Abstract
The HPrK kinase/phosphatase is a common component of the phosphotransferase system (PTS) of gram-positive bacteria and regulates catabolite repression through phosphorylation/dephosphorylation of its substrate, the PTS protein HPr, at a conserved serine residue. Phosphorylation of HPr by HPrK also affects additional phosphorylation of HPr by the PTS enzyme EI at a conserved histidine residue. Sinorhizobium meliloti can live as symbionts inside legume root nodules or as free-living organisms and is one of the relatively rare gram-negative bacteria known to have a gene encoding HPrK. We have constructed S. meliloti mutants that lack HPrK or that lack key amino acids in HPr that are likely phosphorylated by HPrK and EI. Deletion of hprK in S. meliloti enhanced catabolite repression caused by succinate, as did an S53A substitution in HPr. Introduction of an H22A substitution into HPr alleviated the strong catabolite repression phenotypes of strains carrying ΔhprK or hpr(S53A) mutations, demonstrating that HPr-His22-P is needed for strong catabolite repression. Furthermore, strains with a hpr(H22A) allele exhibited relaxed catabolite repression. These results suggest that HPrK phosphorylates HPr at the serine-53 residue, that HPr-Ser53-P inhibits phosphorylation at the histidine-22 residue, and that HPr-His22-P enhances catabolite repression in the presence of succinate. Additional experiments show that ΔhprK mutants overproduce exopolysaccharides and form nodules that do not fix nitrogen.
Sinorhizobium meliloti is a member of the alphaproteobacteria and can grow as free-living organisms, or as intracellular, nitrogen-fixing symbionts of alfalfa and other legumes (5, 13, 35, 54).
S. meliloti is able to utilize a large variety of compounds for growth, but succinate and other C4-dicarboxylic acids play an especially important role in metabolism during both free-living and symbiotic states. C4-dicarboxylic acids are used to fuel and provide reducing equivalents for nitrogen fixation by bacteroids (20, 49). Free-living S. meliloti also utilize succinate and do so in preference to many sugars and other substrates (6, 28, 31, 45, 57).
It has been shown that succinate represses genes needed for utilization of secondary carbon sources such as lactose (31, 57) and that it can prevent the intracellular accumulation of secondary carbon sources, lactose and raffinose, through inducer exclusion (6). This is called succinate-mediated catabolite repression (SMCR) and, though well documented in S. meliloti, the molecular mechanisms of its operation remain obscure. In many other model bacteria where catabolite repression in response to sugars is well understood, sensing of primary carbon sources takes place during their transport through the phosphotransferase system (PTS) (14, 50, 53). The regulatory and physiological responses that constitute catabolite repression are often controlled by the phosphorylation state of the of the PTS proteins HPr and EIIA, which in turn depends on whether or not the PTS is transporting a favored sugar (22, 24, 58, 59). There are key differences between how some gram-negative bacteria, exemplified by Escherichia coli, and some gram-positive bacteria, exemplified by Bacillus subtilis, regulate catabolite repression via the PTS (Fig. 1). In these organisms the PTS delivers a phosphate group to a preferred sugar such as glucose. The phosphate originates from PEP, is transferred to the histidine-15 residue of HPr by enzyme EI, and is then transferred from HPr to EIIA.
FIG. 1.
Mechanisms of catabolite repression in E. coli (A) and B. subtilis (B).
In E. coli and other enteric bacteria, EIIA is the main player in catabolite repression. In the presence of glucose, EIIA is mainly unphosphorylated because the phosphate group it obtains from HPr eventually phosphorylates the incoming sugar. Unphosphorylated EIIA blocks transporters of secondary carbon sources, resulting in inducer exclusion (30, 32, 34). In the absence of glucose, EIIA is mainly in its phosphorylated form, which activates cyclic AMP synthesis by adenylate cyclase. Cyclic AMP binds to the regulator Crp and activates genes necessary for catabolism of secondary carbon sources (7, 23, 51, 56). In contrast, gram-positive bacteria such as B. subtilis rely mainly on HPr for regulation of catabolite repression (48, 58). HPr can be phosphorylated at the serine-46 residue by the HPr kinase/phosphatase HPrK under energy-replete conditions (33, 38, 42, 47). HPr-Ser46-P, after binding to the transcriptional regulator CcpA, can repress genes associated with secondary carbon source utilization (1, 25, 47, 58). Inducer exclusion is also used by some gram-positive organisms for catabolite repression. In these cases the permease blocking is effected by HPr-Ser46-P (41, 58).
S. meliloti possesses genes encoding some of the PTS proteins. Genes for an EI-type protein (SMc02437), HPr (SMc02754), EIIA-type proteins ManX (SMc02753) and EIIANtr (SMc01141), and HPrK (SMc02752) are present in the chromosome. Transport-related PTS proteins EIIB and EIIC are absent, indicating that PTS proteins that have been retained in S. meliloti probably provide critical regulatory functions even though they likely do not catalyze transport (4, 29). In support of this hypothesis, recent work in S. meliloti has shown that the HPr and ManX proteins are involved in regulation of many aspects of cellular physiology, including carbon metabolism, catabolite repression, exopolysaccharide synthesis, and survival in stationary phase (44).
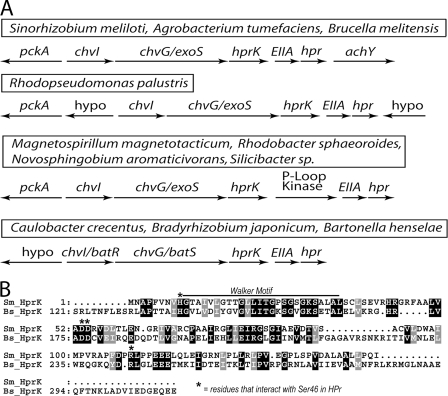
While HPrK is a common regulator in gram-positive bacteria, it is lacking in most gram-negative bacteria, including E. coli and other enteric bacteria. However, it is encoded in the genomes many of alphaproteobacteria (Fig. 2A) (2, 29). In S. meliloti and many other alphaproteobacteria, the gene for hprK is found upstream of the hpr and manX genes (4, 29) (Fig. 2A). The putative S. meliloti HPrK is shorter than HPrK in model gram-positive organisms (see Fig. 2B). In fact, all alphaproteobacteria known to contain a HPrK homolog contain a short version, similar to the one in S. meliloti, rather that the longer version typical of gram-positive organisms (29, 55). In addition, S. meliloti appears to lack the regulator CcpA. Interestingly, an artificially truncated HPrK from Lactobacillus casei, which contained the same regions as the shorter S. meliloti HPrK, exhibited wild-type kinase and phosphatase activities toward HPr (9, 18). This suggests that the S. meliloti HPrK may also have kinase and phosphatase activities toward HPr. Given its unusual presence in S. meliloti and related bacteria and its association with the hpr and manX genes, we generated and characterized hprK mutants as part of an ongoing project to understand the role of the PTS system in regulating SMCR, carbon metabolism, and other aspects of physiology in S. meliloti.
FIG. 2.
Genes encoding HPrK-like proteins in S. meliloti and other alphaproteobacteria. (A) Maps of chromosomal regions containing PTS-like genes from various sequenced alphaproteobacteria. (B) CLUSTALX alignment of HprK from B. subtilis and S. meliloti. Important motifs and residues are indicated. Black boxes indicate identical amino acids and gray boxes conserved amino acids. Note that the S. meliloti protein lacks the N-terminal domain present in the B. subtilis protein.
