Abstract
Enteropathogenic Escherichia coli (EPEC) was the first pathovar of E. coli to be implicated in human disease; however, no EPEC strain has been fully sequenced until now. Strain E2348/69 (serotype O127:H6 belonging to E. coli phylogroup B2) has been used worldwide as a prototype strain to study EPEC biology, genetics, and virulence. Studies of E2348/69 led to the discovery of the locus of enterocyte effacement-encoded type III secretion system (T3SS) and its cognate effectors, which play a vital role in attaching and effacing lesion formation on gut epithelial cells. In this study, we determined the complete genomic sequence of E2348/69 and performed genomic comparisons with other important E. coli strains. We identified 424 E2348/69-specific genes, most of which are carried on mobile genetic elements, and a number of genetic traits specifically conserved in phylogroup B2 strains irrespective of their pathotypes, including the absence of the ETT2-related T3SS, which is present in E. coli strains belonging to all other phylogroups. The genome analysis revealed the entire gene repertoire related to E2348/69 virulence. Interestingly, E2348/69 contains only 21 intact T3SS effector genes, all of which are carried on prophages and integrative elements, compared to over 50 effector genes in enterohemorrhagic E. coli O157. As E2348/69 is the most-studied pathogenic E. coli strain, this study provides a genomic context for the vast amount of existing experimental data. The unexpected simplicity of the E2348/69 T3SS provides the first opportunity to fully dissect the entire virulence strategy of attaching and effacing pathogens in the genomic context.
Escherichia coli is important because it is biology's premier model organism, is a common commensal of the vertebrate gut, and is a versatile pathogen of humans and animals. Molecular epidemiological studies have classified E. coli strains into a number of phylogroups (phylogroups A, B1, B2, D, and E) (13, 42), which are estimated to have diverged in the last 5 to 9 million years (37, 42). Commensal E. coli strains are beneficial to the host and rarely cause disease. However, several clones of E. coli are responsible for a spectrum of diseases, including urinary tract infection, sepsis/meningitis, and diarrhea (for a review, see reference 15). Diarrheagenic E. coli strains are divided into enterotoxigenic E. coli (ETEC), enteroaggregative E. coli, enteroinvasive E. coli, diffusely adhering E. coli, enteropathogenic E. coli (EPEC), and enterohemorrhagic E. coli (EHEC) strains (15), which have different virulence mechanisms.
Whole-genome sequencing approaches have revealed that E. coli has a conserved core of genes common to both commensal and pathogenic strains. The conserved genome framework is decorated with genomic islands and small clusters of genes that have been acquired by horizontal gene transfer and that in pathogenic strains are often associated with virulence (for a review, see reference 32). EPEC strains provide a striking example of a pathovar highly adapted to virulence in the human intestine (8, 15), but until now no EPEC strain has been fully sequenced.
EPEC was the first pathovar of E. coli to be implicated in human disease (4) and remains a leading cause of infantile diarrhea in developing countries (for a review, see reference 6). However, because EPEC strains were found not to invade cells or release diffusible toxins, doubts about their pathogenic potential were raised in the 1960s and 1970s. However, induction of diarrhea in human volunteers (21) provided the decisive evidence that EPEC is a true human pathogen. As a result of this study, one of the strains tested, E2348/69 (serotype O127:H6), isolated in Taunton, United Kingdom, in 1969, became the prototype strain used globally to study EPEC biology and disease. Indeed, E2348/69 is probably the most-studied pathogenic E. coli strain, and until now it was impossible to place the vast amount of biological data in a genomic context.
Typical EPEC strains, which belong to a limited number of O serogroups, contain the EPEC adherence factor plasmid that encodes the bundle-forming pilus (BFP) (10) and also contain the gene regulator locus per (for a review, see reference 6). Typical EPEC strains are further divided into four distinct lineages, EPEC lineages 1 to 4 (18); E2348/69 belongs to EPEC lineage 1 and to the B2 phylogroup.
The hallmark of EPEC infection is formation of distinct attaching and effacing (A/E) lesions, which are characterized by effacement of the brush border microvilli and intimate bacterial attachment (for reviews, see references 6 and 8). The ability to induce A/E lesions is encoded on a pathogenicity island termed the locus of enterocyte effacement (LEE), which is also present in O157 and non-O157 EHEC strains and the mouse pathogen Citrobacter rodentium (24, 25; for a review, see reference 9). The LEE encodes the adhesin intimin, the structural components of a type III secretion system (T3SS) involved in translocation of effector proteins into the mammalian host cell, gene regulators, chaperones, translocators, and seven effector proteins (EspB, EspF, EspG, EspH, EspZ, Map, and Tir) (for a review, see reference 9). Recent studies have shown that the EPEC strain E2348/69 genome encodes several additional non-LEE effectors, including EspJ (23), EspG2, and EspI/NleA, as well as NleB, NleC, NleD, NleE, and NleH (for a review, see reference 9). Additional putative virulence factors include the autotransporter protein EspC (26), lymphostatin (LifA) (17), and several fimbrial operons. Here we report the genome sequence of E2348/69, describe a bioinformatics survey of this strain's virulence factors, and present the results of comprehensive comparative studies performed with commensal and other pathogenic E. coli strains.
