition, we complemented strain 2.4.1ΔrpoHII with a plasmid harboring a 1.8-kb fragment which contained the entire rpoHII and RSP0600 genes, along with 104 bp of the upstream sequence of rpoHII and 14 bp of the downstream region of RSP0600. The fragment was amplified using the oligonucleotides RSP0601_UP_EcoRI and RSP0600_DWN_EcoRI. This fragment was cloned into pRK415 and transferred to strain 2.4.1ΔrpoHII with the same strategy as that used for constructing 2.4.1ΔrpoHII(pRK2.4.1rpoHII) to obtain strain 2.4.1ΔrpoHII(pRK2.4.1rpoHIIRSP0600).
Measurement of sensitivity to 1O2.
For inhibition zone assays, 0.05-ml volumes of exponential-phase R. sphaeroides cultures (optical density at 660 nm of 0.5), grown under semiaerobic conditions, were diluted into 5 ml of prewarmed top agar (0.8% agar) and layered onto minimal malate medium agar plates. Filter paper disks were placed on the surface of the agar, and 5 μl of 10 mM methylene blue solution (Sigma-Aldrich, Seelze, Germany) was applied to the filter disks. Zones of inhibition were measured after incubation for 48 h at 32°C under a fluorescent tube (model NL 36 W/860 daylight; Spectralux Plus, Radium, Wipperfürth, Germany). Inhibition zone assays were also performed in the dark with filter paper disks containing 5 μl of 10 mM methylglyoxal, 200 mM H2O2, and 500 mM paraquat to generate superoxide.
High light and photooxidative-stress conditions.
High light and photooxidative-stress conditions were established as described previously (16). In brief, cultures were grown under semiaerobic conditions overnight to obtain pigmented cells. Cultures were diluted to an optical density at 660 nm of 0.2 and allowed to double once under aerobic growth conditions in darkened flat glass bottles. High light conditions were generated by illumination with 800 W m−2 of white light, and for photooxidative stress, 1O2-producing methylene blue was added to liquid cultures at a final concentration of 0.2 μM.
Radioactive labeling of proteins.
Samples of 7 ml were retrieved from R. sphaeroides cultures, 15 μCi of l-[35S]methionine (GE Healthcare, München, Germany) was added, and incubation was performed for 10 min under the experimental conditions described above. Samples were cooled on ice after incubation, and cells were harvested by centrifugation at 10,000 × g for 10 min at 4°C and stored at −20°C until further processing.
Gel-based proteome analysis.
Proteome analysis performed by two-dimensional (2D) gel electrophoresis followed the procedure described previously (17). In brief, for the extraction of soluble proteins, harvested cells were washed and disrupted by sonication. Intact cells and cell debris were removed by centrifugation, and the supernatant was used for ultracentrifugation at 100,000 × g. The radioactive label was quantified in the colorless supernatant by adding 10-μl aliquots to 1 ml of rotiszint scintillation cocktail (Roth, Karlsruhe, Germany) and measured in a Beckmann LS-6500 scintillation counter (Beckmann Coulter, Fullerton, CA). For 2D gel electrophoresis, protein samples containing 1.5 × 106 cpm were treated with RNase A (Qiagen) and RQ1 DNase I (Promega, Madison, WI) to remove nucleic acids. Proteins were precipitated using trichloroacetic acid, and the protein pellets were dried and then solubilized in sample buffer (17). Then, samples were applied to immobilized pH gradient strips (ReadyStrip; Bio-Rad, Hercules, CA). After isoelectric focusing, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and gels were fixed, dehydrated, dried, and exposed to phosphorimaging screens for 48 h. Phosphorimages were read with a Molecular Imager FX (Bio-Rad) set to a resolution of 100 μm.
Protein spots on digital phosphorimages were compared using Delta2D version 3.3 software as described by the manufacturer (Decodon, Greifswald, Germany).
Extraction of RNA.
Cell samples from growth experiments were rapidly cooled on ice and harvested by centrifugation at 10,000 × g in a cooled centrifuge. For 5′ rapid amplification of cDNA ends (RACE), total RNA was isolated using hot phenol and quantified by photometric analysis at a wavelength of 260 nm. Samples were treated with 1 U of RNase-free DNase I (Invitrogen) per 1 μg RNA to remove contaminating DNA. After DNase I treatment, the RNA was purified by standard procedures using a mixture of phenol-chloroform-isoamyl alcohol and chloroform-isoamyl alcohol before being precipitated with sodium acetate and ethanol. For real-time reverse transcription-PCR (RT-PCR), total RNA was isolated by Total RNA isolation reagent (TRIR; ABGene, Epsom, United Kingdom) as described by the manufacturer. After extraction, DNase I treatment and purification of RNA samples were carried out as described above. Contamination by any remaining DNA was detected by PCR amplification of RNA samples, using primers targeting gloB (RSP0799) (see Table S1 in the supplemental material), as described previously (16).
5′ RACE.
For the determination of 5′ mRNA ends using 5′ RACE, 5 μg of total RNA was reverse transcribed into cDNA by using 20 U of avian myeloblastosis virus reverse transcriptase (Promega) and 12.5 pmol of gene-specific primers (gloA-RACE-1, gst-RACE-1, RSP3075-RACE-1, and 2.4.1rpoZ-B [see Table S1in the supplemental material]). RT was performed for 1 h at 50°C and then 15 min at 60°C with all primers in one reaction mixture. cDNA was then purified with a High Pure PCR product purification kit (Roche). The purified cDNA was poly(A) tailed using 20 U of terminal transferase (Fermentas). Poly(A) tailing was performed for 30 min at 37°C, and the terminal transferase was then heat inactivated at 70°C for 10 min. The poly(A)-tailed cDNAs served as templates for the first of two PCRs. The first PCR was performed using the oligo(dT) anchor primer and a gene-specific RACE-1 primer. For the second PCR, the PCR anchor primer and the gene-specific RACE-2 primer (nested PCR) were used. Resulting PCR products were cloned into the pDrive cloning vector (Qiagen) and sequenced with the respective gene-specific RACE-2 primer.
Real-time RT-PCR.
Primers employed for analyzing the relative expression of target genes, using real-time RT-PCR, are listed in Table S1 in the supplemental material. For normalization of mRNA levels, the rpoZ gene, which encodes the ω subunit of RNA polymerase of R. sphaeroides, was used (39). Conditions for real-time RT-PCR were previously described in detail (16, 17). For real-time RT-PCR, a final concentration of 4 ng μl−1 of total RNA was applied using a one-step RT-PCR kit (Qiagen), and Sybr green I (Sigma-Aldrich) was added at a final dilution of 1:50,000 to the master mixture to detect double-stranded DNA. The relative expression of target genes was calculated relative to the expression of untreated samples and relative to that of rpoZ (40). PCR efficiencies were 2.02 for rpoZ, 1.98 for ggt, 2.31 for gloB, and 2.02 for gpx (16, 17). Additional PCR efficiencies were determined experimentally using serial dilutions of RNA, as follows: 2.04 for rpoE, 2.09 for rpoHII, 2.0 for rpoHI, 2.01 for gloA, 2.02 for groEL, 1.98 for RSP3075, and 2.0 for RSP3537.
