Abstract
Recent advances in the field of prokaryotic N-glycosylation have established a foundation for the pathways and proteins involved in this important posttranslational protein modification process. To continue the study of the Methanococcus voltae N-glycosylation pathway, characteristics of known eukaryotic, bacterial, and archaeal proteins involved in the N-glycosylation process were examined and used to select candidate M. voltae genes for investigation as potential glycosyl transferase and flippase components. The targeted genes were knocked out via linear gene replacement, and the resulting effects on N-glycan assembly were identified through flagellin and surface (S) layer protein glycosylation defects. This study reports the finding that deletion of two putative M. voltae glycosyl transferase genes, designated aglC (for archaeal glycosylation) and aglK, interfered with proper N-glycosylation. This resulted in flagellin and S-layer proteins with significantly reduced apparent molecular masses, loss of flagellar assembly, and absence of glycan attachment. Given previous knowledge of both the N-glycosylation pathway in M. voltae and the general characteristics of N-glycosylation components, it appears that AglC and AglK are involved in the biosynthesis or transfer of diacetylated glucuronic acid within the glycan structure. In addition, a knockout of the putative flippase candidate gene (Mv891) had no effect on N-glycosylation but did result in the production of giant cells with diameters three to four times that of wild-type cells.
It has become widely accepted that glycosylation is an important posttranslational protein modification within all three domains of life. Long recognized and studied in eukaryotes, glycosylation pathways in prokaryotes have received more attention in recent years. Several reviews summarizing the current state of knowledge of protein glycosylation in Archaea (2, 11, 42) and flagellar glycosylation in Bacteria and Archaea (25) attest to the progress that has been made in understanding this important process from a prokaryotic perspective. Specifically, significant advances in comprehending the process of N-linked glycosylation, which is the attachment of polysaccharide structures to specific Asn residues within a conserved Asn-Xaa-Ser/Thr motif (where Xaa is any amino acid except Pro) have occurred. Of note is the N-glycosylation system in Campylobacter jejuni, where the gene products from the pgl locus assemble a branched heptasaccharide on a membrane-bound lipid carrier and then translocate the glycan across the cytoplasmic membrane to facilitate transfer to the appropriate Asn residue of the target protein (24, 34).
The study of archaeal glycosylation in recent years has begun to yield an understanding of how these organisms modify proteins. The first requirement for assembling a glycan of any linkage type is a pool of nucleotide-activated monosaccharide precursors. Several enzymes have been identified from Methanococcus maripaludis that are required for UDP-acetamido sugar synthesis and are predicted to be precursors for flagellin and surface (S) layer protein modifications (29).
With these nucleotide-activated sugars, a set of genes known as the archaeal glycosylation (agl) genes then assemble and attach the desired glycan to its target protein in a stepwise fashion. First, a set of glycosyl transferases construct the glycan on a dolichol phosphate anchor at the cytoplasmic face of the cytoplasmic membrane. In the obligate anaerobic methanogen Methanococcus voltae, at least three glycosyl transferases are predicted to be necessary to assemble the trisaccharide β-ManpNAcA6Thr-(1-4)-β-GlcpNAc3NAcA-(1-3)-β- GlcpNAc, which has been characterized from both the S-layer protein and the flagellins (two major flagellins, FlaB1 and FlaB2, and two minor flagellins, FlaA and FlaB3) (39). In this system, AglH has been proposed as the glycosyl transferase responsible for the attachment of the linking N-acetylglucosamine (33), while AglA is responsible for the terminal sugar attachment (9). In the moderate halophile Haloferax volcanii, a pentasaccharide is found on the S-layer protein that requires the activities of at least five glycosyl transferases, with AglD, AglE, AglF, AglG, and AglI identified as involved so far (1, 3, 4, 41, 42).
The next step in the process is to translocate the glycan from the cytoplasmic to the extracellular side of the membrane. No homologs of either the proposed eukaryotic flippase (Rft1; a unique transporter) (14) or the bacterial flippase (PglK; an ABC transporter) (21) have been detected in either M. voltae or H. volcanii (1, 9). Furthermore, a knockout of the most likely flippase candidate in M. voltae, based on very weak BLAST scores with Rft1 and PglK, had no detectable effect on the flagellin or S-layer glycan (9). This appears to indicate that the protein responsible for glycan translocation is unique in each domain of life and that the archaeal flippase may prove to be an archaeal-specific protein.
The final step in the N-glycosylation pathway is the attachment of the completed glycan to its target protein via the action of an oligosaccharyl transferase. In both M. voltae and H. volcanii, this protein has been identified as AglB (1, 9). Both of the identified proteins are homologs of the eukaryotic Stt3p and the bacterial PglB proteins. A key feature of these proteins is the highly conserved active site, WWDXG, responsible for its transferase activity (40). Interestingly, the soluble domain of AglB from the thermophilic archaeon Pyrococcus furiosus has been crystallized (17) and appears to extend the conserved active site to include a DXXK motif located approximately 60 residues upstream of the WWDXG motif. Significantly, subsequent mutagenesis of the DXXK motif revealed its essential role in the catalytic activity of the yeast STT3 (16). The apparent universality of Stt3p/PglB/AglB to catalyze the N-glycosyl bond within all three domains of life, despite the presence of different linking sugars in different organisms, illustrates the conserved nature of this step in the N-glycosylation process.
The currently proposed glycan assembly model for M. voltae is summarized in Fig. 1. This contribution reports the testing of three genes from M. voltae, two putative glycosyl transferases (GT2 family members) and one flippase candidate, for their possible roles in the N-glycosylation pathway within the organism. Knockouts of the genes encoding the two putative GT2 proteins affected glycan and flagellum assembly, with the flagellins produced migrating as lower-molecular-mass proteins in immunoblots. Additionally, it was shown by mass spectrometry (MS) that the S-layer protein produced in these knockout strains was no longer glycosylated. A knockout of the flippase candidate had no effect on glycosylation of the flagellins but did have a profound effect on cell size.
FIG. 1.
Current model of N-glycosylation of flagellin and S-layer proteins in M. voltae. Steps 1 to 3 diagram the assembly of the trisaccharide via glycosyl transferases onto a lipid carrier at the cytoplasmic face of the cytoplasmic membrane. Step 4 represents the translocation of the glycan to the exterior of the membrane via a flippase enzyme. Finally, step 5 shows the attachment of the complete glycan to the target protein via an STT3 oligosaccharyl transferase. AglH (Mv1751), AglA (Mv151), and AglB (Mv1749) have been previously reported to carry out steps 1, 3, and 5, respectively (8, 28). This study reports the finding that AglC (Mv990) and AglK (Mv991) carry out step 2 in glycan assembly.