MATERIALS AND METHODS
Media, plasmids, and strains.
S. meliloti strain Rm1021 was used as the parent for the construction of all mutants. S. meliloti strains were grown in TY or in M9 mineral salts medium (52) supplemented with 5 ng/ml of cobalt chloride and various carbon sources at the concentrations noted in the text. E. coli XL1B MRF′ (Stratagene) was the host for construction of all plasmids and was grown in LB medium (52) supplemented with antibiotics, IPTG (isopropyl-β-d-thiogalactopyranoside) at 48 μg/ml, and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 40 μg/ml as needed. All other strains and plasmids used are listed in Table 1. Antibiotics were added at the following concentrations as needed: streptomycin at 500 μg/ml, ampicillin at 100 μg/ml, gentamicin at 30 μg/ml (S. meliloti) or 10 μg/ml (E. coli), neomycin at 100 or 200 μg/ml, and streptomycin at 200 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Relevant genotype or markera | Source or reference |
|---|---|---|---|
| Strains | |||
| Rm1021 | S. meliloti, wild type | hpr hprK (wild type) | 37 |
| RB111 | Rm1021, Δhpr | Δhpr | 44 |
| CAP14 | Rm1021, ΔhprK | ΔhprK | This study |
| CAP43 | Rm1021, SMc02324::pCAP77 | hpr hprK (wild type); Nmr | This study |
| CAP49 | RB111, SMc02324::pCAP81 | hpr hprK (wild type); Nmr | 44 |
| CAP60 | RB111, SMc02324::pCAP77 | Δhpr; Nmr | This study |
| CAP62 | CAP14, SMc02324::pCAP83 | ΔhprK/Psma0113::hprK; Nmr | This study |
| CAP78 | Rm1021, Δhpr/ΔhprK | Δhpr ΔhprK | This study |
| CAP92 | CAP78, SMc02324::pCAP81 | ΔhprK | This study |
| CAP93 | RB111, SMc02324::pCAP113 | hprH22A; Nmr | This study |
| CAP94 | CAP78, SMc02324::pCAP113 | ΔhprK/hpr(H22A); Nmr | This study |
| CAP95 | RB111, SMc02324::pCAP114 | hpr(S53A); Nmr | This study |
| CAP97 | RB111, SMc02324::pCAP118 | hpr(H22A/S53A); Nmr | This study |
| Plasmids | |||
| pGEM-T Easy | Cloning vector | Amp | Promega |
| pJQ200 SK | Suicide plasmid with sacB | Gm | 46 |
| pMB439 | Suicide plasmid | Nm | 3 |
| pCAP11 | pMB393, PmelA::gfp | Sp | 44 |
| pDG127 | pGEM-T Easy, hpr | Amp | 44 |
| pCAP31 | pJQ200SK carrying ΔhprK allele | Gm | This study |
| pCAP77 | pMB439 with an internal fragment of SMc02324 | Nm | 44 |
| pCAP81 | pCAP77 with hpr | Nm | 44 |
| pCAP83 | pCAP77 with Psma0113::hprK | Nm | This study |
| PCAP84 | pCAP77 with Psma0113 | Nm | This study |
| pCAP113 | pCAP77 with hpr(H22A) | Nm | This study |
| pCAP114 | pCAP77 with hpr(S53A) | Nm | This study |
| pCAP118 | pCAP77 with hpr(H22A/S53A) | Nm | This study |
Resistance markers; Nm, neomycin; Sp, spectinomycin; Amp, ampicillin; Gm, gentamicin.
Construction of in-frame deletions of hprK.
In-frame deletion of hprK was carried out following the gene replacement approach of Quandt and Hynes (46). For construction of the deletion, the N-terminal coding end and upstream region of hprK were amplified from S. meliloti Rm1021 genomic DNA with the primers 229 and 238. The C-terminal coding end and downstream flanking sequence of hprK were amplified by using primers 242 and 243. The primers are listed in Table 2. The two fragments were separately cloned into pGEM-T Easy, excised with the restriction enzymes SacI-NsiI and SpeI-NsiI, respectively. Fragments were ligated together, eliminating a 243-bp fragment of the gene, and cloned into the sacB-containing suicide vector pJQ200SK, creating plasmid pCAP31. pCAP31 construction was confirmed by PCR and sequencing. The plasmid was transformed into S. meliloti by electroporation, integrating into the chromosome by homologous recombination, yielding strain CAP13. A second recombination resulted in loss of the integrated plasmid and the wild-type copy of hprK and in retention of the hprK in-frame deletion. Isolates that had lost the plasmid were selected on TY-sucrose plates, and the presence of the ΔhprK allele was screened for by PCR. The ΔhprK strain obtained, CAP14, was used in the present study. The same plasmid (pCAP31) was delivered into strain RB111 (Δhpr), yielding strain CAP78(Δhpr/ΔhprK) which carries in-frame, unmarked deletions of hpr and hprK.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) | Description | Notes |
|---|---|---|---|
| 229 | GGACTAGTCGCATCGTCGTCCGGC | Amplification of the N-terminal coding end of hprK | |
| 238 | TGATCATGCATGAAAGCGCGAGCGCCGACTTGCC | Amplification of the N-terminal coding end of hprK | |
| 242 | CGAGCTCATGGCCAGATATCGCAATGCAG | Amplification of the C-terminal coding end of hprK | |
| 243 | CATGCATGATCAGAAATCTGCCGCTGTTGC | Amplification of the C-terminal coding end of hprK | |
| 278 | ATGGCGGCGGCAGCGTGGTC | External primer for site-directed mutagenesis of hpr | |
| 279 | CGCCGGAGGAGGAGTTGCAG | External primer for site-directed mutagenesis of hpr | |
| 462-463 | GTCGGCGGGACCGCCATCATGGGGCTGATGA | Internal primers for hpr(S53A) mutagenesis | Primer 463 is the reverse complement of 462 |
| 464-465 | ACAAACGCGGCCTGGCCGCGCGTGCCTCTG | Internal primer for hpr(H53A) mutagenesis | Primer 465 is the reverse complement of 464 |
Site-directed mutagenesis of hpr.
Site-directed mutants of HPr were constructed by the overlap extension method (52), using two internal primers that were complementary to each other and carried the desired mutation (amino acid substitution), and two external primers downstream and upstream of the gene (Table 2). Briefly, one internal primer and one external primer constituted a primer set used to amplify each one of two segments of the gene. Plasmid pDG127, which contains hpr under its native promoter region, was used as a template. The amplified segments shared an overlapping sequence (carrying the desired mutation), which mixed, denatured, and annealed served as templates for a second round of amplification using only the external primers. The product of this second amplification was the complete gene carrying the desired mutation. The different site-directed mutant genes constructed were hpr(H22A), hpr(S53A), and hpr(H22A/S53A). All were verified by sequencing.