Statistical analysis.
Statistical analysis of the comparison of Cp (crossing point) values obtained for individual genes under different stress conditions, using real-time RT-PCR, was performed with Student's t test using Microsoft Excel 2003 (Microsoft, Redmond, WA). Significance levels (P values of ≤0.1, ≤0.05, and ≤0.01) are indicated in the figure legends. Statistically significant differences in protein synthesis were tested with the Student t test option available in Delta2D software. Proteins affected by 1O2 were included in the analysis if changes in protein synthesis were statistically significant as previously described (16).
RESULTS
RpoHII contributes to the defense of R. sphaeroides against 1O2.
We constructed an rpoHII deletion mutant of R. sphaeroides as described in Materials and Methods. Inhibition zone experiments revealed a significantly higher sensitivity to 1O2 exposure for the rpoHII mutant than for the isogenic wild-type strain 2.4.1. The diameter of the zone of inhibition observed for wild-type strain 2.4.1 was 2.6 ± 0.1 cm (mean ± standard deviation) (n, 3); and it was increased in the rpoHII mutant to 3.2 cm (n, 3) when 10 mM methylene blue was applied in the presence of light for the generation of 1O2. Inhibition zones observed for the wild type and for the rpoHII mutant that harbored plasmid pRK415 were not significantly different from those described above (see Table S2 in the supplemental material). The deletion of rpoHII in strain 2.4.1ΔrpoHII was complemented in trans by the expression of rpoHII on a low-copy-number plasmid yielding strain 2.4.1ΔrpoHII(pRK2.4.1rpoHII). To control for polar effects of the rpoHII deletion on the RSP0600 gene located downstream of rpoHII, a second plasmid containing rpoHII and RSP0600 was used for complementation of the rpoHII mutant, resulting in strain 2.4.1ΔrpoHII(pRK2.4.1rpoHIIRSP0600). Inhibition zones of the two complemented rpoHII mutants formed upon 1O2 exposure were not significantly different from those obtained with the wild type (see Table S2 in the supplemental material). The diameters measured 2.5 ± 0.1 cm (n, 3) for the former and 2.4 ± 0.1 cm (n, 3) for the latter strain. From these observations, we conclude that RpoHII is involved in the response of R. sphaeroides to 1O2 and that the higher sensitivity of the rpoHII mutant is not due to polar effects of the resistance cassette on the downstream gene. Further inhibition zone tests revealed that there were no significant differences in resistance to hydrogen peroxide or superoxide generated by the addition of paraquat between the rpoHII mutant and the wild type (see Table S2 in the supplemental material).
The rpoHII gene is triggered by exposure to 1O2.
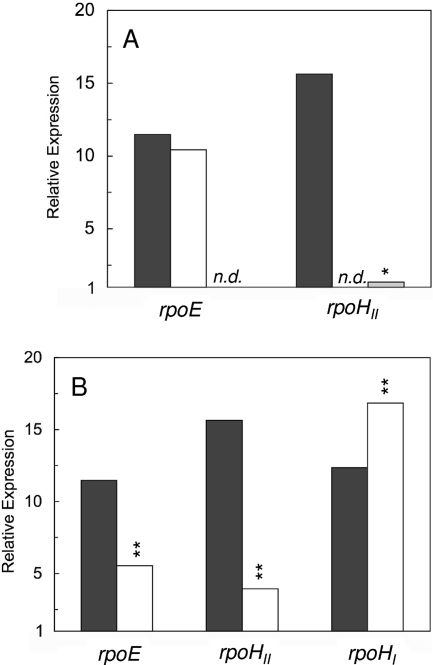
In order to assess in detail the factors that trigger the expression of rpoHII, we used real-time RT-PCR to quantify the relative expression levels of the rpoHII gene in wild-type and mutant strains under different growth conditions. A 15-fold increase in the rpoHII mRNA level was observed 7 min after the onset of 1O2 exposure in the wild type but not in strain TF18, which lacks RpoE and its anti-sigma factor ChrR (Fig. 1A). This result is in agreement with the RpoE-dependent expression of rpoHII (3). Under the same conditions, the mRNA level of rpoE was increased 10- to 12-fold in the wild type and rpoHII mutant, confirming earlier findings that rpoE is triggered by 1O2 exposure (3, 17).
FIG. 1.
Exposure to 1O2 and heat shock affect relative expression levels of sigma factors rpoE and rpoHII. (A) Levels of relative expression are shown for sigma factors rpoE and rpoHII in response to 1O2 exposure in wild-type strain 2.4.1 (black bars), 2.4.1ΔrpoHII (white bars), and rpoE chrR mutant strain TF18 (gray bar). (B) The expression levels of rpoE and rpoHII were compared to that of rpoHI in wild-type strain 2.4.1 under conditions of 1O2 exposure (black bars) and heat shock (white bars). Exposure to 1O2 was performed for 7 min, and heat shock for 30 min at 42°C. Values for relative expression levels represent the increase in gene expression compared to that of the control at time point 0 min and were normalized to mRNA levels determined for rpoZ. Mean values for three independent experiments are shown, and statistically significant differences in expression for mutants relative to that of wild-type cultures were determined using Student's t test. Levels of significance are as follows: *, P ≤ 0.01; **, P ≤ 0.05. n.d., not detectable.
RpoHII is a member of the sigma 32 family of proteins, implying a role in heat shock response. Like RpoHI of R. sphaeroides, RpoHII can complement the temperature-sensitive phenotype of an E. coli rpoH mutant (19). We therefore compared the mRNA levels of rpoHI and rpoHII and also of rpoE after exposure to 1O2 and heat shock (Fig. 1B). To verify that our growth conditions initiated a heat shock response, relative expression levels of the known heat shock gene groEL (25) were monitored. The groEL levels were increased eightfold in both the wild type and the rpoHII mutant after heat shock (data not shown). This result is in agreement with the heat shock response observed previously for R. sphaeroides (25) and with the finding that the rpoHII mutant is not heat sensitive. The rpoE mRNA level increased about 12-fold upon 1O2 exposure and about 5-fold after heat shock (Fig. 1B). Under the same conditions, rpoHII mRNA levels increased by a factor of 15 in response to 1O2 but only by a factor of 4 after heat shock. In comparison, the rpoHI mRNA levels were increased 12- and 17-fold in response to 1O2 and heat shock, respectively. Like the response of rpoHII to 1O2, the response of rpoHII to heat shock was RpoE dependent. These data demonstrate a partial overlap of the 1O2 response and the heat shock response in R. sphaeroides. The pronounced increase in rpoHII mRNA levels under photooxidative-stress conditions over those in response to heat shock hints at rpoHII,s prominent role in response to 1O2 exposure.