MATERIALS AND METHODS
Microbial strains and growth conditions.
The M. voltae PS stock strain, maintained in the Jarrell laboratory since it was obtained from Len Hook in 1983 and used in all our previous publications on M. voltae flagellation (6, 18, 19), was lost due to a freezer malfunction. We obtained a second version of M. voltae PS (hereafter designated strain PS*) from W. B. Whitman, and this strain was used in the generation of knockouts in genes Mv891, Mv990, and Mv991 and for protein analysis in this study. The two strains were grown under identical conditions in Balch medium 3 (5) at 37°C under an atmosphere of CO2/H2 (20:80) and supplemented with 7.5 μg/ml puromycin (MP Biomedicals Inc., Solon, OH) when necessary. Escherichia coli DH5α, used for all cloning steps, was grown in Luria-Bertani medium at 37°C and supplemented with 100 μg/ml ampicillin when necessary.
Construction of knockout plasmids.
Knockout strains were generated using M. voltae PS*. The vector pPAC60 (36) was used as the backbone plasmid for knockout creation via linear gene replacement utilizing a puromycin resistance (purr) cassette. Briefly, 1 kb of sequence upstream and downstream of each gene was amplified via PCR (Table 1), with the addition of restriction enzyme sites, and ligated into pPAC60. One fragment was ligated into the multiple cloning site at HindIII and SpeI or SpeI and NruI sites before the purr cassette, while the other fragment was ligated into the multiple cloning site at NheI and BglII, after the purr cassette. This created a construct with 2 kb of M. voltae sequence, with the targeted gene sequence replaced by the purr cassette. The fragment orientation and sequence were confirmed after transformation into E. coli and screening (Table 1). Each construct was cut by restriction digestion with the outermost restriction enzymes to separate the 2-kb-plus purr cassette fragment from the rest of the plasmid before transformation.
TABLE 1.
PCR primers used in this study
| Primer | Sequencea | |
|---|---|---|
| For linear gene replacement | ||
| Mv990_Up_Bgl | GAAGATCTTTTACATATTTGCAATTACGC | |
| Mv990_Up_Nhe | CTAGCTAGCGATTAAACCAATTTTCGAGAC | |
| Mv990_Down_Spe | GACTAGTTCGAGAACCTTTATACATAGC | |
| Mv990_Down_Hind | CCCAAGCTTGAAACCTAATTCCGATTACG | |
| Mv991_Up_Hind | CCCAAGCTTTTTAAGTTCTGGAGAGAAGC | |
| Mv991_Up_Spe | GACTAGTCCATAATCTAACCAATCTAACC | |
| Mv991_Down_Nhe | CTAGCTAGCGGGCAAAGTTTTGGATTAAA | |
| Mv991_Down_Bgl | GAAGATCTTCCTTTAAAGAATCCATAGG | |
| Mv891_Up_Spe | ATGACTGAAACTAGTGTTAACC | |
| Mv891_Up_Nru | GTTCGCGATAATTTTGAGTAGAGATTTGAC | |
| Mv891_Down_Nhe | CTAGCTAGCTCAATTATCACATTTATTATAGC | |
| Mv891_Down_Bgl | GAAGATCTTGTAAAGCCGAATCCCTTAG | |
| Sequencing primers on pPAC60 | ||
| pSL1180_Up | ATTAAGTTGGGTAACGCCAG | |
| pPAC60_P-sl | ATTTAAAATTACTACCATAATAC | |
| pPAC60_PhmvA | GGTACCGAGCTCGAGTCCAA | |
| pSL1180_Down | TGGGACGTCGACCTGAGGTA | |
Restriction sites used for cloning are underlined.
Generation of M. voltae mutants.
A liposome delivery method first described for Methanosarcina acetivorans (28) was used with minor modifications (37) for all transformations of M. voltae. After transformants were selected on Balch medium 3-plus-puromycin plates, single colonies were picked anaerobically and transferred into Balch medium 3 containing puromycin for subsequent analyses.
Southern hybridization analyses.
Chromosomal DNA was isolated from wild-type and transformant cultures of M. voltae as previously described (13), digested with restriction enzymes, electrophoresed, and transferred to nylon membranes (Roche Molecular Biochemicals) by the capillary transfer method. Digoxigenin (DIG)-labeled probes were generated by PCR amplification of downstream regions (identical to the 1-kb downstream fragment used for cloning) using DIG-dUTP (Roche Molecular Biochemicals) as recommended by the manufacturer. Southern hybridizations were carried out at 55°C, and stringency washes and developing were done as described previously (37).
SDS-PAGE and immunoblotting.
Whole-cell preparations of M. voltae were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (23). The gels were either stained with Coomassie brilliant blue G250 (12) or transferred to Immobilon-P transfer membranes (Millipore, Bedford, MA) as described previously (38). Immunoblots were developed using chicken polyclonal anti-FlaB2 antibodies (shown to react with the major flagellins FlaB1 and FlaB2) (7) as the primary antibody and a horseradish peroxidase-conjugated rabbit anti-chicken immunoglobulin Y (Jackson Immunoresearch Laboratories, West Grove, PA) as the secondary antibody. The blots were developed with a chemiluminescence kit (Roche Molecular Biochemicals) according to the manufacturer's instructions.
MS analysis of flagellins.
Flagellar samples were prepared from both M. voltae PS and M. voltae PS* strains as previously described (20). Each protein sample (50 to 200 μg) was digested overnight with trypsin (Promega, Madison, WI) at a ratio of 30:1 (protein-enzyme [vol/vol]) in 50 mM ammonium bicarbonate at 37°C. The protein digests were analyzed by MS as previously described (39).
MS analysis of S-layer protein.