The DNA fragments carrying the mutations were cloned into pGEM-T Easy and excised with EcoRI, and the resulting fragments were cloned into pCAP77, a suicide vector that contains an internal fragment of the first gene of the rhamnose utilization operon in S. meliloti Rm1021, SMc02324 (36). Suicide plasmids with the mutant alleles were then delivered into the chromosome of strains RB111(Δhpr) and CAP78(Δhpr/ΔhprK). Strain CAP49, a complemented RB111(Δhpr) strain that carries a wild-type copy of hpr in SMc02324, was used as a control strain for experiments with site-directed mutant alleles of hpr integrated in SMc02324. Strain CAP60, an RB111-derived strain that carries an empty vector (pCAP77) in SMc02324, was used as the Δhpr control strain in these experiments. These strains are listed in Table 1.
Complementation of ΔhprK.
A copy of the hprK gene was amplified from Rm1021 genomic DNA and was cloned as a 447-bp fragment into plasmid pCAP84, resulting in pCAP83. Plasmid pCAP84 is a derivative of pCAP77 that contains a weak promoter upstream of the multiple cloning site. The promoter is a 490-bp sequence upstream from the operon comprised of genes sma0113 and sma0114. This region exhibited very weak promoter activity when fused to the reporter gene gfp (P. P. Garcia, D. J. Gage, and C. Arango, unpublished data). Strain CAP14(ΔhprK) was complemented in trans by recombination of pCAP83 in the chromosome at SMc02324. The resulting strain was called CAP83. A control strain, that exhibited wild-type phenotypes, was constructed by recombination of pCAP77 at SMc02324 in Rm1021.
Growth curves and growth rates.
Growth curves and growth rates were obtained as explained previously (44). Concentration of single carbon sources was 0.4%, while concentrations for dual carbon sources were 0.05% for succinate and 0.1% for the secondary carbon source. Yield measurements at different succinate concentrations confirmed that at 0.05% the cultures were limited for carbon when they reached stationary phase. Growth curve cultures were tested for the number of fast-growing suppressors upon reaching stationary phase. Experiments with a fraction of suppressors higher than 10% were not included in the results. Growth experiments for evaluation of gene expression were conducted with strains transformed with plasmid pCAP11, and fluorescence was measured as described previously (44).
β-Galactosidase assays.
Strains were streaked, or spread, onto M9 minimal plates with succinate (0.4%), lactose (0.4%), or succinate plus lactose (0.1 and 0.05%, respectively) as carbon sources. Cells were scraped off the plates, rinsed, resuspended in 15% glycerol, and frozen at −80°C until the time of the assay. Enzyme assays were conducted as described previously (39). For qualitative evaluation of β-galactosidase activity, cells were pregrown on M9 minimal medium with glycerol as carbon source, rinsed, resuspended in no-carbon M9, and plated on M9 minimal plates with succinate (0.1%) plus lactose (0.05%) as carbon sources and X-Gal (40 μg/ml) as an indicator of β-galactosidase activity (SLX plates).
Nodulation assays.
Alfalfa seeds were sterilized and sprouted as described previously (19), with the following modifications. Single seedlings were placed on Nod3 (BNM) agar slants in 18-mm glass tubes and inoculated with 100 μl of a suspension of S. meliloti. Suspensions were made by scraping colonies from M9 plates with glycerol as a carbon source, rinsing, and resuspending in Nod3 medium to a final optical density (OD) of 0.1 at 595 nm. Plant growth tubes were loosely capped and incubated in a growth chamber with 16/8-h light/dark cycle at 26°C. Nodules were counted weekly. At 36 days after inoculation, plant shoots were cut above the cotyledons, dried, and weighed.
Nodule structure microscopy.
Nodules were excised from alfalfa plants, fixed in phosphate-buffered saline with 4% paraformaldehyde, dehydrated in ethanol, and embedded in JB4 embedding resin as described previously (17, 40). Embedded nodules were sliced to give 2-μm-thick sections, dyed with a mixture of DAPI (4′,6′-diamidino-2-phenylindole) and acridine orange, and photographed under epifluorescence illumination using a filter set with a 330- to 380-nm excitation filter and a long-pass emission filter with a 435-nm cutoff.
RESULTS
Deletion of hprK results in greatly reduced growth rates and reduced catabolic gene expression.
Although the main objective of deleting hprK was to investigate whether the protein had a role in catabolite repression, it became evident that the deletion of hprK strongly affected growth. Colonies of strain CAP14(ΔhprK) grown on M9 plates with a variety of carbon sources were significantly smaller than those of strain Rm1021 (wild type) (Fig. 3). Growth rates were measured in minimal medium with succinate, fructose, arabinose, maltose, glycerol, glucose, raffinose, and lactose as the sole carbon sources. The strain lacking hprK exhibited growth rates that were reduced 70 to 80% relative to wild-type growth rates (Table 3). Growth on TY was reduced as well, but not as drastically as in minimal medium. Suppressors readily arose in strain CAP14(ΔhprK), which resulted in faster-growing strains that gave rise to colonies much larger than those of the parental ΔhprK strain (Fig. 3).
FIG. 3.

Colony morphology of S. meliloti containing altered HprK and HPr proteins. (A) Colonies of wild-type strain Rm1021. (B) Colonies of strain CAP14(ΔhprK) (arrows) and ΔhprK suppressor strains (arrowheads). (C) Colonies of control strain CAP43(SMc02324::pCAP77) and substitution mutants CAP93[hpr(H22A)], CAP95[hpr(S53A)], and CAP97[hpr(H22A/S53A)] on M9 plates with 0.4% glycerol. Plates were incubated for 2 weeks (A and B) or for 1 week (C) before being photographed.
TABLE 3.
Growth rate constants of strains Rm1021 (wild type) and CAP14(ΔhprK) in different media (n = 2 to 4)
| Carbon sourcea | Avg growth rate constant k (hr−1) ± SE
|
|
|---|---|---|
| Rm1021 | CAP14(ΔhprK) | |
| Succinate | 0.17 ± 0.015 | 0.05 ± 0.004 |
| Glucose | 0.14 ± 0.002 | 0.04 ± 0.011 |
| Raffinose | 0.11 ± 0.013 | 0.02 ± 0.003 |
| Lactose | 0.09 ± 0.002 | 0.02 ± 0.001 |
| Fructose | 0.19 ± 0.017 | 0.06 ± 0.011 |
| Arabinose | 0.10 ± 0.002 | 0.03 ± 0.001 |
| Maltose | 0.11 ± 0.012 | 0.02 ± 0.0003 |
| Glycerol | 0.21 ± 0.015 | 0.04 ± 0.002 |
| TY (rich medium) | 0.27 ± 0.009 | 0.15 ± 0.0002 |
Single carbon source added to M9 minimal medium. All cultures were grown at 30°C with shaking.
Attempts to alleviate the growth defect by addition of growth factors were unsuccessful (data not shown). The addition of tryptophan or vitamins to M9 plates with lactose as the carbon source did not result in larger colonies for either the wild-type strain or the CAP14(ΔhprK) strain compared to nonamended plates. The addition of Casamino Acids and yeast extract slightly stimulated the growth of wild-type strain Rm1021 but did not fix the growth phenotype of strain CAP14(ΔhprK). These results, together with the slow growth on all types of carbon sources tested, suggest that the altered growth phenotype of strain CAP14(ΔhprK) may be a result of alteration in central metabolic pathways rather than a defect in the biosynthesis of amino acids or cofactors.