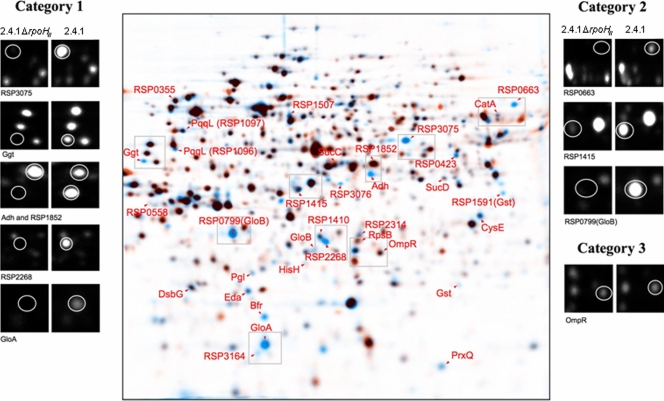
Deletion of rpoHII affects the synthesis of soluble proteins upon 1O2 exposure.
Protein synthesis patterns in strain 2.4.1ΔrpoHII under photooxidative stress were clearly different than the patterns observed in the wild type (Fig. 2; Table 2). Overall, most of the proteins (76%) characterized by the proteome analysis as RpoE dependent (category 1) (17) and conditionally RpoE dependent (category 2; less induction in a mutant lacking RpoE) showed dependency on RpoHII, whereas RpoE-independent proteins (category 3) were not affected by the lack of RpoHII.
FIG. 2.
Superimposed 2D gel electrophoresis images indicating differences in synthesis of soluble proteins between wild-type strain 2.4.1 and strain 2.4.1ΔrpoHII in response to 1O2 exposure. Protein extracts were prepared from cells labeled in vivo with l-[35S]methionine during exponential growth in the presence of methylene blue (0.2 μM) and high light (800 W m−2). Superimposed images were generated by combining the digitalized autoradiograms in Delta 2D software. For both strain 2.4.1 and strain 2.4.1ΔrpoHII, three gels of independent experiments were fused and overlaid. Blue spots represent proteins from strain 2.4.1, and orange spots represent proteins from strain 2.4.1ΔrpoHII. Therefore, in superimposed images, blue spots indicate proteins not synthesized in strain 2.4.1ΔrpoHII. Black spots indicate proteins synthesized similarly in both strains, and orange spots represent proteins exhibiting increased synthesis in strain 2.4.1ΔrpoHII. Protein numbers labeled with RSP prefixes have hypothetical functions.
TABLE 2.
Comparison of relative protein spot intensities after 1O2 exposure
| Category and locus tag | Protein | Function | Relative protein spot intensity (%) ± SDa
|
||
|---|---|---|---|---|---|
| 2.4.1ΔrpoHII | 2.4.1ΔrpoHII (pRK2.4.1rpoHII) | 2.4.1ΔrpoHII (pRK2.4.1rpoHIIRSP0600) | |||
| Category 1 | |||||
| RSP0392† | GloA | Predicted lactoylglutathione lyase: glyoxalase I | 8 ± 4 | 116 ± 12 | 98 ± 11 |
| RSP3272† | Ggt | γ-Glutamyltranspeptidase | 0.0 ± 0.0 | 38 ± 5 | 29 ± 9 |
| RSP3537† | Adh | Zn-containing alcohol dehydrogenase | 0.0 ± 0.0 | 169 ± 25 | 148 ± 2 |
| RSP2314† | Predicted oxidoreductase | 11 ± 5 | 63 ± 16 | 105 ± 7 | |
| RSP3075† | Hypothetical protein | 9 ± 1 | 74 ± 0.7 | 95 ± 2 | |
| RSP3076 | Hypothetical protein | 0.0 ± 1 | 161 ± 11 | 152 ± 9 | |
| RSP1591 | Gst | Predicted glutathioneS-transferase | 0.0 ± 0.0 | 93 ± 2 | 30 ± 10 |
| RSP2645 | Eda | KDPG/KHG bifunctional aldolase | 39 ± 23 | 97 ± 6 | 107 ± 16 |
| RSP2973 | PrxQ | Peroxiredoxin | 20 ± 9 | 159 ± 37 | 91 ± 23 |
| RSP2860 | RpsB | 30S ribosomal protein S2 | 192 ± 44 | 93 ± 9 | 110 ± 5 |
| RSP0423 | Predicted oxidoreductases related to aryl-alcohol dehydrogenases | 30 ± 3 | 111 ± 33 | 90 ± 40 | |
| RSP2268 | Predicted metalloβ-lactamase | 0.0 ± 0.0 | 113 ± 6 | 109 ± 11 | |
| RSP1852 | Hypothetical protein | 147 ± 40 | 108 ± 14 | 117 ± 1 | |
| RSP1464 | DsbG | Putative periplasmic thiol-disulfide interchange protein of the DsbA family | 82 ± 26 | 94 ± 24 | 81 ± 11 |
| Category 2 | |||||
| RSP3164† | Ferredoxin-like protein | 0.0 ± 0.0 | 74 ± 10 | 73 ± 21 | |
| RSP3342 | Bfr | Bacterioferritin | 0.0 ± 0.0 | 147 ± 31 | 160 ± 15 |
| RSP2481 | CysE | Serine acetyltransferase | 35 ± 13 | 62 ± 6 | 59 ± 18 |
| RSP0799 | GloB | Putative hydroxyacylglutathione hydrolase: glyoxalase II | 7 ± 4 | 63 ± 3 | 58 ± 6 |
| RSP2294 | GloB | Putative hydroxyacylglutathione hydrolase: glyoxalase II | 2 ± 1 | 95 ± 1 | 87 ± 11 |
| RSP1397 | Gst | GlutathioneS-transferase | 15 ± 13 | 106 ± 49 | 112 ± 5 |
| RSP2735 | Pgl | Inner membrane subunit of ABC sugar transporter | 32 ± 15 | 112 ± 13 | 104 ± 4 |
| RSP1096 | PqqL | Putative zinc peptidase | 7 ± 5 | 76 ± 8 | 51 ± 0.8 |
| RSP1097 | PqqL | Putative zinc peptidase | 42 ± 12 | 47 ± 19 | 39 ± 0.5 |
| RSP0663 | Formate-tetrahydrofolate ligase | 6 ± 4 | 81 ± 54 | 51 ± 41 | |
| RSP1415 | Putative polysaccharide deacetylase | 0.4 ± 0.0 | 78 ± 0.3 | 79 ± 4 | |
| RSP1507 | NADH-dependent aldehyde dehydrogenase | 4 ± 1 | 35 ± 15 | 49 ± 9 | |
| RSP2779 | CatA | Catalase | 37 ± 17 | 129 ± 55 | 117 ± 49 |
| RSP0355 | Putative serine protease involved in heat shock response, exhibiting chaperone function | 96 ± 25 | 75 ± 36 | 65 ± 3 | |
| RSP0558 | Possible ribosomal protein L11: predicted methyltransferase | 83 ± 43 | 70 ± 0.4 | 65 ± 0.6 | |
| RSP0967 | SucC | Succinyl-coenzyme A synthetase; β subunit | 120 ± 53 | 98 ± 5 | 109 ± 6 |
| RSP0966 | SucD | Succinyl-coenzyme A synthetase; α subunit | 127 ± 59 | 90 ± 9 | 88 ± 25 |
| RSP1410 | Hypothetical protein | 49 ± 46 | 48 ± 3 | 49 ± 2 | |
| RSP2241 | HisH | Phosphoribosyl-ATP pyrophosphatase | 133 ± 58 | 100 ± 0.1 | 100 ± 0.1 |
| Category 3 | |||||
| RSP0847 | OmpR | Two-component transcriptional regulator | 117 ± 66 | 131 ± 32 | 126 ± 3 |
Protein spot intensities ± standard deviations (SD) were normalized to values observed for the wild-type strain 2.4.1 and are shown for the rpoHII mutant strain 2.4.1ΔrpoHII and for the complemented mutants harboring the plasmid pRK2.4.1rpoHII or pRK2.4.1rpoHIIRSP0600. Category 1 includes RpoE-dependent proteins, category 2 contains conditionally RpoE-dependent proteins, and category 3 contains RpoE-independent proteins (17). Boldface proteins depend on RpoHII; †, predicted RpoHII target sequence (6). Mean values of three independent experiments are depicted for the rpoHII mutant and for the two complemented mutants.