S-layer proteins from M. voltae strain PS* and the corresponding aglC and aglK mutant strains were prepared according to the method of Koval and Jarrell (22). Each protein sample (approximately 200 μg) was digested overnight with trypsin (Promega, Madison, WI) at a ratio of 30:1 (protein-enzyme [vol/vol]) in 50 mM ammonium bicarbonate at 37°C. The protein digests were analyzed by nano-liquid chromatography-tandem MS (nano-LC-MS/MS) using a QTOF Ultima hybrid quadrupole time-of-flight mass spectrometer coupled to a Nanoacquity high-performance LC system (Waters, Milford, MA). MS/MS spectra were acquired automatically on doubly, triply, and quadruply charged ions. The tryptic protein digest was also analyzed by nano-LC-MS with alternating low collision energy (CE) (10 V)/high CE (10 V) in the collision cell of the mass spectrometer in order to simultaneously generate intact peptide ions (low CE) as well as their fragment ions (high CE) for each peak. The high-CE spectra were searched for oxonium ions corresponding to β-ManpNAcA6Thr (318 Da; m/z 319) and GlcpNAc3NAcA (258 Da; m/z 259), and the equivalent LC-MS/MS spectra were examined to confirm the identities of the glycopeptides.
Preflagellin peptidase assay.
A preflagellin peptidase assay, previously described (7, 10), was used to generate two versions of nonglycosylated M. voltae FlaB2 flagellin. E. coli membranes containing overexpressed preflagellin FlaB2 were used as substrates, with M. voltae membranes as the source of the preflagellin peptidase FlaK. The unprocessed and processed forms of FlaB2 (with or without the signal peptide) could be distinguished by their small difference in size by immunoblotting.
RT-PCR.
To determine whether Mv991 and Mv990 were cotranscribed, total RNA was isolated from wild-type, Mv991−, and Mv990− cells. The cells were prepared for RNA isolation by subculture and growth for 24 h at 37°C. The cultures were then overpressurized with H2-CO2 (80:20), followed by an additional incubation at 37°C for 1 hour. Total RNA was isolated using the RNeasy Mini Kit (Qiagen), and reverse transcription (RT)-PCR was carried out using the One-Step RT-PCR kit (Qiagen).
Electron microscopy.
Intact M. voltae cells were washed in 50 mM MgSO4 prior to absorption onto Formvar-coated gold grids. Negative staining was done with 2% phosphotungstic acid (pH 7.0). The grids were viewed on a Hitachi H-700 electron microscope operating at 75 kV.
Nucleotide sequence accession numbers.
Sequence data from Mv990 (AglC), Mv991 (AglK), and Mv891 can be found in the GenBank/DDBJ/EMBL databases under the accession numbers EU726231, EU726230, and EU751623, respectively.
RESULTS
Selection of M. voltae genes to target for study.
In an attempt to move beyond direct homology searches and to understand the underlying properties of enzymes involved in N-glycosylation, known N-glycosylation proteins from Saccharomyces cerevisiae, C. jejuni, and M. voltae were examined for two properties, namely, the number of transmembrane domains (TMDs) and types of conserved domains (CDs). TMDs, as predicted by TMpred (15), were deemed an important characteristic to consider because the N-glycosylation process is invariably localized to a membrane, implying that some type of anchoring feature is required. CDs, as recognized by NCBI's Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi; 27), were also considered, since functional similarity does not always translate into sequence (and therefore BLAST) similarity.
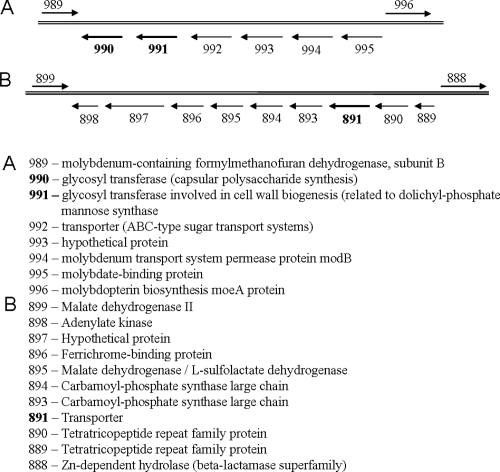
Using this information, several M. voltae gene candidates that were identified as glycosyl transferases or possible transporters were similarly characterized. Genes with the appropriate characteristics, Mv990, Mv991, and Mv891, were chosen for study and are listed in Table 2, and their genomic context is diagramed in Fig. 2. The genomic context was considered important because of its potential to offer clues to a gene's function and also because intergenic regions in M. voltae tend to be quite short, creating the potential for multigene transcripts.
TABLE 2.
Properties of the components of N-linked glycosylation systems
| GI no. | Protein | Functiona | TMDb | CDc |
|---|---|---|---|---|
| S. cerevisiae (Eukaryota) | ||||
| 1370470 | Alg5 | Gt | 2 | Glycos_transf_2 |
| 6325441 | Dpm1 | Gt | 1 | Glycos_transf_2 |
| 536653 | Alg7 | Gt | 11 | Glycos_transf_4 |
| 6321391/6319544 | Alg13/ Alg14 | Gt | 2/2 | Glyco_tran_28_C/ none |
| 536377 | Alg1 | Gt | 3 | Glycos_transf_1 |
| 1322572 | Alg2 | Gt | 4 | Glycos_transf_1 |
| 1301907 | Alg11 | Gt | 3 | Glycos_transf_1 |
| 536015 | Rft1 | Flippase | 12 | Rft protein |
| 586444 | Alg3 | Gt | 10 | ALG3 protein |
| 1302235 | Alg9 | Gt | 10 | Glyco_transf_22 |
| 47678269 | Alg12 | Gt | 13 | Glyco_transf_22 |
| 1420090 | Alg6 | Gt | 13 | Alg6_Alg8 |
| 1420215 | Alg8 | Gt | 14 | Alg6_Alg8 |
| 1706435 | Alg10 | Gt | 11 | DIE2/ALG10 |
| 6321416 | Stt3p | Ot | 14 | STT3 |
| C. jejuni (Bacteria) | ||||
| 5771412 | PglC | Gt | 1 | Bac_transf |
| 5771411 | PglA | Gt | 0 | Glycos_transf_1 |
| 57166812 | PglJ | Gt | 2 | Glycos_transf_1 |
| 57166814 | PglH | Gt | 2 | Glycos_transf_1 |
| 57166813 | PglI | Gt | 1 | Glycos_transf_2 |
| 3413446 | PglK | Flippase | 6 | ABC_ATPase |
| 5771410 | PglB | Ot | 11 | STT3 |
| M. voltae (Archaea) | ||||
| 87045855 | AlgH | Gt | 7 | Glycos_transf_4 |
| Unknown | Unknown | Gt | Unknown | Unknown |
| 87045840 | AglA | Gt | 1 | Glycos_transf_1 |
| Unknown | Unknown | Flippase | Unknown | Unknown |
| 87045854 | AglB | Ot | 13 | STT3 |
| M. voltae genes targeted | ||||
| 190336425 | Mv990 | Gt candidate | 2 | Glycos_transf_2 |
| 190336423 | Mv991 | Gt candidate | 1 | Glycos_transf_2 |
| 190403021 | Mv891 | Flippase candidate | 11 | Polysacc_synt |
Gt, glycosyl transferase; Ot, oligosaccharyl transferase.
