Abstract
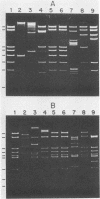
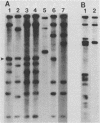
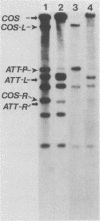
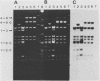
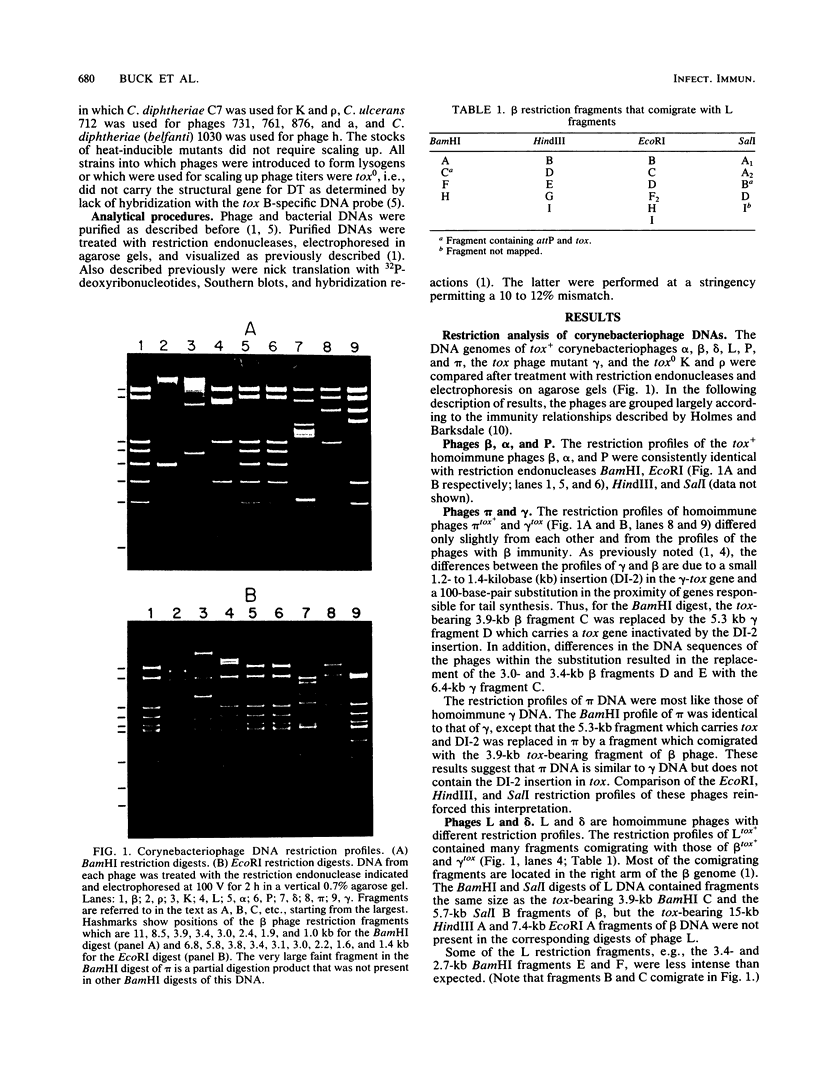
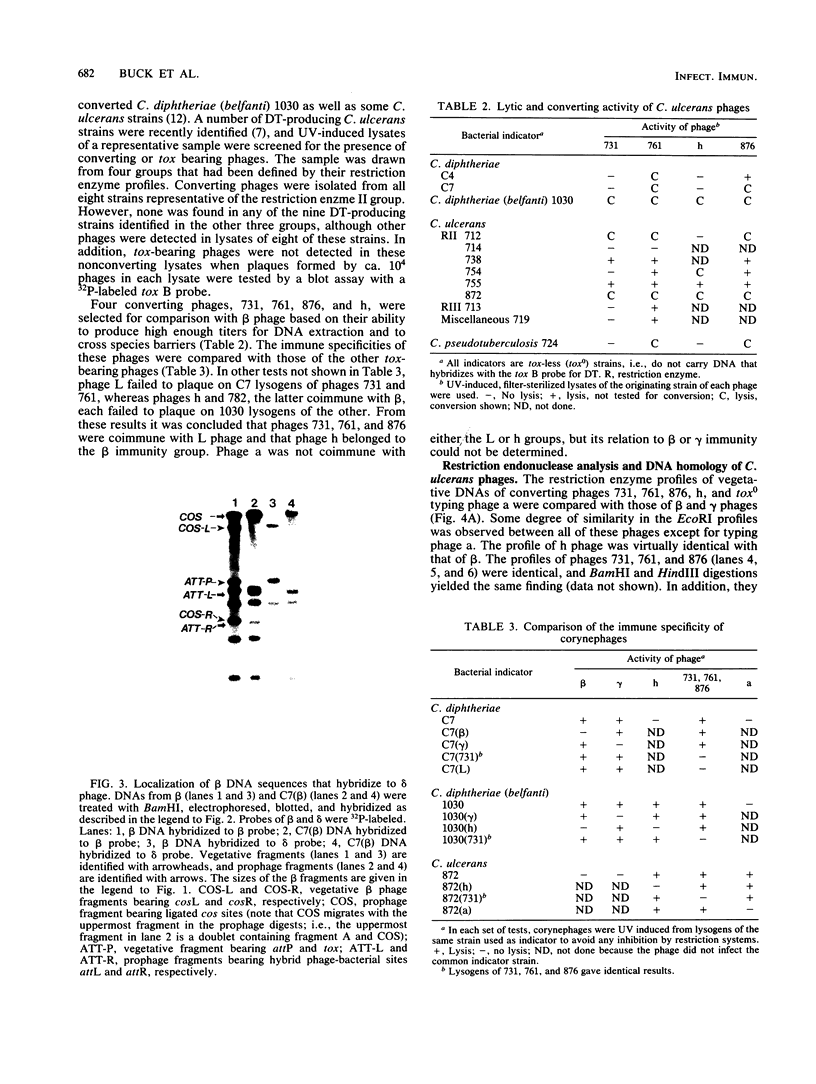
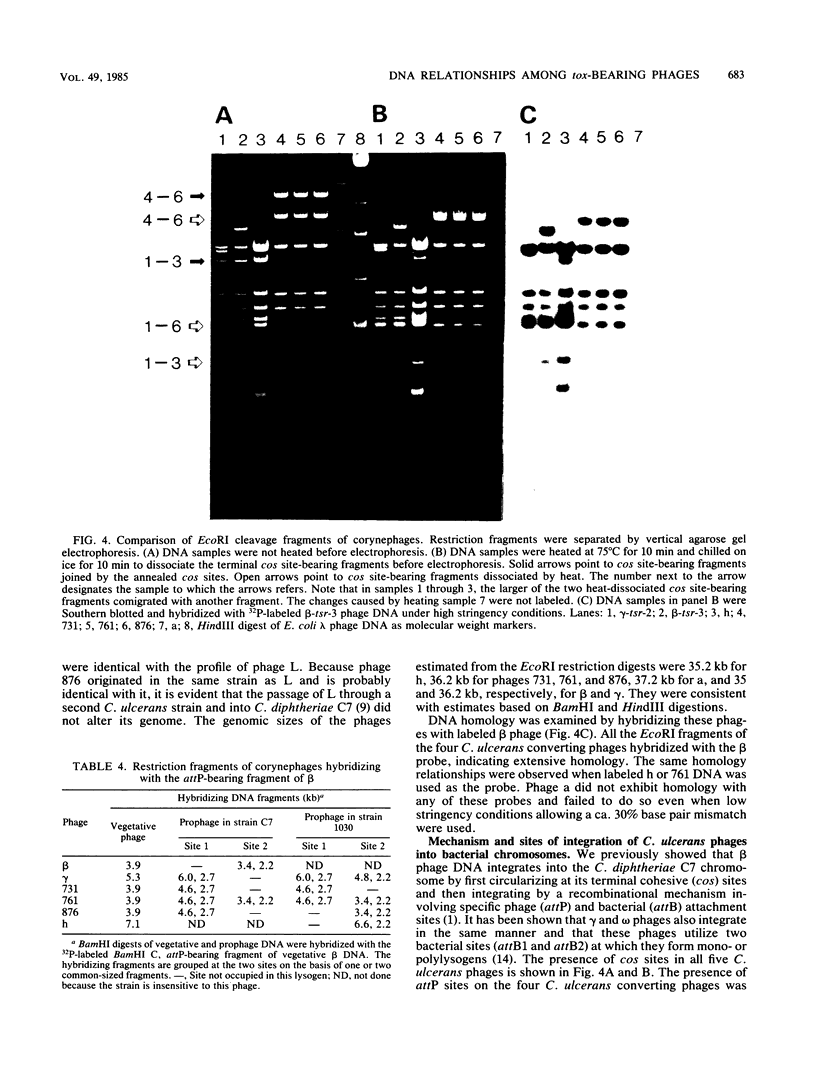
The DNA genomes of a number of tox-bearing, temperate corynebacteriophages isolated from strains of Corynebacterium diphtheriae and Corynebacterium ulcerans were compared. With one exception, these phages displayed similarities in their restriction enzyme digest profiles and extensive homology with prototypic beta converting phage. The exception, phage delta, had a unique restriction profile and exhibited homology with beta over a limited portion of its genome. DNAs of phages from each host contained cohesive ends and integrated as prophage by a mechanism analogous to that employed by coliphage lambda. It is proposed that these tox-bearing phages belong to a common family, the beta family. The role of the beta family in the movement of the tox gene between strains of C. diphtheriae and C. ulcerans is discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buck G. A., Groman N. B. Genetic elements novel for Corynebacterium diphtheriae: specialized transducing elements and transposons. J Bacteriol. 1981 Oct;148(1):143–152. doi: 10.1128/jb.148.1.143-152.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck G. A., Groman N. B. Identification of deoxyribonucleic acid restriction fragments of beta-converting corynebacteriophages that carry the gene for diphtheria toxin. J Bacteriol. 