Products of the genes gloA, ggt, adh, RSP2314, RSP3075, RSP3076, and RSP3164 with a predicted RpoHII target sequence (6) did not show a significant response to 1O2 in the rpoHII mutant (Table 2). In total, 25 proteins previously shown to be dependent or conditionally dependent on the presence of RpoE (17) were synthesized at lower or undetectable levels in the rpoHII mutant, as indicated by blue protein spots in superimposed 2D gels and decreased relative spot intensities (Fig. 2; Table 2). Other proteins depending on RpoHII in category 1 are encoded by the genes gst (RSP1591), eda, prxQ, rpsB, RSP0423, and RSP2268. In category 2, proteins encoded by the genes bfr, cysE, gloB (RSP0799), gloB (RSP2294), gst (RSP1397), pgl, pqqL (RSP1096, RSP1097), RSP0663, RSP1415, RSP1507, and catA were significantly affected by the deletion of rpoHII (Fig. 2; Table 2). In the complemented strain 2.4.1ΔrpoHII(pRK2.4.1rpoHII), most RpoHII-dependent proteins were synthesized in amounts that were similar to that of the wild-type strain 2.4.1. For Ggt, PqqL (RSP1097), and RSP1507, however, the amounts of synthesized proteins were restored to less than 50% of that of the wild type (Table 2). The amounts of proteins synthesized in strain 2.4.1ΔrpoHII(pRK2.4.1rpoHIIRSP0600), which also contains the RSP0600 gene downstream of rpoHII, were similar to those of the wild-type strain, with the exceptions of Gst, RSP2314, and PqqL (RSP1096) (Table 2).
Only one protein that increased in synthesis upon 1O2 exposure was encoded by a gene with a preceding RpoE target sequence: RSP1852, encoding a hypothetical protein related to UV endonucleases (3, 6, 17). A second gene, RSP1464 (dsbG, encoding a periplasmic disulfide isomerase), overlaps with RSP1465, which as yet has no function assigned and is preceded by a putative RpoE target sequence (17). Both proteins were synthesized in the rpoHII mutant when it was exposed to 1O2 (Fig. 2; Table 2), which underlines their regulation in an RpoE-dependent but RpoHII-independent manner.
Additional RpoE-dependent proteins not affected by the deletion of rpoHII were found in category 2 (encoded by RSP0355, RSP0558, RSP1410, sucC, sucD, and hisH). The synthesis of proteins previously classified as RpoE independent (category 3) was also not altered by the deletion of rpoHII. This is reflected by, e.g., the synthesis of OmpR, which is not altered in strain 2.4.1ΔrpoHII, as depicted in superimposed 2D gels (Fig. 2; Table 2).
RpoHII-dependent gene expression.
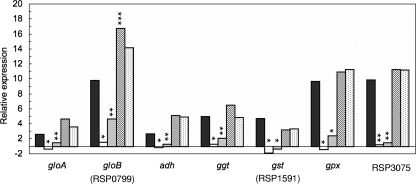
In order to determine the role of RpoHII in the expression of strictly RpoHII-dependent genes, we compared mRNA levels of some of the genes in the rpoHII mutant with those in the rpoE-chrR mutant strain TF18 and in the wild-type strain 2.4.1 by real-time RT-PCR (Fig. 3). We included gpx, for which we did not detect a protein in our proteome analysis. Relative mRNA levels of gpx were increased upon exposure to 1O2 (16) and blue light, and the gene carries a putative RpoHII target sequence in the upstream region (6). Significantly increased mRNA levels (increased by factors of 3 to 10) were observed for the genes gloA, gloB (RSP0799), adh, ggt, gst (RSP1591), gpx, and RSP3075 in the wild type after 7 min of 1O2 exposure (Fig. 3). In contrast, no significant increase in mRNA levels of the above-mentioned genes was observed for the rpoHII mutant. Interestingly, we observed a small increase in mRNA levels in the case of all tested genes, except for gst (RSP1591) in strain TF18. Complementation of the rpoHII mutant with plasmids harboring rpoHII or rpoHII and RSP0600 restored the relative expression, upon 1O2 exposure, of all genes mentioned above. For all of the investigated genes except gst (RSP1591), the relative expression was even higher than that of the wild type, but significantly increased values were observed only for gloB (RSP0799) (Fig. 3).
FIG. 3.
Selected functional genes depending on RpoHII for expression, which contained a putative RpoHII promoter (6). Relative expression levels of gloA, gloB (RSP0799), adh, ggt, gst (RSP1591), gpx, and RSP3075 in strain 2.4.1 (black bars), 2.4.1ΔrpoHII (white bars), rpoE chrR mutant strain TF18 (gray bars), strain 2.4.1ΔrpoHII(pRK2.4.1rpoHII) (hatched bars), and 2.4.1ΔrpoHII(pRK2.4.1rpoHIIRSP0600) (stippled bars) were determined after 7 min of incubation under photooxidative-stress conditions. Relative expression was determined by real-time RT-PCR. Levels of significance are indicated as follows: *, P ≤ 0.01; **, P ≤ 0.05; and ***, P ≤ 0.1.
Because the rpoHII mRNA level was also slightly increased by heat shock (Fig. 1B), mRNA levels of the RpoHII-dependent gene gst (RSP1591) measured under photooxidative stress (Fig. 3) were compared to those in response to heat stress (data not shown). Exposure to heat failed to induce gst (RSP1591) in both the wild-type 2.4.1 and the rpoHII mutant cultures, which supports the specific role of RpoHII-dependent gene expression in response to 1O2 exposure.
Identification of putative RpoHII-specific target sequences.