TMDs were predicted by TMpred.
CDs are from the NCBI Conserved Domain Database.
FIG. 2.
Schematic of the M. voltae genomic regions of interest. Genes in boldface were targeted in this study.
Generation of M. voltae mutants.
The genes Mv990, Mv991, and Mv891 were individually targeted for knockout by removing the genomic copy of the gene and replacing it with a selectable antibiotic marker (puromycin cassette) in M. voltae PS*. The puromycin cassette used in this study was designed with an additional promoter after the antibiotic resistance terminator sequence to allow the restarting of a multigene transcript (36). For Mv891 and Mv991, the puromycin cassette was inserted so that the additional promoter was facing downstream genes. For Mv990, which was the last gene in a directional series of genes (Fig. 2), the puromycin cassette was inserted in the opposite orientation to prevent further transcription. Confirmation of gene replacement was achieved by Southern blot analysis (Fig. 3), in which the expected wild-type and knockout patterns were obtained.
FIG. 3.
Southern blot analysis confirming linear gene replacement. On the left are the Southern blot results of each deletion (the gene is indicated under each blot with the restriction enzyme used). M, λ-HindIII marker representing 23, 9, 6.5, 4.3, 2.3, 2, and 0.5 kb; wt, genomic DNA from wild-type M. voltae; Δ, genomic DNA from the indicated M. voltae deletion strain. On the right are schematics of the genome regions probed, with expected DNA fragment sizes. The targeted genes are dark gray; the purr cassette is represented by black arrows (promoters), a light-gray box (the puromycin gene), and a white block with X inside (the terminator); and the DIG-labeled probe is hatched. HindIII sites are marked by vertical arrows, while XhoI sites are marked by vertical round-topped markers where relevant.
Analysis of flagellin from M. voltae PS*.
Flagellins from the replacement M. voltae PS* strain migrated as slightly larger proteins in immunoblots with respect to the original M. voltae PS strain (data not shown). Therefore, we compared flagellar tryptic peptides from the PS and PS* strains by LC-MS/MS in order to determine if any changes had occurred in the glycan modification structure of the latter. Interestingly, the N-linked glycan observed on the flagellin from the PS* strain was composed of the same trisaccharide that was reported previously (39) but with an additional mass of either 220 Da or 260 Da appended to its nonreducing end. By way of illustration, the MS/MS spectra for the doubly glycosylated tryptic peptide T53-78 (ESTEQVASGLQISQVMGMHNNSNINK) from the FlaB2 proteins from M. voltae strains PS and PS* are presented in Fig. 4b and c, respectively (the two sites of N-linked glycosylation are underlined). The equivalent MS spectrum for each glycopeptide is shown in the insets. In the PS strain, a doubly charged ion was observed at m/z 1,094.5 (Fig. 4b, inset), corresponding to the T53-78 peptide modified with two trisaccharide glycan moieties. MS/MS analysis of this glycopeptide (Fig. 4b) yielded a glycan fragment pattern identical to those presented previously for the strain (39). In contrast, three glycopeptides were observed in the digest of M. voltae PS* flagellin (Fig. 4c, inset), corresponding to the same tryptic peptide with two N-linked glycans additionally modified by 220 and/or 262 Da (220 plus 220 Da, 220 plus 262 Da, and 262 plus 262 Da, respectively). The MS/MS spectrum for the doubly protonated glycopeptide ion at m/z 1,215 additionally modified with one 220- and one 262-Da moiety is presented in Fig. 4c. This MS/MS spectrum shows clear evidence that the original trisaccharide glycan is modified with one or the other moiety, but not both at the same time. Furthermore, the extra moiety is always linked to the β-ManpNAcA6Thr moiety. Structural studies are under way to identify the new modifications, though we suspect that they are carbohydrate in nature. Thus, it appears that although genetically related, the two strains produce unique glycan structures. All further analyses in the current work comparing S-layer and flagellin proteins from wild-type M. voltae to Mv891, Mv990, and Mv991 mutants refer to M. voltae PS* as the parent strain.
FIG. 4.
Nano-LC-MS analysis of the FlaB2 tryptic glycopeptide T53-78 from M. voltae strains PS and PS*. (a) Amino acid sequence of T53-78Y showing the two sites of N-linked glycosylation (underlined). (b) MS/MS spectrum of the quadruply protonated T53-78 glycopeptide ion at m/z 1,094.5 from M. voltae PS flagellin. The corresponding LC-MS spectrum for this glycopeptide is presented in the inset. The lower half of the MS/MS spectrum is dominated by oxonium ions for β-ManpNAcA6Thr (▴, m/z 319) and GlcpNAc3NAcA (•, m/z 259) and their dehydration products. Larger oxonium ions composed of disaccharides (m/z 462.2 and m/z 577.2), as well as the entire trisaccharide glycan (m/z 780.3), were also observed (▪, GlcNAc). The high-mass region of this spectrum is dominated by fragment ions arising from the sequential loss of the components from the trisaccharides attached to both N-linked sites. Rel. int., relative intensity. (c) MS/MS spectrum of the quadruply protonated T53-78 glycopeptide ion at m/z 1,215.0 from M. voltae PS* flagellin. The corresponding LC-MS spectrum (inset) reveals a more complex glycoform profile due to the extension of the two N-linked glycans with either 220-Da (solid pentagon) or 262-Da (solid diamond) moieties (m/z 1,204.5, two 220-Da residues; m/z 1,215.0, one 220- and one 262-Da residue; and m/z 1,225.5, two 262-Da residues). The MS/MS spectrum indicates that the trisaccharide observed in the original M. voltae PS strain is modified here with one additional residue (either 220 or 262 Da) linked to the β-ManpNAcA6Thr (solid triangle) residue. Finally, though less intense than the glycan-related fragment ions, a good b fragment ion series was observed in this MS/MS spectrum, confirming the identity of the peptide (some of the b ions detected are annotated in the spectrum).
Effects on flagellin glycosylation.