Low growth rates on certain carbon sources could be caused by either low expression of the catabolic or transport genes or by low activity of the corresponding proteins. Expression the melA-agp operon, necessary for utilization of α-galactosides such as raffinose and melibiose, was evaluated using the reporter plasmid pCAP11, which carries a PmelA::gfp fusion inducible by α-galactosides. Specific fluorescence of ΔhprK cells at mid-exponential phase grown on raffinose as the sole carbon source was reduced sixfold compared to the wild-type cells [10,700 ± 1,500 fluorescence units/OD units for CAP14(ΔhprK) versus 66,900 ± 2,200 for the wild type, n = 2]. Even when grown in M9 with succinate, a condition that represses the melA-agp operon, the measured levels were significantly lower for strain CAP14(ΔhprK) [900 ± 90 fluorescence units/OD units for CAP14(ΔhprK) versus 4,000 ± 670 for the wild type]. The β-galactosidase activity was also low in strain CAP14(ΔhprK) when measured in cells grown on plates with lactose as the sole carbon source. The enzyme activity in strain Rm1021 was 20.7 ± 0.7 U (average ± the standard error, three independent experiments), while in strain CAP14(ΔhprK) it was 4.3 ± 0.12 U, a fivefold reduction. These results suggest that HPrK is involved, directly or indirectly, in regulation of carbon metabolism, through the regulation of genes or enzymes needed for transport and catabolism.
Loss of HPrK results in severe SMCR.
HPrK regulates catabolite repression in gram-positive organisms via phosphorylation of HPr on a conserved serine residue. Since S. meliloti HPr protein retains this phosphorylatable residue, it seemed likely that HPrK could regulate SMCR in this organism. In a preliminary assessment of SMCR phenotypes, strains Rm1021 (wild type) and CAP14(ΔhprK) were grown on M9 minimal plates with succinate (0.1%) plus lactose (0.05%). The plates also contained X-Gal, which turns blue when cleaved by endogenous β-galactosidase. Colonies from strain Rm1021 were initially white, indicating growth on succinate and repression of β-galactosidase production, but turned blue after succinate was depleted and lactose utilization started. In contrast, strain CAP14(ΔhprK) never turned blue, even after colonies reached the maximum size (Fig. 4). This suggested that strain CAP14(ΔhprK) had a severe alteration of SMCR, leading to very low levels of lactose utilization. If HPrK were generally involved in SMCR, its absence would affect the catabolite repression of other secondary carbon sources, and the effects should be observed in growth curves of the mutant strain on dual carbon sources. The growth of strains Rm1021 and CAP14(ΔhprK) was monitored in M9 medium with succinate plus either raffinose (an α-galactoside) or lactose (a β-galactoside). When grown on succinate plus lactose, the wild-type strain Rm1021 exhibited a short lag of 2 to 3 h after succinate was exhausted, before growth resumed using lactose as a carbon source. This lag was 20 or more hours for the strain CAP14(ΔhprK) (Fig. 5A). An extended diauxic lag was also exhibited by strain CAP14(ΔhprK) in succinate plus raffinose medium (Fig. 5B). The diauxic lag was extended from 5 h in the wild-type strain to approximately 30 h for strain CAP14(ΔhprK). These results indicated that SMCR was stronger in strain CAP14(ΔhprK) than in the wild-type strain, suggesting that HPrK has a role in the process of repression or derepression and that its role is not specific to a single type of sugar.
FIG. 4.
Colony phenotype of S. meliloti containing altered HPrK and HPr proteins on SLX plates. Colonies of the indicated strains were grown for 7 days on succinate (0.1%) plus lactose (0.05%) M9 plates with indicator X-Gal before being photographed. Blue coloration indicates the activity of endogenous β-galactosidase.
FIG. 5.
Growth of ΔhprK mutants on dual carbon sources. The diauxic growth of wild-type strain Rm1021, strain CAP14(ΔhprK), and strain CAP62 (which was complemented with hprK in trans) was examined. Cells were grown in M9 media with 0.1% succinate plus 0.05% lactose (A) or with 0.1% succinate plus 0.05% raffinose (B). Curves were time shifted to align the point of succinate exhaustion.
Epistatic relation between Δhpr and ΔhprK.
To explore the epistatic relation between the hpr deletion and the hprK deletion, a double Δhpr/ΔhprK mutant strain (CAP78) was constructed by introducing a ΔhprK allele into the Δhpr mutant strain RB111. Growth and SMCR phenotypes of strain CAP78(Δhpr/ΔhprK) were similar to those of strain RB111(Δhpr), i.e., relaxed SMCR compared to the wild-type strain, indicating that the phenotypes caused by the deletion of hprK require HPr in one or more of its forms (data not shown, Fig. 4, and Fig. 6A).
FIG. 6.
Growth of ΔhprK and HPr amino acid-substitution mutants on dual carbon sources. (A and B) Diauxic growth of the control strain CAP49, strain CAP92(ΔhprK), strain CAP78(ΔhprK/Δhpr) and strain CAP95[hpr(S53A)]. (C and D) Diauxic growth of the control strain CAP49, strain CAP60(Δhpr), strain CAP93[hpr(H22A)] and strain CAP97[hpr(H22A/S53A)]. Cells were grown in M9 media with 0.1% succinate plus 0.05% lactose (A and C) or with 0.1% succinate plus 0.05% raffinose (B and D). All strains are isogenic and came from the same strain, RB111(Δhpr), into which wild-type hpr or site-directed mutants of hpr, or an empty vector, were introduced at the SMc02324 site. Curves were time shifted to align the point of succinate exhaustion.
Due to the high frequency of secondary mutations that suppress the phenotypes of ΔhprK mutants, it was possible that the strain isolated as (Δhpr/ΔhprK) was one of these suppressor mutants. To verify that this was not the case, plasmid pCAP81 (carrying a copy of hpr) was introduced into the chromosome of CAP78(Δhpr/ΔhprK). The resulting strain (CAP92) exhibited phenotypes typical of strain CAP14(ΔhprK): small colonies on M9 minimal plates with glycerol (data not shown) and white coloration on M9 minimal plates with succinate plus lactose plus X-Gal (SLX plates), confirming that CAP78(Δhpr/ΔhprK) had no hprK suppressor mutations (Fig. 4).
hprK complementation reverses the observed phenotypes of the ΔhprK mutant.
To confirm that the observed phenotypes were a direct result of deleting hprK, a copy of the gene was supplied in trans by inserting it into the SMc02324 gene in the chromosome of strain CAP14(ΔhprK). The gene was supplied in single copy, with its expression controlled from the promoter region of genes sma0113 and sma0114 (P. P. Garcia and D. J. Gage, unpublished results). The resulting strain, CAP62, was evaluated for growth and SMCR on various carbon sources. Introduction of the hprK gene reestablished wild-type phenotypes, fully or partially, in all cases tested (Fig. 5 and data not shown).
To verify that strain CAP62(ΔhprK, SMc02324::hprK) was truly a complemented ΔhprK mutant and did not contain a spontaneous suppressor of ΔhprK, the integrated plasmid containing hprK was exchanged for another suicide plasmid without hprK. Transductants plated on M9 minimal medium with glycerol as the sole carbon source exhibited limited growth (small colonies) compared to the parental strain CAP62(ΔhprK, SMc02324::hprK), confirming that the phenotypes of strain CAP62(ΔhprK, SMc02324::hprK) were due to complementation of ΔhprK in trans and not due to a suppressor mutation (data not shown).