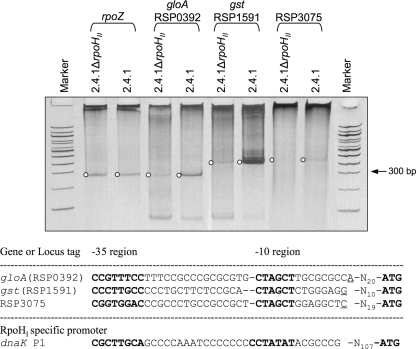
To confirm that transcription initiates at the predicted target sequences for some of the RpoHII-dependent genes, we mapped 5′ ends of the mRNA by 5′ RACE (Fig. 4). For this purpose, we chose gloA, gst (RSP1591), and RSP3075 to investigate genes that strongly depend on RpoHII (Fig. 3).
FIG. 4.
Separation of 5′ RACE products obtained from RNA extracts of wild-type and rpoHII mutant cultures after 10 min of photooxidative stress. PCR products obtained after the second PCR (nested) were separated on a 10% polyacrylamide gel and stained with ethidium bromide. Upstream of the 5′ ends of the sequences corresponding to the depicted DNA bands, RpoHII target sequences were found and are depicted as aligned sequences below the gel image. DNA marker lanes, 100-bp ladder. In the alignment, transcription start sites are underlined and putative −35 and −10 regions are printed in bold letters. The dnaK P1 promoter sequence is shown for comparison and is recognized only by RpoHI in vitro (19).
5′ RACE was performed with RNA extracts from the wild type and strain 2.4.1ΔrpoHII after they were exposed to 1O2. To assess the relative changes in expression, cDNA synthesis was performed with gene-specific primers in the same reaction (see Table S1 in the supplemental material). For all three genes, specific products were PCR amplified from wild-type cultures. Amplification of PCR products obtained from RNA extracts of strain 2.4.1ΔrpoHII was much weaker for gloA and gst (RSP1591) and missing for RSP3075 in comparison to those for the wild type (Fig. 4). A primer specific for rpoZ, a gene used for normalization in the real-time RT-PCR approach described previously (39), yielded PCR products from both strains of very similar intensities (Fig. 4). These data strongly support the RpoHII-dependent expression of genes encoding proteins not synthesized upon 1O2 exposure in strain 2.4.1ΔrpoHII. 5′ ends were determined by sequencing the amplified DNA fragments after cloning them into the pDrive vector. Putative −10 and −35 RpoHII target sequences were found for all three genes upstream of the 5′ end of the respective PCR products (Fig. 4).
Those putative target sequences were similar to those of previously predicted RpoHII-dependent promoters (6) but exhibited differences in the −35 and −10 regions compared to target sequences located upstream of the start codon of several heat shock-triggered genes and recognized by both RpoHI and RpoHII (19). The −35 region putatively recognized by RpoHII during 1O2 exposure contains not only TTG but also TTT or TGG and is flanked by GC-rich positions (Fig. 4). In contrast, the conserved −10 region consists of CTAGCT for all three genes investigated and is separated from the corresponding −35 region by 15 to 16 bp. For comparison, the RpoHI-specific promoter P1 of dnaK is shown, which differs in the −10 region by the exchange of GC with TA (Fig. 4).
Changes in methylglyoxal sensitivity.
The proteome analysis approach indicated that several proteins lacking in the rpoHII mutant upon 1O2 exposure are involved in the degradation of methylglyoxal (Table 2). This toxic nucleophile accumulates during the disorder of sugar metabolism and oxidative stress and is spontaneously formed within cells (24). Differences between the rpoHII mutant and the wild type in sensitivity to methylglyoxal were observed in inhibition zone assays. Inhibition zones observed for the wild type were 3.2 ± 0.1 cm in diameter and were significantly increased to 3.6 ± 0.1 cm for the rpoHII mutant. Strains harboring plasmid pRK415 yielded no significantly different inhibition zone values (see Table S2 in the supplemental material). The inhibition zone diameters yielded by the two complemented rpoHII mutants were not significantly different from that of the wild type and measured 3.2 ± 0.1 cm and 3.3 ± 0.1 cm for strains 2.4.1ΔrpoHII(pRK2.4.1rpoHII) and 2.4.1ΔrpoHII(pRK2.4.1rpoHIIRSP0600, respectively.
DISCUSSION
A functional hierarchy is established by the RpoE-RpoHII sigma factor cascade.
Here we demonstrate that RpoHII acts downstream of RpoE in a sigma factor cascade. This is shown by the lack of rpoHII expression in the rpoE-deficient mutant TF18 under conditions of 1O2 exposure (Fig. 1A). rpoHII was one of the genes identified among the few that are preceded by an RpoE target sequence in the promoter region (3, 6). Interestingly, a rather small number of genes belonging to the RpoE regulon exhibit a conserved RpoE target sequence (3, 6, 17). Their functions are restricted to DNA repair upon UV damage (photolyase, phrA), energy metabolism (cytochrome A, cycA; the cyclopropane/cyclopropene fatty acid synthesis RSP1091 protein), gene regulation (rpoE, rpoHII, and the tspO-like RSP1409 protein), transformation of thiols (RSP3184 and dsbG), and hypothetical functions (RSP1465 and RSP1852). Therefore, the function of proteins encoded by genes exhibiting a conserved RpoE target sequence are rather different than those of the RpoHII regulon (see below), which provides evidence that a functional hierarchy exists in the response to 1O2 exposure.
Our findings show that most of the genes encoding proteins that directly or conditionally depend on RpoE but do not contain a conserved RpoE target sequence are indeed controlled by RpoHII (Fig. 2 to 4; Table 2). Unlike time-oriented sigma factor cascades that regulate, e.g., sporulation, the RpoE-RpoHII cascade controls a functional hierarchy characterized by (i) the rapid induction of RpoHII-dependent protein synthesis upon 1O2 exposure (17) and (ii) the function of proteins encoded by genes preceded by RpoE and RpoHII target sequences (Table 2).
Slightly increased mRNA levels of RpoHII-dependent genes were observed upon 1O2 exposure in the TF18 strain lacking rpoE but not in the rpoHII mutant, except for gloB (RSP0799) (Fig. 3). This finding indicates that the expression of rpoHII may be triggered to a small extent by further molecular factors acting directly on rpoHII that have yet to be identified. Although rpoHI transcript levels were increased by 1O2 exposure up to 12-fold, the synthesis of typical heat shock proteins such as HslO was increased only 2- to 3-fold compared to that of control conditions (17), and spot intensities of additional heat shock proteins such as GroEL and DnaK were not increased (data not shown). In Agrobacterium tumefaciens, the activity of RpoHI is controlled by the chaperone DnaK, which releases RpoHI under heat shock conditions (35). It remains to be elucidated if RpoHI of R. sphaeroides is controlled by a mechanism similar to that in A. tumefaciens.
Analysis of the rpoHII mutant and the complementation with plasmids harboring rpoHII or rpoHII and RSP0600 show that sensitivity to 1O2 and methylglyoxal is caused by the deletion of rpoHII. The amounts of proteins synthesized in the complemented strains are not restored to wild-type levels for all proteins, e.g., those encoded by ggt and gst (Table 2). However, mRNA levels were restored for both genes and were not different for the two complemented strains.
RpoHII controls a functional response to 1O2 exposure.