1981 Oct;148(1):153–162. doi: 10.1128/jb.148.1.153-162.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck G. A., Groman N. B. Physical mapping of beta-converting and gamma-nonconverting corynebacteriophage genomes. J Bacteriol. 1981 Oct;148(1):131–142. doi: 10.1128/jb.148.1.131-142.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck G., Groman N., Falkow S. Relationship between beta converting and gamma non-converting corynebacteriophage DNA. Nature. 1978 Feb 16;271(5646):683–685. doi: 10.1038/271683a0. [DOI] [PubMed] [Google Scholar]
- Groman N. B. Conversion by corynephages and its role in the natural history of diphtheria. J Hyg (Lond) 1984 Dec;93(3):405–417. doi: 10.1017/s0022172400065001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groman N., Cianciotto N., Bjorn M., Rabin M. Detection and expression of DNA homologous to the tox gene in nontoxinogenic isolates of Corynebacterium diphtheriae. Infect Immun. 1983 Oct;42(1):48–56. doi: 10.1128/iai.42.1.48-56.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groman N., Laird W. Heat-inducible mutants of corynebacteriophage. J Virol. 1977 Sep;23(3):587–591. doi: 10.1128/jvi.23.3.587-591.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groman N., Schiller J., Russell J. Corynebacterium ulcerans and Corynebacterium pseudotuberculosis responses to DNA probes derived from corynephage beta and Corynebacterium diphtheriae. Infect Immun. 1984 Aug;45(2):511–517. doi: 10.1128/iai.45.2.511-517.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes R. K., Barksdale L. Comparative studies with tox plus and tox minus corynebacteriophages. J Virol. 