An hpr(S53A) mutant behaves like a mutant devoid of HPrK.
Certain HPr proteins can be phosphorylated on two different residues. In B. subtilis EI phosphorylates HPr at the histidine-15 residue, while HPrK phosphorylates (and dephosphorylates) HPr at the serine-46 (47, 48). The latter form of HPr acts as a corepressor of catabolic genes in response to the presence of favored carbon sources (1, 25, 47, 58). Alignment of S. meliloti HPr to HPr proteins of B. subtilis and other bacteria revealed well-conserved regions around Ser-53 and His-22 in the S. meliloti protein that are similar to regions around phosphorylatable serine and histidine residues in other HPr proteins (4). These results suggested that residues His-22 and Ser-53 may be phosphorylated in S. meliloti HPr. It has been suggested that in S. meliloti HPr-His22-P is involved, directly or indirectly, in repression of secondary carbon source utilization in the presence of succinate (44). This hypothesis does not exclude the possibility that HPr-Ser53-P is also involved in SMCR or in regulating phosphorylation of HPr at the histidine-22 residue. Slow growth on single carbon sources and extended diauxic lags on dual carbon sources in mutants lacking the hprK gene (strains CAP14 and CAP92) suggested that HPr-Ser53-P is important for activation of catabolic genes. To explore the importance of phosphorylation at the two different critical residues, genes coding for HPr-H22A, HPr-S53A, and HPr-H22A/S53A were inserted in single copy in the SMc02324 gene of strain RB111(Δhpr), yielding strains CAP93, CAP95, and CAP97, respectively.
Colonies of CAP95[hpr(S53A)] were smaller than colonies of control strain CAP43 when grown on M9 plates with glycerol as a carbon source (0.4%) (Fig. 3C). Slow growth was also observed in M9 minimal medium with succinate, lactose, or raffinose as carbon sources (data not shown). When grown on SLX plates, colonies of strain CAP95[hpr(S53A)] were white for a longer period of time than the control strains CAP43 and CAP49. The final color of strain CAP95[hpr(S53A)] was pale blue, in contrast to the dark blue colonies of control strains CAP43 and CAP49 (Fig. 4). The levels of β-galactosidase observed for strain CAP95[hpr(S53A)] on SLX plates resembled those of strain CAP14(ΔhprK), although they were slightly less severe (Fig. 4). This suggested that lack of phosphorylation at the serine-53 residue on HPr contributes to the altered phenotypes in both ΔhprK and hpr(S53A) mutants. Enhanced catabolite repression was observed in CAP95[hpr(S53A)] during growth in M9 medium with succinate plus lactose or raffinose (Fig. 6A and B). Once succinate was exhausted, the strain carrying the hpr(S53A) allele exhibited longer diauxic lags than strain CAP49 which carried a wild-type hpr. In summary, the growth and catabolite repression phenotypes observed for strain CAP95[hpr(S53A)] are similar to those observed in strain CAP14(ΔhprK), suggesting that HPrK phosphorylates HPr at the serine-53 residue and that phosphorylation at this residue by HPrK plays a significant role in regulation of carbon utilization.
Phenotypes caused by the lack of phosphorylation of HPr-Ser53 are reversed by an H22A substitution in HPr.
To investigate whether enhanced SMCR in strains unable to phosphorylate serine-53 of HPr was due to the absence of HPr-Ser53-P or was due to an excess of HPr-His22-P, a strain in which both the serine-53 and the histidine-22 residues of HPr were unphosphorylatable was constructed. The resulting strain, CAP97[hpr(H22A/S53A)], was characterized during growth on single and mixed carbon sources. Normal growth on M9 plates with glycerol was restored (Fig. 3C), as was growth in liquid M9 medium with succinate as carbon source (data not shown). Normal catabolite repression phenotype was restored as well in strain CAP97[hpr(H22A/S53A)]. Diauxic lags were shortened to less than the wild-type length in M9 medium with succinate plus lactose or raffinose (Fig. 6C and D). The β-galactosidase levels in strain CAP97[hpr(H22A/S53A)] reverted to wild-type levels, as shown by their darker blue coloration on SLX plates, compared to the pale blue colonies of strain CAP95[hpr(S53A)] (Fig. 4). Similar results were seen in strain CAP94, which has the hpr(H22A) allele in a ΔhprK background (data not shown).
These results are consistent with the idea that inability to phosphorylate the histidine-22 residue of HPr relieves the enhanced catabolite repression imposed by an unphosphorylatable serine-53 residue, suggesting that the accumulation of HPr-His22-P in the ΔhprK and hpr(S53A) mutants slows growth in single carbon sources and enhances repression of lactose and raffinose utilization in the presence of succinate.
Mutants with unphosphorylatable HPr-His22 show partial relief from catabolite repression.
If HPr-His22-P is an effector of SMCR, it would be expected that a strain with unphosphorylatable histidine-22 residue should exhibit a relaxed catabolite repression phenotype. To test this hypothesis, the hpr(H22A) allele was introduced into strain RB111(Δhpr). The resulting strain, CAP93[Δhpr/hpr(H22A)], exhibited normal growth on M9 glycerol plates and in liquid M9 with succinate (Fig. 3C and data not shown). Growth rates on lactose and raffinose were reduced compared to control strains CAP43 and CAP49 (data not shown).
The β-galactosidase levels in strain CAP93[hpr(H22A)] were slightly higher than the wild-type control strains CAP43 and CAP49, as judged from its darker blue color on SLX plates (Fig. 4). Strain CAP93[hpr(H22A)] exhibited a slightly relaxed SMCR phenotype compared to the isogenic control strain CAP49 during diauxic growth in M9 with succinate plus lactose (Fig. 6C) and succinate plus raffinose (Fig. 6D). These results confirm that HPr-His22-P, directly or indirectly, causes repression in the presence of succinate, since both CAP93[hpr(H22A)] and the control strain CAP49 have the ability to produce HPr-Ser53-P but only CAP93 cannot produce HPr-His22-P.
Deletion of hprK alters succinoglycan production and symbiotic properties.
hprK is located immediately downstream of chvG (also known as exoS) in the chromosome of S. meliloti and is likely cotranscribed with chvG, since the two genes overlap. Reports that chvG mutants of S. meliloti exhibit alterations in succinoglycan production and in symbiotic phenotypes (10-12) prompted us to investigate these phenotypes in strain CAP14(ΔhprK). When the ΔhprK strain was plated on TY plates with calcofluor, it exhibited a very bright phenotype starting on the second day of incubation, which lasted for several days. In contrast, strain Rm1021 (wild type) initially presented a bright phenotype that dimmed gradually as a halo developed around the colony (Fig. 7). The bright calcofluor phenotype of CAP14(ΔhprK) indicated that this strain had upregulated production of long-chain succinoglycan. Normal development of a halo suggested that breakdown of long-chain succinoglycan or synthesis of short-chain succinoglycan had occurred and had not been altered by the absence of HPrK.
FIG. 7.
Succinoglycan production in ΔhprK mutants. Strains were pregrown in M9-glycerol medium, rinsed, resuspended, and spotted onto TY plates with 0.02% calcofluor (0.02%). Plates were photographed under UV light to document succinoglycan fluorescence. The exposure time was 0.05 s for all images, except for the day 1 images, which were exposed for 0.1 s.