The role of RpoHII-type sigma factors, which are widely distributed among organisms in the Alphaproteobacteria (19), is largely unknown with respect to environmental response and function of the controlled genes. The proposed function of genes under RpoHII control may explain the adaptive response to photooxidative stress observed for R. sphaeroides (17). 1O2 reacts with a variety of cellular molecules, such as proteins, lipids, DNA, and RNA, and forms partially oxidized reaction products, e.g., protein and lipid peroxides, which are toxic to cells (11, 22). Our data show that several genes encoding proteins involved in GSH-dependent and -independent detoxification of 1O2 reaction products are controlled by RpoHII (Table 2).
The first group of RpoHII-dependent proteins is involved in the GSH-dependent detoxification of reaction products formed from cellular macromolecules interacting with 1O2. Gst (RSP1591 and RSP1397, encoding glutathione S-transferase) catalyzes the detoxification of a wide range of molecules that harbor an electrophilic carbon, sulfur, or nitrogen atom and may reduce organic peroxides (22). Mechanistically, conjugates of the electrophilic substrates are formed with GSH. Those conjugates may be further metabolized by Ggt (RSP3272, encoding γ-glutamyltransferase) and aminopeptidases, which subsequently cleave the glutamyl and glycine residues (22). In R. sphaeroides, Ggt may represent a periplasmic protein that also ensures regeneration of GSH and redox homeostasis. The aminopeptidase function is unclear but may be carried out by the RpoHII-dependent putative Zn-dependent peptidases encoded by RSP1096 and RSP1097, which harbor a PqqL peptidase motif. PQQ was shown to be a factor for survival against oxidative stress in vitro (33) and in a bacterial system such as that of E. coli (27).
The second group of RpoHII-dependent proteins is involved in the degradation of peroxides. The gene product of prxQ (RSP2973, encoding a peroxiredoxin) was shown to reduce H2O2 and cumene hydroperoxide in vitro, and the deletion of prxQ leads to an increased sensitivity to exposure to 1O2 and other reactive oxygen species (48). Gpx (RSP2389, encoding GSH peroxidase) was induced by organic hydroperoxides in Chlamydomonas reinhardtii (15, 30), and bovine Gpx degraded protein peroxides in vitro (10, 34). Although mRNA levels were increased upon 1O2 exposure (16), Gpx was not detected on 2D gels of soluble proteins (Fig. 2) and may therefore be membrane associated in R. sphaeroides. Control of prxQ and gpx expression by RpoHII may be crucial for organic peroxide degradation upon photooxidative stress. Synthesis of CatA (previously called CatE) is decreased in the absence of RpoHII under photooxidative stress. The CatA synthesis induced by RpoHII may reflect (i) the known 1O2 sensitivity of catalases or (ii) the need for increased CatA levels to degrade H2O2 generated from 1O2 reaction products in the cytoplasm.
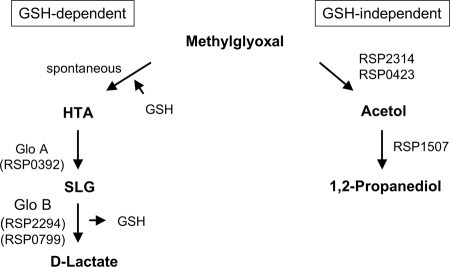
The third group of proteins controlled by RpoHII is involved in methylglyoxal removal. Methylglyoxal is generated under conditions of carbohydrate metabolism disorder (28) and amino acid breakdown and may accumulate under oxidative stress (24). This strong nucleophile rapidly reacts with macromolecules such as proteins and nucleic acids (9). Therefore, intracellular accumulation needs to be avoided to prevent cellular damage (24). Synthesis of proteins involved in GSH-dependent and -independent pathways of methylglyoxal degradation are controlled by RpoHII in R. sphaeroides (Table 2). GSH-dependent degradation of methylglyoxal is carried out by GloA and GloB (RSP0799 and RSP2294, respectively), which represent the glyoxalase system forming d-lactate as an end product (Fig. 5). Degradation of methylglyoxal independently of GSH is performed by methylglyoxal reductase, aldo-keto reductases, and alcohol dehydrogenases, thereby forming lactaldehyde and 1,2-propanediol (Fig. 5). The oxidoreductases YeaE and YghZ found in E. coli have been shown to degrade methylglyoxal in vitro in an NADPH-dependent manner (29). A BLAST search (1) revealed the aldo-keto reductases related to DkgA, YeaE, and YghZ in R. sphaeroides: the RpoHII-dependent RSP0423 and RSP2314 oxidoreductases (27 to 46% identity) (Fig. 5). The acetol formed by those oxidoreductases may be degraded by the predicted RSP1507 aldehyde dehydrogenase to 1,2-propanediol, which may then be further metabolized by the RSP3537 product, a predicted alcohol dehydrogenase (not shown in Fig. 5). The control of methylglyoxal-degrading pathways by RpoHII is supported by an increased sensitivity of the rpoHII mutant to methylglyoxal. Further studies of the functions of those proteins need to be performed to verify their role in the response to 1O2, with respect to methylglyoxal detoxification in R. sphaeroides.
FIG. 5.
Role of selected RpoHII-dependent proteins in GSH-dependent and GSH-independent degradation of methylglyoxal. The putative functions of the depicted proteins were inferred from comparisons to homologous proteins based on BLAST search results. HTA, hemithioacetal; SLG, S-d-lactoylglutathione. For the functions of the depicted proteins, see Table 2.
Our data show that several of the RpoHII-dependent proteins make use of GSH-dependent and -independent reactions to elicit a cellular response to prevent 1O2-caused damages. GSH obviously plays a central role in the defense of R. sphaeroides against 1O2, which exceeds direct scavenging of 1O2 by GSH, as proposed previously (11, 13). Interestingly, 5-methyltetrahydrofolate exhibits the capacity to prevent photosensitizing reactions and may act as a 1O2 scavenger (37). A putative formate-tetrahydrofolate ligase (RSP0663) is under the control of RpoHII. Alternatively, 5-methyltetrahydrofolate would need to be regenerated under photooxidative stress, because it is specifically damaged by 1O2 and acts as a cofactor of, e.g., photolyases and cryptochromes (42).
Additional proteins controlled by RpoHII that are putatively involved in oxidative-stress responses are Bfr (bacterioferritin) and the RSP3164 (ferredoxin-like) protein, which represent iron-binding proteins. The control of intracellular iron concentration is pivotal to preventing the formation of hydroxyl radicals by the reaction of iron(II) with peroxides via the Fenton reaction (18). Ferredoxins are involved in cellular redox reactions and may serve as electron donors for the detoxification of reactive oxygen species. In conclusion, the RpoHII regulon defined by the investigation of the rpoHII mutant by using a proteome approach revealed a functionally sharply defined and newly assigned role for RpoHII in controlling genes in R. sphaeroides in response to 1O2 exposure.