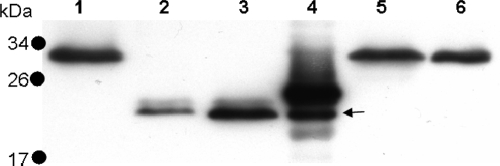
The first indicator proteins examined to determine if any knockouts had an effect on N-glycosylation were the flagellin proteins. M. voltae PS and PS* contain four flagellin proteins (two major flagellins, FlaB1 and FlaB2, and two minor flagellins, FlaA and FlaB3), which are all modified by the same N-linked trisaccharide (PS) or by the trisaccharide with additional 220-Da or 262-Da mass (PS*) (reference 39 and this work). Immunoblotting of whole-cell lysates was used to detect any shifts in the apparent molecular masses of the major flagellins (given the high sequence similarity between FlaB1 and FlaB2, polyclonal antibodies raised to FlaB2 cross-react with FlaB1) (7). Both Mv990− and Mv991− cells had significant reductions in the apparent molecular masses of their flagellins (Fig. 5), indicating an alteration in glycosylation status. To help clarify the extents of the alteration, two forms of M. voltae flagellin were used as controls. M. voltae flagellins, like all archaeal flagellins, are first synthesized as preproteins with a 12-amino-acid leader sequence (35). This signal peptide is cleaved by a dedicated preflagellin peptidase (10) to generate mature flagellins. To indicate the apparent molecular mass of mature, fully glycosylated flagellin (31 to 33 kDa), wild-type whole-cell lysates were used (Fig. 5, lanes 1 and 6). Also, to indicate the apparent molecular mass of unglycosylated flagellin, M. voltae FlaB2 flagellin was overexpressed in E. coli (which alone cannot process or glycosylate archaeal flagellins) (6). Subsequently, FlaB2-containing E. coli membranes were used in an established preflagellin peptidase assay (9) to generate unglycosylated FlaB2 flagellins both with (24 kDa) and without (22 kDa) their signal peptide sequences (Fig. 5, lane 4).
FIG. 5.
Immunoblot of M. voltae flagellins FlaB1/FlaB2 resolved on a 15% acrylamide gel. Lane 1, wild-type M. voltae (32 kDa); lane 2, Mv990 mutant (aglC); lane 3, Mv991 mutant (aglK); lane 4; M. voltae FlaB2 expressed in E. coli with and without (arrow) signal peptide (24 and 22 kDa, respectively); lane 5, Mv891 mutant; lane 6, wild-type M. voltae (32 kDa).
For both Mv990− and Mv991− cells, the FlaB1/FlaB2 flagellins migrate to an apparent molecular mass approximately consistent with completely unglycosylated but N-terminally processed flagellin protein (Fig. 5, compare lanes 2 and 3 with the lower band in lane 4). Conversely, Mv891− flagellin appears to be equivalent in molecular mass to wild-type flagellin, indicating no change in the glycosylation status of the knockout.
Effect on gross cellular appearance and flagellar structure.
To evaluate the effect of each knockout on the overall cell appearance and flagellar function, cells were examined by both phase-contrast microscopy and transmission electron microscopy (TEM). Wild-type M. voltae cells were visibly motile under phase-contrast microscopy and contained many flagella when examined under TEM (Fig. 6A). In contrast, both Mv990− and Mv991− cells appeared nonmotile when viewed under phase-contrast microscopy and, correspondingly, possessed no flagella under TEM examination (Fig. 6B and C). Mv891− cells appeared as motile as wild-type cells under phase-contrast microscopy. However, quite unexpectedly, approximately half the Mv891− cell culture was composed of cells that were three to four times the diameter of wild-type cells (which are usually 1 to 2 μm). This was confirmed by TEM examination, where both giant cells and normal-size cells were clearly observed in the Mv891− culture and cells of both sizes possessed flagella (Fig. 6D).
FIG. 6.
Electron micrographs of M. voltae wild type and mutants. (A) Wild-type M. voltae. (B) Mv990 mutant (aglC). (C) Mv991 mutant (aglK). (D) Mv891 mutant. Scale bar, 1 μm.
Cotranscription of Mv990 and Mv991.
Because of the gene orientation of Mv990 and Mv991 (Fig. 2A) and the short intergenic region between the genes (42 bp), it was thought likely that the genes were cotranscribed. RT-PCR was used to determine the transcription status of Mv990 and Mv991 in wild-type and knockout cells. Figure 7 clearly shows that in wild-type cells, a single RNA transcript exists for both Mv991 and Mv990. Also, in Mv991− cells, the disruption of Mv991 results in the expected loss of RNA transcript for Mv991, and RT-PCR did not detect the region between Mv991 and Mv990 (since the binding site for the forward primer would have been deleted). The additional promoter added at the end of the Mv991 deletion (on the puromycin cassette) restarted transcription, and the transcript for Mv990 was detectable (Fig. 7). Analysis of the RNA content of Mv990− cells revealed the expected loss of RNA for Mv990 and also did not detect the intergenic region (in this case, the binding site for the reverse primer would have been deleted). Transcription of Mv991 was not abolished in the Mv990− mutant. Collectively, these results indicate that the deletion of Mv991 should not have disrupted the translation of Mv990 and vice versa, suggesting that the phenotype obtained from each mutant was the result of that gene deletion alone. However, although these results indicate that gene transcripts could be detected for both Mv990 and Mv991, no attempt was made to determine the transcript levels in either the wild type or the mutant strains.
FIG. 7.
Cotranscriptional analysis of Mv990 and Mv991. The schematic at the top represents the genome region examined and PCR primer set names. *, The 991 primer set used Mv991 RNA, the Connect primer set used wild-type RNA, and the 990 primer set used Mv990 RNA. The RNA was added to the master mix and set on ice for 30 min, followed by 95°C denaturing of the reverse transcriptase.
Effects on S-layer protein glycosylation.
Without flagellar filaments on Mv990− and Mv991− cells available to isolate, further characterization of the exact alteration to the N-glycan in these cells required another indicator protein. Since it had been previously established that the same trisaccharide was present on both the flagellins and the S-layer proteins in M. voltae PS (35) and that apparent molecular-mass shifts of flagellin proteins result in corresponding shifts in S-layer proteins (9), whole-cell lysates separated by SDS-PAGE were examined (data not shown). As expected, Mv990− and Mv991− cells had a significant reduction in the S-layer protein apparent molecular mass compared to the wild type, while Mv891− cells appeared indistinguishable from wild-type cells. There was no other detectable difference in whole- protein profiles between samples.