In planta phenotypes of strain CAP14(ΔhprK) were evaluated in alfalfa (Medicago sativa). Inoculation with the ΔhprK strain induced nodule formation in the plants, and the number of nodules per plant was similar to the plants inoculated with wild-type strain Rm1021. However, the nodules formed were ineffective, as judged by the white coloration of many of them and by the reduced weight of the plant shoots compared to plants inoculated with strain Rm1021 (Fig. 8). Crushed nodules from plants inoculated with both ΔhprK and wild-type strains yielded colonies when spread on TY plates. Colonies recovered from nodules elicited by strain CAP14(ΔhprK) included the ΔhprK mutants (small colonies), as well as larger colonies with suppressor mutations, showing that strain CAP14(ΔhprK) was able to grow in infection threads and occupy the nodules but either failed to transition to the bacteroid state or to fix nitrogen once in that state (data not shown). Nodules occupied by strain Rm1021 show clear meristematic, infection, and nitrogen fixation zones (Fig. 9A) and densely populated bacteroid-infected plant cells (Fig. 9C). Examination of the internal structure of CAP14(ΔhprK) induced nodules revealed low bacteroid numbers and the presence of starch granules in bacteroid-infected plant cells in both white and pink nodules (Fig. 9B and D). There was also a poorly defined infection/nitrogen-fixation zone in white nodules (Fig. 9B), perhaps explaining the low nitrogen fixation efficiency of these nodules.
FIG. 8.
Nodulation phenotype of ΔhprK mutants. Average number of nodules per plant (A), shoot weight (B), and number of pink nodules per plant (C) on alfalfa plants inoculated with S. meliloti wild-type strain Rm1021, strain CAP14(ΔhprK), and the ΔhprK complemented strain CAP62. *, Significantly different from the wild-type strain (P < 0.003, n = 18).
FIG. 9.
Internal structure of nodules induced by wild-type strain and ΔhprK mutant. Images of S. meliloti strain Rm1021 (A and C, gray nodules) or strain CAP14(ΔhprK) (B and D, white nodules) are shown. Panels A and B show the meristematic (M) and nitrogen fixation (N-f) zones. Panels C and D show bacteroids in plant cells from the nitrogen fixation zone. b, bacteroids; n, nucleus; V, vacuole. Arrowheads indicate starch granules which can be seen around the perimeter of nodule cells in panel D. Scale bar, 98 μm in panels A and B and 24 μm in panels C and D.
When strain CAP62(ΔhprK, SMc02324::hprK) was tested for succinoglycan and symbiotic phenotypes, both of the phenotypes were complemented by the introduction of hprK in trans (Fig. 7 and 8). This result indicated that the observed alterations in succinoglycan production and symbiosis in strain CAP14(ΔhprK) were due to the deletion of hprK.
DISCUSSION
Many bacteria do not have a complete PTS and are thought to have retained the partial systems for regulation (4, 8). Possession of a partial PTS is common in the alphaproteobacteria, and is also found in certain gamma-, delta-, and betaproteobacteria; in spirochetes (Treponema denticola, T. pallidium, and Leptospira interrogans); and in Chlorobium tepidum (2, 4, 55).
The PTS of S. meliloti includes the general proteins EI and HPr and the EIIA-type proteins ManX and EIIANtr. Since the permease components EIIB and EIIC are missing, it is unlikely that sugar transport and phosphorylation directs the regulation of catabolite repression in this organism. The EI protein in S. meliloti is similar to EINtr proteins in other bacteria. Although it has not been proved that HPr is phosphorylated by EINtr, it is likely that this is the case, given the lack of a classical EI and the similarity of HPr to NPr (an Ntr-type HPr) (data not shown). In addition, S. meliloti and many other alphaproteobacteria possess HPrK proteins that are shorter than their gram-positive counterparts (4, 55). These contain all of the critical residues involved in both the kinase and the phosphatase activities. This suggests that S. meliloti uses its incomplete PTS to regulate metabolism and may do so through phosphorylation of HPr by HPrK. It has been shown previously that HPr and ManX participate in SMCR in S. meliloti (44) and that research suggested that HPr-His22-P was, directly or indirectly, responsible for exerting repression in the presence of succinate.
HPrK is involved in catabolite repression through phosphorylation of HPr-Ser53.
In B. subtilis and other firmicutes, HPrK phosphorylates HPr on a conserved serine-46 residue. We investigated the role of HPrK and its possible phosphorylation of HPr in SMCR in S. meliloti by construction of mutant strains lacking HPrK and other strains in which potential phosphorylation of HPr at the conserved histidine-22 and serine-53 residues was prevented. A strain devoid of the hprK gene exhibited strengthened SMCR, as evidenced by long diauxic lags. It also showed that downregulation of melA and lac genes was needed for utilization of raffinose and lactose even in the absence of succinate, indicating strong repression of those genes. These phenotypes were replicated by a strain with a mutant HPr that could not be phosphorylated on the conserved serine-53 residue. This indicated that HPrK most likely phosphorylates HPr at the serine-53 residue, the lack of which results in enhanced catabolite repression. We noted that some of the phenotypes observed in the strain with an unphosphorylatable serine-53 in HPr were less severe than those of a strain lacking HPrK. It has been reported that substitution of an alanine for the conserved serine residue can affect phosphorylation at the histidine residue by EI, because the serine residue is part of the HPr-EI interaction interface (15). Thus, the slight differences in phenotype between the ΔhprK and the hpr(S53A) strains may be caused by a lessening of phosphorylation of the HPr-S53A at its histidine-22 site.
Two, not mutually exclusive, hypotheses could explain the enhanced SMCR seen in the ΔhprK strain. Either HPrK is required for activation of catabolic genes or accumulation of HPr-His22-P in the cell results in enhanced SMCR. A mutant strain in which both the serine-53 and the histidine-22 residues of HPr were unphosphorylatable eliminated the strong phenotype caused by an unphosphorylatable serine-53. Also, the strong phenotype of the ΔhprK strain was eliminated when HPr was replaced with HPr-H22A. This suggests that HPr-His22-P mediates the ΔhprK phenotypes and that HPrK may act by slowing down phosphorylation at the histidine-22 residue when it phosphorylates the serine-53 residue. In B. subtilis and other gram-positive organisms, phosphorylation of HPr at the conserved serine residue significantly slows down phosphorylation by EI at the conserved histidine residue (26). In support of the idea that HPr-His22-P is important for exerting SMCR, a mutant devoid of ManX, which likely accumulated HPr-His22-P, exhibited enhanced SMCR (44).
If HPr-His22-P were solely responsible for establishing SMCR, it would be expected that a strain containing only HPr-H22A would exhibit completely relieved SMCR. Strain CAP93[hpr(H22A)] did exhibit relaxed SMCR, but not a complete relief from catabolite repression. A likely explanation for this result is that there is more than one mechanism that operates to establish SMCR. An example of another protein that is involved in regulation of catabolite repression in S. meliloti is the sensor kinase Sma0113 (Garcia and Gage, unpublished).