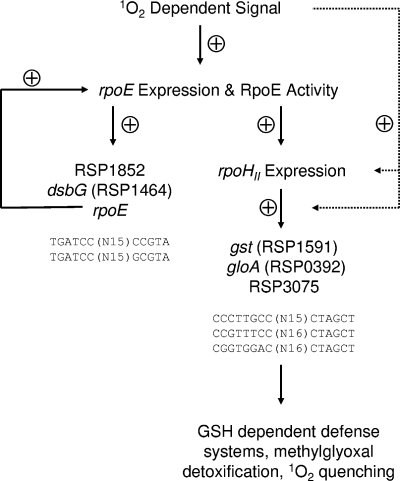
Current model of RpoE-RpoHII-controlled gene expression under photooxidative stress.
Our current model of gene regulation upon photooxidative stress displays the interaction of the rpoE and rpoHII gene products (Fig. 6). The recognition of 1O2 or 1O2-caused damage by R. sphaeroides is not clear to date. However, a recently recognized Zn-binding site in the anti-sigma factor ChrR may be involved in the recognition of 1O2 (7), which may ultimately lead to the dissociation of the RpoE/ChrR complex. The binding of RpoE to specific target sequences triggers the expression of a small number of genes, including rpoE itself and rpoHII (3, 6). Thereby, expression and activity of RpoE are increased and rpoHII-dependent genes are induced. The functions of genes that harbor an RpoE target sequence did not explain the adaptation of R. sphaeroides to photooxidative stress. In contrast, genes that harbor an RpoHII target sequence encode defense systems responding to oxidative stress. GSH-dependent and GSH-independent mechanisms controlled by RpoHII remove organic peroxides generated by the reaction of 1O2 with macromolecules, and accumulation of toxic by-products such as methylglyoxal is prevented. Genes involved in the regeneration or production of 1O2-specific scavengers or metabolites that are especially prone to 1O2 reaction, such as GSH (13) and 5-methyltetrahydrofolate (37), are under the control of RpoHII.
FIG. 6.
The current model displays the role of RpoHII in the RpoE-dependent response to 1O2 in R. sphaeroides. Solid arrows indicate positive effects in the regulatory cascade triggered by 1O2. Dotted arrows indicate the functions of hypothetical expression factors involved in the RpoE-RpoHII-dependent gene induction. Representative target sequences for RpoE (3, 17) and the putative target sequences for RpoHII identified in this study (Fig. 4) are depicted.
RpoHII-like sigma factors have so far been recognized only in the Alphaproteobacteria (19) and may control rather specific functions. In Brucella melitensis, the gene BMEI0280 (rpoH1) represents the R. sphaeroides rpoHII homolog. This gene is not involved in the response to heat stress but may be important for the chronicity of B. melitensis infection (12). In Sinorhizobium meliloti, rpoHII is induced during stationary phase, and an rpoHII mutant showed that the resulting strain is not heat sensitive (38). Because several Alphaproteobacteria have two or even three rpoH homologs, it is reasonable to assume that one of the two heat shock sigma factors was recruited for functions other than defense against heat shock. Our regulon analysis represents the first in vivo study of an RpoHII-type sigma factor by using an rpoHII mutant background and indicates that it is crucial for the regulation of defense systems against photooxidative stress in R. sphaeroides. Furthermore, our data indicate that the 1O2-dependent signal triggers the recognition of RpoHII-specific target sequences and that the RpoHII-dependent response is to some extent independent from RpoE.
Supplementary Material
Acknowledgments
This work was supported by DFG grants Kl563/16 and Kl563/20.
Footnotes
Published ahead of print on 31 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anthony, J. R., J. D. Newman, and T. J. Donohue. 2004. Interactions between the Rhodobacter sphaeroides ECF sigma factor, σE, and its anti-sigma factor, ChrR. J. Mol. Biol. 341345-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony, J. R., K. L. Warczak, and T. J. Donohue. 2005. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc. Natl. Acad. Sci. USA 1026502-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arellano, J. B., Y. A. Yousef, T. B. Melø, S. B. B. Mohamad, R. J. Cogdell, and K. R. Naqvi. 2007. Formation and geminate quenching of singlet oxygen in purple bacterial reaction center. J. Photochem. Photobiol. B 87105-112. [DOI] [PubMed] [Google Scholar]
- 5.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 45827-836. [DOI] [PubMed] [Google Scholar]
- 6.Braatsch, S., O. V. Moskvin, G. Klug, and M. Gomelsky. 2004. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J. Bacteriol. 1867726-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, E. A., R. Greenwell, J. R. Anthony, S. Wang, L. Lim, K. Das, H. J. Sofia, T. J. Donohue, and S. A. Darst. 2007. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell 27793-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cogdell, R. J., T. D. Howard, R. Bittl, E. Schlodder, I. Geisenheimer, and W. Lubitz. 2000. How carotenoids protect bacterial photosynthesis. Philos. Trans. R. Soc. Lond. B 3551345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, R. A. 1984. Metabolism of methylglyoxal in microorganisms. Annu. Rev. Microbiol. 3849-68. [DOI] [PubMed] [Google Scholar]
- 10.Davies, M. J. 2004. Reactive species formed on proteins exposed to singlet oxygen. Photochem. Photobiol. Sci. 317-25. [DOI] [PubMed] [Google Scholar]
- 11.Davies, M. J. 2003. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 305761-770. [DOI] [PubMed] [Google Scholar]
- 12.Delory, M., R. Hallez, J.-J. Letesson, and X. De Bolle. 2006. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J. Bacteriol. 1887707-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Mascio, P., T. P. A. Devasagayam, S. Kaiser, and H. Sies. 1990. Carotenoids, tocopherols and thiols as biological singlet molecular oxygen quenchers. Biochem. Soc. Trans. 181054-1056. [DOI] [PubMed] [Google Scholar]
- 14.Drews, G. 1983. Mikrobiologisches praktikum. Springer Verlag, Heidelberg, Germany.