MATERIALS AND METHODS
Bacterial strain and sequencing.
EPEC serotype O127:H6 strain E2348/69 was isolated in Taunton, United Kingdom, in 1969 during an outbreak of infantile diarrhea. The sequenced strain was obtained from the original stock kept at the Health Protection Agency in Colindale, United Kingdom, and was subjected to minimal laboratory passages.
The whole genome was sequenced to a depth of 8× coverage by using pUC19 (insert size, 2.8 to 5 kb) and pMAQ1b (insert size, 5.5 to 10 kb) small-insert libraries and dye terminator chemistry with ABI3700 automated sequencers. End sequences from larger-insert plasmid (pBACe3.6 [insert size, 20 to 30 kb]) libraries were used as a scaffold. The sequence was assembled and finished as described previously (33).
Gene prediction and annotation and comparative analysis.
Protein-encoding sequences (CDSs) were identified using GeneHacker (43), followed by manual inspection of start codons and ribosome binding sequences of each CDS. Intergenic regions that were >150 bp long were further reviewed to determine the presence of small CDSs encoding proteins with significant homology to known proteins. Functional annotation of the CDSs was performed on the basis of the results of homology searches with the public nonredundant protein database (http://www.ncbi.nlm.nih.gov/) using BLASTP. Genes for tRNAs, transfer-messenger RNA, rRNAs, and other small RNAs were identified by using the Rfam database (11) at the Rfam website (http://www.sanger.ac.uk/Software/Rfam/index.shtml). We also searched the E2348/69 genome for all the RNA genes that have been identified in K-12 and Sakai by using BLASTN. Figure S1 in the supplemental material shows the methods used for cluster analysis of the E2348/69 CDSs and for genomic comparison with eight E. coli genomes.
Nucleotide sequence accession numbers.
The annotated genome sequences of E2348/69 have been deposited in public databases under accession number FM180568 for the complete genome and under accession numbers FM180569 and FM180570 for EPEC strain E2348/69 plasmids pMAR2 and pE2348-2, respectively.
RESULTS AND DISCUSSION
General genomic features of E2348/69.
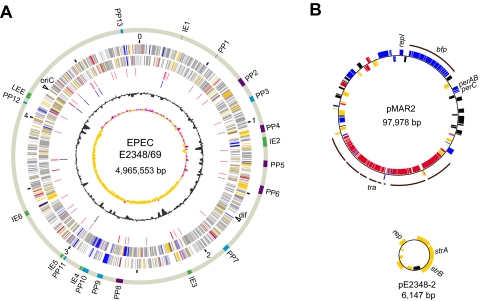
The genome of E2348/69 comprises a circular chromosome (4,965,553 bp), the EPEC adherence factor plasmid (pMAR2; 97,978 bp), and a small drug-resistant plasmid (pE2348-2; 6,147 bp) (Fig. 1 and Table 1; see Table S1 in the supplemental material). The chromosome contains 4,703 predicted protein-encoding genes (including 145 pseudogenes), 92 tRNA genes (including two pseudogenes), and seven rRNA operons. The pMAR2 plasmid is nearly identical to pMAR7, a previously sequenced pMAR2 derivative (5), but we identified three single-nucleotide polymorphisms and two single-base indels in intergenic regions that differentiate these two plasmids (see Fig. S2 in the supplemental material). pMAR2 carries an additional copy of the insertion sequence (IS) element ISEc21, which is not present in pMAR7. pE2348-2 is distantly related to the ColE1 plasmid family and carries the strAB operon encoding streptomycin resistance.
FIG. 1.
Circular maps of the chromosome and plasmids of EPEC strain E2348/69. (A) EPEC strain E2348/69 chromosome. From the outside in, the first circle shows the locations of PPs and IEs (purple, lambda-like PPs; light blue, other PPs; green, IEs and the LEE element), the second circle shows the nucleotide sequence positions (in Mbp), the third and fourth circles show CDSs transcribed clockwise and anticlockwise, respectively (gray, conserved in all eight other sequenced E. coli strains; red, conserved only in the B2 phylogroup; yellow, variable distribution; blue, E2348/69 specific), the fifth circle shows the tRNA genes (red), the sixth circle shows the rRNA operons (blue), the seventh circle shows the G+C content, and the eighth circle shows the GC skew. (B) EPEC strain E2348/69 plasmids. The boxes in the outer and inner circles represent CDSs transcribed clockwise and anticlockwise, respectively. Pseudogenes are indicated by black boxes, and other CDSs are indicated by the colors described above for panel A.
TABLE 1.