Nano-LC-MS/MS characterization of S-layer protein.
We purified the S-layer protein from M. voltae PS* and examined its tryptic peptides by nano-LC-MS/MS for evidence of glycosylation. We confirmed that both S-layer tryptic peptides, which contain N-linked sequons (T64-91 and T92-127), are indeed glycosylated identically to flagellin from this strain (Fig. 8a and d). As with the flagellar glycan, the two S-layer glycopeptides from M. voltae PS* carried the extended glycan, composed of the original trisaccharide with an additional mass of either 220 or 262 Da.
FIG. 8.
Nano-LC-MS analysis of S-layer tryptic peptides T75-91 and T92-127 from M. voltae strain PS* and corresponding aglC and aglK mutant strains. The amino acid sequence for each peptide is presented above panels a and d, and the N-linked site of glycosylation is underlined. The relevant glycopeptide and peptide ions are underlined in all six spectra. The MS spectra show the multiply charged ions (2+ and 3+) for the T75-91 tryptic glycopeptide from the PS* strain (a), the doubly protonated ion at m/z 898.4 corresponding to the unmodified T75-91 peptide from the aglC mutant (b), the same peptide ion in the aglK mutant (c), the multiply protonated glycopeptide ions (3+ and 4+) for the T92-127 tryptic glycopeptide from the PS* strain (d), the triply protonated ion at m/z 1,255.4 corresponding to the T92-127 peptide from the aglC mutant (e), and the same peptide ion in the aglK mutant (f). Rel. int., relative intensity.
In contrast to the parent strain, the corresponding tryptic peptides from a digest of the S-layer proteins from aglC (Fig. 8b and e) and aglK (Fig. 8c and f) mutant strains were no longer glycosylated. Nano-LC-MS/MS analysis confirmed the identity of the doubly protonated ion at m/z 898.4 and the triply protonated ion at m/z 1,255.2 to be unglycosylated T64-91 and T92-127, respectively (data not shown). These ions were not detected in the digest of the parent strain.
DISCUSSION
This report details the continued examination of the M. voltae genome for N-glycosylation pathway genes and the identification of two genes encoding putative glycosyl transferases that affect flagellar glycosylation. In the current study, which revealed that an additional sugar residue is a component of the glycan moiety, it appeared that a fourth glycosyl transferase is required for assembly of the tetrasaccharide found on both S-layer and flagellin proteins. The assembly and attachment portion of the N-glycosylation pathway would therefore contain six steps; four glycosyl transferases to assemble the novel glycan, a flippase to transport the completed glycan across the cytoplasmic membrane, and an oligosaccharyl transferase to attach the glycan to flagellin and S-layer proteins. Previous studies (9, 33) confirmed the identity of the first and third glycosyl transferases (AglH and AglA, respectively) and the oligosaccharyl transferase (AglB). Our current study identified two proteins involved in the biosynthesis/attachment of the diacetylated glucuronic acid second sugar of the glycan, leaving the fourth glycosyl transferase (responsible for attaching the uncharacterized 220- and 262-Da species), as well as the flippase enzyme, unaccounted for.
The two genes targeted in this study as candidates for the second glycosyl transferase were Mv990 and Mv991. Based on the analysis of N-glycosylation genes from all three domains of life (Table 2), it was apparent that glycosyl transferases that have their activities on the cytoplasmic faces of their respective membranes (the glycosyl transferases listed above the flippase enzyme in each system in Table 2) contain few TMDs (generally one to four) and possess a recognizable glycosyl transferase type 1 or 2 domain. The exceptions to this trend appear to be limited to the glycosyl transferase responsible for the attachment of the first nucleotide-activated sugar to the lipid carrier (Alg7, PglC, and AglH) and the unique eukaryotic Alg13/Alg14 complex (in which both proteins must associate to form an active transferase) (8). A search of the M. voltae genome identified Mv990 and Mv991 as putative glycosyl transferase genes that also possessed these traits (Table 2). These genes are located adjacent to each other and at the end of a set of similarly oriented genes on the chromosome (Fig. 2A). Deletions of Mv990 and Mv991 were made, and both resulted in N-glycosylation defects to the flagellin and S-layer proteins (Fig. 5 and 8, respectively). Originally, this result was interpreted to mean that the attempt to restart transcription after the Mv991 deletion had failed and Mv991− actually represented an Mv991/Mv990 double deletion. However, cotranscriptional analysis of these genes in the Mv991− strain (Fig. 7) indicated that RNA for Mv990 was still present. Conversely, Mv990− cells showed no loss of Mv991 transcription (Fig. 7). These results imply that the phenotypes generated by each deletion are the sole result of that single gene loss. One possible explanation is that in the Mv991 mutant (where Mv990 transcription was restarted from the nonnative promoter introduced on the puromycin cassette), the levels of Mv990 transcription were altered significantly from the wild type. This could have affected Mv990 translation and subsequent function. In this case, the Mv991 mutant would indeed represent an Mv990/Mv991 double deletion. Unfortunately, there are no specific antibodies to either Mv990 or Mv991 to confirm either protein's translation status. Also, genetic-system limitations in M. voltae prevent complementation of the deleted genes in trans.
The Mv990/Mv991 results as determined indicate that both genes encode proteins that are involved in flagellar glycosylation. The N-glycosylation pathway in M. voltae PS* is expected to require at least four glycosyl transferases, one for each monosaccharide in the glycan. Genes for the first and third step of this process have already been verified (9, 33), leaving two missing glycosyl transferases to identify. The data from this study identified two putative glycosyl transferases, Mv990 and Mv991, which belong to the large and diverse GT2 family of enzymes. The loss of one is not compensated for by the presence of the other, indicating that they are not redundant in function. While we were unable to determine the precise role of each of the products of these genes in the current study, it is clear that inactivation of either results in failure to glycosylate both flagellin and the S-layer protein. Based on the original study of M. voltae PS, in which a trisaccharide was found to decorate both proteins and mutation of aglA resulted in proteins that were still glycosylated with the disaccharide, it appears unlikely that either of the target genes in the current work is involved in addition of the terminal species (presumed to be a sugar residue). Failure to add this residue to the glycan would not be expected to lead to a complete loss of glycosylation, as cells would still be able to synthesize both the di- and trisaccharide structures on the lipid-linked carrier, which could be transferred to target proteins.