We have provided evidence consistent with a model wherein HPrK phosphorylates HPr at the serine-53 residue, an event that is important in the regulation of SMCR in S. meliloti. Phosphorylation at serine-53 most likely affects SMCR control by slowing down or preventing phosphorylation by EI at the histidine-22 residue. Our results, along with previous results, indicate that HPr-His22-P, directly or indirectly, strengthens SMCR. A simplified model of phosphate transfer and SMCR regulation is shown in Fig. 10. During metabolism, HPr is phosphorylated at the histidine-22 residue by EINtr and transfers the phosphate group to ManX. In the presence of succinate or other preferred carbon sources HPr-Ser53-P levels are low through HPrK phosphatase activity or through a reduction in the synthesis of HPr-S53-P by HPrK. The reduction in HPr-Ser53-P levels allows HPr-His22-P to increase and elicit strengthened SMCR gene regulation and/or inducer exclusion.
FIG. 10.
Proposed model for the effects of HPrK on SMCR in S. meliloti. Phosphate groups enter the PTS by transfer from PEP to the histidine-22 residue of HPr. Phosphatase activity or low kinase activity of HPrK in the presence of succinate allows for accumulation of HPr-His22-P, which elicits SMCR. In the absence of succinate, phosphorylation of the serine-53 residue of HPr prevents HPr-His22-P formation, relieving catabolite repression. Dashed lines indicate phosphorylation events that are assumed to take place based on work in other model systems.
It is interesting that phosphorylation of the conserved serine-53 residue weakens SMCR in S. meliloti because, in B. subtilis and other firmicutes, phosphorylation of the conserved serine strengthens catabolite repression (16, 27). Thus, the regulation of catabolite repression in S. meliloti is mechanistically different from other organisms that possess HPr and HPrK. This difference extends not only to the role of HPr and HPrK but also to how the PTS receives information about carbon status, since succinate is neither imported into the cell by PTS-related proteins nor phosphorylated upon entry.
We have also shown than HPrK is involved in other cellular processes, such as carbon utilization, regulation of succinoglycan production, and symbiosis with alfalfa. It was reported that an S. meliloti strain lacking HPr was also upregulated for succinoglycan production (44). This suggests that the succinoglycan phenotype of the ΔhprK strain is regulated through HPr, as are the SMCR phenotypes. This, however, has yet to be further investigated. Many strains with modified succinoglycan production also exhibit alterations in their symbiotic properties, in particular in their abilities to infect root hairs and construct proper infection threads (11, 21, 43, 60). In contrast, the ΔhprK strain in the present study successfully invaded its host but exhibited a nitrogen fixation defect. The inability to fix nitrogen in strain CAP14(ΔhprK) could be related to its slow-growth phenotype or to failure to differentiate into bacteroids, since the internal structure of alfalfa nodules populated by this strain exhibits a low density of bacteroids.
Acknowledgments
This study was supported by the U.S. Department of Energy contracts DE-FG02-01ER15175 and DE-FG02-06ER15805 to D.J.G.
We thank Preston Garcia for providing the sma0113 promoter, for sharing data, and for useful input.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Asanuma, N., and T. Hino. 2003. Molecular characterization of HPr and related enzymes, and regulation of HPr phosphorylation in the ruminal bacterium Streptococcus bovis. Arch. of Microbiol. 179205-213. [DOI] [PubMed] [Google Scholar]
- 2.Barabote, R. D., and M. H. Saier, Jr. 2005. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69608-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, M. J., V. Oke, and S. R. Long. 2000. New genetic tools for use in the Rhizobiaceae and other bacteria. BioTechniques 29240-245. [DOI] [PubMed] [Google Scholar]
- 4.Boel, G., I. Mijakovic, A. Maze, S. Poncet, M. K. Taha, M. Larribe, E. Darbon, A. Khemiri, A. Galinier, and J. Deutscher. 2003. Transcription regulators potentially controlled by HPr kinase/phosphorylase in gram-negative bacteria. J. Mol. Microbiol. Biotechnol. 5206-215. [DOI] [PubMed] [Google Scholar]
- 5.Brewin, N. J. 1991. Development of the legume root nodule. Annu. Rev. Cell Biol. 7191-226. [DOI] [PubMed] [Google Scholar]
- 6.Bringhurst, R. M., and D. J. Gage. 2002. Control of inducer accumulation plays a key role in succinate-mediated catabolite repression in Sinorhizobium meliloti. J. Bacteriol. 1845385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruckner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209141-148. [DOI] [PubMed] [Google Scholar]
- 8.Cases, I., F. Velazquez, and V. de Lorenzo. 2007. The ancestral role of the phosphoenolpyruvate-carbohydrate phosphotransferase system (PTS) as exposed by comparative genomics. Res. Microbiol. 158666-670. [DOI] [PubMed] [Google Scholar]
- 9.Chaptal, V., F. Vincent, V. Gueguen-Chaignon, V. Monedero, S. Poncet, J. Deutscher, S. Nessler, and S. Morera. 2007. Structural analysis of the bacterial HPr kinase/phosphorylase V267F mutant gives insights into the allosteric regulation mechanism of this bifunctional enzyme. J. Biol. Chem. 28234952-34957. [DOI] [PubMed] [Google Scholar]
- 10.Chen, E. J., E. A. Sabio, and S. R. Long. 2008. The periplasmic regulator ExoR inhibits ExoS/ChvI two-component signaling in Sinorhizobium meliloti. Mol. Microbiol. 691290-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 1805183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 18020-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denarie, J., F. Debelle, and J. C. Prome. 1996. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65503-535. [DOI] [PubMed] [Google Scholar]
- 14.Deutscher, J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 1187-93. [DOI] [PubMed] [Google Scholar]
- 15.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dossonnet, V., V. Monedero, M. Zagorec, A. Galinier, G. Pérez-Martínez, and J. Deutscher. 2000. Phosphorylation of HPr by the bifunctional HPr Kinase/P-Ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion. J. Bacteriol. 1822582-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudley, M. E., T. W. Jacobs, and S. R. Long. 1987. Microscopic studies of cell divisions induced in alfalfa roots by Rhizobium meliloti. Planta 171289-301. [DOI] [PubMed] [Google Scholar]
- 18.Fieulaine, S., S. Morera, S. Poncet, V. Monedero, V. Gueguen-Chaignon, A. Galinier, J. Janin, J. Deutscher, and S. Nessler. 2001. X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain. EMBO J. 203917-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gage, D. J., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 1787159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardiol, A., A. Arias, C. Cervenansky, and G. Martinez-Drets. 1982. Succinate dehydrogenase mutant of Rhizobium meliloti. J. Bacteriol. 1511621-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez, J. E., B. L. Reuhs, and G. C. Walker. 1996. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. USA 938636-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorke, B., and J. Stulke. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6613-624. [DOI] [PubMed] [Google Scholar]