Comparison of general genome features of EPEC strain E2348/69 and eight other sequenced E. coli strains
| Strain | Phylogroup | Pathotype | Serotype | Chromosome
|
Plasmids
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (kb) | G+C content (%) | No. of CDSsa | CDS density (%) | No. of tRNAsa | % Unique region compared with E2348/69 | GenBank accession no. | Size(s) (kb) | GenBank accession no. | ||||
| E2348/69 | B2 | EPEC | O127:H6 | 4,965 | 50.6 | 4,703 (145) | 88.2 | 92 (2) | FM180568 | 97, 6 | FM180569, FM180570 | |
| UTI89 | B2 | UPEC | 5,065 | 50.6 | 5,066 | 91.1 | 88 | 14.0 | CP000243 | 114 | CP000244 | |
| CFT073 | B2 | UPEC | O6:K2:H1 | 5,231 | 50.5 | 5,473 (94) | 91.9 | 89 | 14.9 | AE014075 | ||
| 536 | B2 | UPEC | O6:K15:H31 | 4,938 | 50.5 | 4,685 | 88.7 | 81 | 15.6 | CP000247 | ||
| APEC O1 | B2 | APEC | O1:K1:H7 | 5,082 | 50.6 | 4,467 | 87.5 | 94 | 14.6 | CP000468 | 241, 174, 105, 46 | DQ381420, DQ517526 |
| O157 Sakai | E | EHEC | O157:H7 | 5,498 | 50.5 | 5,361 | 88.1 | 105 (3) | 21.4 | BA000007 | 92, 3 | AB011548, AB011549 |
| MG1655 | A | Commensal | 4,639 | 50.8 | 4,294 (101) | 89.0 | 88 (3) | 23.2 | U00096 | |||
| HS | A | Commensal | O9 | 4,643 | 50.8 | 4,478 (94) | 88.7 | 88 (1) | 24.0 | CP000802 | ||
| E24377A | B1 | ETEC | O139:H28 | 4,979 | 50.6 | 4,873 (118) | 88.6 | 91 (3) | 22.2 | CP000800 | 79, 74, 70, 34, 6, 5 | CP000795 to CP000799, CP000801 |
If available, the numbers of pseudogenes are indicated in parentheses.
Mobile genetic elements.
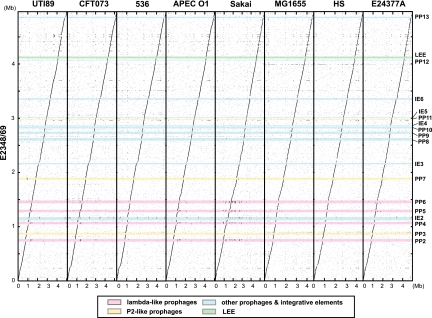
A comparison of the E2348/69 sequence (phylogroup B2) with the genomes of eight sequenced E. coli strains, including K-12 strain MG1655 (phylogroup A), EHEC strain Sakai (phylogroup E), three uropathogenic E. coli (UPEC) strains (UTI89, CFT073, and 536), an avian pathogenic E. coli (APEC) strain (O1) (phylogroup B2), an ETEC strain (E24377A) (phylogroup B1), and a commensal strain (HS) (phylogroup A) (Table 1; see Fig. S3 in the supplemental material), showed that the chromosomes of these organisms are remarkably conserved and display a high degree of overall synteny (Fig. 2). We found no evidence of large chromosome inversions or translocations in E2348/69. However, scattered between the conserved regions in E2348/69 are many strain-specific sequences (about 23 and 21% of the chromosome compared to E. coli K-12 [3] and EHEC strain Sakai [12], respectively [Table 1]), which are mainly mobile genetic elements.
FIG. 2.
Dot plot presentation of DNA sequence homologies between the chromosomes of E2348/69 and eight sequenced E. coli strains. The chromosome sequence of E2348/69 was compared with the chromosome sequences of eight sequenced E. coli strains. The locations of PPs and IEs on the E2348/69 chromosome are indicated.
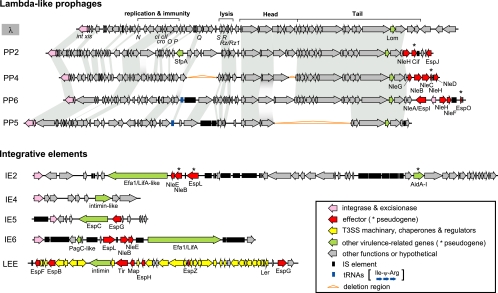
Within the E2348/69-specific sequences we identified 13 prophages (PPs) (PP1 to PP13) and eight integrative elements (IEs) (IE1a, IE1b, IE2 to IE6, and the LEE), which encode an integrase but do not have other phage- and plasmid-related functions (Fig. 1 and 3; see Fig. S4 and Table S2 in the supplemental material). The 13 PPs include four lambda-like phages (PP2, PP4, PP5, and PP6), three P2-like phages (PP3, PP7, and PP 11), a P4-like phage (PP12), an epsilon 15-like phage (PP10), a Mu-like phage (PP9), and a distantly related P22 phage family member (PP8).
FIG. 3.
Genome organizations of four PPs and five IEs carrying E2348/69 virulence-related genes. The four phages are lambda-like phages. Homologous genes in the lambda (accession no. NC_001416) and four PP genomes are indicated by gray shading. T3SS effectors are encoded on three lambda-like phages (PP2, PP4, and PP6) and four IEs (IE2, IE5, IE6, and LEE). Since the NleE family gene on IE2 contains an in-frame 168-bp deletion, it may be nonfunctional.