1970 Jun;5(6):783–784. doi: 10.1128/jvi.5.6.783-794.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes R. K., Barksdale L. Genetic analysis of tox+ and tox- bacteriophages of Corynebacterium diphtheriae. J Virol. 1969 Jun;3(6):586–598. doi: 10.1128/jvi.3.6.586-598.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird W., Groman N. Prophage map of converting corynebacteriophage beta. J Virol. 1976 Jul;19(1):208–219. doi: 10.1128/jvi.19.1.208-219.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximescu P., Oprişan A., Pop A., Potorac E. Further studies on Corynebacterium species capable of producing diphtheria toxin (C. diphtheriae, C. ulcerans, C. ovis). J Gen Microbiol. 1974 May;82(1):49–56. doi: 10.1099/00221287-82-1-49. [DOI] [PubMed] [Google Scholar]
- Rappuoli R. Isolation and characterization of Corynebacterium diphtheriae nontandem double lysogens hyperproducing CRM197. Appl Environ Microbiol. 1983 Sep;46(3):560–564. doi: 10.1128/aem.46.3.560-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R., Michel J. L., Murphy J. R. Integration of corynebacteriophages beta tox+, omega tox+, and gamma tox- into two attachment sites on the Corynebacterium diphtheriae chromosome. J Bacteriol. 1983 Mar;153(3):1202–1210. doi: 10.1128/jb.153.3.1202-1210.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R., Michel J. L., Murphy J. R. Restriction endonuclease map of corynebacteriophage omega ctox+ isolated from the Park-Williams no. 8 strain of Corynebacterium diphtheriae. J Virol. 1983 Feb;45(2):524–530. doi: 10.1128/jvi.45.2.524-530.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]