- 23.Gulati, A., and S. Mahadevan. 2000. Mechanism of catabolite repression in the bgl operon of Escherichia coli: involvement of the anti-terminator BglG, CRP-cAMP and EIIAGlc in mediating glucose effect downstream of transcription initiation. Genes Cells 5239-250. [DOI] [PubMed] [Google Scholar]
- 24.Gunnewijk, M. G., and B. Poolman. 2000. Phosphorylation state of HPr determines the level of expression and the extent of phosphorylation of the lactose transport protein of Streptococcus thermophilus. J. Biol. Chem. 27534073-34079. [DOI] [PubMed] [Google Scholar]
- 25.Halbedel, S., C. Hames, and J. Stulke. 2007. Regulation of carbon metabolism in the mollicutes and its relation to virulence. J. Mol. Microbiol. Biotechnol. 12147-154. [DOI] [PubMed] [Google Scholar]
- 26.Halbedel, S., and J. Stulke. 2005. Dual phosphorylation of Mycoplasma pneumoniae HPr by Enzyme I and HPr kinase suggests an extended phosphoryl group susceptibility of HPr. FEMS Microbiol. Lett. 247193-198. [DOI] [PubMed] [Google Scholar]
- 27.Hanson, K. G., K. Steinhauer, J. Reizer, W. Hillen, and J. Stülke. 2002. HPr kinase/phosphatase of Bacillus subtilis: expression of the gene and effects of mutations on enzyme activity, growth and carbon catabolite repression. Microbiol. 1481805-1811. [DOI] [PubMed] [Google Scholar]
- 28.Hornez, J. P., M. Timinouni, C. Defives, and J. C. Derieux. 1994. Unaffected nodulation and nitrogen-fixation in carbohydrate pleiotropic mutants of Rhizobium meliloti. Curr. Microbiol. 28225-229. [Google Scholar]
- 29.Hu, K.-Y., and M. J. Saier. 2002. Phylogeny of phosphoryl transfer proteins of the phosphoenolpyruvate-dependent sugar-transporting phosphotransferase system. Res. Microbiol. 153405-415. [DOI] [PubMed] [Google Scholar]
- 30.Inada, T., K. Kimata, and H. Aiba. 1996. Mechanism responsible for glucose-lactose diauxie in Escherichia coli: challenge to the cAMP model. Genes Cells 1293-301. [DOI] [PubMed] [Google Scholar]
- 31.Jelesko, J. G., and J. A. Leigh. 1994. Genetic characterization of a Rhizobium meliloti lactose utilization locus. Mol. Microbiol. 11165-173. [DOI] [PubMed] [Google Scholar]
- 32.Kimata, K., H. Takahashi, T. Inada, P. Postma, and H. Aiba. 1997. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc. Natl. Acad. Sci. USA 9412914-12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kravanja, M., R. Engelmann, V. Dossonnet, M. Bluggel, H. E. Meyer, R. Frank, A. Galinier, J. Deutscher, N. Schnell, and W. Hengstenberg. 1999. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol. Microbiol. 3159-66. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda, M., T. H. Wilson, and T. Tsuchiya. 2001. Regulation of galactoside transport by the PTS. J. Mol. Microbiol. Biotechnol. 3381-384. [PubMed] [Google Scholar]
- 35.Long, S. R. 1996. Rhizobium symbiosis: Nod factors in perspective. Plant Cell 81885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mauchline, T. H., J. E. Fowler, A. K. East, A. L. Sartor, R. Zaheer, A. H. Hosie, P. S. Poole, and T. M. Finan. 2006. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. USA 10317933-17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mijakovic, I., S. Poncet, A. Galinier, V. Monedero, S. Fieulaine, J. Janin, S. Nessler, J. A. Marquez, K. Scheffzek, S. Hasenbein, W. Hengstenberg, and J. Deutscher. 2002. Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life? Proc. Natl. Acad. Sci. USA 9913442-13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 40.Monahan-Giovanelli, H., C. A. Pinedo, and D. J. Gage. 2006. Architecture of infection thread networks in developing root nodules induced by the symbiotic bacterium Sinorhizobium meliloti on Medicago truncatula. Plant Physiol. 140661-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monedero, V., O. P. Kuipers, E. Jamet, and J. Deutscher. 2001. Regulatory functions of Serine-46-Phosphorylated HPr in Lactococcus lactis. J. Bacteriol. 1833391-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nessler, S., S. Fieulaine, S. Poncet, A. Galinier, J. Deutscher, and J. Janin. 2003. HPr kinase/phosphorylase, the sensor enzyme of catabolite repression in gram-positive bacteria: structural aspects of the enzyme and the complex with its protein substrate. J. Bacteriol. 1854003-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pellock, B. J., H. P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 1824310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinedo, C. A., R. M. Bringhurst, and D. J. Gage. 2008. Sinorhizobium meliloti mutants lacking phosphotransferase system enzyme HPr or EIIA are altered in diverse processes, including carbon metabolism, cobalt requirements, and succinoglycan production. J. Bacteriol. 1902947-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poole, P., and D. Allaway. 2000. Carbon and nitrogen metabolism in Rhizobium. Adv. Microb. Physiol. 43117-163. [DOI] [PubMed] [Google Scholar]
- 46.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 12715-21. [DOI] [PubMed] [Google Scholar]
- 47.Reizer, J., C. Hoischen, F. Titgemeyer, C. Rivolta, R. Rabus, J. Stulke, D. Karamata, M. H. Saier, Jr., and W. Hillen. 1998. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol. Microbiol. 271157-1169. [DOI] [PubMed] [Google Scholar]
- 48.Reizer, J., S. L. Sutrina, M. H. Saier, G. C. Stewart, A. Peterkofsky, and P. Reddy. 1989. Mechanistic and physiological consequences of HPr(ser) phosphorylation on the activities of the phosphoenolpyruvate:sugar phosphotransferase system in gram-positive bacteria: studies with site-specific mutants of HPr. EMBO J. 82111-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronson, C. W., P. Lyttleton, and J. G. Robertson. 1981. C(4)-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc. Natl. Acad. Sci. USA 784284-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saier, M. H., Jr., and S. Roseman. 1976. Sugar transport. The crr mutation: its effect on repression of enzyme synthesis. J. Biol. Chem. 2516598-6605. [PubMed] [Google Scholar]
- 51.Saier, M. J. 1998. Multiple mechanisms controlling carbon metabolism in bacteria. Biotechnol. Bioeng. 58170-174. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 53.Sauter, T., and E. D. Gilles. 2004. Modeling and experimental validation of the signal transduction via the Escherichia coli sucrose phosphotransferase system. J. Biotechnol. 110181-199. [DOI] [PubMed] [Google Scholar]
- 54.Spaink, H. P. 1995. The molecular basis of infection and nodulation by rhizobia: the ins and outs of sympathogenesis. Annu. Rev. Phytopathol. 33345-368. [DOI] [PubMed] [Google Scholar]
- 55.Stonestrom, A., R. D. Barabote, C. F. Gonzalez, and M. H. Saier, Jr. 2005. Bioinformatic analyses of bacterial HPr kinase/phosphorylase homologues. Res. Microbiol. 156443-451. [DOI] [PubMed] [Google Scholar]
- 56.Stulke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2195-201. [DOI] [PubMed] [Google Scholar]
- 57.Ucker, D. S., and E. R. Signer. 1978. Catabolite-Repression-Like Phenomenon in Rhizobium meliloti. J. Bacteriol. 1361197-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viana, R., V. Monedero, V. Dossonnet, C. Vadeboncoeur, G. Perez-Martinez, and J. Deutscher. 2000. Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol. Microbiol. 36570-584. [DOI] [PubMed] [Google Scholar]
- 59.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao, S. Y., L. Luo, K. J. Har, A. Becker, S. Ruberg, G. Q. Yu, J. B. Zhu, and H. P. Cheng. 2004. Sinorhizobium meliloti ExoR and ExoS proteins regulate both succinoglycan and flagellum production. J. Bacteriol. 1866042-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]