While PP1 appears to be a highly degraded ancestral E. coli phage, corresponding to PP DLP12 of E. coli K-12 (see Fig. S4 and Table S2 in the supplemental material), the other PPs appear to be more recent acquisitions. None of the lambda-like phages of E2348/69 showed high sequence similarity to each other; PP4 is very similar to Sp3, and PP6 is very similar to Sp6, Sp9, Sp10, and Sp12 (lambda-like PPs of EHEC strain Sakai [12]) (see Fig. S5 in the supplemental material). P2-like phages nearly identical to PP3 of E2348/69 are present in UPEC strain CFT073 and commensal strain HS, and a PP7-like phage is present in UPEC strain UTI89 (Fig. 2). Of the 13 PPs identified in E2348/69, only 6 appear to be intact (see Table S2 in the supplemental material).
We also identified a total of 117 IS elements or fragments of IS elements in E2348/69, which were classified into 41 types based on sequence similarity (see Table S3 in the supplemental material). The most abundant IS element is ISEc13 (a total of 30 copies). ISEc21 is a newly identified IS element belonging to the IS110 family. E2348/69 contains six copies of ISEc21.
Genomic comparison with commensal and other pathogenic E. coli strains.
We performed an all-against-all reciprocal BLASTP comparison of the complete gene sets of E2348/69 and the eight previously sequenced E. coli strains. However, large variations in gene predictions confounded this analysis. In order to avoid biases introduced by differences in gene prediction, we compared the E2348/69 gene set with the genomes of the other E. coli strains using one-way comparisons with TBLASTN.
We first identified how many of the E2348/69 genes were unique and how many belonged to paralogous gene families. To this end, we performed a cluster analysis of the 4,656 E2348/69 CDSs (pseudogenes were excluded from this analysis) using BLASTP, which yielded 4,419 unique genes or singlets and 69 gene families containing more than one member. We then performed a TBLASTN analysis using the unique genes and one representative gene from each of the 69 gene families for a comparison with the eight E. coli genomes (see Fig. S1 in the supplemental material).
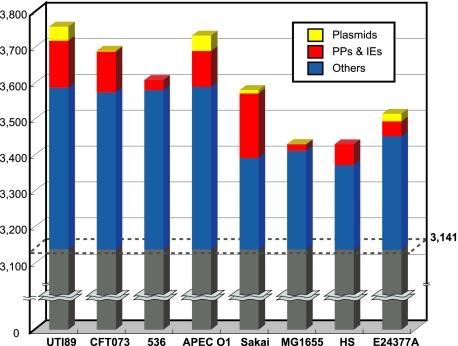
Figure 4 shows that more E2348/69 genes are conserved (as defined by ≥90% identity and ≥60% overlap) in the four strains belonging to phylogroup B2 than in strains belonging to other phylogroups. Of the 4,488 E2348/69 genes or gene families, 3,141 (70%) (which include no IS transposase genes) are conserved in all nine strains examined. With one exception, all of the 3,141 genes were found to be on the chromosomal backbone outside PPs and IEs (E2348_C_1118 was on IE2, which encodes a predicted protein). In contrast, the majority of the E2348/69-specific genes (349/424) were found to be in PPs and IEs (319 genes) or plasmids (30 genes). The remaining 75 E2348/69-specific genes on the chromosome backbone include genes for O127 antigen biosynthesis and two restriction-modification systems, a d-arabitol utilization operon (alt), a retron element, and three fimbrial biosynthesis operons (see Table S4 in the supplemental material). The E2348/69-specific BFP fimbrial operon was found to be in the pMAR2 plasmid. In addition, two E2348/69-specific genes for tRNAAsn (codon AAC) and tRNAThr (codon ACA) were found to be “cargo” on PP8 (see Fig. S4 in the supplemental material).
FIG. 4.
Conservation of E2348/69 genes in the eight sequenced E. coli strains. The numbers of E2348/69 genes conserved in each of the eight sequenced E. coli strains are indicated. Of the 4,492 genes (or multicopy gene families) identified in E2348/69, 3,141 (indicated by gray) were conserved in all the E. coli strains. Other genes were classified into three groups according to their locations in the E2348/69 genome (in plasmids, in PPs and IEs, and in other chromosome regions).
We also identified 98 “phylogroup B2-specific” genes which are conserved in all the phylogroup B2 strains, irrespective of the pathotype. All these phylogroup B2-specific genes are located on the chromosomal backbone, and they include genes encoding sugar transport and utilization and metabolic functions and a gene cluster probably encoding a phylogroup B2-specific di-/tricarboxylate utilization system (see Table S5 in the supplemental material). The rhs (rearrangement hot spot) locus of the phylogroup B2 strains is also unique. While other E. coli strains contain five to seven rhs loci, the phylogroup B2 strains contain one or two such loci. EPEC strain E2348/69 contains only one highly degraded locus, the sequence of which is very different from the sequences of the rhs loci of other E. coli strains, suggesting that it had a distinct origin.