It is likely, therefore, that both Mv990 and Mv991 are involved in synthesis and/or assembly of the second glycan residue, although they clearly have distinct roles, as indicated by the lack of redundancy. There is a proven case in S. cerevisiae in which two gene products are required to dimerize to form a single functional glycosyl transferase (8). The second step in the N-glycosylation pathway in eukaryotes was shown to require both Alg13 and Alg14 for the addition of a nucleotide-activated N-acetylglucosamine. It was found that Alg14 is a membrane-bound protein with no detectable glycosyl transferase activity, while Alg13 has glycosyl transferase activity but remained cytosolic without Alg14. It was determined that Alg14 localizes Alg13 to the membrane for function (8). An alternative explanation for the results presented here for M. voltae is that only one of the genes encodes a protein that functions as the glycosyl transferase, while the other is required for biosynthesis of the GlcpNAc3NAcA. A UDP-GlcNAc 6-dehydrogenase (a WecC homolog), as well as an acetyltransferase, would both be required to produce UDP-GlcNAc3NAcA from a UDP-GlcNAc precursor. It has been shown in related sugar-biosynthetic pathways (29-32) that nucleotide-activated precursors are common substrates for such enzymes and utilize cofactors similar to glycosyl transferases (i.e., NAD/NADP) in their enzymatic reactions. Consequently, these biosynthetic enzymes could carry domains in common with glycosyl transferases and could result in the annotation of these biosynthetic enzymes as glycosyl transferases due to the conservation of particular functional domains. Mutants in either of these genes would produce similar phenotypes: inability to transfer the second sugar to the lipid linked glycan precursor.
Based on the MS analysis of the two S-layer glycopeptides (Fig. 8), it is obvious that both gene products are involved in the N-glycosylation pathway in M. voltae. Although we were unable to precisely define their respective roles, we have renamed Mv990 aglC and Mv991 algK.
Mv891 was investigated as a putative flippase candidate based on its predicted 11 TMDs and polysaccharide synthesis domain (Table 2). However, Mv891 had no effect on the apparent molecular masses of the flagellin or S-layer proteins or on flagellar assembly and function, indicating that the gene does not code for the M. voltae flippase. However, the deletion of Mv891 had a drastic effect on cell size; approximately half the cell population transformed into giant cells with diameters three to four times that of wild-type cells (Fig. 6D). This translates into an exponential increase in cell volume. The only precedent for M. voltae cells with a similar size phenotype comes in the form of a report in which a protein involved in the structural maintenance of chromosomes (smc) was inactivated (26). The result was a mutant population in which ∼20% of the cells contained little or no DNA and a subset of cells (∼2%) had diameters three to four times larger than normal. The authors postulated that these titan cells indicated cell division arrest at a cell cycle checkpoint (26). Examination of the current annotations of the genes from Mv891 to Mv898 (Fig. 2B) revealed no genes annotated as being related to cell division or cell cycle control. Consequently, the reason for this unexpected effect remains unknown.
The overall findings in this study continue to reinforce both the similarities and the differences between the N-glycosylation pathways in Eukarya, Bacteria, and Archaea. A clearer picture is emerging of the characteristics that glycosyl transferases must possess to assemble glycan structures on the cytoplasmic face of a membrane. This offers promise in the future study of N-glycosylation in prokaryotes, where the entire assembly of the glycan appears to occur on the cytoplasmic face of the cytoplasmic membrane. The ability to identify putative glycosylation enzyme candidates by bioinformatics is especially helpful in Archaea, where the genes encoding these pathway components appear to be randomly distributed around the chromosome (9) instead of in a single gene locus, which is the case in Bacteria (34). In addition, the highly conserved nature of the oligosaccharyl transferase STT3p/PglB/AglB across all domains of life reinforces the universality of N-glycosylation in living systems. What has come as a surprise is the apparent divergence of the flippase enzyme in this pathway. The proposed eukaryotic flippase Rft1 is a unique type of transporter (14), while the bacterial flippase PglK falls into the well-defined ABC transporter family (21). Added to this is the fact that the M. voltae proteins similar to Rft1 or PglK appear to have different functions, leaving the archaeal flippase a mystery. Whether the differences in exporting different types of glycan structures through different types of membranes explains this divergence remains to be investigated. What has been established is that examination of the N-glycosylation pathway at the molecular level in Archaea can add to our knowledge of this important protein modification process, not only in this domain, but in general.
Acknowledgments
We thank John Leigh and Barny Whitman for initial genetic analysis of the M. voltae genome. We also acknowledge Wen Ding and Luc Tessier for their assistance with the MS analysis.
B.C. was supported by a Natural Sciences and Engineering Research Council (NSERC) Post-Graduate Scholarship. This work was supported by a Discovery Grant from NSERC (to K.F.J.).