- 15.Fischer, B. B., R. I. L. Eggen, A. Trebst, and A. Krieger-Liszkay. 2006. The glutathione peroxidase homologous gene GpxH in Chlamydomonas reinhardtii is upregulated by singlet oxygen produced in photosystem II. Planta 223583-590. [DOI] [PubMed] [Google Scholar]
- 16.Glaeser, J., and G. Klug. 2005. Photo-oxidative stress in Rhodobacter sphaeroides: protective role of carotenoids and expression of selected genes. Microbiology 1511927-1938. [DOI] [PubMed] [Google Scholar]
- 17.Glaeser, J., M. Zobawa, F. Lottspeich, and G. Klug. 2007. Protein synthesis patterns reveal a complex regulatory response to singlet oxygen in Rhodobacter. J. Proteome Res. 62460-2471. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein, S., D. Meyerstein, and G. Czapski. 1993. The Fenton reagents. Free Radic. Biol. Med. 15435-445. [DOI] [PubMed] [Google Scholar]
- 19.Green, H. A., and T. J. Donohue. 2006. Activity of Rhodobacter sphaeroides RpoHII, a second member of the heat shock sigma factor family. J. Bacteriol. 1885712-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregor, J., and G. Klug. 1999. Regulation of bacterial photosynthesis genes by oxygen and light. FEMS Microbiol. Lett. 1791-9. [DOI] [PubMed] [Google Scholar]
- 21.Happ, H. N., S. Braatsch, V. Broschek, L. Osterloh, and G. Klug. 2005. Light-dependent regulation of photosynthesis genes in Rhodobacter sphaeroides 2.4.1 is coordinately controlled by photosynthetic electron transport via the PrrBA two-component system and the photoreceptor AppA. Mol. Microbiol. 58903-914. [DOI] [PubMed] [Google Scholar]
- 22.Hayes, J. D., J. U. Flanagan, and I. R. Jowsey. 2005. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 4551-88. [DOI] [PubMed] [Google Scholar]
- 23.Hübner, P., J. C. Willison, P. M. Vignais, and T. A. Bickle. 1991. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 1732993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalapos, M. P. 1999. Methylglyoxal in living organisms: chemistry, biochemistry, toxicology and biological implications. Toxicol. Lett. 110145-175. [DOI] [PubMed] [Google Scholar]
- 25.Karls, R. K., J. Brooks, P. Rossmeissl, J. Luedke, and T. J. Donohue. 1998. Metabolic roles of a Rhodobacter sphaeroides member of the sigma 32 family. J. Bacteriol. 18010-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70191-197. [DOI] [PubMed] [Google Scholar]
- 27.Khairnar, N. P., H. S. Misra, and S. K. Apte. 2003. Pyrroloquinoline-quinone synthesized in Escherichia coli by pyrroloquinoline-quinone synthase of Deinococcus radiodurans plays a role beyond mineral phosphate solubilization. Biochem. Biophys. Res. Commun. 312303-308. [DOI] [PubMed] [Google Scholar]
- 28.Kim, I., E. Kim, S. Yoo, D. Shin, B. Min, J. Song, and C. Park. 2004. Ribose utilization with an excess of mutarotase causes cell death due to accumulation of methylglyoxal. J. Bacteriol. 1867229-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko, J., I. Kim, S. Yoo, B. Min, K. Kim, and C. Park. 2005. Conversion of methylglyoxal to acetol by Escherichia coli aldo-keto reductases. J. Bacteriol. 1875782-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leisinger, U., K. Rufenacht, B. Fischer, M. Pesaro, A. Spengler, A. J. Zehnder, and R. I. Eggen. 2001. The glutathione peroxidase homologous gene from Chlamydomonas reinhardtii is transcriptionally up-regulated by singlet oxygen. Plant Mol. Biol. 46395-408. [DOI] [PubMed] [Google Scholar]
- 31.Mackenzie, C., J. M. Eraso, M. Choudhary, J. H. Roh, X. Zeng, P. Bruscella, A. Puskas, and S. Kaplan. 2007. Postgenomic adventures with Rhodobacter sphaeroides. Annu. Rev. Microbiol. 61283-307. [DOI] [PubMed] [Google Scholar]
- 32.Masuda, S., and C. E. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110613-623. [DOI] [PubMed] [Google Scholar]
- 33.Misra, H. S., N. P. Khairnar, A. Barik, K. Indira Priyadarsini, H. Mohan, and S. K. Apte. 2004. Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Lett. 57826-30. [DOI] [PubMed] [Google Scholar]
- 34.Morgan, P. E., R. T. Dean, and M. J. Davies. 2004. Protective mechanisms against peptide and protein peroxides generated by singlet oxygen. Free Radic. Biol. Med. 36484-496. [DOI] [PubMed] [Google Scholar]
- 35.Nakahigashi, K., H. Yanagi, and T. Yura. 2001. DnaK chaperone-mediated control of activity of a σ32 homolog (RpoH) plays a major role in the heat shock response of Agrobacterium tumefaciens. J. Bacteriol. 1835302-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman, J. D., M. J. Falkowski, B. A. Schilke, L. C. Anthony, and T. J. Donohue. 1999. The Rhodobacter sphaeroides ECF sigma factor σE and the target promoters cycA P3 and rpoE P1. J. Mol. Biol. 294307-320. [DOI] [PubMed] [Google Scholar]
- 37.Offer, T., B. N. Ames, S. W. Bailey, E. A. Sabens, M. Nozawa, and J. E. Ayling. 2007. 5-Methyltetrahydrofolate inhibits photosensitization reactions and strand breaks in DNA. FASEB J. 212101-2107. [DOI] [PubMed] [Google Scholar]
- 38.Oke, V., B. G. Rushing, E. J. Fisher, M. Moghadam-Tabrizi, and S. R. Long. 2001. Identification of the heat-shock sigma factor RpoH and a second RpoH-like protein in Sinorhizobium meliloti. Microbiology 1472399-2408. [DOI] [PubMed] [Google Scholar]
- 39.Pappas, C. T., J. Sram, O. V. Moskvin, P. S. Ivanov, R. C. Mackenzie, M. Choudhary, M. L. Land, F. W. Larimer, S. Kaplan, and M. Gomelsky. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 1864748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prentki, P., A. Binda, and A. Epstein. 1991. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene 10317-23. [DOI] [PubMed] [Google Scholar]
- 42.Sancar, A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 1032203-2238. [DOI] [PubMed] [Google Scholar]
- 43.Schilke, B. A., and T. J. Donohue. 1995. ChrR positively regulates transcription of the Rhodobacter sphaeroides cytochrome c2 gene. J. Bacteriol. 1771929-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimada, H., K. Iba, and K.-i. Takamiya. 1992. Blue-light irradiation reduces the expression of puf and puc operons of Rhodobacter sphaeroides under semi-aerobic conditions. Plant Cell Physiol. 33471-475. [Google Scholar]
- 45.Simon, R., M. O'Connell, M. Labes, and A. Puhler. 1986. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other gram-negative bacteria. Methods Enzymol. 118640-659. [DOI] [PubMed] [Google Scholar]
- 46.Uchoa, A., P. Knox, R. Turchielle, N. Seifullina, and M. Baptista. 2008. Singlet oxygen generation in the reaction centers of Rhodobacter sphaeroides. Eur. Biophys. J. 37843-850. [DOI] [PubMed] [Google Scholar]
- 47.van Niel, C. B. 1944. The culture, general physiology, morphology, and classification of the non-sulfur purple and brown bacteria. Bacteriol. Rev. 81-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wakita, M., S. Masuda, K. Motohashi, T. Hisabori, H. Ohta, and K.-I. Takamiya. 2007. The significance of type II and PrxQ peroxiredoxins for antioxidative stress response in the purple bacterium Rhodobacter sphaeroides. J. Biol. Chem. 28227792-27801. [DOI] [PubMed] [Google Scholar]
- 49.Zeilstra-Ryalls, J., M. Gomelsky, J. M. Eraso, A. Yeliseev, J. O'Gara, and S. Kaplan. 1998. Control of photosystem formation in Rhodobacter sphaeroides. J. Bacteriol. 1802801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeilstra-Ryalls, J. H., and S. Kaplan. 2004. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell. Mol. Life Sci. 61417-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.