Although not conserved across the entire B2 phylogroup, 119 genes were found exclusively in strains belonging to this group (see Table S5 in the supplemental material). Since most of these genes are carried on PPs, IEs, or plasmids, they are unlikely to be true orthologues present in the last common phylogroup B2 ancestor. However, there were a few notable exceptions, including the operon for sucrose utilization that occurs in the same chromosomal context in E2348/69 and UPEC strain 536.
Interestingly, of the four non-phylogroup B2 strains, EPEC strain E2348/69 shares the highest number of genes with EHEC strain Sakai (Fig. 4; see Fig. S1 in the supplemental material). Most of the shared genes are carried on PPs and IEs, suggesting that there was independent acquisition through horizontal gene transfer rather than inheritance through vertical descent.
Unexpectedly, we found that all of the genes encoding (pdu) and regulating (pocR) the coenzyme B12-dependent degradation of 1,2-propanediol are present only in E2348/69 and ETEC strain E24377A (see Fig. S6 in the supplemental material). The pdu operon and pocR are found in the same genetic context, alongside the cobTSU genes encoding parts I and III of the cobalamin biosynthetic pathway, as in Salmonella (19). In contrast to Salmonella, the cbi operon, encoding the endogenous biosynthesis of coenzyme B12, has been deleted in the two E. coli strains and the cob-pdu locus is highly divergent in E. coli (see Fig. S6 in the supplemental material). The significance of this diversity is not clear, but the locus was most likely inherited from an E. coli-Salmonella common ancestor and has undergone extensive deletion and rearrangement in multiple lineages of E. coli.
Functional gene loss.
E2348/69 possesses 168 pseudogenes that have frameshifts or premature stop codons or are remnants of genes present in other bacteria (see Table S6 in the supplemental material). Pseudogenes occur about three times more frequently in plasmids, PPs, or IEs (64/869 CDSs) than in the chromosome backbone (101/3,965 CDSs). Pseudogenes found in accessory chromosome regions and in the plasmids were largely remnants of IS element insertion events or were genes related to phage functions (see Table S6 in the supplemental material). However, several of the pseudogenes in PPs are associated with virulence, including genes encoding multiple T3SS effector proteins. Likewise, several of the pseudogenes found in the chromosome backbone are associated with survival in the host, including the genes that disrupt four fimbrial operons and the dmsA gene required for the anaerobic use of dimethyl sulfoxide as a terminal electron acceptor and a remnant of the gene encoding hemolysin E. Interestingly, hlyE is intact in MG1655 and EHEC strain Sakai but is inactivated in the other six E. coli strains compared. The deletion in the E2348/69 hlyE gene is identical to those in the other phylogroup B2 strains. Furthermore, the E2348/69 dsdA gene, encoding d-serine deaminase implicated in d-serine catabolism, which has been shown to be involved in UPEC virulence (39), contains a frameshift mutation. In addition to point mutations and deletions there is also evidence of metabolic streamlining through insertional inactivation; two genes involved in ethanolamine utilization, eutC and eutA, have been inactivated by insertion of Mu-like phage (PP9) and ISEc13, respectively. Likewise, the gene encoding the l-arabinose transporter (araH) and dgoD, required for use of galactonate as a carbon source, have also been disrupted by ISEc13 elements.
Virulence determinants.
Since its discovery (23), it has been apparent that the LEE plays a major role in EPEC pathogenesis. Use of random and targeted discovery programs led to identification of several additional chromosomal and plasmid-encoded virulence factors. When these results were supplemented with the results of homology searches, the genome project data revealed an E2348/69 virulence repertoire that consists of the following elements.
(i) Afimbrial adhesins and outer membrane proteins.
LifA (17) and intimin (14) are major E2348/69 afimbrial adhesins. We also found a second LifA homologue and a Saa autoagglutinating adhesin-like protein. E2348/69 Saa is almost identical to the homologues found in EHEC O157 strain Sakai, UPEC strain UTI89, and APEC (95, 97, and 97% amino acid sequence identity, respectively), but it is only distantly related (30% sequence identity) to Saa found in a LEE-negative Shiga toxigenic E. coli serotype O113:H21 strain (34). E2348/69 also has five intact genes and four pseudogenes encoding afimbrial adhesin homologues or adhesin-like proteins (see Table S7 in the supplemental material). The E2348/69 gene C_2704 encodes an adhesin-like autotransporter that is 70% similar to Salmonella enterica ShdA (16). Upstream of C_2704 is C_2705, which encodes a glycosyltransferase of the type associated with glycosylation of a subset of autotransporter proteins (28). Although thought to be confined to the intestinal lumen, E2348/69 encodes five Lom (also known as Ail, OmpX, or PagC) family proteins, which are outer membrane proteins implicated in cell adhesion, resistance to complement-dependent killing, and survival in macrophages (1), and a homologue of SfpA, a porin implicated in survival of Yersinia enterocolitica during systemic infection (27).
(ii) Fimbrial adhesins.
E2348/69 has eight intact and five incomplete fimbrial operons. The complete fimbrial operon repertoire of E2348/69 in the context of the sequenced E. coli strains is shown in Table S8 in the supplemental material. Among the intact operon products, BFP play a role in microcolony formation in vitro (10) and diarrhea in a human volunteers (2), while long polar fimbriae play no obvious role in cell adhesion in vitro (7). The function of the other fimbrial operons has not been elucidated yet.