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Abu-Qarn, M., and J. Eichler. 2006. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 61511-525. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Qarn, M., J. Eichler, and N. Sharon. 2008. Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. doi: 10.1016/j.sbi.2008.06.010. [DOI] [PubMed]
- 3.Abu-Qarn, M., A. Giordano, F. Battaglia, A. Trauner, H. R. Morris, P. Hitchen, and J. Eichler. 2008. Identification of AglE, a second glycosyltransferase involved in N-glycosylation of Haloferax volcanii S-layer glycoprotein. J. Bacteriol. 1903140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu-Qarn, M., S. Yurist-Doutsch, A. Giordano, A. Trauner, H. R. Morris, P. Hitchen, O. Medalia, A. Dell, and J. Eichler. 2007. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 3741224-1236. [DOI] [PubMed] [Google Scholar]
- 5.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardy, S. L., and K. F. Jarrell. 2003. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 501339-1347. [DOI] [PubMed] [Google Scholar]
- 7.Bayley, D. P., and K. F. Jarrell. 1999. Overexpression of Methanococcus voltae flagellin subunits in Escherichia coli and Pseudomonas aeruginosa: a source of archaeal preflagellin. J. Bacteriol. 1814146-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bickel, T., L. Lehle, M. Schwarz, M. Aebi, and C. A. Jakob. 2005. Biosynthesis of lipid-linked oligosaccharides in Saccharomyces cerevisiae: Alg13p and Alg14p form a complex required for the formation of GlcNAc(2)-PP-dolichol. J. Biol. Chem. 28034500-34506. [DOI] [PubMed] [Google Scholar]
- 9.Chaban, B., S. Voisin, J. Kelly, S. M. Logan, and K. F. Jarrell. 2006. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol. Microbiol. 61259-268. [DOI] [PubMed] [Google Scholar]
- 10.Correia, J. D., and K. F. Jarrell. 2000. Posttranslational processing of Methanococcus voltae preflagellin by preflagellin peptidases of M. voltae and other methanogens. J. Bacteriol. 182855-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichler, J., and M. W. Adams. 2005. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69393-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faguy, D. M., D. P. Bayley, A. S. Kostyukova, N. A. Thomas, and K. F. Jarrell. 1996. Isolation and characterization of flagella and flagellin proteins from the thermoacidophilic archaea Thermoplasma volcanium and Sulfolobus shibatae. J. Bacteriol. 178902-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gernhardt, P., O. Possot, M. Foglino, L. Sibold, and A. Klein. 1990. Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol. Gen. Genet. 221273-279. [DOI] [PubMed] [Google Scholar]
- 14.Helenius, J., D. T. Ng, C. L. Marolda, P. Walter, M. A. Valvano, and M. Aebi. 2002. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415447-450. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374166. [Google Scholar]
- 16.Igura, M., N. Maita, J. Kamishikiryo, M. Yamada, T. Obita, K. Maenaka, and D. Kohda. 2008. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 27234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Igura, M., N. Maita, T. Obita, J. Kamishikiryo, K. Maenaka, and D. Kohda. 2007. Purification, crystallization and preliminary X-ray diffraction studies of the soluble domain of the oligosaccharyltransferase STT3 subunit from the thermophilic archaeon Pyrococcus furiosus. Acta Crystallogr. Sect. F 63798-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarrell, K. F., D. P. Bayley, V. Florian, and A. Klein. 1996. Isolation and characterization of insertional mutations in flagellin genes in the archaeon Methanococcus voltae. Mol. Microbiol. 20657-666. [DOI] [PubMed] [Google Scholar]
- 19.Kalmokoff, M. L., and K. F. Jarrell. 1991. Cloning and sequencing of a multigene family encoding the flagellins of Methanococcus voltae. J. Bacteriol. 1737113-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalmokoff, M. L., K. F. Jarrell, and S. F. Koval. 1988. Isolation of flagella from the archaebacterium Methanococcus voltae by phase separation with Triton X-114. J. Bacteriol. 1701752-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, J., H. Jarrell, L. Millar, L. Tessier, L. M. Fiori, P. C. Lau, B. Allan, and C. M. Szymanski. 2006. Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 1882427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koval, S. F., and K. F. Jarrell. 1987. Ultrastructure and biochemistry of the cell wall of Methanococcus voltae. J. Bacteriol. 1691298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 24.Linton, D., N. Dorrell, P. G. Hitchen, S. Amber, A. V. Karlyshev, H. R. Morris, A. Dell, M. A. Valvano, M. Aebi, and B. W. Wren. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 551695-1703. [DOI] [PubMed] [Google Scholar]
- 25.Logan, S. M. 2006. Flagellar glycosylation—a new component of the motility repertoire? Microbiology 1521249-1262. [DOI] [PubMed] [Google Scholar]
- 26.Long, S. W., and D. M. Faguy. 2004. Anucleate and titan cell phenotypes caused by insertional inactivation of the structural maintenance of chromosomes (smc) gene in the archaeon Methanococcus voltae. Mol. Microbiol. 521567-1577. [DOI] [PubMed] [Google Scholar]
- 27.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. Weese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 942626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Namboori, S. C., and D. E. Graham. 2008. Acetamido sugar biosynthesis in the Euryarchaea. J. Bacteriol. 1902987-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoenhofen, I. C., V. V. Lunin, J. P. Julien, Y. Li, E. Ajamian, A. Matte, M. Cygler, J. R. Brisson, A. Aubry, S. M. Logan, S. Bhatia, W. W. Wakarchuk, and N. M. Young. 2006. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J. Biol. Chem. 2818907-8916. [DOI] [PubMed] [Google Scholar]
- 31.Schoenhofen, I. C., D. J. McNally, J. R. Brisson, and S. M. Logan. 2006. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 168C-14C. [DOI] [PubMed] [Google Scholar]
- 32.Schoenhofen, I. C., D. J. McNally, E. Vinogradov, D. Whitfield, N. M. Young, S. Dick, W. W. Wakarchuk, J. R. Brisson, and S. M. Logan. 2006. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J. Biol. Chem. 281723-732. [DOI] [PubMed] [Google Scholar]
- 33.Shams-Eldin, H., B. Chaban, S. Niehus, R. T. Schwarz, and K. F. Jarrell. 2008. Identification of the archaeal alg7 gene homolog (encoding N-acetylglucosamine-1-phosphate transferase) of the N-linked glycosylation system by cross-domain complementation in Saccharomyces cerevisiae. J. Bacteriol. 1902217-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol. 11233-238. [DOI] [PubMed] [Google Scholar]
- 35.Thomas, N. A., S. L. Bardy, and K. F. Jarrell. 2001. The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25147-174. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, N. A., S. Mueller, A. Klein, and K. F. Jarrell. 2002. Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. Mol. Microbiol. 46879-887. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, N. A., C. T. Pawson, and K. F. Jarrell. 2001. Insertional inactivation of the flaH gene in the archaeon Methanococcus voltae results in non-flagellated cells. Mol. Genet. Genomics 265596-603. [DOI] [PubMed] [Google Scholar]
- 38.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 764350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voisin, S., R. S. Houliston, J. Kelly, J. R. Brisson, D. Watson, S. L. Bardy, K. F. Jarrell, and S. M. Logan. 2005. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 28016586-16593. [DOI] [PubMed] [Google Scholar]
- 40.Yan, Q., and W. J. Lennarz. 2002. Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 27747692-47700. [DOI] [PubMed] [Google Scholar]
- 41.Yurist-Doutsch, S., M. Abu-Qarn, F. Battaglia, H. R. Morris, P. G. Hitchen, A. Dell, and J. Eichler. 2008. aglF, aglG and aglI, novel members of a gene island involved in the N-glycosylation of the Haloferax volcanii S-layer glycoprotein. Mol. Microbiol. 691234-1245. [DOI] [PubMed] [Google Scholar]
- 42.Yurist-Doutsch, S., B. Chaban, D. J. VanDyke, K. F. Jarrell, and J. Eichler. 2008. Sweet to the extreme: protein glycosylation in Archaea. Mol. Microbiol. 681079-1084. [DOI] [PubMed] [Google Scholar]