(iii) T2SS, iron uptake systems, and virulence gene regulators.
The sequenced E. coli strains each encode one or two type II secretion systems (T2SS) (see Fig. S7 in the supplemental material). E2348/69 contains a single T2SS, encoded in the gspM-yghJ locus; although its substrate remains unknown, the heat-labile enterotoxin is secreted by this T2SS in ETEC strain H10407 (40). E2348/69 contains six iron uptake systems (see Table S9 in the supplemental material). These systems include three systems that are not present in K-12 but are largely conserved in UPEC, APEC, and EHEC strain Sakai.
Regulation of virulence genes and coordinate regulation of virulence and housekeeping genes are required for virulence. Major regulators for the E2348/69 virulence genes are encoded on the LEE (Ler, GrlA, and GrlR) and on pMAR2 (Per). In addition, the RcsBCD two-component system (TCS) is known to regulate production of the exopolysaccharide colanic acid, and two TCSs (CxpAR and EvgAS) regulate LEE and BFP gene expression (22, 29, 30). All other TCSs that are widely distributed in E. coli strains are conserved in E2348/69; this includes several TCSs (e.g., QseEF [36]) that have been shown to be involved in virulence in other pathogenic E. coli strains (see Table S10 in the supplemental material).
(iv) T3SSs and their effectors.
E2348/69 encodes 21 T3SS effectors and contains 6 effector pseudogenes, while EHEC strain Sakai encodes 50 effectors and contains 12 effector pseudogenes (41) (Table 2). Recently, we have shown that EPEC lineage 2 strain B171 (serotype O111:NM) encodes at least 28 effectors and contains 12 pseudogenes (31). Twelve of the E2348/69 effectors are encoded by PPs (PP2, PP4, and PP6), seven are encoded by IEs (IE2, IE5, and IE6), and seven are encoded by the LEE (Fig. 3; see Table S2 in the supplemental material). As in EHEC strain Sakai, the three effector-transducing phages are lambda-like, and the effectors are encoded in the regions called exchangeable effector loci downstream of tail fiber genes (Fig. 3).
TABLE 2.
Comparison of T3SSs and effectors of 10 E. coli strains
| T3SS machinery or effector | Presence or no. in strain(s):
|
||||||
|---|---|---|---|---|---|---|---|
| E2348/69 | UTI89, CFT073, 536, APEC O1 | Sakai | MG1655 | HS | E24377A | B171 | |
| T3SS machinery | |||||||
| LEE | + | − | + | − | − | − | + |
| ETT2a | − | − | pa | p | p | p | p |
| Effectors in PPs and IEs | |||||||
| EspB | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| EspF | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| EspG | 2 | 0 | 1 | 0 | 0 | 0 | 1 |
| EspH | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| EspJ | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| EspK | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| EspL | 2 (1)b | 0 | 1 | 0 | 0 | 0 | 1 |
| EspM | 0 | 0 | 2 | 0 | 0 | 0 | 3 (1)c |
| EspN | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| EspO | 1 (1) | 0 | 2 | 0 | 0 | 0 | 0 |
| EspV | 0 | 0 | 1 (1) | 0 | 0 | 0 | 1 (1) |
| EspW | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| EspX | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| EspZ | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| Map | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| NleA/EspI | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| NleB | 3 (1) | 0 | 3 (1) | 0 | 0 | 0 | 3 (2) |
| NleC | 1 | 0 | 1 | 0 | 0 | 0 | 2 (2) |
| NleD | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| NleE | 2 | 0 | 1 | 0 | 0 | 0 | 1 |
| NleF | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| NleG | 1 | 0 | 14 (5) | 0 | 0 | 0 | 6 |
| NleH | 3 (1) | 0 | 2 | 0 | 0 | 0 | 2 (1) |
| TccP | 0 | 0 | 2 (1) | 0 | 0 | 0 | 1 |
| Tir | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| Cif | 1 (1) | 0 | 0 | 0 | 0 | 0 | 1 |
| OspB | 0 | 0 | 0 | 0 | 0 | 0 | 2 (1) |
| Effectors at other loci | |||||||
| EspL | 1 (1) | 1 (1) | 3 (1) | 2 (1) | 2 (2) | 2 (2) | 2 (2) |
| EspR | 0 | 0 | 4 (1) | 2 (2) | 1 | 1 | 1 (1) |
| EspX | 0 | 0 | 6 (1) | 2 (1) | 3 (3) | 3 (1) | 3 (1) |
| EspY | 0 | 0 | 5 (1) | 1 (1) | 0 | 0 | 0 |
| Total | 27 (6) | 1 (1) | 62 (12) | 7 (5) | 6 (5) | 6 (3) | ≥40 (12) |
p, partial.
The numbers in parentheses are the numbers of pseudogenes.
Includes one plasmid-encoded homologue.
While the LEE-encoded effectors are conserved among all the A/E pathogens studied thus far, there is considerable variation in the non-LEE effector protein repertoire between strains. Although EHEC strain Sakai and EPEC strain B171 each encode several strain-specific effectors (Table 2), all of the effector families that can be detected by homology-based approaches in E2348/69 are also found in EHEC strain Sakai. The E2348/69 cif pseudogene is the only exception. Of the intact non-LEE-encoded effectors, only seven families are common to the all three strains: NleA/EspI, NleB, NleE, NleF, NleG, NleH, and EspL. These effectors, together with LEE-encoded effectors, may represent the core set of essential effectors of A/E pathogens. It is noteworthy that E2348/69 encodes only one NleG, while there are 14 homologues in EHEC strain Sakai and 6 homologues in EPEC strain B171. In contrast, E2348/69 contains two intact EspG (encoded on the LEE and IE5) and NleE (encoded on IE2 and IE6) homologues, compared to one copy in both EHEC strain Sakai and EPEC strain B171. Two effector families which are present in both EPEC strain B171 and EHEC strain Sakai but are not present in E2348/69 are the EspM and TccP/TccP2 families (41). In contrast, EspJ, which is involved in inhibition of receptor-mediated phagocytosis (23), is present in E2348/69 and EHEC strain Sakai but is not present in EPEC strain B171. Interestingly, there are no homologues of any of Sakai's non-phage-encoded effectors in Sakai in E2348/69, except a degraded gene for an EspL family protein (Table 2). These effector homologues are also not present in the other phylogroup B2 strains, but they are conserved in B171, as well as MG1655, HS, and E24377A. Consistent with this, the ETT2 gene cluster encoding the second T3SS of E. coli is specifically not present in the phylogroup B2 strains, including E2348/69. These findings suggest that the effector homologues found in the chromosome backbones are largely effectors for the ETT2-encoded T3SS and that, as proposed by Ren et al. (38), this system was acquired by other E. coli lineages after the divergence of the B2 phylogroup, which has been postulated to be the earliest-splitting branch in the E. coli phylogenetic tree (20).
Concluding remarks.
In this study we identified 424 E2348/69-specific genes, most of which are carried on PPs, IEs, or plasmids. We also identified a number of genetic traits that are specific for the phylogroup B2 strains irrespective of the pathotype, including the absence of the ETT2-related T3SS, which is present in E. coli strains belonging to all other phylogroups.
Interestingly, we found that the T3SS of E2348/69 is much simpler than the T3SSs of EHEC strain Sakai and EPEC strain B171. The LEE of E2348/69 is the smallest LEE and consists only of the core 41 CDSs. Moreover, compared with the genomes of EPEC strains B171 (not complete [35]; accession no. AAJX00000000) (31), E22 (not complete [35]; accession no. AAJV00000000), and E110019 (not complete [35]; accession no. AAJW00000000), which encode at least 28 (plus 12 pseudogenes), 40 (plus 6 pseudogenes), and 24 (plus 13 pseudogenes) known effectors, respectively, EPEC E2348/69 has a smaller effector repertoire (only 21 intact effectors) but nonetheless an effector repertoire which is sufficient for colonization and human disease. Importantly, we cannot exclude the possibility that any of the EPEC strains encode novel, yet-to-be-characterized T3SS effectors. Nonetheless, the simplicity of the virulence gene set of E2348/69 provides the first opportunity to fully dissect the entire virulence strategy of A/E pathogens in the genomic context.
Supplementary Material
Acknowledgments
We thank Chris Tang, Centre for Molecular Microbiology and Infection, Imperial College, and Mark Pallen, University of Birmingham, for their assistance in launching the sequencing project and Simon Rutter, Rob Squares, Claire Arrowsmith, and Angela Lord of the Sanger Institute.
This work was supported by The Wellcome Trust and by grants-in-aid for scientific research [Kiban (B) and Priority Areas “Applied Genomics”] from the Ministry of Education, Science, and Technology of Japan.
Footnotes
Published ahead of print on 24 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bartra, S. S., K. L. Styer, D. M. O'Bryant, M. L. Nilles, B. J. Hinnebusch, A. Aballay, and G. V. Plano. 2008. Resistance of Yersinia pestis to complement-dependent killing is mediated by the Ail outer membrane protein. Infect. Immun. 76612-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 2802114-2118. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Bray, J. 1945. Isolation of antigenically homogeneous strains of Bact. coli neopolitanum from summer diarrhoea of infants. J. Pathol. Bacteriol. 57239-247. [Google Scholar]
- 5.Brinkley, C., V. Burland, R. Keller, D. J. Rose, A. T. Boutin, S. A. Klink, F. R. Blattner, and J. B. Kaper. 2006. Nucleotide sequence analysis of the enteropathogenic Escherichia coli adherence factor plasmid pMAR7. Infect. Immun. 745408-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H. D., and G. Frankel. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 2983-98. [DOI] [PubMed] [Google Scholar]
- 7.Fitzhenry, R., S. Dahan, A. G. Torres, Y. Chong, R. Heuschkel, S. H. Murch, M. Thomson, J. B. Kaper, G. Frankel, and A. D. Phillips. 2006. Long polar fimbriae and tissue tropism in Escherichia coli O157:H7. Microbes Infect. 81741-1749. [DOI] [PubMed] [Google Scholar]
- 8.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30911-921. [DOI] [PubMed] [Google Scholar]
- 9.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 732573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girón, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254710-713. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths-Jones, S., A. Bateman, M. Marshall, A. Khanna, and S. R. Eddy. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 811-22. [DOI] [PubMed] [Google Scholar]
- 13.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 1726175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 877839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 16.Kingsley, R. A., R. L. Santos, A. M. Keestra, L. G. Adams, and A. J. Baumler. 2002. Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol. Microbiol. 43895-905. [DOI] [PubMed] [Google Scholar]
- 17.Klapproth, J. M., I. C. Scaletsky, B. P. McNamara, L. C. Lai, C. Malstrom, S. P. James, and M. S. Donnenberg. 2000. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 682148-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacher, D. W., H. Steinsland, T. E. Blank, M. S. Donnenberg, and T. S. Whittam. 2007. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J. Bacteriol. 189342-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence, J. G., and J. R. Roth. 1996. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 14211-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lecointre, G., L. Rachdi, P. Darlu, and E. Denamur. 1998. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 151685-1695. [DOI] [PubMed] [Google Scholar]
- 21.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, and S. Sotman. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i1119-1122. [DOI] [PubMed] [Google Scholar]
- 22.Macritchie, D. M., J. D. Ward, A. Z. Nevesinjac, and T. L. Raivio. 2008. Activation of the Cpx envelope stress response down-regulates expression of several locus of enterocyte effacement-encoded genes in enteropathogenic Escherichia coli. Infect. Immun. 761465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchès, O., V. Covarelli, S. Dahan, C. Cougoule, P. Bhatta, G. Frankel, and E. Caron. 2008. EspJ of enteropathogenic and enterohaemorrhagic Escherichia coli inhibits opsono-phagocytosis. Cell. Microbiol. 101104-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 921664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23399-407. [DOI] [PubMed] [Google Scholar]
- 26.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mildiner-Earley, S., and V. L. Miller. 2006. Characterization of a novel porin involved in systemic Yersinia enterocolitica infection. Infect. Immun. 744361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moormann, C., I. Benz, and M. A. Schmidt. 2002. Functional substitution of the TibC protein of enterotoxigenic Escherichia coli strains for the autotransporter adhesin heptosyltransferase of the AIDA system. Infect. Immun. 702264-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadler, C., Y. Shifrin, S. Nov, S. Kobi, and I. Rosenshine. 2006. Characterization of enteropathogenic Escherichia coli mutants that fail to disrupt host cell spreading and attachment to substratum. Infect. Immun. 74839-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevesinjac, A. Z., and T. L. Raivio. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J. Bacteriol. 187672-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogura, Y., H. Abe, K. Katsura, K. Kurokawa, M. Asadulghani, A. Iguchi, T. Ooka, K. Nakayama, A. Yamashita, M. Hattori, T. Tobe, and T. Hayashi. 2008. Systematic identification and sequence analysis of the genomic islands of the enteropathogenic Escherichia coli strain B171-8 by the combined use of whole-genome PCR scanning and fosmid mapping. J. Bacteriol. 1906948-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pallen, M. J., and B. W. Wren. 2007. Bacterial pathogenomics. Nature 449835-842. [DOI] [PubMed] [Google Scholar]
- 33.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403665-668. [DOI] [PubMed] [Google Scholar]
- 34.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 696999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasko, D. A., M. J. Rosovitz, G. S. Myers, E. F. Mongodin, W. F. Fricke, P. Gajer, J. Crabtree, M. Sebaihia, N. R. Thomson, R. Chaudhuri, I. R. Henderson, V. Sperandio, and J. Ravel. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 1906881-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reading, N. C., A. G. Torres, M. M. Kendall, D. T. Hughes, K. Yamamoto, and V. Sperandio. 2007. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J. Bacteriol. 1892468-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid, S. D., J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 40664-67. [DOI] [PubMed] [Google Scholar]
- 38.Ren, C. P., R. R. Chaudhuri, A. Fivian, C. M. Bailey, M. Antonio, W. M. Barnes, and M. J. Pallen. 2004. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 1863547-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roesch, P. L., P. Redford, S. Batchelet, R. L. Moritz, S. Pellett, B. J. Haugen, F. R. Blattner, and R. A. Welch. 2003. Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. Mol. Microbiol. 4955-67. [DOI] [PubMed] [Google Scholar]
- 40.Tauschek, M., R. J. Gorrell, R. A. Strugnell, and R. M. Robins-Browne. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. USA 997066-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tobe, T., S. A. Beatson, H. Taniguchi, H. Abe, C. M. Bailey, A. Fivian, R. Younis, S. Matthews, O. Marches, G. Frankel, T. Hayashi, and M. J. Pallen. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA 10314941-14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 601136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yada, T., and M. Hirosawa. 1996. Detection of short protein coding regions within the cyanobacterium genome: application of the hidden Markov model. DNA Res. 3355-361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.